Abstract
The current study intended to explore the interaction of the long non-coding RNA (lncRNA), microRNA (miRNA), and messenger RNA (mRNA) under the background of competitive endogenous RNA (ceRNA) network in endometriosis (EMs). The differentially expressed miRNAs (DEmiRs), differentially expressed lncRNA (DELs), and differentially expressed genes (DEGs) between EMs ectopic (EC) and eutopic (EU) endometrium based on three RNA-sequencing datasets (GSE105765, GSE121406, and GSE105764) were identified, which were used for the construction of ceRNA network. Then, DEGs in the ceRNA network were performed with Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and protein-protein interaction (PPI) analysis. Besides, the DEmiRs in the ceRNA network were validated in GSE124010. And the target DELs and DEGs of verified DEmiRs were validated in GSE86534. The correlation of verified DEmiRs, DEGs, and DELs was explored. Moreover, gene set enrichment analysis (GSEA) was applied to investigate the function of verified DEmiRs, DEGs, and DELs. Overall, 1352 DEGs and 595 DELs from GSE105764, along with 27 overlapped DEmiRs between GSE105765 and GSE121406, were obtained. Subsequently, a ceRNA network, including 11 upregulated and 16 downregulated DEmiRs, 7 upregulated and 13 downregulated DELs, 48 upregulated and 46 downregulated DEGs, was constructed. The GO and KEGG pathway analysis showed that this ceRNA network probably was associated with inflammation-related pathways. Furthermore, hsa-miR-182-5p and its target DELs (LINC01018 and SMIM25) and DEGs (BNC2, CHL1, HMCN1, PRDM16) were successfully verified in the validation analysis. Besides, hsa-miR-182-5p was significantly negatively correlated with these target DELs and DEGs. The GSEA analysis implied that high expression of LINC01018, SMIM25, and CHL1, and low expression of hsa-miR-182-5p would activate inflammation-related pathways in endometriosis EU samples.
LINC01018 and SMIM25 might sponge hsa-miR-182-5p to upregulate downstream genes such as CHL1 to promote the development of endometriosis.
Keywords: competing endogenous RNA, endometriosis, inflammation, long non-coding RNA, messenger RNA, microRNA
Introduction
Endometriosis (EMs), characterized by the subsistence of endometrial-like tissue (including stroma and glands) growing outside the uterine cavity, is a common benign gynaecological disorder.1 EMs approximately affects 6% to 10% of women worldwide, mainly during the reproductive age.1 It would induce infertility and various pain, such as pelvic pain, dysmenorrhea, and dyspareunia,2 also a risk of cancerization.3 Of note, there is still no individual theory that can thoroughly explain all the aspects of EMs, even the classic “retrograde menstruation” hypothesis, suggesting viable endometrial debris refluxed through the fallopian tubes into the pelvic cavity to implant.4 Not only such complexity of the disease itself but also the absence of sensitive and specific biomarkers challenged the diagnosis and treatment of EMs. Hence, it is essential to explore the potential molecular mechanisms underlying EMs to deepen our understanding of EMs.
The non-coding RNAs (ncRNAs), transcribed from the DNA-genome but unable to code proteins, function as universal regulators in cellular processes, which could be generally sorted into two types according to their scale: the small long non-coding RNAs (<200 nucleotides in length) and the long non-coding RNAs (⩾200 nucleotides in length).5 The miRNA, one of the most concerned small ncRNAs, has been proved to be dysregulated in EMs, but the specific mechanism remained to clarify,6 particularly in multi-cohorts integrated analysis. The lncRNA, a new star with the advancement of the RNA-sequencing technology, has invoked a research upsurge in recent years, with no exception in Ems.7 Notably, the emerging competing endogenous RNAs (ceRNAs) hypothesis manifested that lncRNAs could serve as a miRNA sponge to regulate the target mRNAs.8 And this hypothesis had been attested in EMs: the first reported lncRNA H19 in EMs sponged miRNA let-7 to regulate its downstream gene IGF1R to impact the proliferation of endometrial stromal cells.9 However, few comprehensive analyses of EMs-associated miRNAs and lncRNAs in the ceRNA network’s milieu have been conducted.
Therefore, we intended to establish an EMs-related ceRNA network to investigate the regulatory role of the lncRNA-miRNA-mRNA axis in EMs (Figure 1). As far as we know, this report represents the first endeavour to construct a lncRNA-associated ceRNA network based on multiple RNA-sequencing datasets in EMs.
Figure 1.
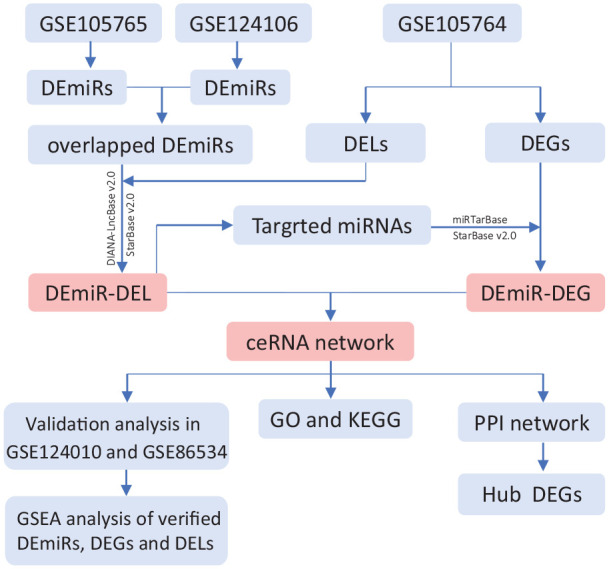
The flowchart of endometriosis-associated ceRNA network analysis.
Materials and methods
Training datasets resources
Two miRNA expression datasets: GSE105765 (eight paired EC and EU endometrium tissue samples),10 and GSE121406 (four paired EC and EU endometrial stromal cells),11 as well as the lncRNA and mRNA expression profile GSE105764 (same eight paired EC and EU endometrium tissue samples in GSE105765),10 were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo). All these datasets were measured by high-throughput RNA-sequencing: GSE105765 was based on platform GPL11154 (Illumina HiSeq 2000), GSE121406 on platform GPL18573 (Illumina NextSeq 500), and GSE105764 on platform GPL20301 (Illumina HiSeq 4000). There was no need for ethical approval or informed consent in this study because the data was publicly available.
