Abstract
Legume plants form nitrogen-fixing nodules in symbiosis with soil bacteria rhizobia. The number of symbiotic nodules is controlled at the whole-plant level with autoregulation of nodulation (AON), which includes a shoot-acting CLV1-like receptor kinase and mobile CLE (CLAVATA3/ENDOSPERM SURROUNDING REGION-related) peptides that are produced in the root in response to rhizobia inoculation. In addition to rhizobia-induced CLE peptides, nitrate-induced CLE genes have been identified in Lotus japonicus and Glycine max, which inhibited nodulation when overexpressed. However, nitrate-induced CLE genes that systemically suppress nodulation in AON-dependent manner have not been identified in Medicago truncatula. Here, we found that MtCLE35 expression is activated by both rhizobia inoculation and nitrate treatment in M. truncatula, similarly to L. japonicus CLE genes. Moreover, we found that MtCLE35 systemically suppresses nodulation in AON-dependent manner, suggesting that MtCLE35 may mediate nitrate-induced inhibition of nodulation in M. truncatula.
Keywords: symbiotic nodules, autoregulation of nodulation, nitrate, CLE, CLV1-like kinase
1. Introduction
The Rhizobium–legume interaction results in the formation of new organs on plant roots—symbiotic nodules, where the process of nitrogen fixation takes place. Nodule number is regulated by the plant systemically, and this process is known as autoregulation of nodulation (AON) [1]. A key component of AON is a CLAVATA 1 (CLV1)-like receptor kinase encoded by SUPER NUMERIC NODULES (MtSUNN) in Medicago truncatula, HYPER NODULATION ABERRANT ROOT FORMATION1 (LjHAR1) in Lotus japonicus, and NODULE AUTOREGULATION RECEPTOR KINASE (GmNARK) in Glycine max [2,3,4]. Legume mutants carrying mutations in these genes exhibit supernodulating phenotype (they form an excessive number of nodules), and some of them are nitrate tolerant (nodulation is not suppressed by high nitrogen concentrations which inhibit nodulation in wild-type plants) [5,6,7]. A grafting experiment demonstrated that the supernodulating phenotype of mutants defective in CLV1-like receptor kinase is determined by the shoot part of the plant [8]. Therefore, a signaling cascade activated by CLV1-like kinase in the shoot suppresses the subsequent nodulation on the roots systemically through a shoot-derived inhibitor, thereby regulating nodule number [1,9,10,11]. CLE (CLAVATA3/ENDOSPERM SURROUNDING REGION-related) peptides represent mobile signal molecules that are produced in the root in response to rhizobia inoculation and trigger AON through shoot-acting CLV1-like receptor kinase [12,13,14]. For L. japonicus CLE peptide, LjCLE-RS2, produced in the root in response to rhizobia inoculation, the presence in xylem sap collected from the shoot and the direct binding to LjHAR1 CLV1-like receptor kinase were shown [15].
In response to AON activation, the expression of cytokinin biosynthesis gene LjIPT3 was increased in the shoot in LjHAR1-dependent manner, and shoot-to-root transported cytokinin was suggested to inhibit nodule initiation in L. japonicus [16]. For the MtIPT3 gene in M. truncatula AON-dependent activation in the shoot was also shown [17]; however, the activation of orthologous gene in soybean, GmIPT5, appeared to be AON-independent [18]. Moreover, the inhibition of shoot-to-root auxin transport was shown to occur downstream of AON activation [19]. Finally, in response to AON the production of miR2111 is reduced in the shoot [10,11]. miR2111 is believed to be a mobile microRNA that is transported from the shoot to the root and increases the competence of the root to rhizobia infection and nodulation via down-regulation of the TML gene, which encodes F-box protein—a negative regulator of symbiosis [10,20]. Overproduction of miR2111 results in hyperinfection and increased nodule number [10]. AON inhibits miR2111 synthesis in the shoot, thereby limiting nodule number produced on the inoculated roots [10,11].
CLE peptides inhibiting nodulation have been identified in different legumes: MtCLE13 and MtCLE12 in M. truncatula [12], LjCLE-RS1, LjCLE-RS2, and LjCLE-RS3 (CLE-ROOT SIGNAL) in L. japonicus [13,21], GmRIC1 and GmRIC2 (RHIZOBIUM INDUCED CLE) in G. max [14]. The expression of these genes is activated in response to rhizobia inoculation, and their overexpression inhibited nodulation in AON-dependent manner [12,13,14]. Nitrate is a well-known regulator of nodulation, since nodule formation takes place at low nitrate level in the soil, whereas high nitrate doses inhibit nodulation [22]. In soybeans and L. japonicus, in addition to rhizobia-induced CLE peptides, nitrate-induced CLE genes have been identified, which are supposed to mediate nitrate-dependent suppression of symbiotic nodule development [13,14,23].
In L. japonicus, the expression of LjCLE-RS2 and LjCLE-RS3 genes is activated by both rhizobia and nitrate treatment [13,21]. However, in soybean in addition to rhizobia-induced CLE peptides that systemically suppress nodulation, nitrate-induced CLE peptides (NIC), GmNIC1 and GmNIC2, have been identified, that are activated in response to nitrate treatment and inhibit nodulation locally in the root [14,23]. GmNIC1 and GmNIC2 overexpression in transgenic roots obtained by Agrobacterium rhizogenes-mediated transformation inhibited nodulation locally, but did not suppress nodulation on non-transgenic roots, which indicates the absence of systemic effect on nodulation [14]. Therefore, in contrast to L. japonicus CLE genes, which are activated by both rhizobia and nitrate and act systemically to suppress nodulation, in soybean there are two groups of CLE-peptides: RIC are induced by rhizobia and systemically suppress nodulation, whereas NIC are activated in response to nitrate treatment and inhibit nodulation locally in the root via root-acting CLE receptor. The mechanism underlying the specific perception of CLE peptide by a shoot and a root-acting receptor are of great interest and remain to be elucidated.
