Abstract
The RNA-dependent RNA polymerase (RdRp) is a key enzyme which regulates the viral replication of SARS-CoV-2. Remdesivir (RDV) is clinically used drug which targets RdRp, however its mechanism of action remains elusive. This study aims to find out the binding dynamics of active Remdesivir-triphosphate (RDV-TP) to RdRp by means of molecular dynamics (MD) simulation. We built a homology model of RdRp along with RNA and manganese ion using RdRp hepatitis C virus and recent SARS-CoV-2 structures. We determined that the model was stable during the 500 ns MD simulations. We then employed the model to study the binding of RDV-TP to RdRp during three independent 500 ns MD simulations. It was revealed that the interactions of protein and template-primer RNA were dominated by salt bridge interactions with phosphate groups of RNA, while interactions with base pairs of template-primer RNA were minimal. The binding of RDV-TP showed that the position of phosphate groups was at the entry of the NTP channel and it was stabilized by the interactions with K551, R553, and K621, while the adenosine group on RDV-TP was pairing with U2 of the template strand. The manganese ion was located close to D618, D760, and D761, and helps in stabilization of the phosphate groups of RDV-TP. Further we identified three hits from the natural product database that pose similar to RDV-TP while having lower binding energies than that of RDV-TP, and that SN00359915 had binding free energy about three times lower than that of RDV-TP.
Keywords: COVID-19, SARS-CoV-2, RdRp, Remdesivir, Coronavirus, Molecular dynamics simulation
Graphical abstract
Highlights
-
•
A homology model of RdRp, RNA, and manganese ion was built.
-
•
Remdesivir was docked to RdRp and was stable for 500 ns MD simulations.
-
•
The binding of remdesivir was stabilized by the interactions with key residues.
-
•
Three hits had lower binding energies than that of remdesivir.
1. Introduction
Coronavirus 2019 (COVID-19) is a disease due to the SARS-CoV-2 virus which presently threatens human life across the globe. It is highly contagious and has affected the lives of millions of people with over one million deaths as of October 2020. There is therefore an urgent need to discover effective therapeutics for eliminating the disease considering its detrimental effects to human life.
SARS-CoV-2 is a positive-strand RNA virus whose genomic structure expresses high similarity with previous severe acute respiratory syndrome coronavirus (SARS-CoV). Like other coronaviruses, the proliferation of the SARS-CoV-2 requires non-structural protein 12 (nsp12) (RNA-dependent RNA polymerase, RdRp), a key enzyme which regulates the viral genome replication and transcription. It is thus the validated target for the development of COVID-19 disease therapeutics. Remdesivir (RDV) is a clinically-used drug which targets RdRp and its efficacy against Ebolavirus (EBOV) infection is well-understood [1,2], SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS) [[3], [4], [5], [6]] has also been previously reported. Due to the high sequence similarity of RdRp structures of SARS-CoV-1 and SARS-CoV-2 (96% sequence identity), it has been therefore approved for emergency use of treating SARS-CoV-2 virus [7].
Remdesivir is a prodrug and nucleoside analogue which functions after biotransformation into its active triphosphate form (RDV-TP). It works as a substrate mimic of ATP, which is a substrate for RdRp for nucleotide addition on the primer RNA strand. However, RDV has been proposed to work differently during chain translocation. Several works proposed that the RDV induced delayed chain termination [8,9], however, its mechanism of action is not completely understood [10,11]. This study aims to elucidate the mechanism of action and dynamics of the RDV binding into RdRp by means of molecular dynamics (MD) simulation. The MD simulation approach has been widely accepted to get invaluable insight into drug binding in a specific target at the molecular level.
Previous computational studies on the interaction between Remdesivir and SARS-CoV-2 RdRp were reported. For instance, studies by Elfiky (2020a, 2020b) [12,13] indicated the strong binding of Remdesivir to RdRp. Kato et al. (2020) described intermolecular interactions between Remdesivir and SARS-CoV-2 RdRp by employing fragment molecular orbital calculations, which indicated that T687, N691, and D760 are critical residues [14]. Meanwhile, a study by Zhang and Zhou (2020) described the inhibitory mechanism of Remdesivir to SARS-CoV-2 RdRp [15]. However, the previous studies lacked of the RNA template-primer in their RdRp complex models, which is crucial in the nucleotide triphosphate binding. Here we reported the Remdesivir interactions with the RdRp complex along with RNA template-primer.
We firstly built the RdRp complex along with the primer RNA, template RNA, and manganese ions prior to docking RDV-TP and ATP to the RdRp model. We then employed the RDV-TP bound to the RdRp model to identify molecular hits of potential RdRp inhibitors from the natural product database through pharmacophore screening. Natural products are continuously showing their importance in drug discovery and development as up to 80% of drugs including those that inhibit viral replication approved by the Food and Drug Administration between 1981 and 2014 are linked to natural products [16]. For instance, catechin and epigallocatechin gallate (EGCG) were reported to have antiviral activity against SARS-CoV and MERS-CoV, respectively [17]. We validated the binding of our natural product hits using 200 ns MD simulation for each compound and performed MM-PBSA energy calculations to evaluate their affinities to RdRp. Interestingly, the three molecular hits had lower binding affinities than that of RDV-TP.
2. Experimental section
2.1. Homology modeling and ligand preparation
The RdRp model was constructed using Maestro's homology modeling tools [18]. The electron microscopy solved structure of SARS-CoV-2 non-structural protein 12 (NSP 12, PDB ID: 6M71) [19] and X-ray solved structure of RNA primer template hepatitis C virus NS5B (4WTG) [20] was employed. The charge state of the RdRp complex was optimized at pH = 7. The complex was further preprocessed, optimized, and minimized with Maestro's protein preparation wizard tool [18].
Using the prepared RdRp structure, a receptor grid file was generated around the RDV-TP binding site and the ionization/tautomeric state of RDV-TP was generated at pH = 7 using Maestro's Epik tool [21,22]. The ligand was then docked to the receptor using the Glide XP docking [23,24] scoring function. The MD simulation was then performed to check the binding dynamics of RDV-TP.
