Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Wastewater, Sewer system, Wastewater-based epidemiology
Abstract
The SARS-CoV-2 virus causing COVID-19 is spread in sewage by the stool of infected individuals, and viral material in sewage can be quantified using molecular tools. This study aimed to monitor the presence of SARS-CoV-2 RNA in sewage in Mexico based on RdRP, S, and N gene analysis. The influent, effluent, and activated sludge from two domestic wastewater treatment plants (WWTP) were evaluated from the early stage of the epidemic to July 2020. Additionally, sampling points in sewer systems were examined, comparing two different RNA-concentration methods: centrifugal ultrafiltration and adsorption-based methods. The adsorption method resulted in RNA titration that was two orders of magnitude higher than with ultrafiltration (up to 3.38 log10 copies RdRP gene/mL of sewage). The surveillance of SARS-CoV-2 RNA in the influent of two WWTP correlated with the cumulative COVID-19 cases in Queretaro city. The higher RNA level in secondary sludge compared to influent suggests that viral RNA becomes concentrated in activated sludge. This result supports SARS-CoV-2 RNA removal in WWTP, where all effluent samples were negative for virus quantification. This work proves that wastewater-based epidemiology is a very valuable tool in developing countries where diagnostic tests for COVID-19 are limited.
1. Introduction
The COVID-19 respiratory pandemic caused by the novel coronavirus has generated more than 30 million confirmed cases in 216 countries (September 19th, 2020), with a fatality rate of 3.3 % [1] Person-to-person transmission is the main route of spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the agent causing COVID-19, which occurs via direct contact or through droplets spread by an infected individual coughing or sneezing [2] or through microdroplets during loud conversations and breathing [3].
In addition, SARS-CoV-2 RNA was found to be present in anal swabs and stools from infected individuals, regardless of intestinal infection signs, with a positive prevalence in a later stage of the disease despite negative analysis using oral sampling [4,5]. Although live viruses exist in stool specimens, current knowledge of possible oral-fecal transmission is limited [6].
Eventually, fecal matter reaches the sewer system, where wastewater treatment plants (WWTP) are potential sampling points representing the communities served by those plants. Recent studies describe the presence of SARS-CoV-2 RNA in WWTP influent with levels of approximately 12 copies mL−1 [7,8]. In this sense, some studies have successfully correlated the SARS-CoV-2 RNA level in wastewater and sludge with the contagious numbers of COVID-19 or even as a potential early warning indicator of spread of the disease in the population [9]. Studies have also shown the potential of water-based epidemiology (WBE) as an outbreak indicator for hepatitis A virus and norovirus in 2013 [10]. In this sense, WBE can be a powerful tool for surveying emerging epidemic hotspots such as COVID-19, with a potential of cost savings compared to medical screening [11,12].
As in many other countries, the surveillance policy of Mexican health authorities focuses on patients with mild to severe symptoms [13], which may underestimate the pandemic spread since up to 43 % of infected people are asymptomatic [14]. To date, Mexico is the fourth country in the world reporting deaths (about 73,000) caused by COVID-19 [1], and there is a current need for more efficient surveillance tools. The WBE can overcome the limitations, economic and practical, of conventional methods [11]. There are few studies using WBE for the surveillance of SARS-CoV-2 in the Latin American region [[15], [16], [17]]. Besides, there exist interest to determine the fate of the SARS-CoV-2 RNA in wastewater treatment plants considering the potential reuse of the wastewater for irrigation purposes [17] and the type of concentration method used to process the sample.
This study aimed to monitor SARS-CoV-2 RNA in a moderately populated city in Mexico from the early stage of the epidemic to July 2020. First, punctual sampling points in sewer systems were analyzed, including a COVID-19 hospital, a quarantine center, a government office, and a complex with a correctional and court facility where two different RNA-concentration methods (centrifugal ultrafiltration and adsorption-based methods) were compared for those samples. Then, the levels of SARS-CoV-2 RNA were monitored in the influent, effluent and activated sludge from two domestic WWTP. The results were correlated with the positive medical cases in the area.