In M. truncatula, two CLE-peptides, MtCLE12 and MtCLE13, were found to be involved in AON [12]. The expression of these two genes was induced in response to rhizobia inoculation, whereas their induction by nitrate has not been shown [12]. Ectopic expression of these genes systemically suppressed nodulation in wild-type plant, but not in MtSUNN mutant lacking a functional CLV1-like receptor kinase, indicating that MtSUNN may be responsible for MtCLE12 and MtCLE13 perception in the shoot. Recently, an additional close homologue of MtCLE12 and MtCLE13 has been identified in M. truncatula genome—the MtCLE35 gene [24,25]. The expression of this gene is induced in rhizobia according to transcriptomic data [25]. However, the role of the MtCLE35 gene in AON has not been investigated. Here, we found that MtCLE35 expression is activated in response to rhizobia inoculation as well as to nitrate treatment, similarly to L. japonicus CLE genes. Moreover, we found that MtCLE35 systemically suppresses nodulation in an AON-dependent manner, suggesting that MtCLE35 is involved in nitrate-induced inhibition of nodulation in M. truncatula.
2. Results
2.1. MtCLE35 Is Expressed during Nodulation and in Response to Nitrate Treatment
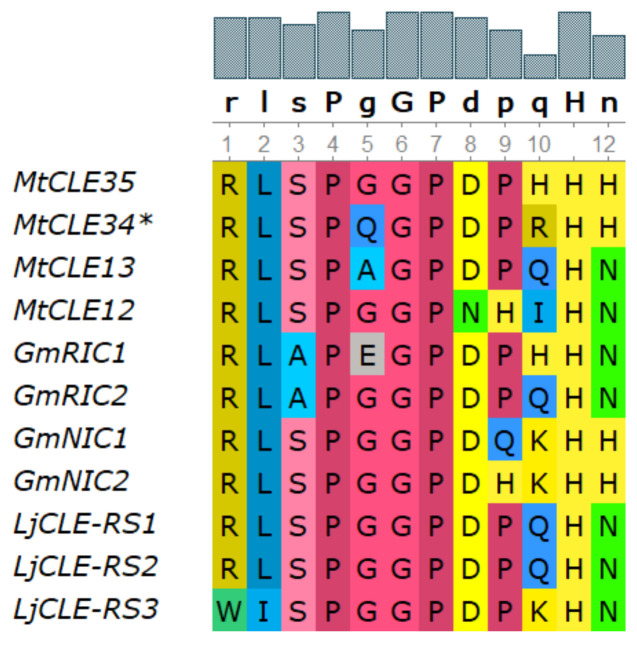
According to phylogenetic analysis, the MtCLE35 gene (Medtr2g091125.1) is closely related to other genes encoding nodulation-suppressing CLE peptides [24] (see Figure S1). The amino acid sequence of CLE domain of MtCLE35 shares high similarity with CLE domain sequences of other nodulation-suppressing CLE peptides, and differs from that of GmNIC1,2 and LjCLE-RS1,2 only at two positions (Figure 1). Moreover, MtCLE35 contains the consensus sequence TLQAR in the signal peptide domain, which was previously found in other nodulation-suppressing CLE peptides [24,26].
Figure 1.
Sequence alignment of CLAVATA3/ENDOSPERM SURROUNDING REGION-related (CLE) domains from Medicago truncatula, Glycine max and Lotus japonicus nodulation-suppressing CLE peptides.
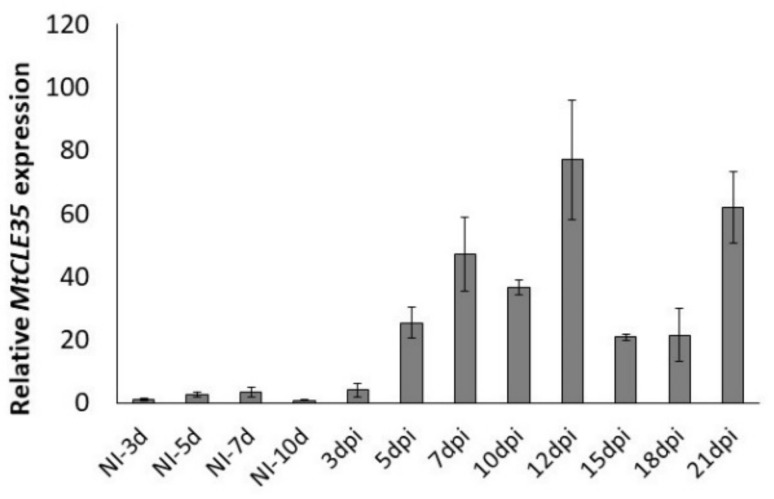
To study the expression dynamics of the MtCLE35 gene during nodule development, the relative transcript levels of MtCLE35 were analyzed at different stages after inoculation using quantitative reverse transcription polymerase chain reaction (qRT-PCR) (from 3 to 21 dpi (days post inoculation)) in comparison to the non-inoculated roots (NI). The induction of MtCLE35 expression was observed at 5 dpi. At later stages of nodulation its expression level increased reaching peak at 10–12 dpi, and after a slight reduction at 15–18 dpi, it was increased again in mature nodules at 21 dpi (Figure 2). This suggests that MtCLE35 acts not only at early stages of nodule development but also at later stages in mature nodules. Similar dynamics of MtCLE35 expression were observed in three independent experiments (see Figure S2 for the data from additional biological repeat).
Figure 2.
The expression levels of the MtCLE35 gene at different days post inoculation (dpi) in comparison to the non-inoculated roots (NI). Results are mean ± standard error of the mean (SEM) of three technical repeats of one biological repeat, representative for three independent experiments.
