Abstract
Hereditary hemorrhagic telangiectasia is a rare disease with autosomal dominant inheritance. More than 80% hereditary hemorrhagic telangiectasia patients carry heterozygous mutations of Endoglin or Activin receptor-like kinase-1 genes. Endoglin plays important roles in vasculogenesis and human vascular disease. In this report, we found a novel missense mutation (c.88T > C) of Endoglin gene in a hereditary hemorrhagic telangiectasia 1 patient. Induced pluripotent stem cells of the patient were generated and differentiated into endothelial cells. The hereditary hemorrhagic telangiectasia-induced pluripotent stem cells have reduced differentiation potential toward vascular endothelial cells and defective angiogenesis with impaired tube formation. Endoplasmic reticulum retention of the mutant Endoglin (Cys30Arg, C30R) causes less functional protein trafficking to cell surface, which contributes to the pathogenesis of hereditary hemorrhagic telangiectasia. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 genetic correction of the c.88T > C mutation in induced pluripotent stem cells revealed that C30R mutation of Endoglin affects bone morphogenetic protein 9 downstream signaling. By establishing a human induced pluripotent stem cell from hereditary hemorrhagic telangiectasia patient peripheral blood mononuclear cells and autologous correction on mutant hereditary hemorrhagic telangiectasia-induced pluripotent stem cells, we were able to identify a new disease-causing mutation, which facilitates us to understand the roles of Endoglin in vascular development and pathogenesis of related vascular diseases.
Keywords: hereditary hemorrhagic telangiectasia (HHT), induced pluripotent stem cell (iPSC) differentiated endothelial cells, Endoglin
Introduction
Hereditary hemorrhagic telangiectasia (HHT) is an autosomal dominant inheritance rare disease affecting approximately 1 in 5000 people. Typical clinical symptoms of HHT are spontaneous and recurrent epistaxis, gastrointestinal bleeding, telangiectasia of skin and mucosa, and arteriovenous malformations (AVMs) of brain and lung.1–3 The diagnosis can be made depending on the presence of four criteria: spontaneous recurrent epistaxis; multiple telangiectasias (especially on nose, lips, fingers, and gastrointestinal tract); AVMs in lung, liver, and brain; and the familial inheritance. A patient was diagnosed with HHT when he/she met at least three of the four criteria. Endoglin (ENG), Activin receptor-like kinase-1 (ALK1), SMAD4, or bone morphogenetic protein (BMP9) mutations are responsible for more than 85% of HHT,4–8 they also account for about 25% of disease-causing mutations in pulmonary arterial hypertension (PAH). Most mutation genes reported so far in HHT and PAH are the components of BMP pathway that is essential for angiogenesis. Moreover, PAH is a potential complication of HHT.9,10 After the Fifth World Symposium in 2013, HHT-induced PAH is classified as group 1.2 PAH.11 Previous studies showed a stronger correlation between PAH and HHT2 (ALK1) than correlation between PAH and HHT1 (ENG).12 However, recent studies have shown that there are no significant differences in the prevalence of PAH between HHT1 (12%) and HHT2 (19%) patients undergoing pulmonary AVM embolization.13 So the understanding of the molecular pathology of HHT1 would also provide insight into PAH.
About 61% of HHT patients carry ENG mutation, these cases are classified as HHT1.14 According to the online database (http://arup.utah.edu/database/ENG/ENG_display.php), more than 505 variants of ENG were found in human genetic database, with 65% of which are pathogenic mutations. Nonsense and missense mutations accounted for 30% of the total pathogenic mutations. Heterozygous mutations result in protein truncation or protein degradation or intracellular retention, which eventually contributes to the reduced functional ENG protein in HHT1. ENG is a type 1 membrane glycoprotein located on cell surfaces and highly expressed on human endothelial cells (ECs).15 It has a large extracellular domain composed of two orphan region (OR) and bipartite zona pellucida module, a hydrophobic transmembrane domain, and a short cytoplasmic tail domain. The OR is located at outermost extracellular region and is the part that binds ligands such as BMP9.16,17 It contains two pairs of conserved cysteines, C30–C207 and C53–C182, which form disulfide bridges in OR1 and OR2, respectively.17 The crucial role of ENG in angiogenesis was proved by different models. ENG knockout mice die at gestational day 10.0–10.5 due to defects in vessel and heart development.18–20 Moreover, conditional knockout mice with ENG specific deletion in vascular cell precursors die at E10.5–12.5 due to vascular defects with misdirection of intersegmental vessels and dilatation of dorsal aortic.21 Banerjee et al. found that ENG promotes EC specification in the mouse embryonic stem cell (ESC) model.22 One recent work revealed abnormal formation of intersegmental blood vessels and dorsal longitudinal anastomotic vessel in ENG knock out zebrafish, further provided the evidence that ENG plays roles in vasculogenesis.23 So far, molecular pathogenic study of HHT are mainly based on the mouse or zebrafish models, and the generation of new models, such as humanized model, will help us further understand the pathogenesis of the disease.
Previous study revealed that ECs from HHT1 and HHT2 have decreased TGF-β signaling, including activins and BMPs.24–26 Recent structural studies showed that BMP9 directly interacted with ENG through its N-terminal orphan domain, and many HHT1-associated mutations caused misfolded ENG protein,17 some of which compromised BMP signaling as a result from the defects in BMP9 binding or trafficking to the cell surface.27
In this study, we identified a novel mutation at ENG gene of a HHT1 patient, and established the patient peripheral blood mononuclear cells (PBMNCs) derived induced pluripotent stem cells (iPSCs), which were further differentiated into ECs to evaluate the effect of C30R ENG mutation on early vascular development in HHT. At the same time, we assessed the function of the ECs by tube formation assay and subcellular co-localization detection by fluorescent microscopy. The endoplasmic reticulum (ER) retention of the mutant ENG protein leads to the reduction of functional ENG protein trafficking to cell surface, thereby affecting the downstream of BMP signaling, which contributes to the pathogenesis of HHT.