2. Materials and methods
2.1. Sewage samples
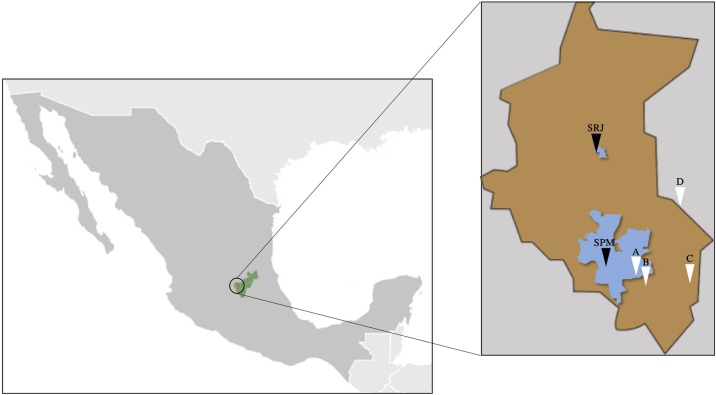
Two WWTP (denoted SPM and SRJ) located in the city of Santiago de Queretaro (1.2 million inhabitants in the metropolitan area), Queretaro State, Mexico, were selected for this study (Fig. 1 ). These plants treat approximately 60 % of the wastewater generated in the city (Table 1 ). Three sampling points were considered in the WWTP: influent (after coarse and fine screening), secondary sludge (from sludge recirculation system), and effluent (after the disinfection process, where available). Both WWTP operate with secondary treatment. At the sampling period, only the large plant chlorinates the effluent. To validate the SARS-CoV-2 quantification methods, four sample points were selected from the street sewer systems at different facilities where positive and negative COVID-19 cases were reported (Fig. 1, Table 2 ). Grab samples were collected in 2020 from April 23rd to July 3rd from the WWTP and on July 14th and 20th from the sewer systems. Five hundred mL were taken for the sewage and effluent samples, and 15 mL was taken for the sludge samples. All samples were collected in the morning between 10 and 11 a.m. and were transported to the laboratory at 4 °C, accordingly [8,9]. The samples from the WWTP were stored at −70 °C for later use; the samples from the other sewer systems were processed the same day of sampling for RNA isolation. Personal protective equipment (e.g., coverall, gloves, and face, eye, and respiratory protection) was used during the sampling and further processing steps. Total suspended solids (TSS) from the sewer system samples were determined according to standard methods [18].
Fig. 1.
Sampling sites. Green, brown and blue polygons denote Queretaro State, Santiago de Queretaro and the urban area served by each sampled WWTP (SPM and SRJ). Letters denote the sampling location for the different street sewer systems: A) water utility company; B) COVID-19 hospital; C) quarantine center and D) correctional and court facility.
Table 1.
Data on the population and operating characteristics of WWTP.
| WWTP | Capacity (L/s) |
Process | Served population (inhabitants) |
|
|---|---|---|---|---|
| Designed | Treated | |||
| SPM | 750 | 635.3 | Dual (biofilter/activated sludge). Chlorinated effluent | 320,545 |
| SRJ | 30 | 26.9 | Activated sludge. Non chlorinated effluent | 13,535 |
SPM, San Pedro Martir area; SRJ, Santa Rosa Jauregui area.
Table 2.
Data of sampling points from sewer systems.
| Sampling point | Total population (Patients and staff) |
% COVID-19 positivity reporteda |
|---|---|---|
| Hospital | 176 | 33 |
| Quarantine center | 82 | 35 |
| Water utility company | 53 | 0 |
| Correctional and court facility | 2100 | 0 |
According to clinical diagnosis reported by local authorities.
2.2. Concentration of genetic material
Before RNA isolation, the genetic material from the sewer system samples was concentrated by two different methods: centrifugal ultrafiltration and adsorption onto a membrane. For practical purposes, the adsorption method was selected to concentrate the influent and effluent samples from the WWTP. The concentration step was not applied for sludge samples from the WWTP.
2.2.1. Virus concentration by centrifugal ultrafiltration
A total of 120 mL of raw sewage was filtered through a 0.2-μm membrane of polyethersulfone (Millipore, Netherlands) to remove bacterial cells and debris. Subsequently, the viral material was concentrated from the filtered water using a centrifugal filter device (Amicon Ultra-15 10 K, Millipore, Netherlands) by centrifugation at 2200 x g for 20 min [7]. The concentrate was recovered with a sterile pipette and stored at −20 °C until use.
2.2.2. Virus concentration by adsorption onto a negatively charged membrane
Between 30 and 100 mL of raw sewage or effluent was used. The pH of the sample was adjusted to 3.5 with 2 N HCl, and the sample was filtered through a 0.45-μm pore and 47-mm diameter negatively charged nitrocellulose membrane (Millipore, Netherlands) via a glass funnel and base (Millipore, Netherlands) [19]. The membrane was stored at −20 °C until use. Subsequently, the membrane was cut and used directly in the RNA extraction procedure.