The activation of MtCLE35 expression in developing nodules is consistent with transcriptomic data obtained by LCM (Laser Capture Microdissection)-RNA-seq (https://iant.toulouse.inra.fr/symbimics/) for M. truncatula [27] (Figure S3A). Moreover, MtCLE35 expression was also increased in developing nodules according to Small Secreted Peptide Gene Expression Atlas (SSP-GEA) available in the Medicago truncatula Small Secreted Peptide Database (https://mtsspdb.noble.org/ [28]) (Figure S3B).
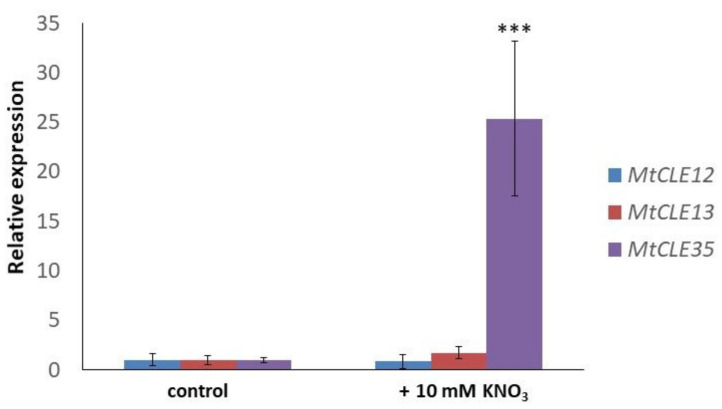
Next, we checked the effect of nitrate treatment on the MtCLE35 gene expression. After 24 h of nitrate treatment (10 mM KNO3), the expression level of the MtCLE35 gene was 25-fold increased (Figure 3). However, no increase of MtCLE12 and MtCLE13 in response to nitrate has been revealed. This suggests that MtCLE35 is a nitrate-responsive gene.
Figure 3.
The expression levels of MtCLE12, MtCLE13, and MtCLE35 in response to nitrate treatment (10 mM KNO3). Asterisks indicate statistically significant differences in MtCLE35 expression compared with control (p < 0.001).
2.2. Overexpression of MtCLE35 Suppresses Nodulation in Wild-Type Plants
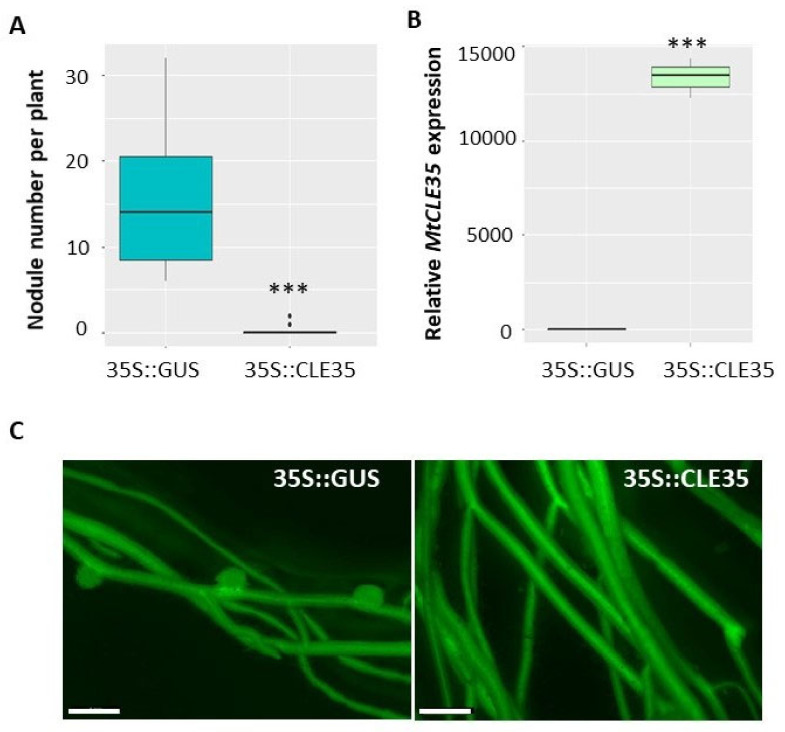
To explore the role of the MtCLE35 gene in nodulation we have overexpressed this gene under 35S promoter in M. truncatula roots (35S::MtCLE35). β-glucuronidase (GUS)-overexpressing roots were used as a control (35S::GUS). green fluorescent protein (GFP) fluorescence was used as a positive marker to select transgenic roots, where MtCLE35 overexpression was confirmed by qPCR analysis (Figure 4). Nodules were counted on transgenic roots at 21 dpi. In wild type A17 line, overexpression of MtCLE35 resulted in a significant reduction of nodulation. In the control GUS-overexpressing roots, on average 16 nodules were counted per transgenic root, whereas in MtCLE35-overexpressing roots in most cases nodulation was completely inhibited, and only two out of 15 plants carried up to two nodules per transgenic root in each biological experiment (Figure 4).
Figure 4.
MtCLE35 overexpression inhibits nodulation in wild-type plants. (A) Nodule number in wild-type (A17) A green fluorescent protein (GFP)-positive transgenic roots transformed with the 35S::GUS (control) or 35S::MtCLE35 construct. Asterisks indicate statistically significant differences (Mann-Whitney U test, p < 0.001). (B) The expression level of the MtCLE35 gene in GFP-positive transgenic roots transformed with the 35S::GUS (GUS_OE, control) or 35S::MtCLE35 construct. Asterisks indicate statistically significant differences p < 0.001. Box plots are presented with median values. (C) Examples of the nodulation phenotype of GFP-positive transgenic roots carrying the 35S::GUS (control) or 35S::MtCLE35 construct on wild-type plants. Bars = 1000 µm.