For comparison and validation of both our constructed RdRp complex and modeling methods for accuracy and consistency, we utilized the recently deposited crystal complex of RNA polymerase nsp12-nsp7-nsp8 with bound template-primer RNA and RDV-TP (PDB ID: 7BV2) [25]. After docking, we performed a sequence alignment of our newly bound polymerase-RDV-TP complex onto 7BV2, including the RNA template-primer and RDV-TP models and corresponding metal ions for best comparison. Figure S1 shows that our model is consistent with the geometry of 7BV2 in regards to partial protein conformations, RNA template-primer shape and RDV-TP binding poses. This information encouraged us to further continue with this model in the future simulations.
Pharmacophore screening was performed using active Remdesivir (Figure S2) against 325,187 natural product molecules [26] by means of Ligand Scout 4.3 [27]. In our pharmacophore model, we defined seven pharmacophore features including two negative ionizable, one aromatic ring and four H-bond acceptors and their corresponding positions. The natural origin compounds were our greatest priority in the present study since they exhibited inhibition to the RdRp of other RNA viruses, high bioavailability, low toxicity, and received greater importance for their antiviral applications [28,29]. Before docking, the ionization/tautomeric states of each compound were generated at pH = 7 using Maestro's Epik tool [21,22]. The lowest ionization/tautomeric states were selected, and then their geometries were minimized to their most energetically favorable states. To check if our docked ligand poses were reasonable, we compared the docked poses of the 65 ligands with the RDV-TP pose. Clearly, the 65 ligands bind similarly to the binding pocket of RDV-TP but with some differences. This provided a reasonable starting pose for future MD simulations which could further refine the binding pose given the full conformation flexibility from the simulations.
2.2. MD system setup
Several MD simulations were performed: one 500 ns simulation for the apo-form of RdRp, three 500 ns simulations for the RdRp-RDV-TP complex, three 500 ns simulations for the ATP-RdRp complex, and a 200 ns simulation for each complex of SN00166900, SN00303170, and SN00359915. MD simulations were conducted by using AMBER16 software [30]. The AMBER ff14SB [31], RNA.OL3 [32,33], and GAFF2 [34] forcefields were applied for protein, RNA and ligands, respectively. A salt concentration of 0.15 M was applied to each system and were placed in a truncated octahedron TIP3P water box with a 10 Å radius. Each system underwent a 10000 step minimization three times, which consisted of 500 steps of steepest descents and 9500 conjugate gradients each. Then, minimizations were applied to each system to relax ion and water molecular positioning, with second and third minimizations being conducted with and without protein backbone atoms restraint, respectively. Each system was heated from 0 to 100, 100–200, and 200–300K, with each undergoing 50 ps under NVT ensemble with a time step of 0.0005 ps and a 5 kcal mol−1 A−2 restraint. Each system was then allowed three steps of relaxation under NPT ensemble, each with and without 3 kcal mol−1 A−2, 1 kcal mol−1 A−2 restraints, respectively.
The Langevin thermostat with a collision rate of 1.0 ps−1 was applied to all covalent bonds involving hydrogen atoms and were restrained using SHAKE algorithm [35]. The PME method was employed to treat long-range electrostatic interactions with an integration step of 2 fs [36]. The long-range non-bonded interactions were calculated with a cutoff distance of 9.0 Å by applying periodic boundary conditions.
To check the convergence of MD simulations, we calculated the RMSD values of protein Cα, RNA backbone, and ligand main atoms for each trajectory by employing the CPPTRAJ module [37]. In addition, the root mean square fluctuations (RMSF) of each individual residue of the protein, RNA and individual atoms of the ligands were calculated to characterize the local structural fluctuations in each complex.
Clustering analysis was employed to group similar RdRp-ligand structures with ions and solvent molecules being omitted. The backbone RMSD matrix was used as the metric for similarity, and the clustering method was accessed by DBSCAN [38] via CPPTRAJ module. The distance cutoff (epsilon) was 2.5 and minpoints 15, and the most abundant clusters were used for analysis. For the RDV-TP complex, the last 200 ns of simulation data were used, while for complex of SN00166900, SN00303170, and SN00359915, the last 100 ns of simulation data were used for clustering analysis.
The binding energy of ligands were calculated using the Molecular Mechanics-Poisson Boltzmann solvent accessible surface area (MM-PBSA) method [39] via the MMPBSA.py module. Trajectories of 200 snapshots of the last 100 ns of each complex was used for calculations.
3. Results and discussion
3.1. Homology model of RdRp complex
Before constructing the RdRp complex model, sequence alignment between SARS-CoV-2 non-structural protein 12 (NSP 12, PDB ID: 6M71) and RNA primer template hepatitis C virus NS5B (4WTG) was conducted. Superposition of the modelled and crystallographic structures is depicted (Figure S3-A), which shows that both structures align well with each other, particularly for amino acid residues 1–541. The interactions of protein and template-primer RNA mostly originated from contributions in the RNA phosphate group. Hydrogen-bonding interactions occurred with S501, R569, S592, Y595, and Y689, while salt bridge interactions occurred with K500, K577, and R583 (Figure S3–B). The template-primer RNA base pair interactions with the protein occurred between Uracil and S682 (H-bond) and K500 (pi-cation) (Figure S3, B and C). This indicated that the elongation step of RdRp function is not sequence specific which was also implied in the previous study [25].
During the MD simulations, we monitored the H-bond occupancy between RNA and protein during the 500 ns timescale (Table S2). It showed that the high H-bond occupancies occurred between U5 (U554) primer strand and U2 (U543) of the template strand with S814 (S445) and S682 (S313) occupancies of 96.87% and 82.53%, respectively. The modest occupancies occurred in many H-bonds interactions; for instance, those between A3 (A544) of the template strand with D684 (D315) (69.16%), A4 (A545) with R569 (R200) (66.03%), and U8 (U549) of template strand with Y595 (Y226) (58.40%). In addition, various H-bond interactions between protein and RNA and between RNA bases were observed to have low occupancies (Table S2).