Identification of DEmiRs, DEGs, and DELs
The “DESeq2” R package12 was applied to analyze the differentially expressed microRNAs (DEmiRs) with a threshold of |log2 fold change (FC)| ⩾ 2 and adjust P-value < 0.01; the differentially expressed genes (DEGs) and differentially expressed lncRNAs (DELs) with a threshold of |log2 FC| ⩾ 3 and adjust P-value < 0.01. Moreover, intersection analysis was conducted to detect the shared DEmiRs between GSE105765 and GSE121406.
CeRNA network construction
In the light of the ceRNA hypothesis, screened DEGs, DELs, and overlapped DEmiRs were applied to build the lncRNA–miRNA–mRNA regulatory network. The predicted lncRNAs interacted with overlapped DEmiRs were mined in downloaded databases StarBase v2.013 and DIANA-LncBase v2.0,14 both of which provided the experimentally validated miRNA-lncRNA interactive information. Next, these predicted lncRNAs were further intersected with the identified DELs in GSE105764. Additionally, the overlapped DEmiRs-targeted mRNAs were predicted from the miRTarBase15 and StarBase v2.013 databases and later were intersected with the identified DEGs in GSE105764. Finally, the filtered DEmiR-DEL and DEmiR-DEG interactive pairs were employed to build a ceRNA regulatory network, which was visualized in software Cytoscape 3.6.1.16
GO and KEGG pathway enrichment analysis
The DEGs included in the established ceRNA network were performed with GO and KEGG pathway enrichment analysis by Enrichr (http://amp.pharm.mssm.edu/Enrichr/), a useful online tool for querying functional annotation and biological information of genes. Retrieved GO terms and KEGG pathways with a P-value < 0.05 were supposed to be significantly enriched.
PPI network establishment
To further explore the potential interplay of DEGs in the ceRNA network, the Search Tool for the Retrieval of Interacting Genes database (STRING-Version 10.0, http://stringdb.org) was adopted to create a PPI network with the interaction score > 0.4. Then, the PPI network was carried into Cytoscape 3.6.116 for visualization, and the degree score of nodes was analyzed by the plugin NetworkAnalyzer. Moreover, 10 hub genes were determined by the “Degree” method in the plugin CytoHubba.
Validation analysis of DEmiRs, DEGs, and DELs in the ceRNA network
The validation analysis of all DEmiRs in the ceRNA network was performed in GSE124010 (based on platform GPL25134).17 This dataset contained 3 normal endometria (NM) from healthy candidates and 3 EU samples from EMs patients. It would be interesting to know whether the candidate DEmiRs in EU vs. EC in training datasets were also changed in the EU versus NM in the validation dataset. After the positively verified DEmiRs were acquired, their target DEGs and DELs were further chosen to validate in GSE86534, which profiled the mRNA and lncRNA expression in four paired EU and EC tissue samples from EMs patients grounded on platform GPL20115.18 The p-value < 0.05 was considered significant.
Correlation analysis of the verified DEmiRs, DEGs, and DELs
To explore the correlation between verified DEmiRs, and their target DEGs and DELs, the miRNA profile in GSE105765 and mRNA-lncRNA profile in GSE105764 examined on the same samples were combined to perform the Spearman correlation analysis. Due to the lack of validation datasets detecting the miRNA and mRNA-lncRNA profile in the same samples, we only validated the correlation between target DEGs and DELs in GSE86534. The P-value < 0.05 was considered significant.
Gene set enrichment analysis (GSEA) of the verified DEmiRs, DEGs, and DELs
GSEA is a computational method to evaluate whether a defined gene set exerts a significant difference between two biological phenotypes.19 Since the EU samples might play a fundamental role in the pathogenesis of Ems,20 we investigated the function of the verified DEmiRs, DEGs, and DELs in EU samples by GSEA analysis. According to the median expression of the verified DEmiRs, DEGs and DELs, the EU samples were respectively divided into two groups: the high- and low-expression groups, and the file “h.all.v7.0.symbols.gmt” in GSEA websites (https://www.gsea-msigdb.org/gsea/index.jsp) was used as the reference gene set. The analysis was performed in GSEA software, and the statistical threshold was FDR q-value < 0.25. Then, top-ranking results were visualized in R software.
Results
Differentially expressed DEGs, DELs, and DEmiRs
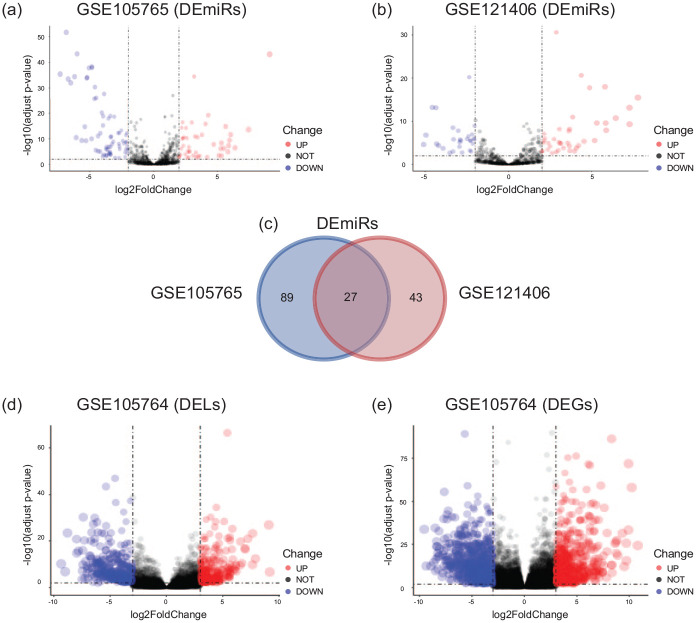
With the criteria of |log2 FC| ⩾ 2 and adjust P-value < 0.01, 116 DEmiRs (47 upregulated and 69 downregulated) were obtained from GSE105765, along with 70 DEmiRs (40 upregulated and 30 downregulated) from GSE121406 (Figure 2a and b). The intersection analysis showed 27 common DEmiRs (11 upregulated and 16 downregulated) between GSE105765 and GSE121406 (Figure 2c). Additionally, with the criteria of |log2 FC| ⩾ 3 and adjust P-value < 0.01, 1352 DEGs (693 upregulated and 659 downregulated) and 595 DELs (278 upregulated and 317 downregulated) were identified from GSE105764 (Figure 2d and e).