2.3. RNA extraction and quantification
RNA was extracted from the concentrated, negatively charged membranes or sludge samples (600 μL) using the RNeasy Power Microbiome kit (Qiagen, Germany) according to the manufacturer's instructions. Fifty microliters of RNA extract were recovered from each sample.
2.3.1. Quantification of RNA SARS-CoV-2
As a first attempt to detect and quantify SARS-CoV-2, the S protein and N protein genes were evaluated in some of the recovered sewage and sludge RNA extracts from the WWTP using TaqMan 2019-nCoV Assay Kit v1 (Applied Biosystems, USA) according to the manufacturer’s instructions. Synthetic sequences of the target genes (TaqMan 2019-nCoV Control Kit v1, Applied Biosystems, USA) were used as positive controls. Although the assay kit is designed for qualitative analysis, quantification of the N and S genes was performed using three 10-fold dilutions (from 1.0 E + 02 to 1.0 E + 04 copies per reaction) of the control kit as a calibration suspension. The reverse transcription and amplification of the genes were performed with SuperScript III Platinum One-Step qRT-PCR Kit (Invitrogen, USA). Internal (human RNase P RPPH1), positive and negative controls were performed for each qPCR assay.
The RdRP gene specific for SARS-CoV-2 was quantified in all RNA samples as previously reported [20]. All primers and probes were synthesized by Sigma-Aldrich (USA). Quantification of the RdRP gene was performed using four 10-fold dilutions (from 1.0 E + 02 to 1.0 E + 05 copies per microliter) of the Wuhan coronavirus 2019 RdRP gene control as a calibration suspension (European Virus Archive Global, Germany). All RT-qPCR assays were performed in duplicate using a Real-Time PCR System (Applied Biosystems StepOne™, USA). Reactions were considered positive if the threshold cycle (CT) was obtained before 40 cycles [7].
3. Results and discussion
Three SARS-CoV-2 genes, RdRP, N protein and S protein, from two WWTP in Santiago de Queretaro were evaluated in influent, effluent and secondary sludge from April 13th to July 3rd. Additionally, the RdRP gene was examined in sewage samples from the sewer system at different relevant points, representing confirmed COVID-19 cases and other points without confirmed cases, using two different RNA concentration methods. For N and S protein gene quantification, the RN-ase P gene was evaluated as an internal control, with a CT average value of 29.8 ± 4.3, indicating no inhibitory effects on RT-PCR by wastewater. Positive control samples showed average CT values of 18.5, 19.9, and 23.5 for the N, S and RdRP genes, respectively, which were not detected in the negative control samples. The slopes of the standard curves for the quantification of the evaluated genes were -3.35 for RdRP, -3.68 for N, and −3.78 for S. Y-intercept values were 46.93, 36.23, and 41.12 and amplification efficiencies 93 %, 87 %, 84 % for RdRP, N, and S gene quantification, with correlation coefficients of 0.98, 0.98, and 0.99, respectively.
3.1. Comparison of methods
Four sampling points were considered to evaluate the feasibility of two different concentration methods. These samples were composed of two positive sampling points for SARS-CoV-2 (sewer system from a COVID-19 hospital and a COVID-19 quarantine center), and the other two were presumably negative for SARS-CoV-2 (a government office and a correctional and court facility). Both methods were able to detect the RdRP gene in all samples collected on July 14th, 2020. The adsorption method generated higher values (up to two orders of magnitude) than the centrifugal ultrafiltration method (Table 3 ). For the second sampling day (July 20th), only the adsorption method was positive for RdRP quantification.
Table 3.
RdRP gene quantification (log10 copies/mL) results using different RNA concentration methods from sewer systems at two different sampling dates.
| Sampling point | 14/07/2020 |
20/07/2020 |
||
|---|---|---|---|---|
| Ultrafiltration | Adsorption | Ultrafiltration | Adsorption | |
| Hospital | 1.9830 | 2.3210 | negative | 2.5951 |
| Quarantine center | 1.9185 | 3.3781 | negative | 2.5808 |
| Water utility company | negative | negative | negative | negative |
| Correctional facility | 1.3019 | 2.7713 | negative | negative |
Such variation between detection methods was recently addressed by Ahmed et al. [21] using murine hepatitis virus as a surrogate of SARS-CoV-2. Those authors found that the adsorption-extraction methods with MgCl2 pre-treatment using mixed cellulose esters membranes was more efficient than ultracentrifugation. Conversely, Sherchan et al. [22] reported better performances with the ultrafiltration method than those obtained with adsorption and elution, though the N1 and N2 genes were evaluated. To the best of our knowledge, only one study [23] has assessed the adsorption method followed by RNA extraction directly from the membrane, similar to this work. Nevertheless, those authors reported contradictory results between the ultrafiltration and adsorption methods, with only one positive sample for each. Considering the different conclusions of studies comparing viral concentration methods for SARS-CoV-2 [[21], [22], [23]], more studies and interlaboratory validations are needed to prompt a wider use of WBE studies for COVID-19.