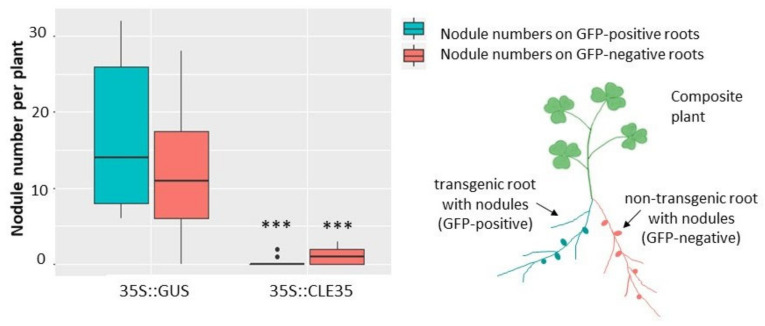
2.3. The Inhibitory Effect of MtCLE35 Overexpression on Nodulation Is Systemic
To check if such inhibitory effect of MtCLE35 overexpression on nodulation is systemic, we counted nodule numbers not only on transgenic roots, overexpressing MtCLE35, but also on non-transgenic root which emerged together with transgenic roots in composite plants after A. rhizogenes-mediated transformation. In addition to MtCLE35 coding DNA sequence) under 35S promoter, the genetic construct used for plant transformation also contains GFP cassette (enhanced green fluorescent protein (eGFP)) under rolD promoter) to select transgenic roots. In this system, GFP-positive roots represent transgenic roots, which overexpress MtCLE35 (or GUS in control plants), whereas GFP-negative roots (that do not demonstrate GFP fluorescence) represent non-transgenic ones (Figure 5). A significant reduction of nodule number was found in both transgenic (GFP-positive, MtCLE35-overexpressing) and GFP-negative non-transgenic roots which do not have the 35S::MtCLE35 insert (Figure 5). Representative images of composite wild-type plants containing both transgenic MtCLE35-overexpressing roots and non-transgenic roots are presented inFigure S4. Only very few nodules were found on non-transgenic roots in composite plants containing MtCLE35-overexpressing roots. This fact suggests a systemic nature of MtCLE35 inhibitory effect, in which a long-distance transport of MtCLE35 gene product seems to be involved.
Figure 5.
MtCLE35 overexpression systemically inhibits nodulation in wild-type plants. Nodule number in wild-type (A17) composite plants on green fluorescent protein (GFP)-positive transgenic roots (cyan) carrying the 35S::GUS (control) or 35S::MtCLE35 construct and on GFP-negative non-transgenic roots (coral) are represented. Box plots are presented with median values. Asterisks indicate statistically significant differences (Mann-Whitney U test, p < 0.001).
2.4. The Effect of MtCLE35 on Nodulation Depends on MtSUNN Receptor Kinase
Next, to check if MtCLE35 action on nodulation depends on MtSUNN receptor kinase, we analyzed the effect of MtCLE35 overexpression on nodulation in sunn-4 mutant plants. In contrast to the wild type, MtCLE35 overexpression had no obvious effect on nodulation in sunn-4 mutant (Figure 6, Figure S5). Both GUS-overexpressing control roots and MtCLE35-overexpressing roots of sunn-4 mutant plants demonstrated a supernodulating phenotype. No statistically significant difference in nodule numbers was found between control (GUS-overexpressing roots) and MtCLE35-overexpressing roots, indicating that the inhibitory effect of MtCLE35 overexpression on nodulation was abolished in sunn-4 mutant (Figure 6).
Figure 6.
MtCLE35 overexpression does not affect nodulation phenotype in sunn-4 mutant. (A) Nodule number on green fluorescent protein (GFP)-positive transgenic roots of sunn-4 plants transformed with the 35S:GUS (control) or 35S::MtCLE35 construct. Box plots are presented with median values. No statistically significant differences were found between nodule number in 35S:MtCLE35 and 35S:GUS (control) overexpressing plants. (B) Examples of the nodulation phenotype of GFP-positive transgenic roots carrying the 35S::GUS (control) or 35S::MtCLE35 construct on sunn-4 mutant plants. Bars = 1000 µm.
Therefore, functional MtSUNN gene is required for MtCLE35-dependent inhibition of nodulation in M. truncatula.
3. Discussion
Here, we found that MtCLE35 expression is induced both by rhizobia and by nitrate treatment. In contrast, the MtCLE12 and MtCLE13 genes that were previously found to be AON-dependent inhibitors of nodulation induced by rhizobia [12] did not exhibit nitrate responsiveness. Moreover, we showed that MtCLE35 overexpression in transgenic roots inhibited nodulation in wild-type plants, both in transgenic roots and in non-transgenic roots of composite plants bearing MtCLE35-overexpressing roots, indicating that MtCLE35 suppresses nodulation systemically. Our results indicate that MtCLE35 systemically inhibits nodulation in an AON-dependent manner, since MtCLE35 inhibitory effect on nodulation was observed in wild-type plants, but not in sunn-4 supernodulating mutant defective in AON. Therefore, MtCLE35 peptide is involved in AON, and MtSUNN receptor kinase may act as its receptor in the shoot. Collectively, our data suggest that MtCLE35 is a mediator of nitrate-dependent inhibition of nodulation in M. truncatula (Figure 7).
Figure 7.