3.2. The binding of remdesivir and ATP to RdRp complex
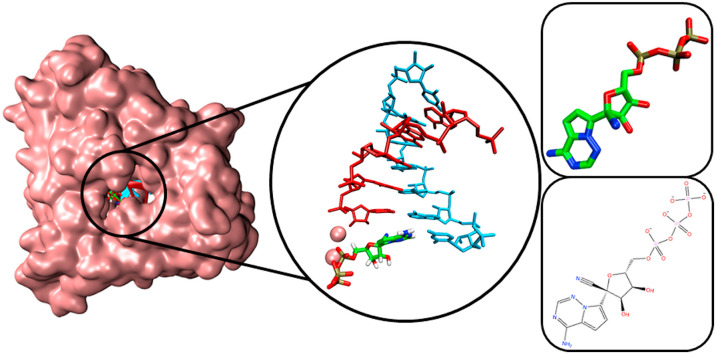
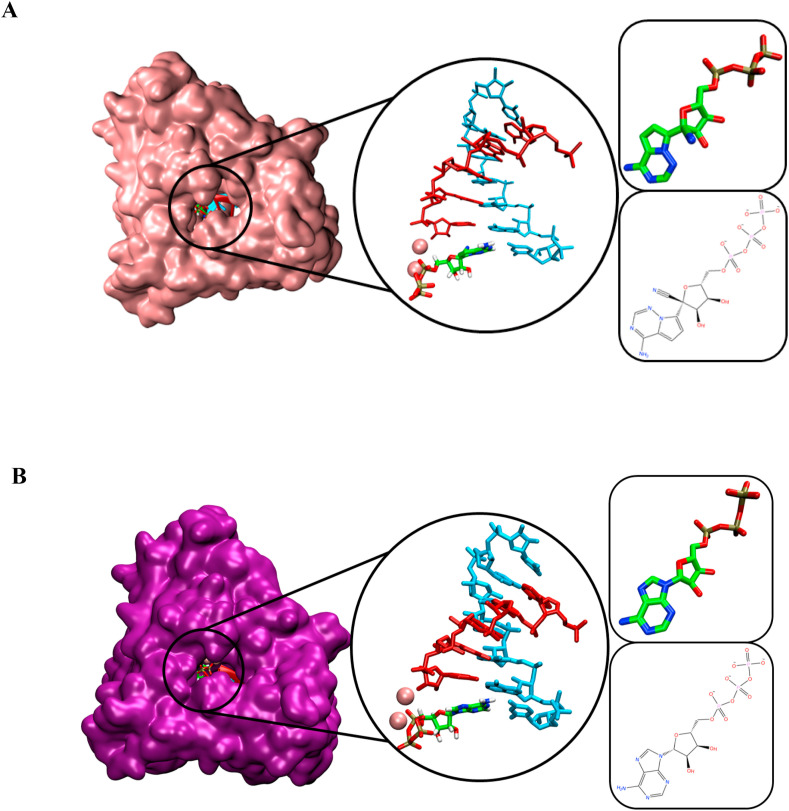
RDV-TP was docked at the i position against RdRp SARS-CoV-2 model which contains 6 bases in the primer strand, 8 bases in the template strand, and a manganese ion (Fig. 1 A). The conformation of RDV-TP was identical with the ATP pose (Fig. 1B), with both RDV-TP and ATP were located at the catalytic site of RdRp. The position of the phosphate group at the NTP entrance channel may reflect its role in inhibiting the entry of NTP to the active site.
Fig. 1.
The 3D poses of RDV-TP (A) and ATP (B) in the RdRp SARS-CoV-2 model. Primer and template strands are colored red and cyan colors, respectively.
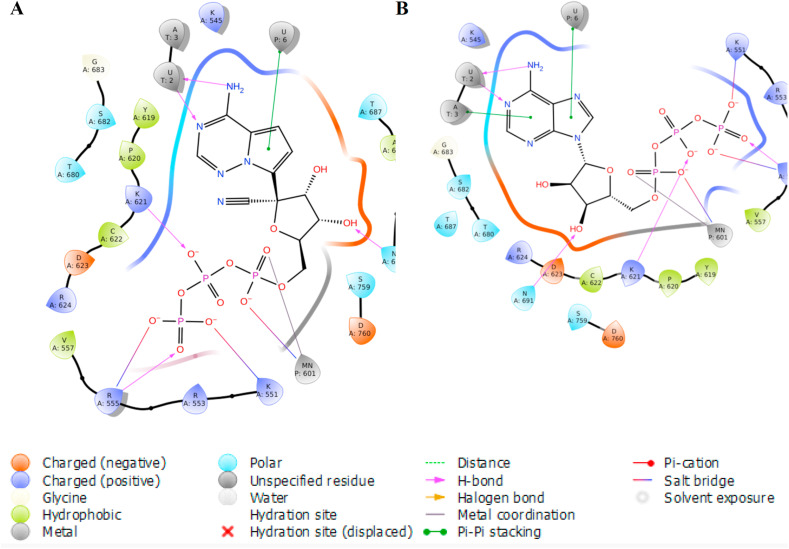
The detailed interactions of docked RDV-TP and ATP with RdRp are depicted (Fig. 2 ). Both phosphate groups of RDV-TP and ATP formed H-bond interactions with positively charged amino acid residues K621 and R555, while salt bridge interactions formed with K551 and R555. The phosphate groups of both ligands were additionally stabilized by manganese ions through metal coordination and salt bridge interactions. The adenosine motifs of both RDV-TP and ATP established H-bond interactions with U2 of template strands and pi-pi stacking interactions with U6 of primer strands. Additional pi-pi stacking interaction were formed with A3 of the template strand in the ATP conformation. Interactions with N691 were formed in both RDV-TP and ATP through their 3′-OH groups. In both complexes, D623 and D760 were located close to phosphate and manganese ions, while the nitrile group of RDV-TP was surrounded by T687 and A688. Clearly, the docked poses of both RDV-TP and ATP were almost identical.
Fig. 2.
The detailed interaction of docked RDV-TP (A) and ATP (B) with RdRp.