Materials and methods
DNA extraction and mutation analysis
Human genomic DNA was prepared from peripheral blood (PB) according to manufacturer's instructions of RelaxGene Blood DNA System (TIANGEN Biotech, Beijing, PR China). The complete coding exons and adjacent introns of ENG and ALK1 genes were amplified for sequencing. Mutation analyses were performed by Chromas and Vector NTI software.
PBMNCs isolation and expansion
PBMNCs were isolated by Ficoll-Paque Premium (GE Healthcare, Little Chalfont, UK) according to the method published by Bin-Kuan Chou et al.28 In brief, mononuclear cells (MNCs) were isolated by loading 20 mL diluted PB onto a layer of 15ml of Filcoll-Paque Premium, then the layer of MNCs was harvested, washed, and resuspended in mononuclear cell (MNC) medium, which consisted of 50% Iscove's Modified Dulbecco's Medium (Gibco, Grand Isle, NY, USA), 50% Ham's F12 (Gibco), Insulin-Transferrin-Selenium-Ethanolamine (Gibco), lipid concentrate (Gibco), 5 mg/mL bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO, USA), 50 µg/mL of L-ascorbic acid (Sigma), 2 mM Glutamax (Gibco), 200 µM 1-thioglycerol (Sigma), supplemented with 10 ng/mL interleukin-3 (PeproTech, Rocky Hill, NJ, USA), 100 ng/mL human stem cell factor (PeproTech), 40 ng/mL insulin-like growth factor 1 (PeproTech), 2 U/mL erythropoietin (R&D Systems, Minneapolis, USA), 100 mg/mL human holo-transferrin (R&D Systems), and 20 mM dexamethasone (Sigma). Cells were seeded at low-attachment six-well plates and maintained in MNC medium for 8–12 days before reprograming, and culture medium was changed every other day.
Reprograming and iPSCs cell culture
Reprograming was performed according to the previously published method28 and modified. CF1 mouse embryonic fibroblasts (MEFs) were prepared in-house using standard procedure. Human ESC medium consists of Dulbecco's Modified Eagle Media: Nutrient Mixture F-12 (Gibco), 20% knockout serum replacement (Gibco), Non-Essential Amino Acids Solution (Gibco), 55 µM beta-mercaptoethanol (Sigma), 2 mM Glutamax, and 20 ng/mL basic fibroblast growth factor (bFGF, R&D Systems). Three episomal vectors including pEB-C5, pEB-Tg, and pEB-P53shRNA for reprograming were from Professor Feng Ma. In brief, 2 × 106 cells were nucleofected with a total of 10 µg of three episomal vectors using Nucleofector II (Lonza, Walkersville, USA) and maintained in PBMNCs medium. Twenty-four hours later, the cells were transferred into a six-well plate coated with irradiated MEFs, and then maintained in 50% MNC medium and 50% human ESC medium supplemented with 10 µM Y27632 (Sigma). The culture medium was refreshed every other day with complete human ESC medium supplemented with 250 µM sodium butyrate (Sigma) until manually picking colonies. Colonies with iPSC morphology were directly transferred into a new plate coated with Matrigel (BD Biosciences, Heidelberg, Germany) and maintained in mTeSR1 medium (Stem Cell Technologies, Vancouver, Canada) for 14–18 days after electroporation and expanded. For maintenance of self-renewal, cells were fed daily and passaged using ethylene diamine tetraacetic acid (Gibco) at a dilution of 1:10 every 3–5 days.
Quantitative PCR
RNA extraction was performed using trizol reagent (Sigma); 2 µg of total RNAs were reverse transcribed into complementary DNA (cDNA) templates by using TransScript One-Step genomic DNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, PR China) according to the instructions. The quantitative PCR were performed by using TransStart Tip Green qPCR SuperMix (TransGen Biotech). The expression levels of genes were analyzed by using CFX Connect TM Real-Time PCR Detection System (BioRad, Hercules, CA, USA) and CFX Manager Software. The primers sequences of specific genes for qPCR are listed below:
| OCT4 | Forward: 5′-TGGGCTCGAGAAGGATGTG-3′; Reverse: 5′-GCATAGTCGCTGCTTGATCG-3′ |
| NANOG | Forward: 5′-CATGAGTGTGGATCCAGCTTG-3′; Reverse: 5′-CCTGAATAAGCAGATCCATGG-3′ |
| SOX2 | Forward: 5′-GAAAAACGAGGGAAATGGG-3'′ Reverse: 5′-TTTGCGTGAGTGTGGATGG-3′ |
| CD31 | Forward: 5′-CCAAGGTGGGATCGTGAGG-3′; Reverse: 5′-TCGGAAGGATAAAACGCGGTC-3′ |
| Actin | Forward: 5′-GCACCACACCTTCTACAATGA-3′; Reverse: 5′-GTCATCTTCTCGCGGTTGGC-3′ |
Immunostaining
Immunostaining analysis was performed as follows: cells growing on cover slips were washed with phosphate buffered saline (PBS) and fixed with 4% paraformaldehyde for 10 min at room temperature (RT). Cells were permeabilized with 0.1% Triton X-100 and blocked with 2% BSA in Dulbecco's phosphate buffered saline, then incubated with primary antibodies for octamer-binding transcription factor-4 (OCT4, Abcam, Cambridge, UK), NANOG (Abcam), SOX2 (Abcam), Calnexin (Cell Signaling Technology, Danvers, USA), and ENG (Santa Cruz Biotechnology, Dallas, TX, USA) overnight at 4 ℃. For cell surface antigens staining, primary antibodies for SSEA4 (Abcam), CD31 (Abcam), CD144 (Cell Signaling Technology), and ENG were applied on fixed cells overnight at 4 ℃ after blocking. After washing, cells were treated with AlexaFluor 488 or AlexaFluor 594 conjugated secondary antibody (Life Technologies, Carlsbad, CA, USA) for 1 h in the dark at RT. For F-actin staining, cells were washed and fixed, then treated with Phalloidin-fluoresceine isothiocyanate (FITC, Yeasen, Shanghai, China) for 30 min at RT. Nuclei were stained with DAPI (Sigma). Images were photographed by light microscopy (Nikon Eclipse Ti-S, Japan) and captured using NIS-Elements BR 4.30.01 software.