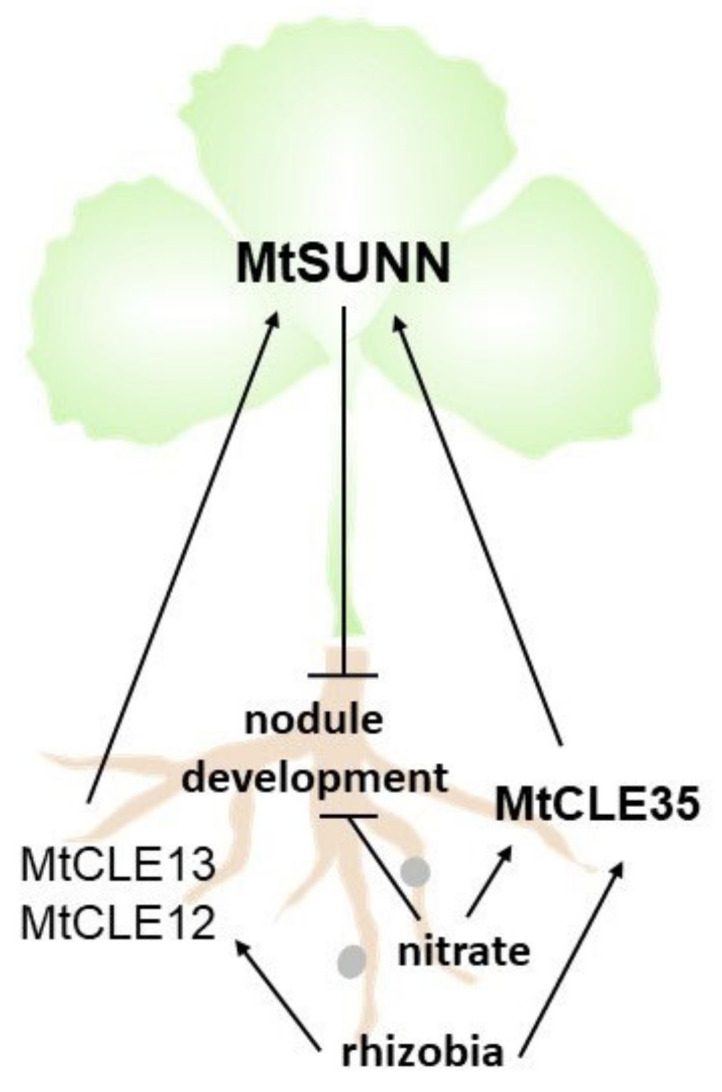
Model of MtCLE35 action in autoregulation of nodulation (AON). The MtCLE12 and MtCLE13 genes are induced in response to rhizobia inoculation, whereas the expression of the MtCLE35 gene is induced by both rhizobia inoculation and nitrate treatment. The CLAVATA3/ENDOSPERM SURROUNDING REGION-related (CLE) peptides encoded by the MtCLE12, MtCLE13, and MtCLE35 genes systemically inhibit nodulation via shoot-acting MtSUNN receptor kinase. MtCLE35 mediates nitrate-dependent inhibition of nodulation.
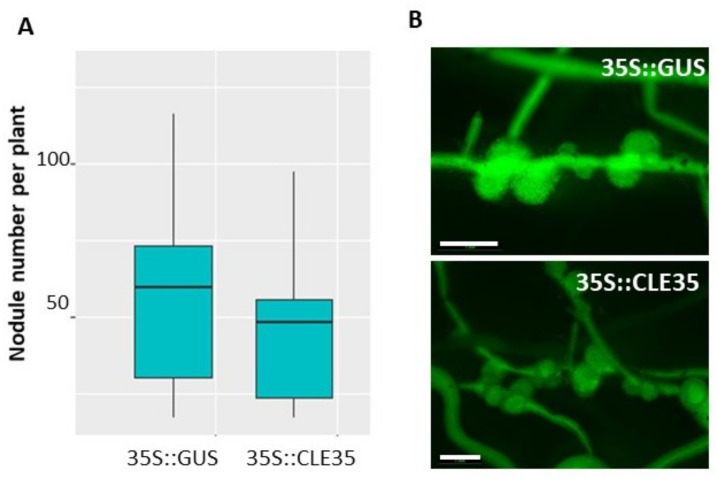
The nitrate-induced MtCLE35 expression was also described in a recent preprint paper available at bioRxiv [29]. In this paper, the inhibitory effect of MtCLE35 on nodulation was also reported, although it was less pronounced according to the presented data (on average, approximately 1.5 nodules were formed on MtCLE35-overexpressing plants compared to approximately three nodules formed in the control plants), and the systemic AON-dependent MtCLE35 action was not investigated in this study.
Nitrate-induced CLE peptides that suppress nodulation when overexpressed have been previously characterized in L. japonicus and G. max. In L. japonicus two CLE peptides, LjCLE-RS2 and LjCLE-RS3, are induced by both nitrate treatment and rhizobia inoculation [13,21], whereas in soybean there are two groups of CLE genes induced by different factors: rhizobia-induced CLE (RIC) and nitrate-induced CLE (NIC) [14,23]. The MtCLE35 regulation seems to be similar to that of L. japonicus LjCLE-RS2 and LjCLE-RS3, and this could be due to the similarity of their regulatory sequences that can be activated by both nitrate- and nodulation-responsive factors. In soybean, two distinct groups of CLE peptide have evolved: RIC peptides that inhibit nodulation systemically in AON-dependent way, and NIC peptide that act locally in the root to suppress nodulation in response to nitrate. Therefore, in legume plants multiple CLE genes have been evolved to control nodulation. Some of them are induced by rhizobia-activated signaling cascade alone, other CLE genes are regulated by both rhizobia and nitrate signaling, whereas the other group of CLE genes are only nitrate-responsive. A detailed comparative analysis of the regulatory sequences of these CLE genes should help to elucidate reasons for such differences in their regulation.
All CLE peptides suppressing nodulation in legumes were shown to act through CLV1-like kinase, and for LjCLE-RS2 the direct binding with LjHAR1 was confirmed [15]. Moreover, recently it was found that MtCLE33 and MtCLE53 peptides are activated by phosphate and arbuscular mycorrhizal fungi, respectively, and regulate mycorrhization in MtSUNN-dependent manner [30]. Therefore, MtSUNN/LjHAR1 should be responsible for the reception of multiple CLE peptides, activating by diverse factors—microsymbionts and nutrients. It is of great interest to study if there are additional receptor proteins that could help MtSUNN/LjHAR1 to distinguish between different CLE peptides and to elucidate how the specific action of the downstream signaling pathway induced by MtSUNN/LjHAR1 is achieved.