During the 500 ns simulations, several H-bond interactions were conserved. The RDV-TP H-bond interactions occurring with U2 (U543) of template strand were retained at high occupancy (75.39%), while that occurring with D760 (D391) was maintained at modest occupancy (48.5%). In contrast, the H-bond with R555 (R186) was maintained at low occupancy (23.62%). Numerous H-bond interactions between RDV-TP and RdRp complex occurred with low occupancies (Table S3). In the case of ATP, the H-bond occupancies observed in the docked conformation were low such as those occurring between ATP and R555 (R186) (33.87%) and K551 (K182) (12.91%). Clearly, the RDV-TP was more capable at maintaining base pairing particularly with U2 (U543) of template strand. RDV-TP had more intense H-bond interactions during the MD simulation as compared with its ATP counterpart. The H-bond occupancies of ATP complex during 500 ns MD simulation are tabulated in Table S4.
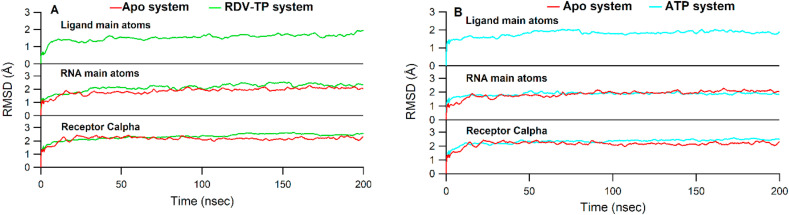
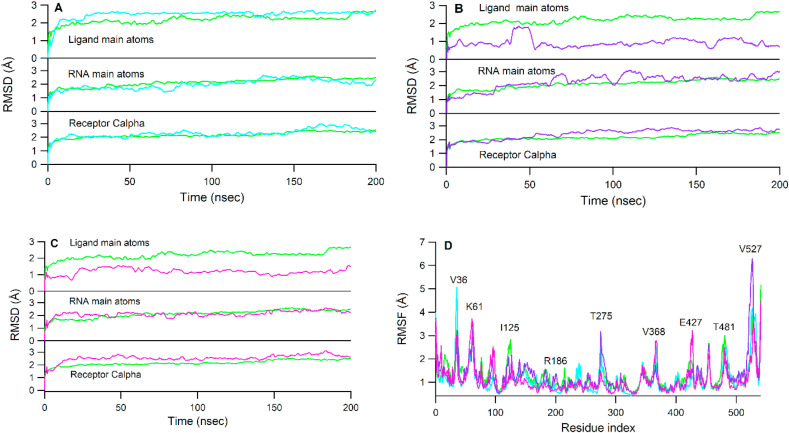
The trajectory convergence during MD simulation was checked through the RMSD values averaged over three independent 500 ns MD simulation for backbone receptor Cα, RNA and ligand main atoms for apo-RdRp, ATP, and RDV-TP complexes (Fig. 3 ). It is clear in Fig. 3 that the receptor backbone quickly reached stability before 100 ns and remained stable throughout the simulation time for both RDV-TP and ATP systems. The RMSD of RNA main atoms showed greater fluctuations and reached stability at ~280 ns in both RDV-TP and ATP, while the average fluctuation during the whole simulation time was ~2 Å. The ligand RMSD of RDV-TP reached stability at ~50 ns, while those of the ATP system occurred at an earlier simulation time (~100 ns). The RMSD values of both RDV-TP and ATP systems were higher than that of the apo system, which indicated that the binding of RDV-TP and ATP induced greater fluctuations in RdRp complex during 500 ns simulation time. The RMSD values for the first, second and third 500 ns MD simulations of ATP and RDV-TP show similar patterns (Figure S4).
Fig. 3.
RMSD values for the receptor Cα, RNA main atoms and ligand main atoms averaged over three 500 ns runs for apo RdRp and RDV-TP (A) and apo RdRp and ATP (B). Apo, RDV-TP, and ATP were colored red, green, and cyan colors, respectively.
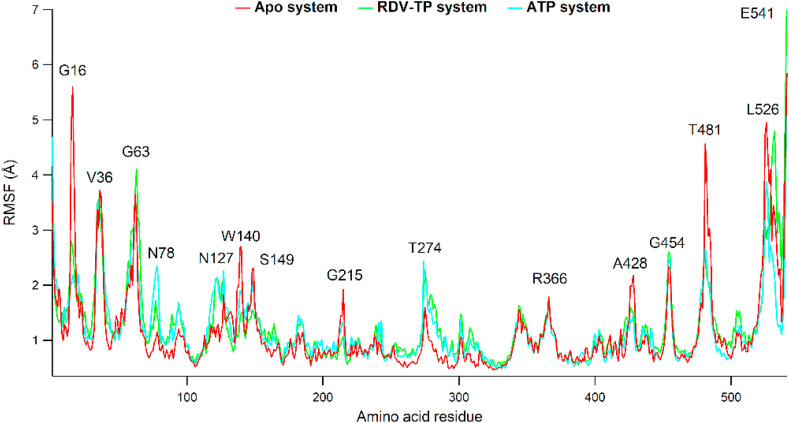
To observe the fluctuations of amino acid residues, RNA residues, ATP and RDV-TP atoms during MD simulation, the root mean square fluctuation (RMSF) was recorded (Fig. 4 ). As shown in Fig. 4, the high peaks were observed at G16 (G385), V36 (V405), G63 (G432), N78 (N447), N127 (N496), W140 (W509), S149 (S518), G215 (G584), T274 (T644), R366 (R735), A428 (797), G454 (G823), T481 (T850), which corresponded to loop regions of the protein, while L526 (L895) and E541 (E919) corresponded to the C-terminals of the protein. Residues interacting with RDV-TP such as K551 (K182), R555 (R186), D623 (D254), N691 (N322), and D760 (D391) were observed to be stable.
Fig. 4.
RMSF plot of protein, in which apo RdRp, RDV-TP, and ATP complexes are colored red, green, and cyan colors, respectively.