Figure 2.
Identification of DEmiRs, DELs, and DEGs in endometriosis. (a) Volcano plots for DEmiRs between ectopic (EC) and eutopic (EU) endometrium in GSE105764. (b) Volcano plots for DEmiRs between EC and EU endometrium in GSE121406. (c) Venn diagram for the overlapping DEmiRs between GSE105764 and GSE121406. (d) Volcano plots for DELs between EC and EU endometrium in GSE105765. (e) Volcano plots for DEGs between EC and EU endometrium in GSE105765. DEmiRs, differentially expressed microRNAs; DEGs, differentially expressed genes; DELs: differentially expressed long non-coding RNAs; EC, ectopic endometrium; EU, eutopic endometrium.
CeRNA network construction
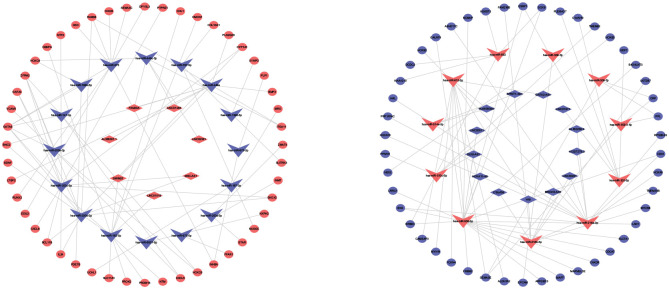
Based on the filtered DEmiR-DEL and DEmiR-DEG interactive pairs, the EMs-related ceRNA network was established, including 11 upregulated and 16 downregulated DEmiRs, 7 upregulated and 13 downregulated DELs, 48 upregulated and 46 downregulated DEGs (Figure 3).
Figure 3.
Competing endogenous RNA (ceRNA) network in endometriosis. The red indicates the upregulated RNAs in EC compared to EU samples, and the blue indicates the downregulated RNAs in EC compared to EU samples. The v-shape represents DEmiRs, the diamond represents DELs, and ellipse represents DEGs. DEmiRs, differentially expressed microRNAs; DELs, differentially expressed long noncoding RNAs; DEGs, differentially expressed genes. EC, ectopic endometrium; EU, eutopic endometrium.
GO and KEGG pathway enrichment analysis
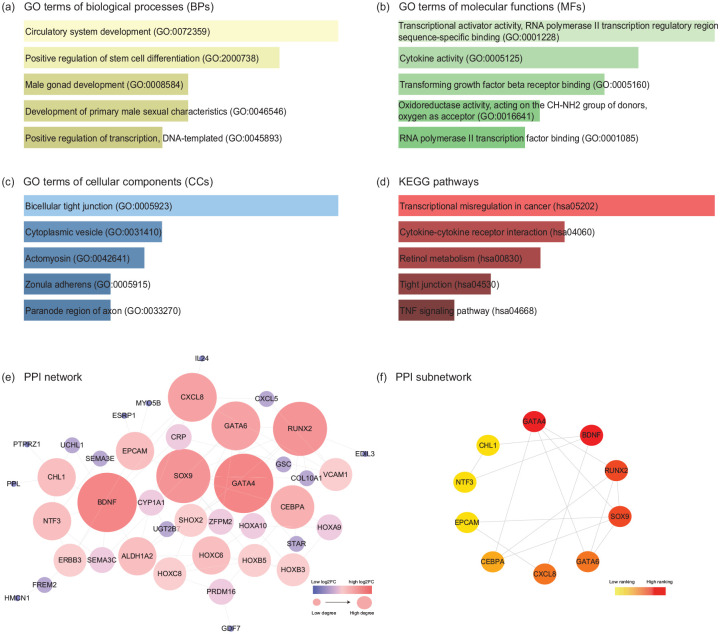
A total of 94 DEGs in the ceRNA network were processed with functional enrichment analysis by website Enrichr. The GO analysis revealed that the top five significantly enriched biological processes (BPs) were Circulatory system development, Positive regulation of stem cell differentiation, Male gonad development, Development of primary male sexual characteristics and Positive regulation of transcription (Figure 4a); the top five molecular functions (MFs) were Transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding, Cytokine activity, Transforming growth factor-beta receptor binding, Oxidoreductase activity, Acting on the CH-NH2 group of donors and RNA polymerase II transcription factor binding (Figure 4b); the top five cellular components (CCs) were Bicellular tight junction, Cytoplasmic vesicle, Actomyosin, Zonula adherens and Paranode region of axon (Figure 4c). Moreover, KEGG pathway analysis indicated that these DEGs were primarily concentrated in Transcriptional misregulation in cancer, Cytokine-cytokine receptor interaction, Retinol metabolism, Tight junction and TNF signaling pathway (Figure 4d).
Figure 4.
GO, KEGG pathway, and PPI network analyses of DEGs in the EMs-related ceRNA network. The top 5 enriched (a) biological processes, (b) cellular components, (c) molecular functions, and (d) KEGG pathways of DEGs in the ceRNA network. The horizontal axis represents the number of genes, and the vertical axis represents GO terms or KEGG pathway names. All entries were ranked by p-value in the ascending order. (e) PPI network constructed by the DEGs in the ceRNA network. According to the degree score calculated by plugin NetworkAnalyzer, the node colour changes gradually from blue to red and the node sizes from small to large in the ascending order. (f) 10 hub DEGs in the PPI network analyzed by the “Degree” method in plugin CytoHubba. The node colour changes gradually from yellow to red in the ascending order according to the degree score. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction; DEGs, differentially expressed genes.