Generation and expansion of ECs from PBMNCs-derived iPSCs
ECs induction of PBMNCs-derived iPSCs were performed according to the previously published protocol.29 In brief, iPSCs were passaged at 1:50–100 and cultured with mTeSR1 for four days. Then, iPSCs culture medium were refreshed with BSA polyvinylalcohol essential lipids (BEL) medium supplemented with Activin A (25 ng/mL, R&D), BMP4 (30 ng/mL, R&D), VEGF165 (50 ng/mL, R&D), and CHIR99021 (1.5 µM, Tocris Bioscience, Bristol, UK) for three days to generate mesoderm cells. For vascular cells specification, mesoderm induction medium were substituted by BEL medium supplemented with VEGF165 (50 ng/mL), SB431542 (10 µM, Millipore, Billerica, MA, USA) for four days. To expand vascular ECs, cells were treated with the same medium as vascular specification for 3–4 more days. Next, mature vascular endothelial (VE) cells were purified by using CD31-dynabeads (Miltenyi Biotec, Bergisch Gladbach, Germany). The purified VE cells were maintained and expanded in EC-serum-free media (SFM, Gibco) containing VEGF165 (30 ng/mL), bFGF (30 ng/mL, R&D), and fetal bovine serum (FBS, 1%, Gibco).
Flow cytometry
Differentiated cells were dissociated into single cells by using 1 mg/mL Accutase (Stem Cell Technologies) and suspended in PBS containing 2% BSA, then the cells were incubated with FITC-conjugated CD31 antibody (BD Pharmingen, San Diego, CA) for 1 h at 4 ℃ in the dark. After washing three times with pre-cold PBS, the cells were suspended in 500uL PBS and kept in the dark on ice until analysis on a FACScalibur flow cytometer. Flow cytometry was performed using ACCURI C6 (BD Biosciences) with data analyzed by using CFlow Plus software (BD Biosciences).
Endothelial tube formation
ECs derived from iPSCs were maintained in EC (FULL)-SFM medium. Before the treatment, on the day, single cells were prepared by Accutase and seeded at 1 × 104 cells per well in a 96-well plate coated with 10 mg/mL Matrigel at 37 ℃ for 1 h. Tube formation of ECs were observed 3 h after seeding. Images were photographed by light microscopy (Nikon E100) with NIS-Elements BR 4.30.01 software. The number of tube junctions and tube branches were assessed by online analysis tool WimTube (https://www.wimasis.com/en/WimTube).
Plasmids
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/Cas9 construct containing a single guide RNA (gRNA) was generated as described previously.30 Briefly, the guide RNA (gRNA, 5′-ACCACTAGCCAGGTCTCGAA-3′; score 92) was designed using online “CRISPR Design” software (http://crispr.mit.edu/) and was inserted into the BbsI sites of CRISPR/Cas vector px458 (Addgene) according to the protocol provided by Zhang.31 For donor plasmids, 1500bp DNA fragments including exon2 were obtained using normal human genomic DNA as template and cloned into piggyBac plasmid (pPB –puDtk, a gift from Dr. Huang Yue) to generate donor plasmids.
CRISPR–Cas9 correction of C30R–iPSCs
C30R–iPSCs were cultured with mTeSR1 for five days, then dispersed to single-cell suspensions by Accutase (Gibco), 1 × 106 cells were nucleofected with a total of 8 µg of donor vectors and 2 µg of CRISPR/cas9 vectors using Nucleofector II (Lonza, Walkersville, USA) and maintained in mTeSR medium supplemented with 10 µM Y27632 for 24 h. Then cells were treated with 0.5 µg/mL puromycin for five days; 8–10 days later, colonies were picked and expanded for PCR and sequencing verification.
Western blot
ECs were grown to confluence in six-well plates and serum-restricted in EC-SFM with 0.1% FBS for 22 h, and then treated without or with 0.5 ng/mL BMP9 for 2 h. The cells were then harvested for 2D-PAGE analysis as described previously.30 Briefly, cells were immediately frozen on dry ice, and then lysed in radio immunoprecipitation assay buffer (Sigma) containing protease and phosphatase inhibitors (ThermoFisher Scientific, California, USA) on ice. Total protein lysates (10 µg) were separated as described previously.30 Membranes were incubated overnight at 4 ℃ with primary antibodies including anti-glyceraldehyde-3-phosphate dehydrogenase (TransGen Biotech), anti-Id1 (BioCheck, Foster City, USA), and anti-p-Smad1/5 (Cell Signaling Technology, Danvers, USA). Quantification of bands was performed by Quantity one software (Bio-Rad Laboratories, Inc).
Statistical analysis
Statistics comparison was performed by one-way ANOVA followed by Dunnett's post hoc test (GraphPad Prism 6). A value of P < 0.05 was considered statistically significant.
