Abstract
Background:
PARPBP (PARP1 binding protein) is an important suppressor of homologous recombination during DNA repair, but the expression and function of PARPBP in breast cancer remain unclear.
Methods:
PARPBP expression was analyzed in breast cancer patient samples and public datasets for its correlation with clinical outcome. The function of PARPBP in breast cancer cell proliferation and anthracycline treatment response were studied both in vitro and in vivo.
Results:
PARPBP was upregulated significantly at both mRNA and protein levels in breast cancer tissues compared with normal breast tissues. PARPBP high expression group had poorer overall survival (OS) than the PARPBP low expression group. Knockdown of PARPBP suppressed breast cancer cell proliferation and colony formation while overexpression of PARPBP did the opposite. We found that transcription factor forkhead box M1 (FOXM1) could activate PARPBP expression by directly binding to the promoter of PARPBP. In addition, high expression of PARPBP related with anthracycline resistance in breast cancer. Depletion of PARPBP increased breast cancer cell apoptosis and DNA damage caused by epirubicin. Moreover, tumor xenograft experiments further demonstrated that PARPBP was involved in breast cancer anthracycline resistance.
Conclusion:
Taken together, our results highlight that PARPBP is a prognostic marker and confers anthracycline resistance on breast cancer.
Keywords: biomarker, breast cancer, chemotherapy, PARPBP, prognosis
Introduction
Breast cancer is a devastating disease whose management is complicated by its high molecular heterogeneity.1,2 Chemotherapy is a conventional and essential treatment that has reduced the death rate for breast cancer patients.3 However, resistance to chemotherapeutic agents is a major obstacle for the effective treatment of breast cancer. Chemotherapy-refractory breast cancer patients recur within months to years after treatment, leading finally to subsequent death. Therefore, there is a critical need to elucidate the mechanisms of resistance to chemotherapy and to develop new chemosensitizers.
Induction of DNA damage is a predominant anti-tumor mechanism for many chemotherapy drugs.4 Anthracycline antibiotics cause DNA damage by embedding between the DNA double-stranded bases. Platinum-based drugs induce DNA damage by binding to DNA, thereby creating inter- or intra-strand cross links. However, some tumor cells can develop drug resistance through repair mechanisms that counteract the DNA damage. Research has shown that enhancing the DNA repair capability of tumor cells results in intrinsic and therapy-induced chemoresistance. Anthracyclines are currently the cornerstone drugs used widely in breast cancer chemotherapy.5 In contrast, the scope of application of platinum drugs is relatively limited, mainly for triple-negative breast cancer (TNBC) or breast cancer patients with BRCA1/2 mutations.6,7 Therefore, we focus only on anthracycline-based resistance in this study.
PARPBP (PARP1 binding protein), also named PARI or C12orf48, is an important homologous recombination inhibitor of human cells during DNA repair.8 Previous studies have shown that PARPBP is expressed abnormally in a variety of tumors, and interacts directly with some regulators of DNA repair, including PARP-1, PCNA, and RAD51.9,10 Downregulation of PARPBP could preserve genomic stability and improve homologous recombination. A previous study by Pitroda et al. developed a Recombination Proficiency Score (RPS), which was calculated based on the expression levels for four genes including PARPBP.11 They showed that RPS provided predictive characterization of individual breast cancers. Low RPS breast tumors exhibit a heightened sensitivity to DNA-damaging therapy.12 However, the function of PARPBP in breast cancer remains unclear.
In this study, we performed in vitro and in vivo experiments to evaluate the effects of PARPBP in breast cancer anthracycline resistance. A better understanding of breast cancer chemoresistance could aid the development of new therapeutic approaches and provide targets for breast cancer patients.
Materials and methods
Bioinformatics analysis of public datasets
PARPBP mRNA expression data in breast cancer and various types of cancers and normal tissues were from Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancerpku.cn/index.html) and Oncomine database (www.oncomine.org).13,14 The threshold of differential expression was set at a 1.5-fold difference in transcripts per million (TPM) between cancers and normal tissues with a P value < 0.0001. The Cancer Genome Atlas (TCGA) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) datasets were analyzed for co-expression.15,16 Figures were generated using Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancerpku.cn/index.html), the cBio Cancer Genomics Portal (http://cbioportal.org) and the KM Plotter Online Tool (http://www.kmplot.com).2,13,17–19 TCGA publication guidelines were followed for the use of all TCGA data in this manuscript.
Patients and tissue specimens
In total, 162 breast cancer tissues (cohorts 1, 2) were obtained from the Sun Yat-Sen University Cancer Center and prepared as paraffin blocks. The Ethics Committee of Sun Yat-Sen University Cancer Center Health Authority approved this study (GZR2017-163). All patients provided written informed consent for this research. All samples were collected in accordance with Health Insurance Portability and Accountability Act guidelines and the Declaration of Helsinki. Cohort 1: a total of 137 breast cancer tissues used for survival analysis were collected between March 2005 and September 2011. Using the formalin-fixed paraffin-embedded (FFPE) method, a tissue microarray was built for immunohistochemistry (IHC) analysis. Cohort 2: a total of 25 patients received anthracycline-based neoadjuvant chemotherapy. The response criteria were determined by the Response Evaluation Criteria in Solid Tumors (RECIST, version 1.1) and postoperative pathology. Tissue specimens were collected between March 2017 and September 2018 before patients received neoadjuvant chemotherapy. Progressive disease or stable disease was considered as chemo resistant (resistance group), while partial response or pathological complete response (pCR) as chemo sensitive (non-resistance group). Pathological complete response was defined as no residual invasive carcinoma in both breast and lymph nodes.
IHC analysis
Immunohistochemistry staining was performed as described previously.20 Both staining intensity (0, negative staining; 1, weak staining; 2, moderate staining; 3, strong staining) and extent of staining as the percentage of positive cells (1, 0–25%; 2, 26–50%; 3, 51–75%; 4, 76–100%) were scored. The final quantitation of each staining was obtained by multiplying the above two scores. PARPBP expression was classified into two groups: high expression group (final score was higher than 2.0) and low expression group (2.0 or less).
Cell lines
Human breast cancer cell lines (MDA-MB-231, BT549, HCC38, T47D, MDA-MB-468, BT474, Skbr-3, and MCF-7) and normal mammary epithelial cell lines (MCF-10A) were obtained from the American Type Culture Collection (Manassas, VA, USA). The drug resistance cell subline of MCF-7 (MCF-7/EPI) was derived from the parental cells by using the low concentration epirubicin (EPI) stepwise incremental method.21 All cell lines used in this study had been cultured for less than 6 months and tested mycoplasma free when the experiments were performed.