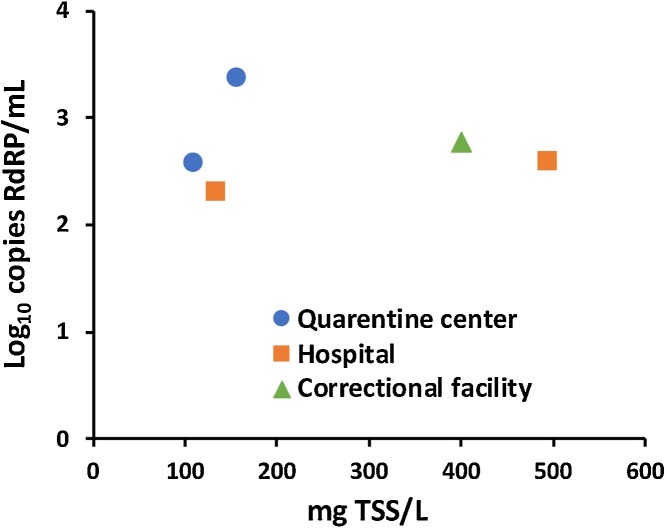
The coronavirus's affinity to suspended solids and organic matter present in the water due to the viral envelope's hydrophobicity can explain the better results obtained with the adsorption method [24]. In addition, the ultrafiltration method only recovers the genetic material present in the liquid fraction of sewage. The relationship of virus RNA copies to TSS content showed that not a clear correlation exists, except for those obtained from the quarantine center, where the correlation was higher than 3.54 log10 copies per mg of TSS. In average, the solids correlation among positive samples in sewer system was 3.4 ± 0.5 log10 copies per mg of TSS (Fig. 2 ). The duration of viral shedding in the feces can explain the higher correlation in sewage from the quarantine center than the COVID-19 hospital. The quarantine facility aims to treat COVID-19 patients in recovery after hospitalization, or with mild illness severity, with an average stay length of 15 days. In this sense, a high number of positive samples for SARS‐CoV‐2 RNA in feces has been reported than for oral swab at a later stage of infection [5]. Unlike pharyngeal analysis (median of positivity at 6.5 days), the viral RNA's positivity in feces is 7–13 days after onset of symptoms. After the first day of laboratory confirmation, the viral shedding last in feces is in the range of 8–14 days, regardless of the illness severity [4]. Although the solid content correlation results are conservative, considering the limited sampling dates from the sewer system- in this work, a recent study analyzing viral RNA in primary sludge from different WWTP, showed the potential use of the solid matrix in wastewater as an indicator of the SARS-CoV-2 prevalence [9]. In addition, the positive result from the sewage of the correctional and court facility highlights the effectivity of the method, detecting the COVID-19 prevalence before reporting any positive case by authorities, possibly due to the lack of medical testing. Thus, SARS-CoV-2 RNA sewage surveillance is a useful tool for authorities in isolated locations, such as office buildings and industrial and correctional facilities.
Fig. 2.
SARS-CoV-2 RNA versus the TSS concentration from different sewer systems and using the adsorption method.
3.2. Detection of SARS-CoV-2 in WWTP influent, effluent and activated sludge
Sixty-six samples were collected from two WWTP in Santiago de Queretaro, comprising influent, secondary sludge, and effluent (22 samples for each point), and 36 % of the influent samples were positive for at least one of the SARS-CoV-2 analyzed genes (Table 4 ). A similar positivity rate and quantification have been reported in sewage, ranging from 0.12 to 4 and 0.37–73 copies/mL, for the N gene using adsorption-direct RNA extraction [23,25] and the electronegative membrane-vortex (EMV) method [26], respectively.
Table 4.