4. Materials and Methods
4.1. Plant Material, Bacterial Strains, and Growth Conditions
M. truncatula A17 and sunn-4 seeds were sterilized with sulfuric acid and washed several times with sterile distilled water, then transferred to the plates with 1% agar for germination. Plates were held at +4 °C for one day, then seeds were transferred to the room temperature and a dark place for 48 h. For expression analysis, plants were grown first on Fahraeus medium [31] for one week and then transferred to vermiculite (Sludyanaya Fabrika, Saint-Petersburg, Russia)-containing pots (3–4 plants per a pot) which were also moistened with nitrogen free Fahraeus medium in the growth chambers under a 16 h photoperiod at 21 °C (75% relative humidity). Ten days after germination, each plant was inoculated with 1 mL of Sinorhizobium meliloti strain Sm2011 liquid culture (grown in YEM (Yeast Extract Mannitol) [32] medium up to OD600 = 0.7) was used. Non-inoculated control roots together with the inoculated roots were harvested and used for RNA extraction.
Agrobacterium rhizogenes-mediated plant transformation was carried out according to our previous description [33].
Nitrate treatment of M. truncatula plants was performed in a hydroponic system. First, plants were grown on plates containing Fahraeus medium [31] for one week and then transferred to the hydroponic system, containing nitrogen-free liquid Fahraeus medium. After 4 days, KNO3 was added to the medium to a final concentration of 10 mM. Plant roots were harvested in 24 h after KNO3 treatment for gene expression analysis, and untreated plants was used as a control.
4.2. RNA Extraction and cDNA Synthesis
RNA extraction was performed using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. DNase treatment was conducted using Rapid Out DNA Removal Kit (Thermo Fisher Scientific, Waltham, MA, USA). The concentration and quality of extracted RNA was measured using a NanoDrop 2000c ultraviolet-visible (UV-Vis) Spectrophotometer (Thermo Scientific, Waltham, MA, USA) and equal amount of RNA was used for cDNA synthesis. cDNA synthesis was carried out using a Revert Aid Reverse Transcriptase kit (Thermo Fisher Scientific, Waltham, MA, USA).
4.3. Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) Analysis
qRT-PCR analysis was performed using CFX-96 real-time PCR detection system with a C1000 thermal cycler (Bio-Rad Laboratories, Alfred Nobel Drive Hercules, CA, USA) with SYBR Green intercalating dyes (Sintol, Moscow, Russia). The data were analyzed by the CFX Manager software (Bio-Rad Laboratories, Alfred Nobel Drive Hercules, CA, USA) with the 2−ΔΔ Ct method [34]. Actin (Medtr7g026230) and ubiquitin (Medtr4g091580) genes were used as reference genes. All the qRT-PCRs were undertaken in three technical repeats. Primers were designed using primer3 (http://bioinfo.ut.ee/primer3-0.4.0/) and Vector NTI Advance 10 program (InforMax, http://www.informaxinc.com, Thermo Fisher Scientific, Waltham, MA, USA), and were synthesized by Evrogen (http://www.evrogen.com, Moscow, Russia). Primers used for qRT-PCR are listed in Table S1. Dissociation curves (55–95 °C) were used to confirm the specificity of PCR amplification.
4.4. Molecular Cloning
The CDS sequence of MtCLE35 (Medtr2g091125.1) was amplified using high fidelity Phusion polymerase (Thermo Fisher Scientific, Waltham, MA, USA) with adding attB site (underlined) to the forward and reverse primers (MtCLE35_CDS_FOR: 5’-AAAAAAGCAGGCTTCATGGCAAACACACAAATAACTATATTT-3’ and MtCLE35_CDS_REV: 5’-CAAGAAAGCTGGGTTCTACTTGTTTTGTGGACCTGCA-3’) and then cloned into the entry vector pDONR221 (Thermo Fisher Scientific, Waltham, MA, USA). Then the CDS was cloned from entry vector to the destination vector pB7WG2D (containing 35S promoter for overexpression and GFP cassette to select transgenic plants under fluorescence) using LR Clonase™ II enzyme (Thermo Fisher Scientific, Waltham, MA, USA) The construct was verified by sequencing. The resulting vector was transformed to Agrobacterium rhizogenes strain MSU440.
4.5. GFP (Green Fluorescent Protein) Fluorescence Detection
GFP detection and imaging was performed using Leica M205 FA fluorescence stereo microscope (www.leica-microsystems.com, Leica Microsystems, Wetzlar, Germany). The images were processed using the ImageJ program (National Institutes of Health, Bethesda, MD, USA) [35].
4.6. Statistical Methods and Computer Software
In the gene expression assay during nodulation at each time points four plants were used in each biological repeat (both for inoculated and non-inoculated plants). One-way analysis of variance (ANOVA) and Student’s t-test were used to compare gene expression levels. The box plot for nodule number in MtCLE35-overexpressing and control (GUS-overexpressing) plants was drawn in RStudio (https://rstudio.com/, Boston, MA, USA). A Mann–Whitney U test was used to compare nodule numbers in MtCLE35-overexpressing and control (GUS-overexpressing) plants; in each group from 15 to 25 plants have been analyzed in each biological repeat. At least three independent biological repeats were done for each experiment.
Multiple alignment of protein sequences was performed using UGENE software (http://ugene.net/ru/, Novosibirsk, Russia) [36] with Clustal W algorithm [37]. For phylogenetic analysis, protein sequences were retrieved from Phytozome v12.1 for Medicago truncatula and from Genbank National Center for Biotechnological Information (NCBI) database2 for Lotus japonicus and Glycine max. Sequences were aligned using the MEGAX program (https://www.megasoftware.net/) with the Clustal W algorithm, and the phylogenetic tree was generated using Maximum Likelihood method with 1000 bootstrap replicates.
5. Conclusions
The MtCLE35 gene is induced by both rhizobia inoculation and nitrate treatment in Medicago truncatula. MtCLE35 systemically suppresses nodulation in an AON-dependent manner, mediating the nitrate-induced inhibition of nodulation in M. truncatula.