RNA interference and overexpression
Sequences of siRNAs were listed in Supplemental Table S2. Overexpression and knockdown constructs pcDNA3.1-PARPBP-HA, pcDNA3.1-FOXM1, and shFOXM1 were obtained from GeneCopoeia (Rockville, MD, USA). The lentivirus for knocking down PARPBP (shPARPBP) was packaged and purchased from GenePharma (Shanghai, China) using siPARPBP1 corresponding sequences. Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative reverse transcription polymerase chain reaction
Total RNA was isolated using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The Nano Drop ND-1000 Spectrophotometer (Nano Drop, Waltham, MA, USA) was used to evaluate RNA quality. Complementary DNA was synthesized using the PrimeScript RT reagent kit (Takara Bio Inc., Dalian, China). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using SYBR Premix Ex Taq (Takara Bio Inc., Dalian, China). Each reaction was performed in triplicate. Primer sequences are given in Supplemental Table S1. Values were normalized to internal controls, and fold changes were calculated through relative quantification (2−ΔΔCt).
Cell counting kit-8 assay
Cell viability was assessed by cell counting kit-8 assay (CCK8; Dojindo Laboratories, Kumamoto, Japan). Cells (1 × 103) were seeded into 96-well plates. After 2 h of CCK8 solution (10 μl) incubation at 37°C, the absorbance at 450 nM was measured using a microtiter plate reader (Bio-Tek EPOCH2, BioTek Instruments, Inc., Winooski, VT, USA).22
Colony formation assay
At 48 h after transfection, cells were cultured with or without EPI at the indicated concentrations for 3 h. The cells were then harvested, seeded 500 cells per well into six-well plates and cultured for an additional 2 weeks. For scoring the colony-forming units, plates were stained with crystal violet (crystal violet 0.5%, ethanol 2%) and photographed.
Western blot analysis
Western blot analysis was performed as previously described.23 The following primary antibodies were used in western blot analysis: anti-PARPBP (1:500, Abcam, Cambridge, MA, USA), anti-FOXM1 (1:1000, Cell Signaling Technology, MA, USA) and β-actin (1:5000, Cell Signaling Technology, MA, USA). The membranes were further incubated with secondary antibody (1:5000 dilution) and ECL reagents (New England Biolabs, Ipswich, MA, USA) were used to detect the protein.
Apoptosis assay
Cell apoptosis was detected using the Andy Fluor 488 Annexin V and PI Apoptosis Kit (GeneCopoeia). After treatment with or without EPI at the indicated concentrations for 48 h, cells were harvested and resuspended in 400 ml of binding buffer. Next, 5 μl of Annexin V-FITC and 2 μl of propidium iodide (PI) were added to the suspensions, and the cells were incubated in the dark at 4°C for 15 min before being analyzed through a fluorescence cytometer; 10,000 events were considered for the analysis.
Promoter reporters and dual-luciferase assay
The PARPBP promoter region (−1500, +76) was amplified and the resulting fragment cloned into the luciferase reporter plasmids pGL3-basic vector (Promega, Madison, WI, USA), designated as pGL3-PARPBP. Mutant construct pGL3-PARPBP-MU was generated by site-directed mutagenesis. Luciferase assay was performed as described previously.23 Each experimental analysis was repeated three times.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed using the Zymo-Spin ChIP kit (Zymo Research, Irvine, CA, USA) according to the manufacturer’s instructions. Chromatin was sheared mechanically by sonication after cells were collected, and cross-linked by formaldehyde. Protein–DNA complexes were precipitated by control immunoglobulin G, anti-histone H3, and anti-FOXM1 antibodies respectively, followed by eluting the complex from the antibodies. The amount of DNA was assessed further by quantitative real-time PCR, using the primers specific for PARPBP promoter and SYBR Select Master Mix (Applied Biosystems, Grand Island, NY, USA).
Tumor xenograft experiments
Animal experimentation was conducted in accordance with the guidelines of the local institutional animal care and use committee. All animal studies were approved by the Ethics Committee (GDREC2019402A). PARPBP-overexpressed (PARPBP/231) and control cells (Ctrl/231) were collected and suspended in 200 μl of PBS at a concentration of 5 × 106 cells per ml, then injected into the mammary fat pads of 6-week-old female BALB/c nude mice (five in each group). At 10 days after injection, the mice were injected intraperitoneally with 5 mg/kg EPI or normal saline (NS) (once per 2 days) for another 2 weeks. Dynamic tumor growth status was monitored weekly using an IVIS imaging system (PerkinElmer, Waltham, MA, USA).23 The tumor size was measured every 4 days. Tumor volumes were determined according to the following formula: A × B2/2, where A is the largest diameter, and B is the diameter perpendicular to A.24 The xenograft tumors were harvested after 4 weeks and the tumors were weighed. Mice were sacrificed under anesthesia (10% chloral hydrate, peritoneal injection), and all efforts were made to minimize discomfort and pain. Tumor tissues were also processed and sectioned for histological evaluation.
Statistical analysis
Statistical analyses were performed using SPSS 22.0 software (SPSS, Chicago, IL, USA). Student’s t test was used to make a statistical comparison between groups. The Chi-squared test and Fisher’s exact test were used to investigate the significance of the correlation between PARPBP expression and clinicopathological features in breast cancer patients. Survival curves were calculated by the Kaplan–Meier method and compared with the log-rank test. A p value <0.05 was considered significant.