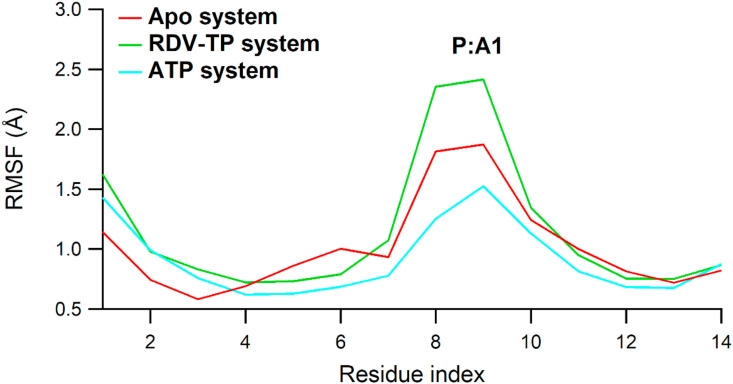
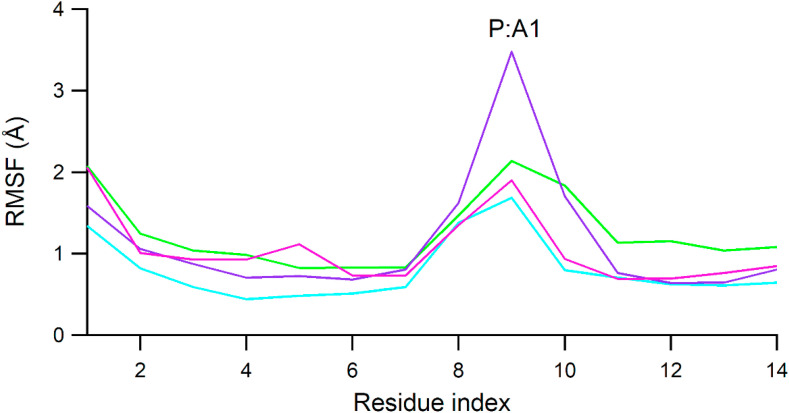
Meanwhile, RMSF of the RNA main atoms (Fig. 5 ) was highest at A1 (~2.5 Å) which is the first sequence of primer strand and did not interact directly with both RDV-TP and ATP. It is worth noting that the RMSF of U2 and A3 of template strands as well as U6 of primer stands in both RDV-TP and ATP systems were observed to be stable around 1 Å.
Fig. 5.
The RMSF plot of RNA main atoms. Apo RdRp, RDV-TP, and ATP complexes are colored red, green, and cyan colors, respectively.
The RMSF of ligand atoms of both RDV and ATP system are depicted (Figure S5). The high peaks were recorded at O9, O10, and O11 (i.e. the oxygen atoms at γ phosphate group of RDV-TP and ATP). Atoms of the adenosine motif of RDV-TP fluctuated less than those of ATP system, which may indicate that base pairing of RDV-TP occurred with more intense interaction compared with the ATP system. Figures S6, S7, and S8 show the RMSF values for each three independent 500 ns MD simulation.
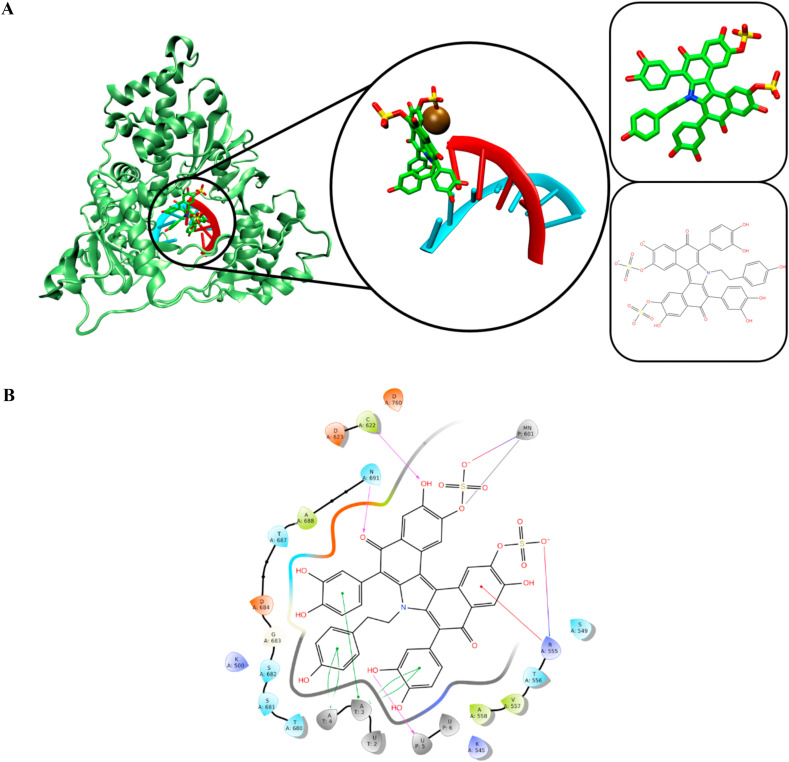
We then performed clustering analysis to generate structural representatives of the most abundant clusters for the 500 ns simulations. The most abundant clusters of RDV-TP and ATP complexes had 100% occupancy. The adenosine motif of RDV-TP interacted with U2 (U543) of template strand via H-bonding, which is absent in ATP interactions. The phosphate groups of RDV-TP and ATP interacted with positively charged residues such as K182 (K551), R184 (R553), and K252 (K621). The manganese ions were also located in the vicinity of the phosphate groups of RDV-TP and ATP, which indicated its role in ligand stabilization. Additionally, the manganese ions were also in close proximity with aspartate residues such as D249 (D618), D391 (D760), and D392 (D761). The nitrile group of RDV-TP was observed to be located in the empty space between C253 (C622), P251 (P620), and V188 (V557).
Our study is partly consistent with several previous studies. For instance, Hillen et al. (2020) implied that N691, S682 and D623 may recognize the 2′-OH group of NTP, thereby rendering the RdRp specific for the synthesis of RNA rather than DNA [40]. Yin et al. (2020) indicated that K545, R555, S682, N691, and D760 supported Remdesivir binding [25], while Zhang and Zhou (2020) implied that D618, S549, and R555 are major contributors to Remdesivir binding [15].