PPI network establishment
By searching those 94 DEGs in the ceRNA network in the STRING database, with the interaction score > 0.4, a PPI network consisting of 41 nodes and 74 edges was constructed (Figure 4e). Furthermore, according to the degree scores, 10 hub DEGs in the PPI network were selected out: GATA4, BDNF, RUNX2, SOX9, GATA6, CXCL8, CEBPA, EPCAM, NTF3, CHL1 (Figure 4f).
Validation analysis of DEmiRs, DEGs, and DELs in the ceRNA network
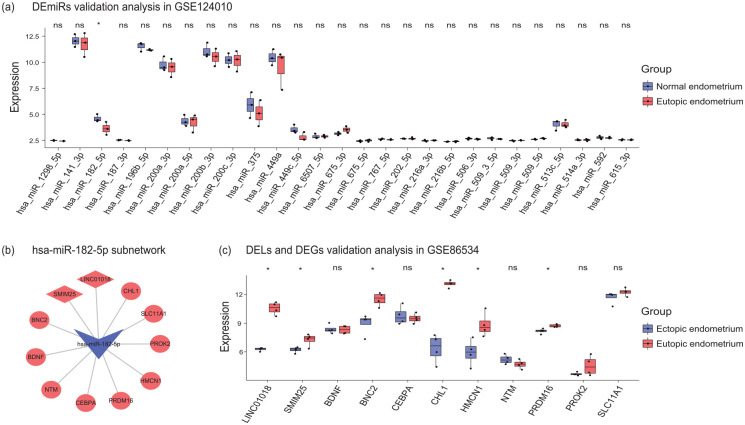
All DEmiRs in the ceRNA network were validated in GSE124010. Probably due to the limited sample size, we only found hsa-miR-182-5p were significantly low-expressed in EU samples when compared to NM samples in GSE124010 (Figure 5a). Since hsa-miR-182-5p was down-regulated in EC vs. EU in training datasets and EU vs. NM in the validation dataset, it might be a constant dysregulated miRNA in EMs development. Hence, the target DEGs and DELs of hsa-miR-182-5p were chosen to validated in GSE86534 (Figure 5b). The results showed that two lncRNAs LINC01018 and SMIM25 along with four genes BNC2, CHL1, HMCN1, and PRDM16 were significantly upregulated in EC compared to EU samples in GSE86534 (Figure 5c).
Figure 5.
Validation analysis of DEmiRs, DEGs, and DELs in the ceRNA network. (a) All DEmiRs in the ceRNA network were validated in GSE124010. (b) The target DEGs and DELs of hsa-miR-182-5p in the ceRNA network. (c) The target DEGs and DELs of hsa-miR-182-5p were validated in GSE86534. DEmiRs, differentially expressed microRNAs; DELs, differentially expressed long noncoding RNAs; DEGs, differentially expressed genes. *P-value < 0.05.
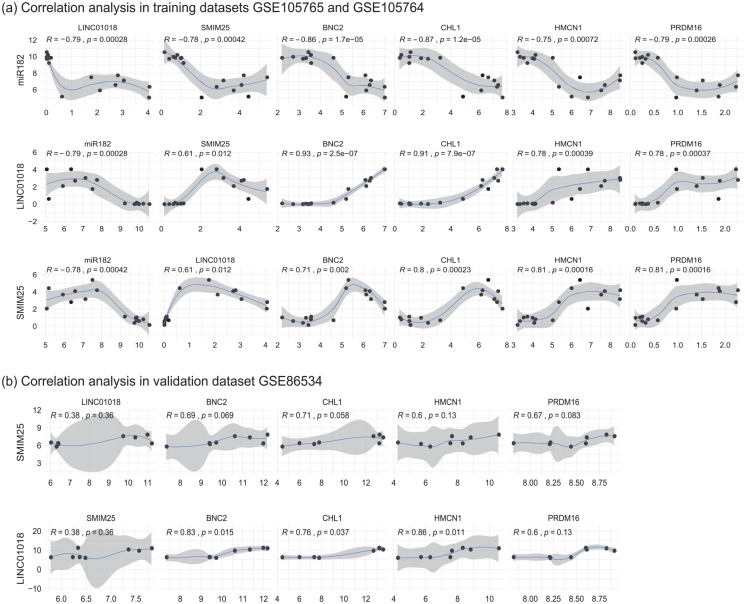
The relationship between hsa-miR-182-5p and its target DEGs and DELs
The correlation analysis in combined data of training datasets GSE105764 and GSE105765 indicated that hsa-miR-182-5p was significantly negatively associated with its target DELs (LINC01018 and SMIM25) and DEGs (BNC2, CHL1, HMCN1, PRDM16). Moreover, those target DELs (LINC01018 and SMIM25) were significantly positively associated with the target DEGs (BNC2, CHL1, HMCN1, PRDM16) (Figure 6a). In the validation dataset GSE86534, LINC01018 and SMIM25 were also proved to be positively correlated with BNC2, CHL1, HMCN1, and PRDM16, respectively, although the P-value was not always lower than 0.05 probably due to the small sample size (Figure 6b). Noticeably, LINC01018 and CHL1 were respectively the most up-regulated DEL and DEG both in the training and validation datasets (supplement Tables S1 and S2). Moreover, CHL1 was also identified as the hub nodes in the PPI network. Hence, we selected CHL1 as the representative DEG in subsequent GSEA analysis.
Figure 6.
The relationship between hsa-miR-182-5p and its target DEGs and DELs. (a) The Spearman correlation analysis of hsa-miR-182-5p and its target DEGs and DELs in combined data of training datasets GSE105764 and GSE105765. (b) The Spearman correlation analysis of the target DEGs and DELs of hsa-miR-182-5p in the validation dataset GSE86534. DELs, differentially expressed long noncoding RNAs; DEGs, differentially expressed genes. *P-value < 0.05.