RdRP, N and S genes quantification (log10 copies/mL) results from influent, sludge and effluent from two different WWTP during the sampling period.
| Plant | Gene | 23/04/2020 | 30/04/2020 | 07/05/2020 | 14/05/2020 | 21/05/2020 | 29/05/2020 | 04/06/2020 | 11/06/2020 | 18/06/2020 | 25/06/2020 | 03/07/2020 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Influent | ||||||||||||
| SPM | RdRP | negative | 1.6212 | negative | negative | negative | negative | negative | negative | 5.1758 | negative | negative |
| N | na | na | na | na | na | na | na | negative | na | 3.4892 | na | |
| S | negative | na | na | negative | na | negative | na | na | na | na | na | |
| SRJ | RdRP | negative | negative | negative | negative | 1.5375 | negative | 2.5258 | negative | negative | 2.3348 | 3.8905 |
| N | na | na | na | na | 1.8602 | na | na | negative | na | 3.7557 | na | |
| S | negative | na | na | 1.3483 | na | negative | na | na | na | na | na | |
| Effluent | ||||||||||||
| SPM | RdRP | negative | negative | negative | negative | negative | negative | negative | negative | negative | negative | negative |
| N | na | na | na | na | na | na | na | negative | na | negative | na | |
| S | negative | na | na | negative | na | negative | na | na | na | na | na | |
| SRJ | RdRP | negative | negative | negative | negative | negative | negative | negative | negative | negative | negative | negative |
| N | na | na | na | na | negative | na | na | negative | na | negative | na | |
| S | negative | na | na | negative | na | negative | na | na | na | na | na | |
| Activated sludge | ||||||||||||
| SPM | RdRP | negative | negative | negative | negative | negative | negative | negative | 10.0065 | 10.2384 | 9.2912 | 10.5631 |
| N | na | na | na | na | na | na | na | 3.2680 | na | na | na | |
| S | negative | na | na | negative | na | negative | na | na | na | na | na | |
| SRJ | RdRP | negative | negative | negative | negative | 9.5295 | negative | 10.6740 | negative | 10.7534 | negative | 10.5631 |
| N | na | na | na | na | 1.4401 | na | na | negative | na | na | na | |
| S | 3.5244 | na | na | 3.2595 | na | 5.2862 | na | na | na | na | na | |
na: not analyzed.
Of the secondary sludge samples, 45 % were positive for at least one of the analyzed SARS-CoV-2 genes (Table 4). Values for the RdRp and S genes in sludge were higher than those in influent. A similar range of values was reported in primary sludge from a WWTP at New Haven, Connecticut, USA, with close to 106 copies of N1 and N2 SARS-CoV-2 genes/sludge mL [9]. In our study, the secondary sludge presented a higher level of genes by up to two orders of magnitude (Table 4). The hydrophobicity of the viral envelope can play an important role in explaining such concentration of the solid matrix in the wastewater treatment process [24]. The higher levels for sludge than those obtained for sewage and the fact that RNA can be directly isolated from sludge without a concentration step suggest that the solid matrix in the WWTP is a feasible indicator for SARS-CoV-2 surveillance.
Variations on the positivity detection among SARS-CoV-2 genes on time has also been reported by other surveillance studies in wastewater [7,8]. Such variation was associated with the limit of detection (LOD) for the different genes, being 3.6, 10 and 10 copies per qRT-PCR assay for RdRP, N and S genes. N and S genes were reported for commercial kits similar to this study [[20], [21], [22], [23], [24], [25], [26], [27]]. However, for wastewater samples analysis, LOD's determination needs to consider the concentration and extraction step, already discussed (section 3.1). The LOD was determined for the nucleocapsid region at 4.45 log10 copies per liter of wastewater [8], but no similar studies have been conducted for RdRp and S genes.
Regarding effluent samples, none of them were positive for the SARS-CoV-2 genes analyzed, in agreement with previous work in which most of the results for secondary treatment effluent were negative for different N genes [8]. Notably, the effluent from WWTP SRJ was not disinfected because of operational problems in the chlorination unit during the sampling period. Nevertheless, no residual SARS-CoV-2 RNA was detected, indicating that the gene fragments were removed during the wastewater treatment process.
3.3. Surveillance of SARS-CoV-2 in WWTP
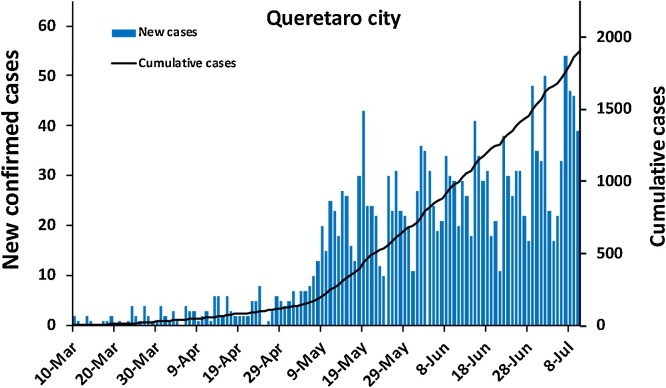
The first COVID-19 case in Santiago de Queretaro city was confirmed on March 10th; 44 days later, sampling at the two WWTP in the city for this work began. To evaluate the correlation between SAR-CoV-2 concentration in wastewater and COVID-19 cumulative cases, a weekly sampling for 70 days was carried out, matching with the exponential COVID-19 spread in the city (Fig. 3 ). In the first three sampling days, only two samples resulted in positive quantification of the RdRP and S genes: an influent and a sludge sample. From May 14th onwards, positive and increasing quantification of the S and N and RdRP genes was observed in influent (Table 4).