In addition, previous reports by Nudler (2009) and Burton et al. (2005) indicated that the substrate was pairing with the RNA template after entering through the main channel of the RdRp complex [41,42]. Its structural resemblance to natural ATP enables it to compete with ATP during viral synthesis [43]. We confirmed the interaction of U6 of primer strand at i site and RDV-TP at i+1 site and hence we speculated that the interaction would lead to delayed chain termination in i+3 or i+5 sites. Study by Gordon et al. (2020) and Tchesnokov et al. (2019) implied that the inhibition of the viral nucleic acid synthesis occurred through the delayed chain termination mechanism, in which following incorporation of RDV-TP, several more NTP molecules could still be added to RNA [6,8]. In brief, our study provides the details at an atomistic level on how Remdesivir behaves during its interaction with RdRp, which may be useful for finding more potent compounds for treating COVID-19 through RdRp targeting.
We then applied pharmacophoric screening against the natural database using pharmacophore features. Screening against natural product database using seven pharmacophoric features, i.e. two negative ionizable, one aromatic ring, and four H-bond acceptors (Figure S2), retrieved 56 molecular hits. We proceeded to select the three best hits for a 200 ns molecular dynamics simulation for each. The three best docked hits are subjected for 200 ns MD simulation, i.e. SN00166900, SN00303170, and SN00359915.
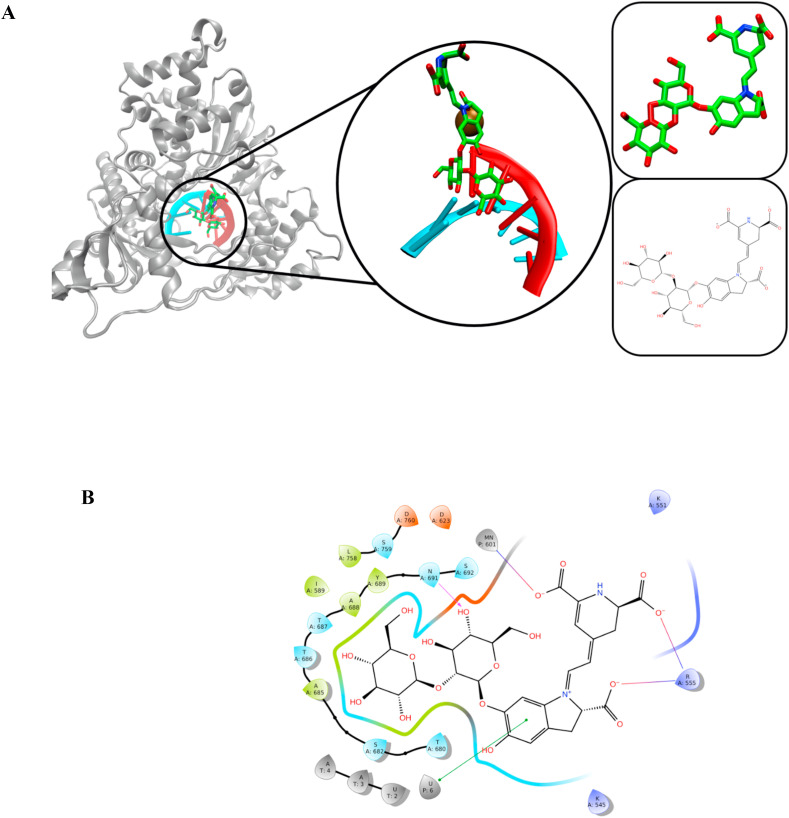
The best docked hit molecules show the similar poses with RDV-TP. The oxane group of SN00166900 was seen to sit after U6 of primer strand and its phenyl ring established pi-pi interactions with U6. More occurrences with N691 and R555 were seen through H-bond and salt bridge interactions. Additionally, the manganese ion interacted with OH group of ligand through salt bridge interactions. Fig. 6 displays the interaction of SN00166900 with RdRp.
Fig. 6.
The interaction of SN00166900 with RdRp complex. Primer and template strands are colored red and cyan colors, respectively.
Meanwhile, the SN00303170 pose was also located in the U6 of primer strand and the pi-pi interaction was recorded between its phenyl ring and U6. The residues R555, N691, and manganese ion were also observed to interact with SN00303170 through H-bond and salt bridge interactions. Fig. 7 depicts the conformation of SN00303170 in the RdRp complex.
Fig. 7.
The interaction of SN00303170 with RdRp complex. Primer and template strands are colored red and cyan colors, respectively.
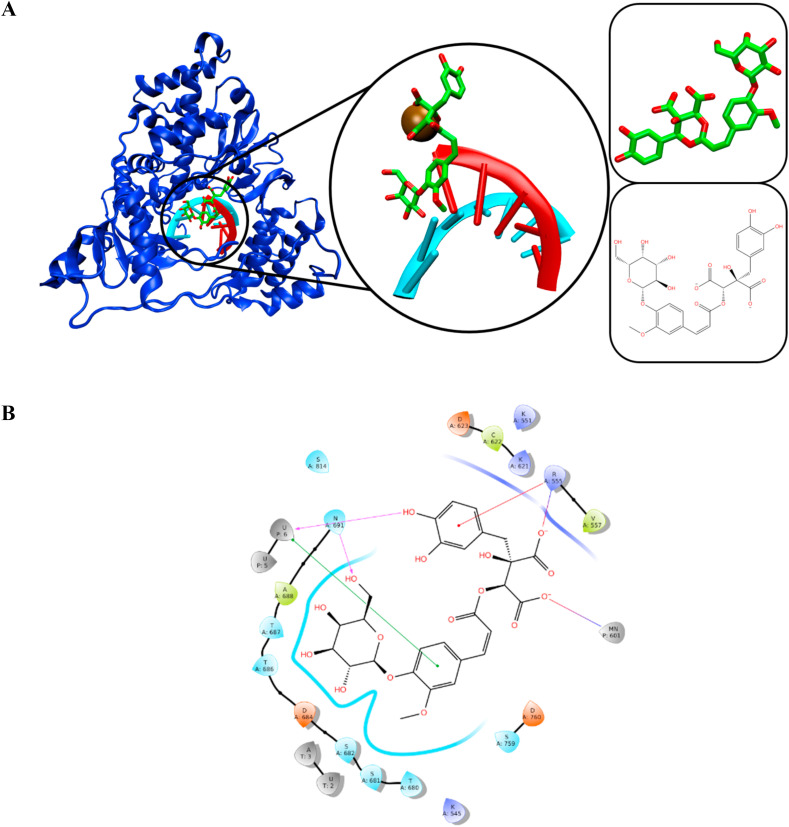
The docked pose of SN00359915 was similar to other ligands, in which it occupied the space above the U6 primer strand. The pi-pi stacking interaction in SN00359915 binding occurred at A3 and A4 of the template strand. The H-bond interactions were recorded between the ligand with R555, N691, and C622. The manganese ion also formed a salt bridge interaction with SN00359915. Fig. 8 depicts the conformation of SN00359915 in the RdRp complex.