Results
PARPBP expression is upregulated in breast cancer and correlates with prognosis of breast cancer patients
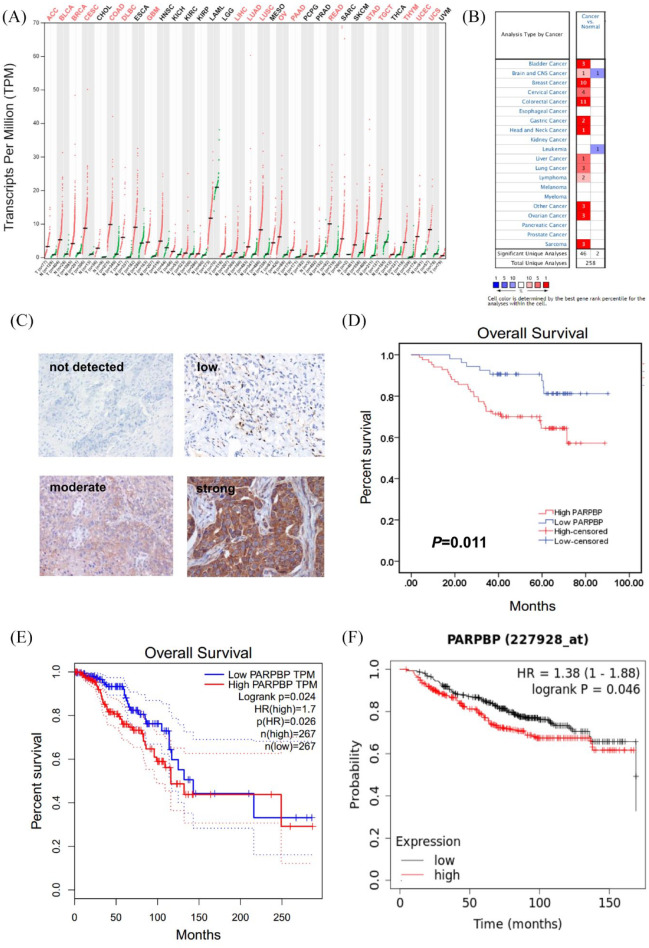
We first explored PARPBP mRNA level in common types of human cancer and normal tissues from public databases. Analysis of data from the GEPIA database showed that PARPBP mRNA expression was upregulated significantly in breast invasive carcinoma (BRCA) and 17 other common types of cancer tissues compared with adjacent normal tissues (Figure 1A). Oncomine database analysis also revealed that PARPBP mRNA expression of breast cancer increased in 10 data sets compared with normal tissues (Figure 1B). Then, we validated expression of PARPBP at the protein level by IHC of breast cancer tissue microarrays (all subtypes, n = 137). While PARPBP staining in adjacent normal breast tissues was usually undetected, a high proportion of breast cancer tissues displayed strong (47/137), moderate (37/137), or low (41/137) PARPBP staining, and only 12 patients had undetectable staining (Figure 1C). These results suggested that PARPBP was upregulated significantly at both mRNA and protein levels in breast cancer tissues compared with normal breast tissues.
Figure 1.
PARPBP expression is up-regulated in breast cancer and correlates with prognosis of breast cancer patients. PARPBP mRNA expression in different normal human tissues and cancer tissues from (A) GEPIA (red represents cancer tissues; green represents normal human tissues) and (B) oncomine database. (C) Representative IHC images of PARPBP in breast cancer tissues. (D, E) High levels of PARPBP correlate with poor prognosis. The OS curves for our cohort1 (D), TCGA (E), and KM Plotter datasets (F).
GEPIA, Gene Expression Profiling Interactive Analysis; HR, hazard ratio; IHC, immunohistochemistry; OS, overall survival; PARBP, PARP1 binding protein; TCGA, The Cancer Genome Atlas; TPM, transcripts per million.
Based on the final quantitation of each breast cancer tissue IHC staining (multiplying the staining intensity score and the extent of staining score), 137 patients were classified into two groups: PARPBP high expression group (score >2.0; n = 84) and PARPBP low expression group (score ⩽2.0; n = 53). As shown in Table 1, the expression of PARPBP was correlated positively with the tumor status, lymph node status, and TNM stage of breast cancer patients (p < 0.001). Furthermore, we found that the PARPBP high expression group had poorer overall survival (OS) compared with the PARPBP low expression group (Figure 1D). Similar OS results were also observed for breast cancer patients in the TCGA (Figure 1E) and KM Plotter tool datasets (Figure 1F) based on relative mRNA expression of PARPBP.
Table 1.
Relationship between PARPBP expression and clinicopathologic factors of breast cancer patients.
| Variables | n = 137 | PARPBP |
p value | |||
|---|---|---|---|---|---|---|
| High | No. (%) | Low | No. (%) | |||
| Age (years) | 1 | |||||
| <50 | 75 | 46 | 33.58% | 29 | 21.17% | |
| ⩾50 | 62 | 38 | 27.74% | 24 | 17.52% | |
| Menopause | 0.857 | |||||
| Yes | 52 | 31 | 22.63% | 21 | 15.33% | |
| No | 85 | 53 | 38.69% | 32 | 23.36% | |
| Tumor status (T) | <0.001* | |||||
| T1 | 35 | 11 | 8.03% | 24 | 17.52% | |
| T2 + T3 + T4 | 102 | 73 | 53.28% | 29 | 21.17% | |
| Lymph node status (N) | <0.001* | |||||
| N0 | 53 | 18 | 13.14% | 35 | 25.55% | |
| N1 | 37 | 22 | 16.06% | 15 | 10.95% | |
| N2 | 22 | 20 | 14.60% | 2 | 1.46% | |
| N3 | 25 | 24 | 17.52% | 1 | 0.73% | |
| Histological grade | 0.295 | |||||
| G1 + G2 | 106 | 62 | 45.26% | 44 | 32.12% | |
| G3 | 31 | 22 | 16.06% | 9 | 6.57% | |
| TNM stage | <0.001* | |||||
| I–II | 87 | 38 | 27.74% | 49 | 35.77% | |
| III–IV | 50 | 46 | 33.58% | 4 | 2.92% | |
| Subtype | 0.353 | |||||
| HR+/HER2+ | 18 | 9 | 6.57% | 9 | 6.57% | |
| HR+/HER2− | 72 | 42 | 30.66% | 30 | 21.90% | |
| HR−/HER2+ | 24 | 18 | 13.14% | 6 | 4.38% | |
| TNBC | 23 | 15 | 10.95% | 8 | 5.84% | |
Significant at p < 0.001.
HER2, human epidermal growth factor receptor 2; HR, hormone receptor; PARPBP, PARP1 binding protein; TNBC, triple negative breast cancer; TNM, tumor, node, metastasis.