The GSEA analysis of hsa-miR-182-5p and its targets in EU samples
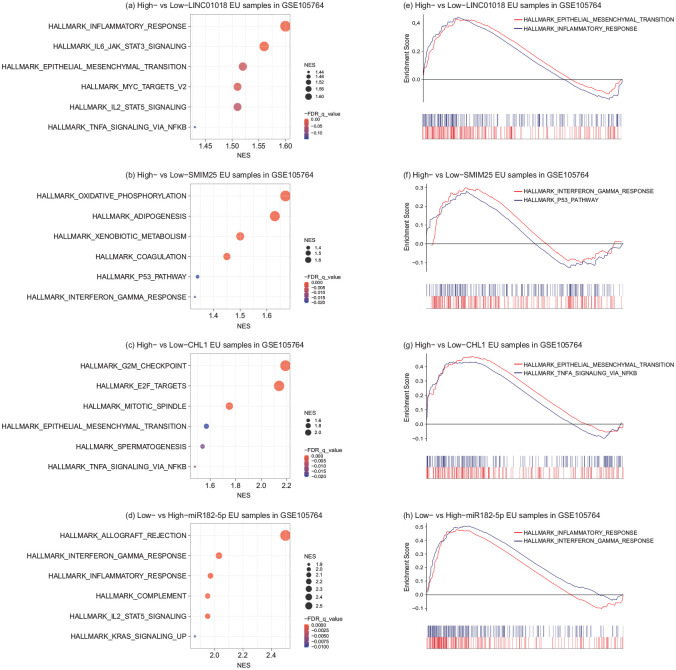
To investigated the function of hsa-miR-182-5p and its targets in EU samples, the GSEA analysis was performed. The EU samples in training datasets were divided into high- and low-expression groups according to the median expression of hsa-miR-182-5p, LINC01018, SMIM25, and CHL1, respectively. The results showed that the pathway “INFLAMMATORY_RESPONSE” was activated in high-expressed LINC01018 and low-expressed hsa-miR-182-5p EU samples compared to respective control samples. Besides, the pathway “INTERFERON_GAMMA_RESPONSE” and “TNFA_SIGNALING_VIA_NFKB” were also respectively triggered in high-expressed SMIM25 and CHL1 EU samples compared to low-expression controls. Interestingly, high expression of LINC01018 and low expression of hsa-miR-182-5p were also associated with activation of the pathway “EPITHELIAL_MESENCHYMAL_TRANSITION,” a well-described pathological process in EMs (Figure 7).
Figure 7.
The GSEA analysis of hsa-miR-182-5p and its targets in EU samples. The top 6 activated pathways in high-expressed LINC01018 (a), SMIM25 (b), CHL1 (c), and low-expressed hsa-miR-182-5p (d) EU samples compared to respective controls. And representative pathways were displayed in classic GSEA plots (e), (f), (g), (h). GSEA, gene set enrichment analysis; EU, eutopic endometrium. NES, normalized enrichment score.
Discussion
Endometriosis (EMs) is a heterogeneous disorder because of the diverse implanting locations with different depth of infiltration and non-specific clinical symptoms.1 The pathological mechanism of EMs remains enigmatic. Progressively accumulating evidence declared that the dysregulation of lncRNA affected miRNA activity, such as the ceRNA hypothesis, which was probably involved in the pathology of Ems.5,6 However, the lncRNA-associated ceRNA network based on multiple RNA-sequencing datasets remains unexplored in EMs.
To address this challenge, we established an EMs-associated ceRNA network comprised of 11 upregulated and 16 downregulated DEmiRs, 7 upregulated and 13 downregulated DELs, 48 upregulated and 46 downregulated DEGs. The GO and KEGG pathway analysis indicated that this ceRNA network was related to inflammation-related pathways, such as “Cytokine-cytokine receptor interaction” and “TNF signalling pathway.” And the inflammatory response is the central link of the genesis of Ems.21 The validation analysis revealed that hsa-miR-182-5p was not only downregulated in EC vs. EU samples but also downregulated in the EU versus NM samples. Besides, the target DELs (LINC01018 and SMIM25) and DEGs (BNC2, CHL1, HMCN1, PRDM16) of hsa-miR-182-5p were proved to be upregulated in EC versus EU samples. The negative correlation of hsa-miR-182-5p and these target DELs and DEGs was proved in training datasets. LINC01018 and SMIM25 were found positively correlated with BNC2, CHL1, HMCN1, PRDM16 in training and validation datasets. The GSEA analysis showed that high expression of LINC01018, SMIM25, and CHL1 (the DEG with the maximum log2FC) and low expression of hsa-miR-182-5p would activate inflammation-related pathways in EU samples in EMs. Hence, we supposed that LINC01018 and SMIM25 might sponge hsa-miR-182-5p to upregulate downstream genes such as CHL1 to promote the development of EMs.
To the best of our knowledge, the lncRNA LINC01018 and SMIM25 in our established ceRNA network are firstly reported in EMs. Wang et al. reported that LINC01018 was downregulated in hepatocellular carcinoma (HCC) tissues, and the over-expression of LINC01018 inhibited proliferation and promoted apoptosis of HCC cells via the up-regulation of FOXO1 by sponging hsa-miR-182-5p.22 Notably, a certain degree of proliferation and reduced apoptosis were the key features of Ems.21 However, the expression trend of LINC01018 in HCC in Wang et al.’s study22 was contrary to our findings in EMs. We supposed that LINC01018 might have tissue-specific expression and affect cell proliferation and apoptosis in EMs via specific mechanisms different from those in HCC. Moreover, the upregulation of LINC01018 would be induced by fasting in humanized livers.23 And the genome-wide association study (GWAS) indicated that the expression of LINC01018 in the liver was associated with the body mass index (BMI).23 Interestingly, women with EMs were reported with lower BMI24 and dysregulated lipid metabolism.25 Although our GSEA analysis indicated LINC01018 related to inflammatory response, it would be interesting to know whether LINC01018 affects lipid metabolism in EMs in future studies.