Fig. 3.
New and confirmed COVID-19 cases in Santiago de Queretaro since the first confirmed case on March 10th. Data retrieved from the COVID-19 dashboard from Mexican health authorities (https://coronavirus.gob.mx/datos/#DownZCSV).
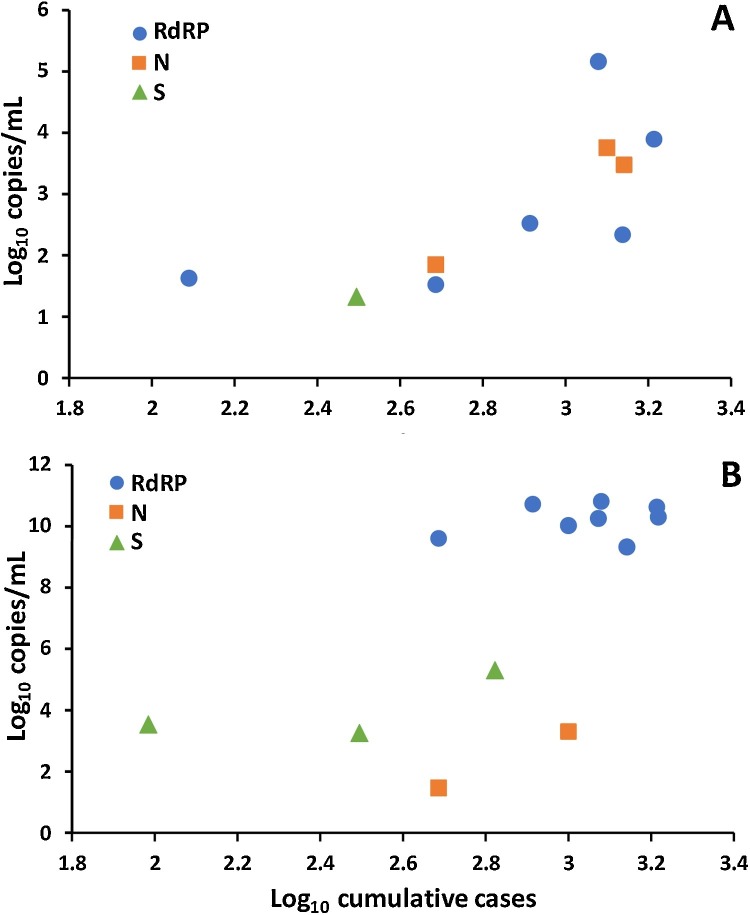
A correlation analysis showed a precise match of increments in different gene concentrations in influent to the cumulative COVID-19 cases reported by health authorities (Fig. 4 A). Similar positive correlations between SAR-CoV-2 RNA in sewage and COVID-19 cases have been reported in other countries, such as the Netherlands and USA [7,26]. No correlation between SARS-CoV-2 RNA in secondary sludge and COVID-19 confirmed cases was observed; the gene concentration was similar regardless of the cumulative cases (Fig. 4B). This sustained RNA detection in sludge is assumed to be due to migration of the genetic material among different matrices during the wastewater treatment process, in this case, virus affinity to solids. A previous study analyzing human coronavirus in sewage concludes that the organic matter and suspended solids protect viruses; the hydrophobicity of the coronaviruses’ envelope makes them more readily adhere to solids. Then, this absorption mechanism can be a removal route of viruses by solids settling [24]. In this regard, our study suggests that influent is the most suitable matrix to quantify the RNA of SARS-CoV-2 for surveillance studies, unlike secondary sludge. But analyzing secondary sludge can help to elucidate the mechanism of virus removal in WWTP.
Fig. 4.
SARS-CoV-2 RNA quantification (log10 copies/mL) as a function of cumulative COVID-19 confirmed cases (log10 cumulative cases) in Santiago de Queretaro. A) WWTP influent and B) secondary sludge.