PARPBP promotes breast cancer cell proliferation
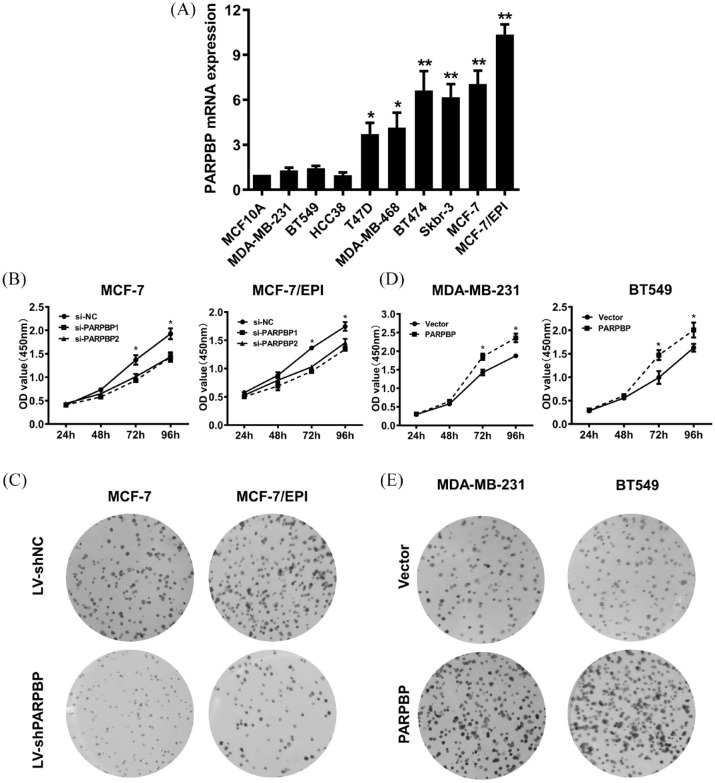
To assess the effects of PARPBP in breast cancer cell proliferation, we first selected suitable cell lines by examining PARPBP mRNA levels in normal mammary epithelial cell line MCF-10A and nine human breast cancer cell lines. As shown in Figure 2A, PARPBP expression is relatively high in MCF-7, MCF-7/EPI, T47D, MDA-MB-468, BT474, and Skbr-3 cell lines, but low in MDA-MB-231, BT549, Hce 38, and MCF-10A. Two specific small interference RNAs against PARPBP mRNA and a negative control were transfected transiently into MCF-7 or MCF-7/EPI. The efficiency of PARPBP knockdown was analyzed by qRT-PCR. In cell viability assays, both MCF-7 and MCF-7/EPI cells transfected with si-PARPBP1/2 slowed cell growth and proliferation compared with cells transfected with the negative control (Figure 2B). In addition, lentivirus particles containing shNC or shPARPB were infected into MCF-7 and MCF-7/EPI cells, and PARPBP knockdown also decreased the ability of colony formation of breast cancer cells (Figure 2C). Conversely, the proliferation and colony formation ability of PARPBP-overexpressed MDA-MB-231 and BT549 cells increased significantly relative to controls (Figure 2D, E). These observations suggested that PARPBP has a positive effect on breast cancer cell growth in vitro.
Figure 2.
PARPBP promotes breast cancer cell proliferation. (A) PARPBP mRNA levels determined by qRT-PCR in MCF-10A and nine human breast cancer cell lines. (B) The growth of MCF-7 and MCF-7/EPI cells infected with si-PARPBP1/2 or si-NC was assayed by CCK8. *p < 0.05; **p < 0.01. (C) Colony formation assays performed on MCF-7 and MCF-7/EPI cells transfected with shPARPB or shNC. (D) The growth of MDA-MB-231 and BT549 cells transfected with PARPBP overexpressing or control vector was assayed by CCK8. *p < 0.05. Colony formation assays performed on MDA-MB-231 and BT549 cells transfected with PARPBP overexpressing or control vector.
CCK8, cell counting kit-8 assay; PARBP, PARP1 binding protein; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
High expression of PARPBP correlates with anthracycline-based resistance
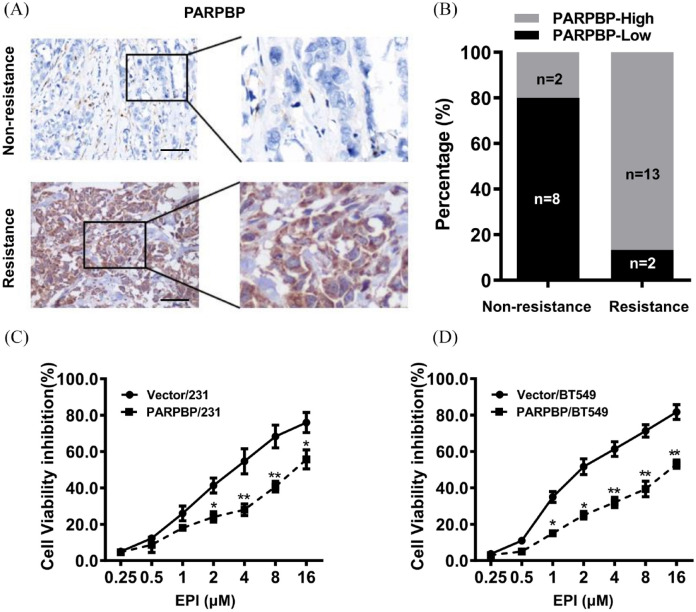
To assess the possible role of PARPBP in anthracycline-based resistance, we performed IHC staining for PARPBP in 25 cases of human breast cancer who later received anthracyclines-based neoadjuvant chemotherapy. The results showed that many more patients had high expression of PARPBP in the chemoresistant group (13/15) than in the nonresistant group (2/10) (Figure 3A, B). In vitro, while EPI treatment at concentration from 1 to 16 µM caused dramatic decrease in cell viability in MDA-MB-231 and BT549 cells, overexpression of PARPBP in these cells recovered cell growth rate markedly (Figure 3C, D). These results indicated that PARPBP might attenuate the sensitivity of breast cancer cell to anthracycline-based chemotherapy drugs.
Figure 3.
High expression of PARPBP correlates with anthracycline resistance in breast cancer. (A) Representative images of immunohistochemical staining of PARPBP in chemo-resistant and non-resistant tumors are shown. Scale bars, 50 μm. (B) Expression of PARPBP in chemoresistant tumors were significantly higher than that in non-resistant breast tumors. (C) PARPBP/231 and Vector/231 cells were treated with various concentrations of EPI for 48 h. (D) PARPBP/BT549 and Vector/BT549 cells were treated with various concentrations of EPI for 48 h. *p < 0.05, **p < 0.01.
EPI, epirubicin; PARBP, PARP1 binding protein.