Results
The diagnosis and genetic test to identify an unknown mutation in HHT
The HHT patient referred in this research is a 62-year-old female patient with recurrent epistaxis for 40 years, visible telangiectasia in nasopharyngeal mucosa and fingertips and digestive tract, moderate anemia, gastrointestinal bleeding, pulmonary AVMs (Fig. 1a), and enhanced chest CT showed a round mass in the right lower lung, considering hemangioma. Echocardiography further confirmed the diagnosis of pulmonary arteriovenous fistula. Her mother has a history of epistaxis, and her nephew had intermittent nasal bleeding (Fig. 1b). DNA analysis of this patient in all exon of ENG and ALK1 genes revealed a missense mutation in position c.88T > C of ENG exon 2 (Fig. 1c), and this mutation has not been reported before based on the database supplied online (http://arup.utah.edu/database/ENG/ENG_display.php). The mutation results in amino acid substitution C30R in the OR1 region of ENG (Fig. 1d). The C30* of ENG was considered to be a pathogenic one,4,32 implying the functional importance of the site.
Fig. 1.
Pedigree, clinical diagnostic and genetic analysis of HHT1 patient. (a) The photo of computed tomographic pulmonary angiography of patient. Pulmonary arteriovenous aneurysm is marked by red arrow; (b) pedigree of the patient and her normal relatives, the patient is marked by black arrow; (c) genetic analysis of the patient. The nucleotide substitution c.88T > C of ENG was found in exon2. The mutation is marked by red arrow; (d) schematic representation of extracellular domain of ENG. Two pairs of disulfide bonds and C30R substitution at the OR domain of ENG were marked.
AVM: arteriovenous malformations.
Reprograming of HHT1 patient (ENG, C30R) PBMNCs into iPSC
The roles of ENG gene mutation in human vascular development remains to be elucidated. To study the dynamic changes during vascular development, we established a human cell model by generating patient iPSCs and investigated the impact of ENG mutation on vascular development by in vitro differentiation assay. The reprograming procedure is displayed in Fig. 2a, reprogramed iPSCs from patient PBMNCs have the morphology of human pluripotent stem cells (Fig. 2b and c). The generated iPSC line have the same mRNA expression level of pluripotency marker genes (OCT4, NANOG, SOX2) as human embryonic stem cells H9 (Fig. 2d). It was consistent with protein staining of the makers (OCT4, SOX2, SSEA4, and NANOG) (Fig. 2e). The iPSCs at passage 10 have no exogenous episomal vector DNA and are ready for further analysis (Fig. 2f). Karyotype analysis demonstrates a normal female karyotype (46, XX) of iPSCs derived from patient PBMNCs (Fig. 2g). Its pluripotency was further confirmed by in vivo teratoma formation experiment, three germ layers including ectoderm, mesoderm, and endoderm were found in teratoma (Fig. 2h).
Fig. 2.
Reprograming of HHT1-patient (ENG, C30R) PBMNCs into iPSC. (a) The procedure of HHT1 patient-derived PBMNCs preparation and reprograming; (b) morphological change of cells from PBMNCs expansion to reprogramed iPSCs. The left panel represents PBMNCs, the middle panel represents colonies emerge in feeder-dependent culture medium, and the right panel represents iPSCs maintained in feeder-free system, scale bars, 500 µm; (c) alkaline phosphatase staining of C30R–iPSCs, scale bar, 500 µm; (d) the mRNA expression of pluripotency marker genes including OCT4, SOX2, and NANOG in iPSCs derived from patient (C30R–iPSC) and health donor (Wt iPSC), human embryonic stem cells H9 used as control, normalized to Actin (n = 3 independent experiments); (e) immunostaining of pluripotency markers including OCT4, SOX2, SSEA4, and NANOG in iPSCs. (f) PCR results of episomal vector-specific DNA in iPSCs at passage 10 with vector DNA as positive control and H9 ESCs as negative control. (g) Karyotype analysis of C30R–iPSCs demonstrates a normal female karyotype (46, XX); (h) in vivo pluripotency detection for three germ layers formation by teratoma test, ectoderm (nervous tissue), mesoderm (cartilage tissues), and endoderm (glandular tissue) formation were indicated by hematoxylin and eosin staining, scale bars, 200 µm in ectoderm and mesoderm, 50 µm in endoderm.
PBMNC: peripheral blood mononuclear cell.
Deficiencies in differentiation and tube formation of CD31+ ECs from HHT1 patients
In order to investigate whether the C30R iPSCs have defects in vascular EC induction, iPSCs derived from the patient (C30R–iPSCs) and a healthy donor (Wt-iPSCs) were differentiated into ECs. CD31+ cells were checked by flow cytometry at vascular specification stage (day 7) and VE expansion stage (day 10). Compared with Wt-iPSCs, the number of CD31+ early vascular cells at day 7 (Fig. 3a) were decreased in C30R line. Wt-iPSCs showed about 64% EC induction, while C30R–iPSCs displayed about 33.4% EC induction at day 7 (Fig. 3a). In corresponding to the protein expression pattern, the mRNA level of CD31 in C30R was also significantly decreased (Fig. 3b). After expansion, the percentage of CD31+ mature vascular ECs from C30R–iPSCs (32.4 ± 1.3%) was much lower than that from Wt-iPSCs (52.4 ± 2.5%) (Fig. 3c). The results indicated that iPSCs derived from HHT patient have less potential to differentiate into vascular ECs (CD31+).
Fig. 3.
Deficiencies in differentiation and tube formation of CD31+ endothelial cells from HHT1 patients. Flow cytometry analysis of the percentage of CD31+ cells demonstrated the number of the positively stained cells were greatly reduced in C30R–iPSCs at EC differentiation day 7 (a) and day10 (c), Wt-iPSCs (ENG, Wt) differentiation was performed as positive control; (b) quantitative PCR analysis revealed the reduced expression of CD31 at day7 in C30R cells, normalized to Actin (n = 3 independent experiments), * P < 0.05; (d) immunostaining analysis revealed that enriched C30R-ECs and Wt-ECs expressed CD31 and CD144, scale bars, 100 µm. (e) C30R VEs displayed impaired tube formation, scale bars, 500 µm; (f) the junction number and branch number of each assay were evaluated by WimTube (n = 3 independent experiments). * P < 0.05.