Fig. 8.
The interaction of SN00359915 with RdRp complex. Primer and template strands are colored red and cyan colors, respectively.
3.3. MD simulation of molecular hits
To get the binding dynamics of each ligand, RMSD values for 200 ns were monitored. Fig. 9 shows the RMSD values for the receptor Cα, RNA and ligand main atoms of each complex for the 200 ns simulation.
Fig. 9.
The RMSD values of RDV-TP and SN00166900 (A), RDV-TP and SN00303170 (B), RDV-TP and SN00359915 (C), and RMSF (D) values for each complex. The RDV-TP, SN00166900, SN00303170, and SN00359915 are colored green, cyan, purple, and pink colors, respectively.
The RMSD of protein Cα, RNA and ligand main atoms of each complex was stable during 200 ns. The stability of protein Cα was reached rapidly in the first 25 ns and for the rest of the simulation time for each SN00166900, SN00303170, and SN00359915 system. It was observed that the RMSD values of protein Cα of RDV-TP complex was lower than those of SN00166900, SN00303170, and SN00359915 complexes. Meanwhile, the RMSD values of RNA main atoms were slightly lower in SN00166900 and SN00359915 systems and was higher in SN00303170 than those of the RDV-TP complex. However, the ligand RMSD values were lower in SN00303170 and SN00359915 than the RDV-TP system and was higher in the SN00166900 system. Meanwhile, RMSF values of the protein were also monitored during 200 ns. High peaks were observed at V36 (V405), K61 (K430), T275 (T644), V368 (V737), T481 (T850), which corresponded to loop regions of the protein and V527 (V905) and E541 (E919), which was the carboxyl ends. While those residues interacting with ligands such as R555 (R186) and N691 (N322) were observed to be stable. Fig. 9D depicts the RMSF of protein residues for each complex.
Similar to the RMSF plot of RDV-TP, RMSF of RNA residues (Fig. 10 ) was high at A1 of the primer strand, which is the first sequence of the primer strand and was not involved in direct ligand interaction. The RMSF of U6 of primer strand and A3 and A4 of the template strand, which involved in the ligand binding, were observed to be stable.
Fig. 10.
Plot of RNA residue fluctuation was recorded as RMSF values as RDV-TP (green) SN00166900 (cyan), SN00303170 (purple), and SN00359915 (pink).
Clustering analysis shows the most dominant structure for each complex during the 200 ns simulations. The ligand SN00166900 maintained H-bond interactions with U553 (U4) and U554 (U5) of the primer strand in addition to D83 (D452), R184 (R553), and R186 (R555). The manganese ion was stabilized through interaction with the ligand OH group. Binding of SN00303170 was preserved through interactions with D249 (D618), T187 (T556), A393 (A762), and H441 (H810). While, SN00359915 interacted with D83 (D452), R184 (R553), R186 (R555), T187 (T556), C253 (C622), S312 (S681), D315 (D684), and N322 (N691).
3.4. Binding free energy calculation
The total binding free energy was calculated from electrostatic (ΔE ELE), van der Waals (ΔE VDW), non-polar term of solvation energy (ΔE PB), and polar term of solvation energy (ΔE PBSUR). The contribution of each energy term was depicted in Table 1 . The total binding energy (ΔE PBTOT) was −18.90 ± 7.59 kcal mol−1 for RDV-TP, −16.62 ± 10.34 for ATP, −35.33 ± 7.56 for SN00166900, −45.23 ± 5.53 for SN00303170, and −64.86 ± 4.66 for SN00359915. It is clear that the SN00359915 had lowest total binding free energy out of the four ligands, and it is around three times as negative than that of RDV-TP. The large contribution of SN00359915 binding to RdRp was originated from van der Waals energy term.
Table 1.
Binding energy predicted by MM-PBSA protocol calculated for 200 snapshots at last 100 ns trajectory.
| Ligand | ΔEELE (kcal/mol) | ΔEVDW (kcal/mol | ΔEPB (kcal/mol) | ΔEPBSUR (kcal/mol) | ΔEPBTOT (kcal/mol) |
|---|---|---|---|---|---|
| SN00166900 | −61.92 ± 6.39 | −36.41 ± 8.57 | 69.72 ± 5.39 | −6.72 ± 0.18 | −35.33 ± 7.56 |
| SN00303170 | −26.81 ± 4.47 | −52.53 ± 6.28 | 40.36 ± 4.09 | −6.26 ± 0.27 | −45.23 ± 5.53 |
| SN00359915 | −9.02 ± 4.88 | −71.15 ± 5.54 | 22.35 ± 3.78 | −7.04 ± 0.21 | −64.86 ± 4.66 |
| RDV-TP | −52.48 ± 30.14 | −18.19 ± 6.16 | 56.19 ± 26.08 | −4.42 ± 0.23 | −18.90 ± 7.59 |
| ATP | −40.99 ± 32.16 | −27.11 ± 6.32 | 56.00 ± 25.73 | −4.52 ± 0.13 | −16.62 ± 10.34 |
In addition, the three hits identified in this study showed higher predicted binding energies than RDV-TP. The superiority of SN00359915 binding, for instance, which were supported by more diverse interactions with key residues such as those with R555 and N691, while was additionally corroborated by favorable interactions with D452, R553, T556, C622, S681, and D684.