Depletion of PARPBP increases breast cancer cell apoptosis and DNA damage
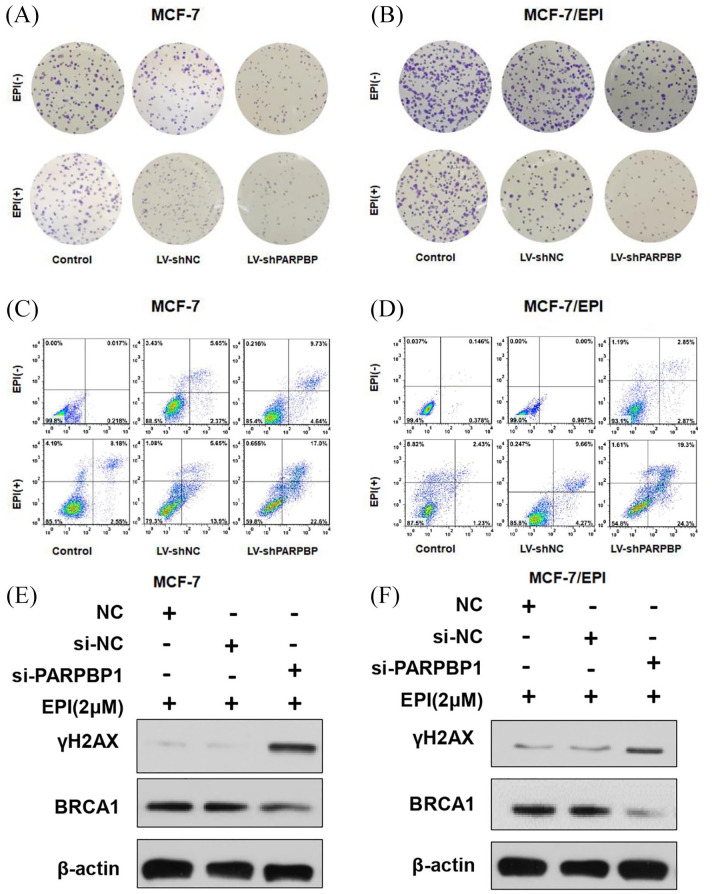
Anthracycline-based chemotherapy drugs such as EPI function by inducing cell apoptosis and DNA damage. Therefore, we examined the effect of PARPBP in cell apoptosis and DNA damage induced by EPI. We found that EPI treatment at 2 µM had minimal effect on colony formation of MCF-7 and MCF-7/EPI cells, but resulted in drastic decrease of colony formation when PARPBP was knocked down (Figure 4A, B). After EPI treatment, the apoptosis proportion was significantly increased in cells transfected with PARPBP shRNA compared with cells transfected with negative control or cells without transfections (Figure 4C, D). These results indicated that depletion of PARPBP could increase breast cancer cell apoptosis caused by EPI. Meanwhile, PARPBP depletion increased significantly the EPI-induced level of γH2AX but decreased BRCA1 protein levels, indicators of DNA damage, and DNA repair capability (Figure 4E, F).
Figure 4.
Depletion of PARPBP increases breast cancer cell apoptosis and DNA damage caused by epirubicin. (A, B) Colony-forming ability of the control, LV-shNC- and LV-shPARPBP-transfected MCF-7 (A) and MCF-7/EPI cells (B) in the absence or presence of EPI (2 μM) for 48 h. (C, D) The apoptotic rates of MCF-7 (C) and MCF-7/EPI cells (D) transfected with LV-shNC and LV-shPARPBP in the absence or presence of EPI (2 μM) for 48 h were visualized by flow cytometry. (E, F) Western blot analysis of γH2AX and BRCA1 expression in MCF-7 (E) and MCF-7/EPI cells (F) with or without transfection of LV-shNC or LV-shPARPBP after EPI treatment (2 μM) removal.
EPI, epirubicin; PARBP, PARP1 binding protein.
PARPBP promotes breast cancer anthracycline-based resistance in vivo
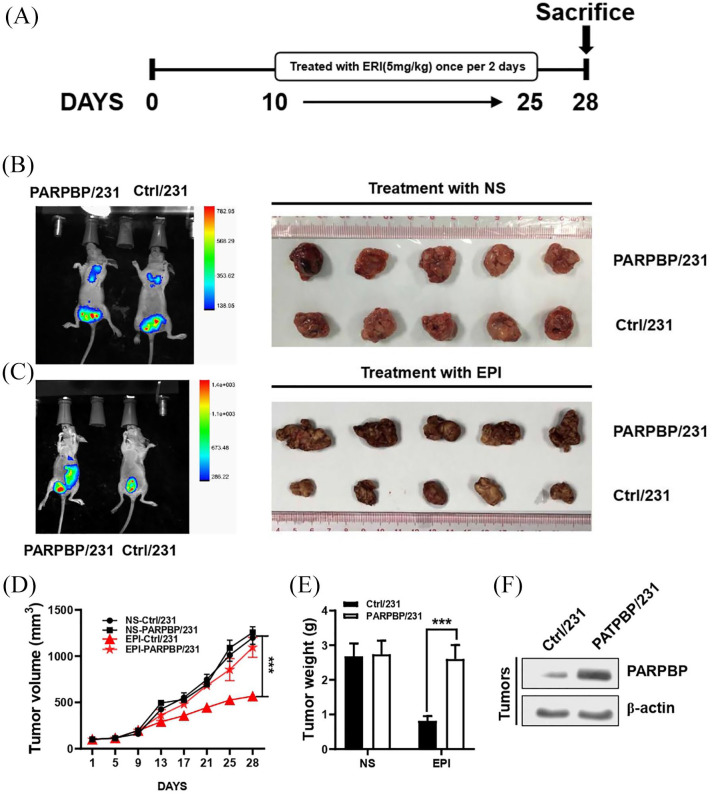
To investigate the functional role of PARPBP in regulating the drug resistance of breast cancer in vivo, PARPBP-overexpressing and control cells were injected into the mammary fat pads of female BALB/c nude mice followed by EPI treatment (Figure 5A). The volume and weight of tumors in PARPBP-overexpressed group (PARPBP/231) were significantly higher than those in the control group, but similar to those without EPI treatment (Figure 5B–F). That means EPI effectively inhibited tumor growth in mice with control tumors but not in mice with PARPBP-overexpressing tumors, suggesting that PARPBP is important for chemoresistance in breast cancer.
Figure 5.
PARPBP promotes breast cancer anthracycline-based resistance in vivo. (A) PARPBP/231 and Ctrl/231 cells were injected into the mammary fat pads of nude mice (n = 5). Tumor development was allowed for 10 days, and then the mice were injected intraperitoneally with 5 mg/kg EPI or normal saline for another 2 weeks. (B, C) Tumors from PARPBP/231 and Ctrl/231 mice treated with NS (B) or EPI (C) are shown. (D) The growth curves of the tumors are plotted. (E) The weights of the xenograft tumors are summarized. ***p < 0.001 versus respective control in Student’s t test. (F) Expression of PARPBP in breast cancer xenografts was examined by western blotting.