EC: endothelial cell; iPSC: induced pluripotent stem cell.
We have shown that VE differentiation rate was reduced in patient-derived iPSCs. To examine the functional change in C30R cells, tube formation experiment was performed. Although no significant difference was found in the expression of VE cells markers CD31 and CD144 between purified Wt-VEs and C30R-VEs (Fig. 3d), the tube formation ability was apparently decreased in enriched C30R VEs (Fig. 3e). These results were further confirmed by junction number and branch number counting C30R VEs (Fig. 3f). These observations suggested an impaired tube formation of VE cells from the HHT patient.
The ER retention of mutant ENG in C30R iPSCs-EC
Next, we wonder whether the amino acid mutation of ENG carried by HHT patient would affect the structure and subcellular localization of ENG protein. Subcellular localization analysis by immunostaining revealed that wild-type ENG localized in the cell membrane, while C30R mutant protein aggregated around the nucleus (Fig. 4a). Further analysis by co-staining demonstrated that the mutant protein was co-localized with ER marker Calnexin (Fig. 4b). Thus, the C30R mutation of ENG affects its cell surface localization. The heterozygous mutation of ENG may reduce the amount of functional ENG protein to traffic to cell surface.
Fig. 4.
C30R mutation in HHT1 patient causes protein ER retention. (a) Subcellular localization analysis of the ENG by immunostaining on either non-permeabilized cells or permeabilized cells. C30R ECs displayed ER retention of the protein on permeabilized cells, scale bars, 10 µm; (b) immunostaining analysis by confocal microscopy revealed the co-localization of ER marker Calnexin and ENG (C30R) on permeabilized cells, scale bars, 10 µm. Merging shows co-localization of both proteins in yellow. Nuclei were stained with DAPI. ER retention of ENG (C30R) are marked by white arrow.
ENG: Endoglin; EC: endothelial cell.
Autologous correction of the c.88T > C mutation in iPSCs derived from the HHT1 patient by CRISPR/Cas9
To correct the mutation of c.88T > C at ENG in the HHT patient-derived iPSCs, we constructed a donor plasmid by inserting 1500bp fragments of the genomic sequence including exon2 into piggyBac vector. The piggyBac vector contains a puro gene for positive selection (Fig. 5a). The fragments were amplified from normal human genomic DNA. Three gRNAs were designed and chemically synthesized, and inserted into PX458 vector which could express both of Cas9 and gRNA. These constructs were individually transfected into 293T cells. The efficincy of these gRNAs to cleave ENG were determined by the T7 endonuclease 1 assay (data not shown). The HHT patient-derived iPSCs were co-transfected with the donor plasmid and px458 containing gRNA to generate corrected clones. Corrected clones were identified by genome DNA fragments sequencing and further confirmed by RT-PCR products sequencing (Fig. 5b). Analyses of the potential off targets site showed that no mutation were found in the corrected iPSCs (data not shown). The corrected iPSCs have the same mRNA expression level of pluripotency marker genes (OCT4, NANOG, SOX2) as iPSCs from HHT patient (Fig. 5c).
Fig. 5.
CRISPR–Cas9 correction of the c.88T > C mutation in iPSCs derived from the HHT1 patient. (a) Schematic representation of our CRISPR–Cas9/HDR strategy for the correction of the c.88T > C (middle, E2). A donor plasmid (top) containing 1500bp of wild-type sequence including exon 2. Tested gRNAs and PAM (orange) are located downstream of the mutation site. Repaired allele (bottom). (b) Sequencing of representative RT-PCR products. The mutation is marked by red arrow. (c) The mRNA expression of pluripotency marker genes including OCT4, SOX2, and NANOG in iPSCs, normalized to Actin (n = 3 independent experiments).
Note: ENG mutation, iPSC-derived ECs from the HHT patient; Correction, CRISPR-corrected iPSC-derived ECs.
ENG: Endoglin.
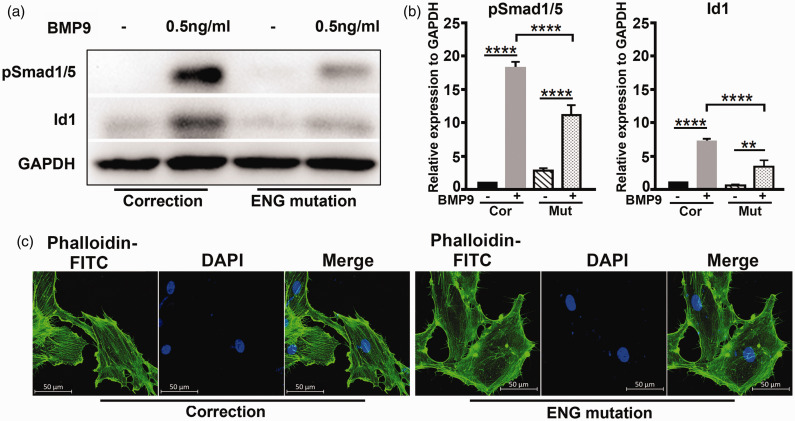
C30R mutation of ENG affects BMP9 downstream signaling
Other ENG HHT pathogenic mutations have been reported, many of which lead to the decrease of BMP9 signal.27 C30 forms a disulfide bond with C207; C30R mutant failed to localize to the cell surface as C207R mutant, which had no response to BMP9.27 We wonder whether C30R heter-mutants carried by the HHT patient may cause reduced BMP9 signaling. To address the question, we used the CRISPR/cas9 genetic correction on mutant HHT-iPSCs to perform autologous comparison. As shown in Fig. 6a and b, in cells bearing ENG mutation and after correction, BMP9 treatment results in increased level of BMP downstream Smad1/5 phosphorylation and Id1 expression, the fold change of which were much higher in corrected cells, indicating the mutation of ENG affects BMP9 downstream signaling. We further examined whether the cytoskeleton of ECs was affected in ECs with defective BMP/ENG signaling. We have compared the cytoskeleton of iPSC-derived ECs with ENG mutation and after correction by F-actin staining (F-actin was labeled with phalloidin-FITC). As shown in Fig. 6c, ENG mutation cells showed a disorganized cytoskeleton, and after correction, the cells formed a highly organized cytoskeleton. These results are consistent with previous reports that HHT1 BOECs showed a disorganized and depolymerized cytoskeleton.24
Fig. 6.