4. Conclusion
In this study, RdRp in complex with RNA and manganese ions were built using homology modeling. The RdRp model was stable during 500 ns MD simulation. The binding of RDV-TP to RdRp showed that the position of the phosphate group was at the entry of NTP channel, which may reflect its role in inhibiting the entry of NTP to the active site. Additionally, the phosphate group established interactions with K551, R553, and K621, while the adenosine motif formed base pairing with U2 of template strand. The metal ions stabilized the phosphate group of RDV-TP and were located close to D618, D760, and D761. Using pharmacophore models, the three natural product hits SN00166900, SN00303170 and SN00359915 were observed to have were identified which had lower binding energies than that of RDV-TP.
Author contributions
MA performed simulation, analyzed data, and wrote the manuscript. STW, DJB and NP performed simulation. CW designed the work, analysis data, and supervised the work. All authors approved the manuscript.
Acknowledgment
MA wish to thank Ministry of Research and Technology Republic of Indonesia for partially funding this research. CW thanks the support from the National Science Foundation of USA under Grants NSF RUI-1904797/ACI-1429467 and XSEDE MCB 170088.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2020.104156.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Mulangu S. A randomized, controlled trial of ebola virus disease therapeutics. N. Engl. J. Med. 2019;381(24):2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoenen T., Groseth A., Feldmann H. Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Microbiol. 2019;17(10):593–606. doi: 10.1038/s41579-019-0233-2. [DOI] [PubMed] [Google Scholar]
- 3.Lu R. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int. J. Biol. Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A.C. Tructure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tchesnokov E.P. Mechanism of inhibition of ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses-Basel. 2019;11(4) doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhai P. The epidemiology, diagnosis and treatment of COVID-19. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon C.J. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295(15):4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tchesnokov E.P. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 2008;283(49):34218–34228. doi: 10.1074/jbc.M806797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko W.-C. Arguments in favour of remdesivir for treating SARS-CoV-2 infections. Int. J. Antimicrob. Agents. 2020;55(4) doi: 10.1016/j.ijantimicag.2020.105933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elfiky A.A. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: an in silico perspective. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1761882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato K., Honma T., Fukuzawa K. Intermolecular interaction among Remdesivir, RNA and RNA-dependent RNA polymerase of SARS-CoV-2 analyzed by fragment molecular orbital calculation. J. Mol. Graph. Model. 2020;100 doi: 10.1016/j.jmgm.2020.107695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L., Zhou R. Structural basis of the potential binding mechanism of remdesivir to SARS-CoV-2 RNA-dependent RNA polymerase. J. Phys. Chem. B. 2020;124(32):6955–6962. doi: 10.1021/acs.jpcb.0c04198. [DOI] [PubMed] [Google Scholar]
- 16.Newman D.J., Cragg G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016;79(3):629–661. doi: 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- 17.Nascimento Junior J.A.C. SARS, MERS and SARS-CoV-2 (COVID-19) treatment: a patent review. Expert Opin. Ther. Pat. 2020:1–13. doi: 10.1080/13543776.2020.1772231. [DOI] [PubMed] [Google Scholar]
- 18.Madhavi Sastry G. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368(6492):779. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Appleby T.C. Tructural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015;347(6223):771. doi: 10.1126/science.1259210. [DOI] [PubMed] [Google Scholar]
- 21.Greenwood J.R. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput. Aided Mol. Des. 2010;24(6):591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 22.Shelley J.C. Epik: a software program for pKaprediction and protonation state generation for drug-like molecules. J. Comput. Aided Mol. Des. 2007;21(12):681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 23.Friesner R.A. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 24.Friesner R.A. Extra precision Glide: docking and scoring incorporating a model of hydrophobic enclosure for Protein−Ligand complexes. J. Med. Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 25.Yin W. Tructural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368(6498):1499. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Banerjee P. Super Natural II--a database of natural products. Nucleic Acids Res. 2015;43:D935–D939. doi: 10.1093/nar/gku886. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolber G., Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005;45(1):160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- 28.Naithani R. Ntiviral activity of phytochemicals: a comprehensive review. Mini Rev. Med. Chem. 2008;8(11):1106–1133. doi: 10.2174/138955708785909943. [DOI] [PubMed] [Google Scholar]
- 29.Lin L.-T., Hsu W.-C., Lin C.-C. Antiviral natural products and herbal medicines. J. Traditional Complementary Med. 2014;4(1):24–35. doi: 10.4103/2225-4110.124335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salomon-Ferrer R. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theor. Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 31.Maier J.A. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11(8):3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zgarbová M. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theor. Comput. 2011;7(9):2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banáš P. Performance of molecular Mechanics force fields for RNA simulations: stability of UUCG and GNRA hairpins. J. Chem. Theor. Comput. 2010;6(12):3836–3849. doi: 10.1021/ct100481h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J. Development and testing of a general amber force field. J. Comput. Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 35.Ryckaert J.-P., Ciccotti G., Berendsen H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23(3):327–341. [Google Scholar]
- 36.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- 37.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theor. Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 38.Shao J. Clustering molecular dynamics trajectories: 1. Characterizing the performance of different clustering algorithms. J. Chem. Theor. Comput. 2007;3(6):2312–2334. doi: 10.1021/ct700119m. [DOI] [PubMed] [Google Scholar]
- 39.Kollman P.A. Calculating structures and free energies of complex Molecules: combining molecular Mechanics and continuum models. Acc. Chem. Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 40.Hillen H.S. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- 41.Nudler E. RNA polymerase active center: the molecular engine of transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton Z.F. NTP-driven translocation and regulation of downstream template opening by multi-subunit RNA polymerases. Biochem. Cell. Biol. 2005;83(4):486–496. doi: 10.1139/o05-059. [DOI] [PubMed] [Google Scholar]
- 43.Saha A. Probable molecular mechanism of remdesivir for the treatment of COVID-19: need to know more. Arch. Med. Res. 2020;51(6):585–586. doi: 10.1016/j.arcmed.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.