EPI, epirubicin; NS, normal saline; PARBP, PARP1 binding protein.
FOXM1 binds to the PARPBP promoter directly and regulates its activity
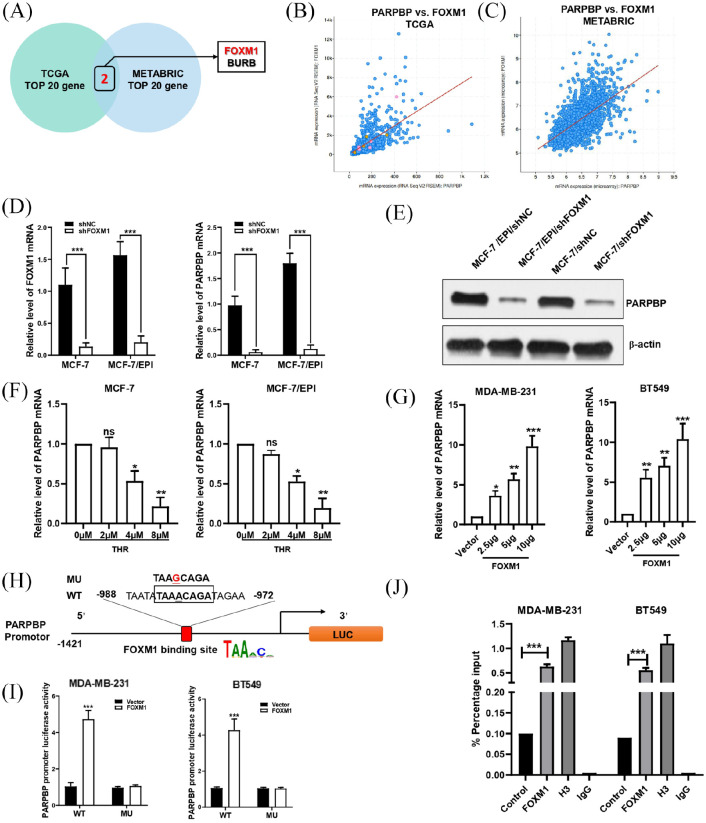
We used TCGA and METABRIC datasets to identify the putative co-expression genes of PARPBP in human breast cancer. FOXM1 and BURB are the intersection of the top 20 co-expressed genes in the two datasets (Figure 6A–C). Next, we focused on FOXM1, which is a transcriptional activator and a critical mediator of epirubicin and paclitaxel resistance in breast cancer. We transfected FOXM1 shRNA into MCF-7 and MCF-7/EPI cells (Figure 6D). The results indicated that both the mRNA and protein levels of PARPBP were decreased significantly with FOXM1 knockdown (Figure 6D, E). Similarly, MCF-7 and MCF-7/EPI cells treated with FoxM1 inhibitor thiostrepton (THR) at concentrations of 4 and 8 µM depressed PARPBP levels (Figure 6F). Conversely, the mRNA level of PARPBP in MDA-MB-231 and BT549 cells transfected with FoxM1 increased with increasing dosage of FoxM1 expression construct transfected (Figure 6G). All these results suggested strongly that expression of PARPBP was activated by FoxM1.
Figure 6.
FOXM1 directly binds to the PARPBP promoter and regulates its activity. (A) Venn diagram shows that FOXM1 and BURB are the intersection of the top 20 PARPBP co-expressed genes in the TCGA and METABRIC datasets. (B, C) Correlation between FOXM1 and PARPBP in TCGA (B) and METABRIC (C). (D, E) Downregulation of FOXM1 by transfecting shFOXM1 reduced PARPBP expression by qRT-PCR (D) and western blotting (E). (F) MCF-7 and MCF-7/EPI cells treated with FoxM1 inhibitor THR at concentrations of 0, 2, 4, and 8 μM. (G) Overexpression of FOXM1 by transfecting FOXM1 expression plasmid in MDA-MB-231 and BT549 cells increased the mRNA level of PARPBP. (H) Schematic of the PARPBP promoter reporter and its putative FOXM1-binding site. (I) Luciferase reporter assay in MDA-MB-231 and BT549 cells (with FOXM1 expression plasmid or empty vector) transfected with luciferase reporter constructs containing WT or MU PARPBP promoter. Data represent means ± SD of at least three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001. (J) ChIP assay showing the binding of FOXM1 to PARPBP promotor. Chromatins were isolated from MDA-MB-231 and BT549 cells, and specific primers for PARPBP promotor was used to DNA quantification. The enrichment percentage = 2% × 2[CT (input sample) − CT (IP sample)]. Normal IgG and histone H3 were used as negative and positive controls, respectively.
ChIP, chromatin immunoprecipitation; GEPIA, Gene Expression Profiling Interactive Analysis; IgG, immunoglobulin G; METABRIC, Molecular Taxonomy of Breast Cancer International Consortium; MU, mutant; OS, overall survival; PARBP, PARP1 binding protein; SD, standard deviation; THR, thiostrepton; TCGA, The Cancer Genome Atlas; TPM, transcripts per million; WT, wild type.
To explore whether FOXM1 regulates PARPBP promoter activity, we scanned the PARPBP gene promoter region with the canonical binding DNA motifs of FOXM1 (5′-TAAaCa-3′) and found such a motif between −988 and −972 upstream of the transcription start site (Figure 6H). Then, we generated a luciferase reporter construct with wild-type or FOXM1 binding motif-mutated PARPBP promoter (Figure 6H), and tested the effect of FOXM1 expression on PARPBP promoter activity in MDA-MB-231 and BT549 cells. The expression of FOXM1 increased dramatically luciferase activity driven by the wild-type but not the mutant PARPBP promoter (Figure 6I). In addition, we performed ChIP assay and verified the binding of FOXM1 to the endogenous PARPBP promoter region (Figure 6J). Collectively, our findings supported the view that FoxM1 could transcriptionally activate PARPBP by interacting with predicted binding sites.
Discussion
Breast cancer persists as a leading cause of cancer death in women worldwide.25 Although systemic chemotherapy is effective in early and advanced breast cancer, the high rates of recurrence and resistance are still the major challenges in breast cancer treatment. In the present study, we revealed roles for PARPBP in breast cancer prognosis and anthracycline resistance.