C30R mutation of ENG affects BMP9 downstream signaling. Western blot (a) and densitometric (b) analysis of Smad1/5 phosphorylation and Id1 in ENG mutation and correction cultured without or with 0.5 ng/mL BMP9 for 2 h (n = 3 independent experiments). ****P < 0.0001, **P < 0.01. (c) Immunostaining analysis of F-actin in ENG mutation and correction by confocal microscopy, ENG mutation showed a disorganized cytoskeleton, while correction a highly organized cytoskeleton, scale bars, 50 µm.
Cor, correction; Mut, ENG mutation; BMP: bone morphogenetic protein; ENG: Endoglin.
Discussion
In this study, a novel mutation of ENG (c.88T > C) was identified in HHT patient, this mutation results in a substitute of Cys by Arg at position 30 of translated amino acid sequence. The crystal structure of ENG protein revealed that c30-c207 and c53-c182 are two pairs of conserved disulfide bonds. C207R, which resulted in disruption of ENG folding by interfering with correct disulfide bond formation, was reported as a pathogenic mutation.17 Thus, the C30R mutation may affect the folding of ENG in the same way as C207R mutation. C30* was reported as a pathogenic mutation with premature termination of ENG translation.4,32 Autologous correction demonstrated the difference of BMP signaling and cytoskeleton organization before and after correction. Taken together, these reports suggested that the mutation identified in this research may be a pathogenic mutation. For this study, we sequenced whole exons of ENG and ALK1 genes in patient samples. But we know that HHT-related mutations were found in at least four genes, including ENG, ALK1, Smad4, and BMP9,4–6 and mutations were also founded at non-coding regions, promoters, and regulatory regions.7,8 The deep sequencing may provide more information about other pathogenic mutation.
Although it has been known that HHT is a vascular disease with deficient angiogenesis and vascular remodeling, its molecular mechanism is still unclear. Most studies on the pathogenesis of HHT are based on mouse or zebrafish models, which are generated by knocking out/down of ENG or ALK1 or Smad4 or BMP9 genes. As far as we know, there are no humanized models to study the pathogenesis of the disease. To obtain patient-derived cells is necessary for establishing humanized disease models. The vascular cells, such as arterial smooth muscle cells and ECs, can be directly obtained from organ donors or patients undergoing vascular surgical procedures. However, they are not readily available in patients with HHT because the majority of patients do not require surgery. On the other hand, the proliferative potential of primary cultured vascular cells obtained from human tissues is limited. The advantages of using pluripotent stem cells include that they have self-renew capacity with unlimited passage and can be differentiated into nearly any cell type for different research purpose. Establishment of patient-specific or disease-specific pluripotent stem cells is useful for studying the cellular and molecular pathological mechanisms of diseases when the patient sample is not available. Here, we use 10 mL of peripheral blood from patient for reprograming PBMNCs to iPSCs and further differentiated into EC lineage. This amount of peripheral blood is sufficient to separate PBMNCs for reprograming. The episomal iPSC reprograming vectors were employed in this study for producing transgene-free, virus-free iPSCs. The differentiation rate study demonstrated that the efficiency of differentiating HHT iPSCs toward the VE was reduced compared with iPSCs derived from healthy donor. Previous studies revealed that eng−/− mice die around gestational day 10.5 primarily due to the defects in vascular and heart development.18–20 Our results further confirmed the role of ENG in vascular cells by using human iPSCs.
Heterozygous germline mutation of ENG has been reported as a major cause of HHT1. Mallet et al. revealed that various mechanisms contributed to ENG loss-of-function, including less responsiveness to BMP9 stimulation and incorrected subcellular localization.27 In our study, we suspected C30R mutation may affect the disulfide bridge formation with C207 in OR1 of ENG. Structural analysis revealed that ENG binds to BMP9 through its conserved hydrophobic groove rather than disulfide bridges in OR1.17,27 Thus, C30R mutation of ENG may cause misfolding but not affect binding affinity with BMP9. Current studies of cellular mechanism in HHT are mainly based on transfection with mutated ENG gene.27,33–35 The iPSCs generated in this study allow us to detect the endogenous proteins in derived ECs, which reflects the expression pattern of ENG in HHT patient. Our study showed that C30R mutation resulted in the mislocalization of ENG to the ER. It suggested that heterozygous mutation of ENG (c.88T > C) caused misfolding of the protein, which was trapped in the ER and cannot traffic to cell surface as wild-type. The ER retention was also found in C207R (cannot form disulfide bond with C30 in OR1) mutated ENG.27 In similar mechanism, C53R cannot form disulfide bond with C182 in OR2, and ENG (C53R) displayed ER accumulation. These results indicated that disulfide bonds in OR are important for the correct folding of proteins. Ali et al. revealed that ER retention was a potential pathogenic mechanism of HHT1 and HHT2.34,36 One possible treatment could be using trafficking reagent to help misfolded but functional ENG/ALK1 relocate to cell membrane from ER.