The SMIM25, also known as LINC01272, was upregulated gastric cancer (GC), and the over-expression of SMIM25 promoted the migration and invasion ability of GC cells by activating the epithelial-mesenchymal transition (EMT) process.26 The EMT defines a process by which epithelial cells lose their cell polarity and cell-to-cell adhesion and acquire the migratory and invasive properties to become mesenchymal cells.27 These changes are supposed to contribute to the establishment of endometriotic lesions in Ems.27 Moreover, the upregulation of SMIM25 might be an indicator of inflammatory bowel disease (IBD) and Crohn disease.28,29 More recently, Hung et al. reported that SMIM25 was upregulated in unstable plaque and highly monocyte- and macrophage-specific.30 And the knockdown of SMIM25 significantly reduced the phagocytosis.30 Hence, this study renamed the SMIM25 as PELATON (plaque enriched lncRNA in atherosclerotic and inflammatory bowel macrophage regulation). Notably, peritoneal macrophages’ impaired phagocytic ability was found in women with EMs, which might contribute to the failure to eradicate aberrant ectopic cells.31 Additionally, aberrant SMIM25 expression might influence the endometrial receptivity via the inflammation reaction.32 Considering the crucial role of inflammation and abnormal immunity in EMs, we speculated possible involvement of SMIM25 in the pathogenesis of endometriosis.
The verified DEmiR hsa-miR-182-5p belonged to the miR-183/96/182 family, which might adopt a critical role in the process of apoptosis, DNA repair, lipid metabolism, and immune signalling.33 By RNA-sequencing, microarray profiling, and qRT-PCR validation, the down-regulation of hsa-miR-182-5p was observed in EC compared to EU samples.10,34 Meanwhile, the dysregulation of hsa-miR-182-5p was also found in the plasma of EMs patients.35 Similarly, has-miR-183 was also reported downregulated in EC versus EU samples and EU versus NM samples, thus promoting invasion and suppressing apoptosis of endometrial stromal cells by targeting ITGB1P.36,37 It has been reported that hsa-miR-182-5p was significantly decreased in atherosclerosis models, and the over-expression of hsa-miR-182-5p inhibited the oxidative stress and macrophage apoptosis by targeting Toll-like receptor 4 (TRL4).38 Quite a few oxidative stress biomarkers had been found significantly higher in women with EMs than healthy controls.39 Continued oxidative stress would contribute to chronic inflammation,40 which provides a favourable condition for the implantation and growth of endometriotic cells. Besides, the macrophages are abundant in ectopic lesions, in the peritoneal cavity and peritoneal fluid of women with EMs compared to controls.31 And two phenotypes of macrophages: “classically activated” macrophages and “alternatively activated” macrophages, collectively contributed to the mixed pro- and anti-inflammatory microenvironment for the establishment of ectopic lesions.31 Furthermore, hsa-miR-182-5p was decreased in metastatic non-small cell lung cancer (NSCLC) tissues compared to primary tumour tissues.41 And it inhibited the metastasis of lung cancer cells via suppressing the EMT process,41 a well-known precondition for the initial implantation of endometriotic lesions.27
Our GSEA analysis revealed that high expression of CHL1 (cell adhesion molecule L1 Like), the target genes of has-miR-182-5p with the maximum log2FC, would activate the EMT process. CHL1 is a member of the L1 gene family of neural cell adhesion molecules (L1-CAMs), which involved developing the nervous system and a series of morphogenic events, such as cell migration and adhesion.42 As the homology of CHL1, L1CAM was upregulated in atypical EMs compared to typical EMs, aggravating pain in EMs by promoting nerve growth.43 Similarly, CHL1 was also found over-expressed in EMs,44 although it was reported under-expressed in cervical cancer,42 breast cancer,45 nasopharyngeal cancer,46 and papillary thyroid cancer.47 The overexpression of CHL1 inhibited the motility of nasopharyngeal cancer cells by the suppression of EMT.46 And the silencing of has-miR-182 promoted the expression of CHL1, thus suppressing the growth and invasion of papillary thyroid carcinoma cells.47 Notably, enhanced invasion and proliferation and the activated EMT were the key features of EMs.21,27 Thus, CHL1 might act in specific ways to influence these processes in EMs.
Nevertheless, three other target genes of has-miR-182 were seldom reported in EMs. BNC2 (Basonuclin 2) is fundamental for the proliferation of craniofacial mesenchymal cells during embryogenesis.48 The polymorphisms in the BNC2 gene were associated with ovarian cancer but not with EMs, indicating EMs is mediated by BNC2 in other ways.49 HMCN1 (Hemicentin 1) participates in the architecture of adhesive and flexible epithelial cell junctions.50 The upregulation of HMCN1 was found in ovarian cancer (OC) fibroblasts, thus promoting the invasion of OC fibroblasts.50 PRDM16 (PR Domain Containing 16) was involved in adipose biology and also maintenance of hematopoietic and neuronal stem cells.51 The deletion of PRDM16 in mice contributed to increased apoptosis of hematopoietic stem cells (HSCs).52 And a steady flow of HSCs would facilitate the angiogenesis and inflammation in EMs ectopic lesions.53
However, our analysis has some limitations. Firstly, due to the scarcity of available lncRNA and miRNA datasets of EMs, the sample size in the available training and validation datasets is small. Expanding the sample size would enhance the reliability of the results. Secondly, the datasets are expected to include normal endometrium (NM) from healthy women as the normal control to explore the molecular changes in EU samples. Besides, the expression of target genes of hsa-miR-182-5p was only analyzed in the mRNA level, which would be improved by validation in the protein level. Moreover, functional experiments need to be performed to explain the detailed regulatory mechanism of hsa-miR-182-5p in a ceRNA manner in EMs.
Conclusion
In conclusion, we firstly constructed the lncRNA-associated ceRNA network based on multiple RNA-sequencing datasets in endometriosis. Our study revealed that the LINC01018 and SMIM25 sponged miR-182-5p to upregulate downstream genes such as CHL1 to promote the development of endometriosis, which would provide new insights into the roles of non-coding RNAs in the pathogenesis of endometriosis.
Supplemental Material
Supplemental material, sj-xlsx-1-iji-10.1177_2058738420976309 for LINC01018 and SMIM25 sponged miR-182-5p in endometriosis revealed by the ceRNA network construction by Li Jiang, Mengmeng Zhang, Sixue Wang, Yuzhen Xiao, Jingni Wu, Yuxin Zhou and Xiaoling Fang in International Journal of Immunopathology and Pharmacology
Supplemental material, sj-xlsx-2-iji-10.1177_2058738420976309 for LINC01018 and SMIM25 sponged miR-182-5p in endometriosis revealed by the ceRNA network construction by Li Jiang, Mengmeng Zhang, Sixue Wang, Yuzhen Xiao, Jingni Wu, Yuxin Zhou and Xiaoling Fang in International Journal of Immunopathology and Pharmacology
Acknowledgments
The authors would like to thank the GEO training course of Helix-Life as well as Biotrainee for the training of bioinformatics analysis, and Dr. Jianming Zeng (CEO of Biotrainee) and Dr. Guozi (Chongqing Medical University) for generously sharing their experience and codes.