Previous studies have reported that PARPBP overexpression was associated with hyperproliferation and severe clinical outcomes in lung, gastric, pancreatic, cervical cancers, hepatocellular carcinoma, and myeloid leukemia.26–29 Here, we found that PARPBP was upregulated significantly in breast cancer tissues at both mRNA and protein levels compared with normal breast tissues. The expression of PARPBP was correlated positively with tumor status, lymph node status, and tumor, node, metastasis (TNM) stage of breast cancer patients. In addition, high expression of PARPBP was associated with poor prognosis in breast cancer patients, which suggested that PARPBP may be a promising prognostic biomarker.
In this study, we found that high expression of PARPBP promotes cell growth of breast cancer. PARPBP knockdown decreased cell proliferation and colony formation in breast cancer. Conversely, the proliferation and colony formation ability of PARPBP-overexpressed breast cancer cells were increased significantly relative to the control group. The positive role of PARPBP in cell growth of breast cancer cells agrees with its adverse prognosis for breast cancer. We observed that PARPBP was upregulated in breast cancer. In particular, it had a relatively high expression in estrogen receptor (ER)-positive (MCF7,MCF7/EPI and T47D cells) and a relatively low expression in TNBC cells (MDA-MB-231 and BT549 cells). High PARPB expression in ER-positive breast cancers correlated directly with resistance to chemotherapy. In fact, we found that the expression of PARPBP in anthracycline-based resistant tumors was significantly higher than that in nonresistant tumors. Therefore, we hypothesized that PARPBP contributes to anthracycline resistance.
EPI – a representative anthracycline antibiotic – is the mainstay chemotherapy drug in the treatment of breast cancer. Epirubicin intercalates into DNA, which might cause DNA damage and apoptosis in cancer cells. The histone variant H2AX is a principal component of chromatin involved in the detection, signaling, and repair of DNA double-strand breaks.30,31 Usually, γH2AX levels are used as an indicator of the degree of DNA damage.32 In this study, we found knockdown of PARPBP not only increased apoptosis of breast cancer cells caused by EPI, but also attenuated intracellular γH2AX level in cancer cells treated with EPI. These results indicate that PARPBP might protect breast cancers from DNA damage caused by chemotherapy. Using tumor xenograft experiments, we also found PARPBP enhanced breast cancer cell chemoresistance.
DNA repair capacity is critical for survival of cancer cells upon therapeutic DNA damage, and thus is an important determinant of susceptibility to chemotherapy in cancer patients.33 Many studies have demonstrated that targeted therapies such as poly(ADP ribose) polymerase (PARP) inhibitors could sensitize tumor cells to DNA-damaging chemotherapies. By inhibiting PARP-mediated repair of DNA lesions created by chemotherapies, greater potency might be achieved.34 But determining the optimal use of PARP inhibitors within drug combination approaches is still challenging. In previous studies, similar synthetic lethal interactions have been demonstrated between inhibitors of PARP and some other genes related to DNA repair and the DNA damage response, including ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related pathways.35,36 PARPBP is an element of the homologous recombination (HR) pathway of DNA repair. PARPBP downregulation improves genomic stability and HR in the HR-deficient Fanconi anemia/BRCA pathway in activated cancer cells.9 Therefore, the effect of combining PARPBP and PARP inhibition deserves to be explored intensively. In addition, O’Connor’s study investigated the roles of PARPBP in pancreatic cancer and found PARPBP has been shown to activate other pathways of DNA repair such as translesion synthesis (TLS) and non-homologous end joining (NHEJ), which are error-prone DNA repair pathways that enable cells to replicate when HR is inhibited.27 TLS and NHEJ activities often result in point mutations, deletions, translocations, and chromosomal rearrangements. It might increase tumor cell sensitivity to DNA crosslinking agent, platinum-based compounds, and radiotherapy. As we know, in general ER+ breast cancer does not respond well to platinum. In future studies, it will be interesting to investigate whether the PARPBP overexpressing ER+ breast cancers are sensitive to platinum-based compounds and/or radiotherapy.
Through analysis of the TCGA and METABRIC databases, we found that the expression of PARPBP was correlated positively with the expression of FOXM1. FOXM1 is a proliferation-associated transcription factor with important functions also in cell proliferation, differentiation, and apoptosis.37 FOXM1 is generally highly expressed in several aggressive human carcinomas and related to oncogenesis in many tissue types, including breast cancer.38 Investigators have also shown that FOXM1 might be involved in resistance to cisplatin, paclitaxel, and epirubicin.39,40 Here, we found a putative binding site of FOXM1 in the PARPBP promoter and confirmed that PARPBP transcription is modulated directly by FOXM1. Our findings additionally identified an important role of FOXM1 in breast cancer chemoresistance by regulating PARPBP expression.
In conclusion, we show that PARPBP is upregulated in breast cancer and that it might be regulated by FOXM1. High PARPBP expression levels are associated with poor OS in breast cancer patients. We also demonstrate that downregulation of PARPBP can effectively promote EPI sensitivity in breast cancer cells. Thus, we propose that detecting the expression of PARPBP in breast cancer patients treated with anthracycline-based chemotherapy may help oncologist to judge whether there is the need to use small molecule inhibitors of PARPBP as novel targeted therapy for patients harboring PARPBP-overexpressing breast cancer. PARPBP might be an attractive candidate target for the treatment of breast tumors that are resistant to anthracycline treatment.
Supplemental Material
Supplemental material, sj-pdf-1-tam-10.1177_1758835920974212 for PARPBP is a prognostic marker and confers anthracycline resistance to breast cancer by Bo Chen, Jianguo Lai, Danian Dai, Rong Chen, Ning Liao, Guanfeng Gao and Hailin Tang in Therapeutic Advances in Medical Oncology
Acknowledgments
We thank all the patients and their families for participation.
Footnotes
Conflict of interest statement: The authors declare that there is no conflict of interest.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by funds from the National Natural Science Foundation of China (81902828, Bo Chen); High-level Hospital Construction Project (DFJH201921, Bo Chen); the Fundamental Research Funds for the Central Universities (y2syD2192230, Bo Chen); and Medical Scientific Research Foundation of Guangdong Province (B2019039, Bo Chen)
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Bo Chen, Department of Breast Cancer, Cancer Center, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, 510080, China.
Jianguo Lai, Department of Breast Cancer, Cancer Center, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, China.
Danian Dai, Department of Breast Cancer, Cancer Center, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, China.
Rong Chen, Department of Breast Oncology, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China.