Previous reports have shown that BMP pathway plays a key role in vascular diseases, such as HHT and PAH.37,38 In our study, we found that ECs from HHT patient had less sensitivity to BMP9, which may lead to the abnormal tube formation and disorganized cytoskeleton. A disorganized cytoskeleton in vascular cells affects their response to flow stress.24,26 Previous reports have showed that vascular tone response to blood flow play a role in the formation of AVM.23,39 We hypothesize that the mutation of ENG (C30R) results in defective BMP/ENG signal, which may lead to the disorganized cytoskeleton of ECs, and eventually contributes to the generation of AVM.
In 2014, about 13.8% adult hospitalizations with HHT had a concurrent diagnosis of PH in the United States.40 From a genetic point of view, both diseases seem to be intrinsically linked with the mutations occurred in BMP signaling pathway: 70% of HPAH patients and 20% of IPAH patients carry BMPR2 heterozygous mutations, while 80% of HHT patients carry ENG or ALK1 heterozygous mutations. However, why the defects of different components in same signaling pathway lead to different phenotypes? For example, HHT manifests dilated arteriovenous teratoma, while PAH is characterized with occlusive pulmonary vascular remodeling. The underline mechanism needs to be further studied and clarified.
In conclusion, we found a novel missense mutation of ENG in HHT patient, and established a human stem cell model of HHT by generating patient iPSCs. The iPSCs showed decreased differentiation efficiency toward EC. The ER retention of mutant ENG reduced its cell surface localization to affect BMP signaling. Autologous mutation correction further provides evidence of the cytoskeleton defects in ECs in HHT.
Acknowledgements
We thank Xiaoxi Yang for providing help on iPSCs generation, and Yonggang Zhang for providing related reagents. We also thank Hongxian Liu for performing part of the experiments.
Footnotes
Author contributions: JY participated in study design and revised the manuscript. ZF participated in the design of the study, data acquisition, statistical analysis, and drafting of the manuscript. XZ conducted the genetic screening, XL and YL participated in cell culture and data collection, FM provided the technique support on iPSC generation, and BL provided clinical advices and the material of HHT patient.
Conflict of interest: The author(s) declare that there is no conflict of interest.
Ethical approval: The patients’ information was recorded according to Peking Union Medical College (PUMC) regulation together with the related application approved by the ethics committee of IBMS, Chinese Academy of Medical Sciences, and School of Basic Medicine. Informed consent was obtained from the patient.
Funding: This work was supported by grants from National Key Research and Development Program of China stem cell and translational research (No: 2016YFA0102300), the Natural Science Foundation of China (NSFC, No. 81870051), CAMS Innovation Fund for Medical Sciences (CIFMS 2016-I2M-4-003), and PUMC Youth Found and the Fundamental Research Funds for the Central Universities (2017310013).
ORCID iDs
References
- 1.Giordano P, Nigro A, Lenato GM, et al. Screening for children from families with Rendu-Osler-Weber disease: from geneticist to clinician. J Thromb Haemost 2006; 4: 1237–1245. [DOI] [PubMed] [Google Scholar]
- 2.Kjeldsen AD, Oxhoj H, Andersen PE, et al. Prevalence of pulmonary arteriovenous malformations (PAVMs) and occurrence of neurological symptoms in patients with hereditary haemorrhagic telangiectasia (HHT). J Intern Med 2000; 248: 255–262. [DOI] [PubMed] [Google Scholar]
- 3.Sabba C, Pasculli G, Lenato GM, et al. Hereditary hemorrhagic telangiectasia: clinical features in ENG and ALK1 mutation carriers. J Thromb Haemost 2007; 5: 1149–1157. [DOI] [PubMed] [Google Scholar]
- 4.McAllister KA, Grogg KM, Johnson DW, et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet 1994; 8: 345–351. [DOI] [PubMed] [Google Scholar]
- 5.Johnson DW, Berg JN, Gallione CJ, et al. A second locus for hereditary hemorrhagic telangiectasia maps to chromosome 12. Genome Res 1995; 5: 21–28. [DOI] [PubMed] [Google Scholar]
- 6.Gallione CJ, Repetto GM, Legius E, et al. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4). Lancet 2004; 363: 852–859. [DOI] [PubMed] [Google Scholar]
- 7.Johnson DW, Berg JN, Baldwin MA, et al. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet 1996; 13: 189–195. [DOI] [PubMed] [Google Scholar]
- 8.Gallione CJ, Richards JA, Letteboer TG, et al. SMAD4 mutations found in unselected HHT patients. J Med Genet 2006; 43: 793–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olivieri C, Lanzarini L, Pagella F, et al. Echocardiographic screening discloses increased values of pulmonary artery systolic pressure in 9 of 68 unselected patients affected with hereditary hemorrhagic telangiectasia. Genet Med 2006; 8: 183–190. [DOI] [PubMed] [Google Scholar]
- 10.Sopena B, Perez-Rodriguez MT, Portela D, et al. High prevalence of pulmonary hypertension in patients with hereditary hemorrhagic telangiectasia. Eur J Intern Med 2013; 24: e30–e34. [DOI] [PubMed] [Google Scholar]
- 11.Simonneau G, Gatzoulis MA, Adatia I, et al. [Updated clinical classification of pulmonary hypertension]. Turk Kardiyoloji Dernegi arsivi: Turk Kardiyoloji Derneginin yayin organidir 2014; 42(Suppl 1): 45–54. [PubMed] [Google Scholar]
- 12.Harrison RE, Flanagan JA, Sankelo M, et al. Molecular and functional analysis identifies ALK-1 as the predominant cause of pulmonary hypertension related to hereditary haemorrhagic telangiectasia. J Med Genet 2003; 40: 865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chizinga M, Rudkovskaia AA, Henderson K, et al. Pulmonary hypertension prevalence and prognosis in a cohort of patients with hereditary hemorrhagic telangiectasia undergoing embolization of pulmonary arteriovenous malformations. Am J Respir Crit Care Med 2017; 196: 1353–1356. [DOI] [PubMed] [Google Scholar]