The presence of RNA of SARS-CoV-2 in secondary sludge leads to new concerns. According to WHO, the potential infectious disease risks of excreta, including the possible presence of SARS-CoV-2, sewage and sludge, must be treated in well-designed and managed treatment plants. Also, it is suggested that there is no evidence on the survival of infectious SARS-CoV-2 viruses in wastewater; therefore, conventional wastewater treatment processes should inactivate enveloped viruses, including SARS-CoV-2 [28]. However, recently Bivins et al. [29] reported the persistence of infectivity of inoculated SARS-CoV-2 in wastewater samples. They estimated that the time for a 90 % reduction (T90) of viable SARS-CoV-2 virus in wastewater at room temperature was 1.5 days. Even though viral genetic material is more persistent in wastewater (T90 = 3.3 days), the authors suggest that the detection of SARS-CoV-2 RNA in wastewater alone does not justify the risk of infection, considering that the study has the limitation that was conducted in wastewater samples previously frozen and thawed and then inoculated with relative high virus titers. The same authors suggest complementary studies to evaluate the influence of pH, solids content, and mixing conditions, factors that may affect the viability of the virus. Further studies are needed to elucidate the migration and transport of virus fragments from the liquid phase to secondary sludge, and the removal mechanisms.
Among medical diagnostic testing for COVID-19 in OECD countries, Mexico has the lowest rate (0.6 tests per 1000 population), which is much lower than the average of 27.7, though testing is identified as a tool for reducing the risk for a new outbreak [30]. Recently, a second COVID-19 outbreak has been reported in Europe. In countries such as Spain and France, the number of reported new cases in late August is similar to that during the first pandemic peak in April [1]. This scenario is expected to occur in the Americas. Considering this scenario, proving the feasibility of SARS-CoV-2 surveillance in wastewater for developing countries such as Mexico is a valuable contribution as a sensitive tool for assessing virus spread in the population, with cost savings compared to medical testing [11].
4. Conclusions
This work is the first report proving the use of wastewater-based epidemiology in Mexico as a feasible tool to detect the prevalence of COVID-19 using the RdRP, N, and S genes of SARS-CoV-2. Comparing two different RNA concentration methods, ultrafiltration and adsorption based, the latter resulted in higher RNA levels, as explained by the affinity of the virus material to the solid fraction from sewage. The method had sufficient sensitivity to detect SARS-CoV-2 RNA from the sewer system in locations with reported COVID-19 cases, such as a COVID-19 hospital and quarantine center, and presumably isolated locations such as correctional and court facilities. Additionally, the surveillance of SARS-CoV-2 RNA in the influent of two WWTP showed a significant matching to the cumulative confirmed cases in Queretaro city. The secondary sludge from the WWTP resulted in RNA levels eight orders of magnitude higher than those in influent, suggesting migration of the genetic material from the liquid to solid matrix in the wastewater treatment process. All the effluent samples were negative for virus quantification indicating that SARS-CoV-2 RNA is removed by WWTP.
CRediT authorship contribution statement
Julián Carrillo-Reyes: Conceptualization, Methodology, Validation, Formal analysis, Writing - original draft, Writing - review & editing. Martín Barragán-Trinidad: Methodology, Formal analysis, Investigation, Writing - review & editing. Germán Buitrón: Conceptualization, Methodology, Validation, Investigation, Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare no conflict of interest to disclose.
Acknowledgments
This work was supported by the Institute of Engineering of the Universidad Nacional Autónoma de México [grant number 0406]. We acknowledge the Water Council from Queretaro for their support during sampling in the sewer system. The authors of this paper acknowledge the European Virus Archive Global (EVA-GLOBAL), that has received funding from the European Union’s Horizon 2020 (No 871029), for providing SARS-CoV-2 gene controls.