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (81671437, 81801425, 81771558) and the Natural Science Foundation of Hunan Provincial, China (2016JC2049). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
ORCID iD: Li Jiang  https://orcid.org/0000-0002-4422-567X
https://orcid.org/0000-0002-4422-567X
Data Availability Statement: Data is available at NCBI GEO, accession numbers: GSE105765, GSE121406, GSE105764, GSE124010, GSE86534.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Giudice LC. (2010) Endometriosis. New England Journal of Medicine 362: 2389–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berkley KJ, Rapkin AJ, Papka RE. (2005) The pains of endometriosis. Science 308: 1587–1589. [DOI] [PubMed] [Google Scholar]
- 3. Nezhat F, Datta MS, Hanson V, et al. (2008) The relationship of endometriosis and ovarian malignancy: A review. Fertility and Sterility 90: 1559–1570. [DOI] [PubMed] [Google Scholar]
- 4. Sampson JA. (1927) Peritoneal endometriosis due to the menstrual dissemination of endometrial tissue into the peritoneal cavity. American Journal of Obstetrics & Gynecology 14: 422–469. [Google Scholar]
- 5. Beermann J, Piccoli M-T, Viereck J, et al. (2016) Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiological Reviews 96: 1297–1325. [DOI] [PubMed] [Google Scholar]
- 6. Saare M, Rekker K, Laisk-Podar T, et al. (2017) Challenges in endometriosis miRNA studies - From tissue heterogeneity to disease specific miRNAs. Biochimica et Biophysica Acta - Molecular Basis of Disease 1863: 2282–2292. [DOI] [PubMed] [Google Scholar]
- 7. Panir K, Schjenken JE, Robertson SA, et al. (2018) Non-coding RNAs in endometriosis: A narrative review. Human Reproduction Update 24: 497–515. [DOI] [PubMed] [Google Scholar]
- 8. Salmena L, Poliseno L, Tay Y, et al. (2011) A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 146: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ghazal S, McKinnon B, Zhou J, et al. (2015) H19 lncRNA alters stromal cell growth via IGF signaling in the endometrium of women with endometriosis. EMBO Molecular Medicine 7: 996–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao L, Gu C, Ye M, et al. (2018) Integration analysis of microRNA and mRNA paired expression profiling identifies deregulated microRNA-transcription factor-gene regulatory networks in ovarian endometriosis. Reproductive Biology and Endocrinology 16: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rekker K, Tasa T, Saare M, et al. (2018) Differentially-expressed miRNAs in ectopic stromal cells contribute to endometriosis development: The plausible role of miR-139-5p and miR-375. International Journal of Molecular Sciences 19: 3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Love MI, Huber W, Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li J-H, Liu S, Zhou H, et al. (2014) starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Research 42: D92−D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. (2016) DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Research 44: D231−D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chou C-H, Shrestha S, Yang C-D, et al. (2018) miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Research 46: D296–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shannon P, Markiel A, Ozier O, et al. (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou C-F, Liu M-J, Wang W, et al. (2019) miR-205-5p inhibits human endometriosis progression by targeting ANGPT2 in endometrial stromal cells. Stem Cell Research & Therapy 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang C, Wu W, Zhu H, et al. (2019) Knockdown of long noncoding RNA CCDC144NL-AS1 attenuates migration and invasion phenotypes in endometrial stromal cells from endometriosis†. Biology of Reproduction 100: 939–949. [DOI] [PubMed] [Google Scholar]
- 19. Subramanian A, Tamayo P, Mootha VK, et al. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu H, Lang JH. (2011) Is abnormal eutopic endometrium the cause of endometriosis? The role of eutopic endometrium in pathogenesis of endometriosis. Medical Science Monitor 17: RA92–RA99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bulun SE, Yilmaz BD, Sison C, et al. (2019) Endometriosis. Endocrine Reviews 40: 1048–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang S, Xu M, Sun Z, et al. (2019) LINC01018 confers a novel tumor suppressor role in hepatocellular carcinoma through sponging microRNA-182-5p. American Journal of Physiology-Gastrointestinal and Liver Physiology 317: G116–G126. [DOI] [PubMed] [Google Scholar]
- 23. Ruan X, Li P, Chen Y, et al. (2020) In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nature Communications 11: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferrero S, Anserini P, Remorgida V, et al. (2005) Body mass index in endometriosis. European Journal of Obstetrics & Gynecology and Reproductive Biology 121: 94–98. [DOI] [PubMed] [Google Scholar]
- 25. Melo AS, Rosa-e-Silva JC, de Rosa-e-Silva ACJS, et al. (2010) Unfavorable lipid profile in women with endometriosis. Fertility and Sterility 93: 2433–2436. [DOI] [PubMed] [Google Scholar]
- 26. Leng X, Liu G, Wang S, et al. (2020) LINC01272 Promotes migration and invasion of gastric cancer cells via EMT. OncoTargets and Therapy 13: 3401–3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang Y-M, Yang W-X. (2017) Epithelial-to-mesenchymal transition in the development of endometriosis. Oncotarget 8: 41679–41689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang S, Hou Y, Chen W, et al. (2018) KIF9‑AS1, LINC01272 and DIO3OS lncRNAs as novel biomarkers for inflammatory bowel disease. Molecular Medicine Reports 17: 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haberman Y, BenShoshan M, Di Segni A, et al. (2018) Long ncRNA landscape in the ileum of treatment-naive early-onset Crohn disease. Inflammatory Bowel Disease 24: 346–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hung J, Scanlon JP, Mahmoud AD, et al. (2020) Novel plaque enriched long noncoding RNA in atherosclerotic macrophage regulation (PELATON). Arteriosclerosis, Thrombosis, and Vascular Biology 40: 697–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vallvé-Juanico J, Houshdaran S, Giudice LC. (2019) The endometrial immune environment of women with endometriosis. Human Reproduction Update 25: 564–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xu H, Zhou M, Cao Y, et al. (2019) Genome-wide analysis of long noncoding RNAs, microRNAs, and mRNAs forming a competing endogenous RNA network in repeated implantation failure. Gene 720: 144056. [DOI] [PubMed] [Google Scholar]