Acknowledgments
The authors thank Sofie Goormachtig (Ghent University, VIB, Ghent, Belgium) for the seeds of M. truncatula sunn-4 mutant. We thank the Research Resource Center for Molecular and Cell Technologies of Saint-Petersburg State University for the equipment used in this study, and Kirill Demchenko (Komarov Botanical Institute, Russian Academy of Sciences) for the assistance with root imaging.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/11/1456/s1. Table S1: Primers used for the expression analysis; Figure S1: Phylogenetic tree based on the protein sequences of CLE genes of Medicago truncatula, Lotus japonicus, and Glycine max. The tree was generated using Maximum likelihood algorithm with 1000 bootstrap replicates; Figure S2: The expression levels of the MtCLE35 gene at different days post inoculation (dpi) in comparison to the non-inoculated roots (NI). Results are mean ± SEM of three technical repeats of one biological repeat, representative for three independent experiments; Figure S3: The expression levels of the MtCLE35 gene in nodules in comparison with the root according to transcriptomic data obtained by LCM (Laser Capture Microdissection)-RNA-seq for M. truncatula (https://iant.toulouse.inra.fr/symbimics, [27]) (A), MtCLE35 expression at 0, 4, 10, 14 and 28 dpi according to theSmall Secreted Peptide Gene Expression Atlas (SSP-GEA) available in The Medicago truncatula Small Secreted Peptide Database (https://mtsspdb.noble.org [28] (B); Figure S4: Examples of nodulation phenotypes of composite wild-type plants containing both transgenic GFP-positive control (β-glucuronidase (GUS))-overexpressing) (A and B) and MtCLE35-overexpressing (C–F) roots and non-transgenic GFP-negative roots. White arrows indicate non-transgenic GFP-negative roots exhibiting faint autofluorescence. Red arrows point at nodules on GFP-positive transgenic roots, yellow arrows point at nodules on GFP-negative non-transgenic roots; Figure S5: Examples of nodulation phenotypes of composite sunn-4 mutant plants containing both transgenic GFP-positive control (GUS-overexpressing) (A and B) and MtCLE35-overexpressing (C–F) roots and non-transgenic GFP-negative roots. White arrows indicate non-transgenic GFP-negative roots exhibiting faint autofluorescence. Red arrows point at nodules on GFP-positive transgenic roots, yellow arrows point at nodules on GFP-negative non-transgenic roots.
Author Contributions
Conceptualization, M.L. and L.L.; validation, Y.Y.; formal analysis, M.L., M.A., and Y.Y.; investigation, M.L. and M.A.; data curation, M.L.; writing—original draft preparation, M.L. and M.A.; writing—review and editing, M.L. and M.A.; funding acquisition, M.L. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Russian Scientific Foundation project no. 16-16-10011, Russian Foundation for Basic Research (RFBR) projects 20-016-00129, 19-016-00177, and grant from Saint-Petersburg State University ID 60256785.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reid D.E., Ferguson B.J., Hayashi S., Lin Y.H., Gresshoff P.M. Molecular mechanisms controlling legume autoregulation of nodulation. Ann. Bot. 2011;108:789–795. doi: 10.1093/aob/mcr205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishimura R., Hayashi M., Wu G.J., Kouchi H., Imaizumi-Anraku H., Murakami Y., Kawasaki S., Akao S., Ohmori M., Nagasawa M., et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature. 2002;420:426–429. doi: 10.1038/nature01231. [DOI] [PubMed] [Google Scholar]
- 3.Searle I.R., Men A.E., Laniya T.S., Buzas D.M., Iturbe-Ormaetxe I., Carroll B.J., Gresshoff P.M. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299:109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- 4.Schnabel E., Journet E.P., de Carvalho-Niebel F., Duc G., Frugoli J. The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 2005;58:809–822. doi: 10.1007/s11103-005-8102-y. [DOI] [PubMed] [Google Scholar]
- 5.Carroll B.J., McNeil D.L., Gresshoff P.M. Isolation and properties of soybean [Glycine max (L.) Merr.] mutants that nodulate in the presence of high nitrate concentrations. Proc. Natl. Acad. Sci. USA. 1985;82:4162–4166. doi: 10.1073/pnas.82.12.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day D.A., Lambers H., Bateman J., Carroll B.J., Gresshoff P.M. Growth comparisons of a supernodulating soybean (Glycine max) mutant and its wild-type parent. Physiol. Plant. 1986;68:375–382. doi: 10.1111/j.1399-3054.1986.tb03368.x. [DOI] [Google Scholar]
- 7.Okamoto S., Kawaguchi M. Shoot HAR1 mediates nitrate inhibition of nodulation in Lotus japonicus. Plant Signal. Behav. 2015;10:e1000138. doi: 10.1080/15592324.2014.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delves A.C., Mathews A., Day D.A., Carter A.S., Carroll B.J., Gresshoff P.M. Regulation of the soybean-Rhizobium nodule symbiosis by shoot and root factors. Plant Physiol. 1986;92:588. doi: 10.1104/pp.82.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soyano T., Hirakawa H., Sato S., Hayashi M., Kawaguchi M. Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci. USA. 2014;111:14607–14612. doi: 10.1073/pnas.1412716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsikou D., Yan Z., Holt D.B., Abel N.B., Reid D.E., Madsen L.H., Bhasin H., Sexauer M., Stougaard J., Markmann K. Systemic control of legume susceptibility to rhizobial infection by a mobile microRNA. Science. 2018;362:233–236. doi: 10.1126/science.aat6907. [DOI] [PubMed] [Google Scholar]
- 11.Gautrat P., Laffont C., Frugier F. Compact Root Architecture 2 Promotes Root Competence for Nodulation through the miR2111 Systemic Effector. Curr. Biol. 2020;30:1339–1345. doi: 10.1016/j.cub.2020.01.084. [DOI] [PubMed] [Google Scholar]