References
- 1.WHO . 2020. WHO Coronavirus Disease (COVID-19) Dashboard | WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/table accessed 24 July. [Google Scholar]
- 2.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ningthoujam R. COVID 19 can spread through breathing, talking, study estimates. Curr. Med. Res. Pract. 2020;10:132–133. doi: 10.1016/j.cmrp.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C., Yang J., Ye G., Cheng Z. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amirian E.S. Potential fecal transmission of SARS-CoV-2: current evidence and implications for public health. Int. J. Infect. Dis. 2020;95:363–370. doi: 10.1016/j.ijid.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 8.Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellmér M., Paxéus N., Magnius L., Enache L., Arnholm B., Johansson A., Bergström T., Norder H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis a virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014;80:6771–6781. doi: 10.1128/AEM.01981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Tot. Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lesimple A., Jasim S.Y., Johnson D.J., Hilal N. The role of wastewater treatment plants as tools for SARS-CoV-2 early detection and removal. J. Water Process Eng. 2020;38 doi: 10.1016/j.jwpe.2020.101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SS . 2020. Lineamiento Estandarizado Para La Vigilancia Epidemiológica Y Por Laboratorio De La Enfermedad Respiratoria Viral.http://www.gob.mx/salud/documentos/lineamiento-estandarizado-para-la-vigilancia-epidemiologica-y-por-laboratorio-de-la-enfermedad-respiratoria-viral [Google Scholar]
- 14.Gudbjartsson D.F., Helgason A., Jonsson H., Magnusson O.T., Melsted P., Norddahl G.L., Saemundsdottir J., Sigurdsson A., Sulem P., Agustsdottir A.B., Eiriksdottir B., Fridriksdottir R., Gardarsdottir E.E., Georgsson G., Gretarsdottir O.S., Gudmundsson K.R., Gunnarsdottir T.R., Gylfason A., Holm H., Jensson B.O., Jonasdottir A., Jonsson F., Josefsdottir K.S., Kristjansson T., Magnusdottir D.N., le Roux L., Sigmundsdottir G., Sveinbjornsson G., Sveinsdottir K.E., Sveinsdottir M., Thorarensen E.A., Thorbjornsson B., Löve A., Masson G., Jonsdottir I., Möller A.D., Gudnason T., Kristinsson K.G., Thorsteinsdottir U., Stefansson K. Spread of SARS-CoV-2 in the icelandic population. N. Engl. J. Med. 2020;382:2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ampuero M., Valenzuela S., Valiente-Echeverria F., Soto-Rifo R., Barriga G.P., Chnaiderman J., Rojas C., Guajardo-Leiva S., Diez B., Gaggero A. SARS-CoV-2 detection in sewage in Santiago, chile - preliminary results. MedRxiv. 2020 doi: 10.1101/2020.07.02.20145177. 2020.07.02.20145177. [DOI] [Google Scholar]
- 16.Fongaro G., Stoco P.H., Souza D.S.M., Grisard E.C., Magri M.E., Rogovski P., Schorner M.A., Barazzetti F.H., Christoff A.P., de Oliveira L.F.V., Bazzo M.L., Wagner G., Hernandez M., Rodriguez-Lazaro D. SARS-CoV-2 in human sewage in Santa Catalina, Brazil, November 2019. MedRxiv. 2020 doi: 10.1101/2020.06.26.20140731. 2020.06.26.20140731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerrero-Latorre L., Ballesteros I., Villacrés-Granda I., Granda M.G., Freire-Paspuel B., Ríos-Touma B. SARS-CoV-2 in river water: implications in low sanitation countries. Sci. Tot. Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.APHA, AWWA, WFE . 21st ed. American Public Health Association; 2005. Standard Methods for the Examination of Water & Wastewater. [Google Scholar]
- 19.Ahmed W., Harwood V.J., Gyawali P., Sidhu J.P.S., Toze S. Comparison of concentration methods for quantitative detection of sewage-associated viral markers in environmental waters. Appl. Environ. Microbiol. 2015;81:2042–2049. doi: 10.1128/AEM.03851-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Tot. Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherchan S.P., Shahin S., Ward L.M., Tandukar S., Aw T.G., Schmitz B., Ahmed W., Kitajima M. First detection of SARS-CoV-2 RNA in wastewater in North America: a study in Louisiana, USA. Sci. Tot. Environ. 2020;743 doi: 10.1016/j.scitotenv.2020.140621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Tot. Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gundy P.M., Gerba C.P., Pepper I.L. Survival of coronaviruses in water and wastewater. Food Environ. Virol. 2008;1:10. doi: 10.1007/s12560-008-9001-6. [DOI] [Google Scholar]
- 25.Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Tot. Environ. 2020;737 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J. Advanced Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.WHO . 2020. Water, Sanitation, Hygiene, and Waste Management for SARS-CoV-2, the Virus That Causes COVID-19.https://www.who.int/publications-detail-redirect/WHO-2019-nCoV-IPC-WASH-2020.4 [Google Scholar]
- 29.Bivins A., Greaves J., Fischer R., Yinda K.C., Ahmed W., Kitajima M., Munster V.J., Bibby K. Persistence of SARS-CoV-2 in water and wastewater. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.OECD . 2020. Testing for COVID-19: a Way to Lift Confinement Restrictions.https://www.oecd.org/coronavirus/policy-responses/testing-for-covid-19-a-way-to-lift-confinement-restrictions-89756248/ [Google Scholar]