- 33. Dambal S, Shah M, Mihelich B, et al. (2015) The microRNA-183 cluster: The family that plays together stays together. Nucleic Acids Research 43: 7173–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Filigheddu N, Gregnanin I, Porporato PE, et al. (2010) Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. Journal of Biomedicine and Biotechnology 2010: 369549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vanhie A, Peterse OD, Beckers D, et al. (2019) Plasma miRNAs as biomarkers for endometriosis. Human Reproduction 34: 1650–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen J, Gu L, Ni J, et al. (2015) MiR-183 regulates ITGB1P expression and promotes invasion of endometrial stromal cells. BioMed Research International 2015: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi X-Y, Gu L, Chen J, et al. (2014) Downregulation of miR-183 inhibits apoptosis and enhances the invasive potential of endometrial stromal cells in endometriosis. International Journal of Molecular Medicine 33: 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qin S-B, Peng D-Y, Lu J-M, et al. (2018) MiR-182-5p inhibited oxidative stress and apoptosis triggered by oxidized low-density lipoprotein via targeting toll-like receptor 4. Journal of Cellular Physiology 233: 6630–6637. [DOI] [PubMed] [Google Scholar]
- 39. Carvalho LFP, Samadder AN, Agarwal A, et al. (2012) Oxidative stress biomarkers in patients with endometriosis: Systematic review. Archives of Gynecology and Obstetrics 286: 1033–1040. [DOI] [PubMed] [Google Scholar]
- 40. Reuter S, Gupta SC, Chaturvedi MM, et al. (2010) Oxidative stress, inflammation, and cancer: How are they linked? Free Radical Biology and Medicine 49: 1603–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li Y, Zhang H, Li Y, et al. (2018) MiR-182 inhibits the epithelial to mesenchymal transition and metastasis of lung cancer cells by targeting the Met gene. Molecular Carcinogenesis 57: 125–136. [DOI] [PubMed] [Google Scholar]
- 42. Schmid RS, Maness PF. (2008) L1 and NCAM adhesion molecules as signaling coreceptors in neuronal migration and process outgrowth. Current Opinion in Neurobiology 18: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Finas D, Huszar M, Agic A, et al. (2008) L1 cell adhesion molecule (L1CAM) as a pathogenetic factor in endometriosis. Human Reproduction 23: 1053–1062. [DOI] [PubMed] [Google Scholar]
- 44. Zhang C, Wu W, Ye X, et al. (2019) Aberrant expression of CHL1 gene and long non-coding RNA CHL1-AS1, CHL1-AS2 in ovarian endometriosis. European Journal of Obstetrics & Gynecology and Reproductive Biology 236: 177–182. [DOI] [PubMed] [Google Scholar]
- 45. He L-H, Ma Q, Shi Y-H, et al. (2013) CHL1 is involved in human breast tumorigenesis and progression. Biochemical and Biophysical Research Communications 438: 433–438. [DOI] [PubMed] [Google Scholar]
- 46. Chen J, Jiang C, Fu L, et al. (2019) CHL1 suppresses tumor growth and metastasis in nasopharyngeal carcinoma by repressing PI3K/AKT signaling pathway via interaction with Integrin β1 and Merlin. International Journal of Biological Sciences 15: 1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu H, Fang J, Zhang J, et al. (2014) miR-182 targets CHL1 and controls tumor growth and invasion in papillary thyroid carcinoma. Biochemical and Biophysical Research Communications 450: 857–862. [DOI] [PubMed] [Google Scholar]
- 48. Vanhoutteghem A, Maciejewski-Duval A, Bouche C, et al. (2009) Basonuclin 2 has a function in the multiplication of embryonic craniofacial mesenchymal cells and is orthologous to disco proteins. Proceedings of the National Academy of Sciences 106(34): 14432−14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cesaratto L. (2016) BNC2 is a putative tumor suppressor gene in high-grade serous ovarian carcinoma and impacts cell survival after oxidative stress. Cell Death and Disease 7(12): e2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu C, Pan H, Torng P, et al. (2019) SRPX and HMCN1 regulate cancer‑associated fibroblasts to promote the invasiveness of ovarian carcinoma. Oncology Reports. Epub ahead of print 17 October 2019. DOI: 10.3892/or.2019.7379. [DOI] [PubMed] [Google Scholar]
- 51. Chi J, Cohen P. (2016) The multifaceted roles of PRDM16: Adipose biology and beyond. Trends in Endocrinology & Metabolism 27: 11–23. [DOI] [PubMed] [Google Scholar]
- 52. Aguilo F, Avagyan S, Labar A, et al. (2011) Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood 117: 5057–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Figueira PGM, Abrão MS, Krikun G, et al. (2011) Stem cells in endometrium and their role in the pathogenesis of endometriosis: Stem cells, the endometrium, and endometriosis. Annals of the New York Academy of Sciences 1221: 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-xlsx-1-iji-10.1177_2058738420976309 for LINC01018 and SMIM25 sponged miR-182-5p in endometriosis revealed by the ceRNA network construction by Li Jiang, Mengmeng Zhang, Sixue Wang, Yuzhen Xiao, Jingni Wu, Yuxin Zhou and Xiaoling Fang in International Journal of Immunopathology and Pharmacology
Supplemental material, sj-xlsx-2-iji-10.1177_2058738420976309 for LINC01018 and SMIM25 sponged miR-182-5p in endometriosis revealed by the ceRNA network construction by Li Jiang, Mengmeng Zhang, Sixue Wang, Yuzhen Xiao, Jingni Wu, Yuxin Zhou and Xiaoling Fang in International Journal of Immunopathology and Pharmacology