- 12.Mortier V., Den Herder G., Whitford R., Van de Velde W., Rombauts S., D’haeseleer K., Holsters M., Goormachtig S. CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 2010;153:222–237. doi: 10.1104/pp.110.153718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okamoto S., Ohnishi E., Sato S., Takahashi H., Nakazono M., Tabata S., Kawaguchi M. Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 2009;50:67–77. doi: 10.1093/pcp/pcn194. [DOI] [PubMed] [Google Scholar]
- 14.Reid D.E., Ferguson B.J., Gresshoff P.M. Inoculation- and nitrate-induced CLE peptides of soybean control NARK-dependent nodule formation. Mol. Plant Microbe Interact. 2011;24:606–618. doi: 10.1094/MPMI-09-10-0207. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto S., Shinohara H., Mori T., Matsubayashi Y., Kawaguchi M. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nat. Commun. 2013;4:2191. doi: 10.1038/ncomms3191. [DOI] [PubMed] [Google Scholar]
- 16.Sasaki T., Suzaki T., Soyano T., Kojima M., Sakakibara H., Kawaguchi M. Shoot-derived cytokinins systemically regulate root nodulation. Nat. Commun. 2014;5:4983. doi: 10.1038/ncomms5983. [DOI] [PubMed] [Google Scholar]
- 17.Azarakhsh M., Lebedeva M.A., Lutova L.A. Identification and Expression Analysis of Medicago truncatula Isopentenyl Transferase Genes (IPTs) Involved in Local and Systemic Control of Nodulation. Front. Plant Sci. 2018;9:304. doi: 10.3389/fpls.2018.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mens C., Li D., Haaima L.E., Gresshoff P.M., Ferguson B.J. Local and Systemic Effect of Cytokinins on Soybean Nodulation and Regulation of Their Isopentenyl Transferase (IPT) Biosynthesis Genes Following Rhizobia Inoculation. Front. Plant Sci. 2018;9:1150. doi: 10.3389/fpls.2018.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Noorden G.E., Ross J.J., Reid J.B., Rolfe B.G., Mathesius U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 2006;14:1494. doi: 10.1104/pp.105.075879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magori S., Oka-Kira E., Shibata S., Umehara Y., Kouchi H., Hase Y., Tanaka A., Sato S., Tabata S., Kawaguchi M. Too much love, a root regulator associated with the long-distance control of nodulation in Lotus japonicus. Mol. Plant Microbe Interact. 2009;22:259–268. doi: 10.1094/MPMI-22-3-0259. [DOI] [PubMed] [Google Scholar]
- 21.Nishida H., Handa Y., Tanaka S., Suzaki T., Kawaguchi M. Expression of the CLE-RS3 gene suppresses root nodulation in Lotus japonicus. J. Plant Res. 2016;129:909–919. doi: 10.1007/s10265-016-0842-z. [DOI] [PubMed] [Google Scholar]
- 22.Nishida H., Suzaki T. Nitrate-mediated control of root nodule symbiosis. Curr. Opin. Plant Biol. 2018;44:129. doi: 10.1016/j.pbi.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Lim C.W., Lee Y.W., Lee S.C., Hwang C.H. Nitrate inhibits soybean nodulation by regulating expression of CLE genes. Plant Sci. 2014;229:1–9. doi: 10.1016/j.plantsci.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Hastwell A.H., de Bang T.C., Gresshoff P.M., Ferguson B.J. CLE peptide-encoding gene families in Medicago truncatula and Lotus japonicus, compared with those of soybean, common bean and Arabidopsis. Sci. Rep. 2017;7:9384. doi: 10.1038/s41598-017-09296-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Bang T.C., Lundquist P.K., Dai X., Boschiero C., Zhuang Z., Pant P., Torres-Jerez I., Roy S., Nogales J., Veerappan V., et al. Genome-Wide Identification of Medicago Peptides Involved in Macronutrient Responses and Nodulation. Plant Physiol. 2017;175:1669–1689. doi: 10.1104/pp.17.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hastwell A.H., Gresshoff P.M., Ferguson B.J. The structure and activity of nodulation-suppressing CLE peptide hormones of legumes. Funct. Plant Biol. 2015;42:229–238. doi: 10.1071/FP14222. [DOI] [PubMed] [Google Scholar]
- 27.Roux B., Rodde N., Jardinaud M.F., Timmers T., Sauviac L., Cottret L., Carrère S., Sallet E., Courcelle E., Moreau S., et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014;77:817–837. doi: 10.1111/tpj.12442. [DOI] [PubMed] [Google Scholar]
- 28.Boschiero C., Dai X., Lundquist P.K., Roy S., de Bang T.C., Zhang S., Zhuang Z., Torres-Jerez I., Udvardi M.K., Scheible W.R., et al. MtSSPdb: The Medicago truncatula Small Secreted Peptide Database. Plant Physiol. 2020;183:399–413. doi: 10.1104/pp.19.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mens C., April H., Su H., Gresshoff P.M., Mathesius U., Ferguson B. Characterisation of Medicago truncatula CLE34 and CLE35 in nodulation control. bioRxiv. 2020:231605. doi: 10.1101/2020.07.31.231605. [DOI] [PubMed] [Google Scholar]
- 30.Müller L.M., Flokova K., Schnabel E., Sun X., Fei Z., Frugoli J., Bouwmeester H.J., Harrison M.J. A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat. Plants. 2019;5:933. doi: 10.1038/s41477-019-0501-1. [DOI] [PubMed] [Google Scholar]
- 31.Fahraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- 32.Vincent J.M. A Manual for the Practical Study of the Root-Nodule Bacteria. Blackwell Scientific Publications; Oxford, UK: 1970. IBP Handbook No. 15. [Google Scholar]
- 33.Azarakhsh M., Rumyantsev A.M., Lebedeva M.A., Lutova L.A. Cytokinin biosynthesis genes expressed during nodule organogenesis are directly regulated by the KNOX3 protein in Medicago truncatula. PLoS ONE. 2020;15:e0232352. doi: 10.1371/journal.pone.0232352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Schindelin J., Rueden C.T., Hiner M.C., Eliceiri K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okonechnikov K., Golosova O., Fursov M., UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 37.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.