- 14.Kuhnel T, Wirsching K, Wohlgemuth W, et al. Hereditary hemorrhagic telangiectasia. Otolaryngol Clin North Am 2018; 51: 237–254. [DOI] [PubMed] [Google Scholar]
- 15.Gougos A, Letarte M. Identification of a human endothelial cell antigen with monoclonal antibody 44G4 produced against a pre-B leukemic cell line. J Immunol 1988; 141: 1925–1933. [PubMed] [Google Scholar]
- 16.Castonguay R, Werner ED, Matthews RG, et al. Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. J Biol Chem 2011; 286: 30034–30046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito T, Bokhove M, Croci R, et al. Structural basis of the human endoglin-BMP9 interaction: insights into BMP signaling and HHT1. Cell Rep 2017; 19: 1917–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourdeau A, Dumont DJ, Letarte M. A murine model of hereditary hemorrhagic telangiectasia. J Clin Invest 1999; 104: 1343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li DY, Sorensen LK, Brooke BS, et al. Defective angiogenesis in mice lacking endoglin. Science 1999; 284: 1534–1537. [DOI] [PubMed] [Google Scholar]
- 20.Arthur HM, Ure J, Smith AJ, et al. Endoglin, an ancillary TGFbeta receptor, is required for extraembryonic angiogenesis and plays a key role in heart development. Dev Biol 2000; 217: 42–53. [DOI] [PubMed] [Google Scholar]
- 21.Young K, Krebs LT, Tweedie E, et al. Endoglin is required in Pax3-derived cells for embryonic blood vessel formation. Dev Biol 2016; 409: 95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee S, Dhara SK, Bacanamwo M. Endoglin is a novel endothelial cell specification gene. Stem Cell Res 2012; 8: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sugden WW, Meissner R, Aegerter-Wilmsen T, et al. Endoglin controls blood vessel diameter through endothelial cell shape changes in response to haemodynamic cues. Nature Cell Biol 2017; 19: 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez LA, Sanz-Rodriguez F, Zarrabeitia R, et al. Blood outgrowth endothelial cells from Hereditary Haemorrhagic Telangiectasia patients reveal abnormalities compatible with vascular lesions. Cardiovas Res 2005; 68: 235–248. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez LA, Garrido-Martin EM, Sanz-Rodriguez F, et al. Gene expression fingerprinting for human hereditary hemorrhagic telangiectasia. Hum Mol Genet 2007; 16: 1515–1533. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez LA, Sanz-Rodriguez F, Blanco FJ, et al. Hereditary hemorrhagic telangiectasia, a vascular dysplasia affecting the TGF-beta signaling pathway. Clin Med Res 2006; 4: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallet C, Lamribet K, Giraud S, et al. Functional analysis of endoglin mutations from hereditary hemorrhagic telangiectasia type 1 patients reveals different mechanisms for endoglin loss of function. Hum Mol Genet 2015; 24: 1142–1154. [DOI] [PubMed] [Google Scholar]
- 28.Chou BK, Gu H, Gao Y, et al. A facile method to establish human induced pluripotent stem cells from adult blood cells under feeder-free and xeno-free culture conditions: a clinically compliant approach. Stem Cells Transl Med 2015; 4: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlova VV, van den Hil FE, Petrus-Reurer S, et al. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat Protoc 2014; 9: 1514–1531. [DOI] [PubMed] [Google Scholar]
- 30.Xing Y, Zhao S, Wei Q, et al. A novel piperidine identified by stem cell-based screening attenuates pulmonary arterial hypertension by regulating BMP2 and PTGS2 levels. Eur Respir J 2018; 51: pii: 1702229. [DOI] [PubMed] [Google Scholar]
- 31.Ran FA, Hsu PD, Wright J, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013; 8: 2281–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berg J, Porteous M, Reinhardt D, et al. Hereditary haemorrhagic telangiectasia: a questionnaire based study to delineate the different phenotypes caused by endoglin and ALK1 mutations. J Med Genet 2003; 40: 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pece-Barbara N, Cymerman U, Vera S, et al. Expression analysis of four endoglin missense mutations suggests that haploinsufficiency is the predominant mechanism for hereditary hemorrhagic telangiectasia type 1. Hum Mol Genet 1999; 8: 2171–2181. [DOI] [PubMed] [Google Scholar]
- 34.Ali BR, Ben-Rebeh I, John A, et al. Endoplasmic reticulum quality control is involved in the mechanism of endoglin-mediated hereditary haemorrhagic telangiectasia. PloS One 2011; 6: e26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Forg T, Hafner M, Lux A. Investigation of endoglin wild-type and missense mutant protein heterodimerisation using fluorescence microscopy based IF, BiFC and FRET analyses. PloS One 2014; 9: e102998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hume AN, John A, Akawi NA, et al. Retention in the endoplasmic reticulum is the underlying mechanism of some hereditary haemorrhagic telangiectasia type 2 ALK1 missense mutations. Mol Cell Biochem 2013; 373: 247–257. [DOI] [PubMed] [Google Scholar]
- 37.Cai J, Pardali E, Sanchez-Duffhues G, et al. BMP signaling in vascular diseases. FEBS Lett 2012; 586: 1993–2002. [DOI] [PubMed] [Google Scholar]
- 38.Dyer LA, Pi X, Patterson C. The role of BMPs in endothelial cell function and dysfunction. Trends Endocrinol Metab 2014; 25: 472–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin Y, Muhl L, Burmakin M, et al. Endoglin prevents vascular malformation by regulating flow-induced cell migration and specification through VEGFR2 signalling. Nat Cell Biol 2017; 19: 639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harder EM, Fares WH. Hospitalizations with hereditary hemorrhagic telangiectasia and pulmonary hypertension in the United States from 2000 to 2014. Respir Med 2019; 147: 26–30. [DOI] [PubMed] [Google Scholar]