Ning Liao, Department of Breast Cancer, Cancer Center, Guangdong Provincial People’s Hospital, Guangdong Academy of Medical Sciences, Guangzhou, Guangdong, 510080, China.
Guanfeng Gao, Department of Breast Oncology, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China.
Hailin Tang, Department of Breast Oncology, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, China.
References
- 1. Chen B, Zhang G, Wei G, et al. Heterogeneity of genomic profile in patients with HER2-positive breast cancer. Endocr Relat Cancer 2020; 27: 153–162. [DOI] [PubMed] [Google Scholar]
- 2. Chen B, Tang H, Chen X, et al. Transcriptomic analyses identify key differentially expressed genes and clinical outcomes between triple-negative and non-triple-negative breast cancer. Cancer Manag Res 2019; 11: 179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waks AG, Winer EP. Breast cancer treatment: a review. JAMA 2019; 321: 288–300. [DOI] [PubMed] [Google Scholar]
- 4. Holohan C, Van Schaeybroeck S, Longley DB, et al. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013; 13: 714–726. [DOI] [PubMed] [Google Scholar]
- 5. Turner N, Biganzoli L, Di Leo A. Continued value of adjuvant anthracyclines as treatment for early breast cancer. Lancet Oncol 2015; 16: e362–e369. [DOI] [PubMed] [Google Scholar]
- 6. Pandy JGP, Balolong-Garcia JC, Cruz-Ordinario MVB, et al. Triple negative breast cancer and platinum-based systemic treatment: a meta-analysis and systematic review. BMC Cancer 2019; 19: 1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang N, Li K, Huang W, et al. Efficacy of platinum in advanced triple-negative breast cancer with germline BRCA mutation determined by next generation sequencing. Chin J Cancer Res 2020; 32: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mochizuki AL, Katanaya A, Hayashi E, et al. PARI regulates stalled replication fork processing to maintain genome stability upon replication stress in mice. Mol Cell Biol 2017; 37: e00117–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moldovan GL, Dejsuphong D, Petalcorin MI, et al. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol Cell 2012; 45: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burkovics P, Dome L, Juhasz S, et al. The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic Acids Res 2016; 44: 3176–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pitroda SP, Pashtan IM, Logan HL, et al. DNA repair pathway gene expression score correlates with repair proficiency and tumor sensitivity to chemotherapy. Sci Transl Med 2014; 6: 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pitroda SP, Bao R, Andrade J, et al. Low recombination proficiency score (RPS) predicts heightened sensitivity to DNA-damaging chemotherapy in breast cancer. Clin Cancer Res 2017; 23: 4493–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45: W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 9: 166–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun 2016; 7: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731. [DOI] [PubMed] [Google Scholar]
- 20. Dai DN, Li Y, Chen B, et al. Elevated expression of CST1 promotes breast cancer progression and predicts a poor prognosis. J Mol Med 2017; 95: 873–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Braunstein M, Liao L, Lyttle N, et al. Downregulation of histone H2A and H2B pathways is associated with anthracycline sensitivity in breast cancer. Breast Cancer Res 2016; 18: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen B, Wei W, Huang X, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics 2018; 8: 4003–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang H, Chen B, Liu P, et al. SOX8 acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Carcinogenesis 2019; 40: 1278–1287. [DOI] [PubMed] [Google Scholar]
- 24. Liu P, Tang H, Chen B, et al. miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett 2015; 357: 384–392. [DOI] [PubMed] [Google Scholar]
- 25. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 26. Piao L, Nakagawa H, Ueda K, et al. C12orf48, termed PARP-1 binding protein, enhances poly (ADP-ribose) polymerase-1 (PARP-1) activity and protects pancreatic cancer cells from DNA damage. Genes Chromosomes Cancer 2011; 50: 13–24. [DOI] [PubMed] [Google Scholar]
- 27. O’Connor KW, Dejsuphong D, Park E, et al. PARI overexpression promotes genomic instability and pancreatic tumorigenesis. Cancer Res 2013; 73: 2529–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu B, Ding Y, Liao X, et al. Overexpression of PARPBP correlates with tumor progression and poor prognosis in hepatocellular carcinoma. Dig Dis Sci 2019; 64: 2878–2892. [DOI] [PubMed] [Google Scholar]
- 29. Nicolae CM, O’Connor MJ, Schleicher EM, et al. PARI (PARPBP) suppresses replication stress-induced myeloid differentiation in leukemia cells. Oncogene 2019; 38: 5530–5540. [DOI] [PubMed] [Google Scholar]
- 30. Celeste A, Fernandez-Capetillo O, Kruhlak MJ, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 2003; 5: 675–679. [DOI] [PubMed] [Google Scholar]
- 31. Chen WT, Alpert A, Leiter C, et al. Systematic identification of functional residues in mammalian histone H2AX. Mol Cell Biol 2013; 33: 111–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gruosso T, Mieulet V, Cardon M, et al. Chronic oxidative stress promotes H2AX protein degradation and enhances chemosensitivity in breast cancer patients. EMBO Mol Med 2016; 8: 527–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ka NL, Na TY, Na H, et al. NR1D1 recruitment to sites of DNA damage inhibits repair and is associated with chemosensitivity of breast cancer. Cancer Res 2017; 77: 2453–2463. [DOI] [PubMed] [Google Scholar]
- 34. Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science 2017; 355: 1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kubota E, Williamson CT, Ye R, et al. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle 2014; 13: 2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Southgate HED, Chen L, Tweddle DA, et al. ATR inhibition potentiates PARP inhibitor cytotoxicity in high risk neuroblastoma cell lines by multiple mechanisms. Cancers 2020; 12: 1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Song L, Wang X, Feng Z. Overexpression of FOXM1 as a target for malignant progression of esophageal squamous cell carcinoma. Oncol Lett 2018; 15: 5910–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu L, Wu J, Guo Y, et al. Overexpression of FoxM1 predicts poor prognosis of intrahepatic cholangiocarcinoma. Aging 2018; 10: 4120–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kwok JM, Peck B, Monteiro LJ, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res 2010; 8: 24–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park YY, Jung SY, Jennings NB, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis 2012; 33: 1843–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-tam-10.1177_1758835920974212 for PARPBP is a prognostic marker and confers anthracycline resistance to breast cancer by Bo Chen, Jianguo Lai, Danian Dai, Rong Chen, Ning Liao, Guanfeng Gao and Hailin Tang in Therapeutic Advances in Medical Oncology