Abstract
Differentiation of human pluripotent stem cells (hPSCs) into ectoderm provides neurons and glia useful for research, disease modeling, drug discovery, and potential cell therapies. In current protocols, hPSCs are traditionally differentiated into an obligate rostro‐dorsal ectodermal fate expressing PAX6 after 6 to 12 days in vitro when protected from mesendoderm inducers. This rate‐limiting step has performed a long‐standing role in hindering the development of rapid differentiation protocols for ectoderm‐derived cell types, as any protocol requires 6 to 10 days in vitro to simply initiate. Here, we report efficient differentiation of hPSCs into a naive early ectodermal intermediate within 24 hours using combined inhibition of bone morphogenic protein and fibroblast growth factor signaling. The induced population responds immediately to morphogen gradients to upregulate rostro‐caudal neurodevelopmental landmark gene expression in a generally accelerated fashion. This method can serve as a new platform for the development of novel, rapid, and efficient protocols for the manufacture of hPSC‐derived neural lineages.
Keywords: differentiation, induced pluripotent stem cells, neural differentiation, neural induction, pluripotent stem cells
Efficient conversion into primal ectoderm by treatment with BGJ398 and LDN193189 accelerates neural induction in human pluripotent stem cells.
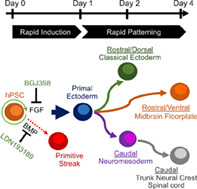
Significance statement.
This article reports a major advance in the ability to direct the differentiation of human pluripotent stem cells (hPSCs) into neural precursors in culture. This study describes that inhibiting selected signaling pathways enables the immediate exit from pluripotency and converts the stem cells within 24 hours into precursor cells that can be rapidly directed to generate neural populations desirable for research and potential cell therapies. This work removes a historical bottleneck that has prevented improving the control and speed of making neural cells from hPSCs and will likely change the way human neurons are generated in the future.
1. INTRODUCTION
Neurodevelopment is possibly the most complex developmental system. As many as 10 000 unique neuronal subtypes have been hypothesized to exist, whose embryonic origins derive from spatiotemporally fate‐restricted ectodermal subpopulations created by morphogen gradients in vivo. Undirected differentiation of human pluripotent stem cells (hPSCs) 1 , 2 , 3 recapitulates these complex neurodevelopmental dynamics in vitro, favoring production of heterogenous subpopulations of neuronal subtypes in the absence of exogenously provided developmental cues. Substantial effort has been expended to combat the emergence of heterogenous populations through directed differentiation using neurodevelopmental guidance cues such as morphogens. 4 , 5 , 6 , 7 , 8 However, even the most advanced current directed differentiation protocols to manufacture specific neuronal subpopulations are only partially successful. 9 , 10 , 11 , 12 , 13 , 14
All differentiation protocols for the production of neural lineages from hPSCs are lengthy, transpiring across timescales that mimic those seen during development in vivo. Aberrant differentiation into heterogenous populations may be an intrinsic property of long‐duration protocols, due to numerous factors including cell growth, cell‐cell interactions, cell‐substrate interactions, pH drift, and nutrient fluctuation; collectively defined here as “stochastic drift.” Lengthy protocols also reduce the ability to optimize due to prolonged experimental feedback cycles.
Accelerating differentiation protocols could improve purities as a byproduct of reducing stochastic drift and optimization turn‐around. This has proven difficult, however, especially for ectoderm formation. Premature neurogenesis has been demonstrated in rodent models 15 , 16 ; however, suggesting differentiation kinetics can be modulated. These phenotypes often arise due to disrupted self‐renewal within stem cell niches, which provides a potential mechanism for developing more rapid and efficient differentiation protocols.
2. ONLINE METHODS
2.1. Human induced pluripotent stem cell culture
A panel of human induced pluripotent stem cells (hiPSCs) derived using nonintegrating Sendai virus vectors 17 were generated in‐house 18 , 19 , 20 and adapted to culture as previously described. 21 Briefly, human fibroblasts or conjunctival biopsies were cultured and then reprogrammed with CytoTune‐iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific A16517). Cultures were maintained on Essential 8 Medium (Thermo Fisher Scientific A1517001) on recombinant human vitronectin (Peprotech AF‐140‐09) and propagated with hypertonic citrate solution. Lyophilized recombinant human vitronectin was reconstituted into a 500 μg/mL stock solution using Dulbecco's phosphate‐buffered saline plus calcium and magnesium (DPBS, Thermo Fisher Scientific 14 040‐133) and human serum albumin (0.1 %wt/vol MilliporeSigma A9731‐5G). This stock solution was further diluted 1:100 into DPBS for coating tissue culture plastic (coating volume 0.1 mL/cm2) for at least 1 hour at room temperature. hPSCs were grown in Essential 8 Medium with daily media exchanges (volume 0.25 mL/cm2). Every 3 to 4 days when 50% to 70% confluence was achieved, hPSCs were chelation passaged with hypertonic citrate buffer (4.4 g/L sodium citrate dibasic monohydrate, 24 g/L potassium chloride dissolved into ultra‐distilled water and filtered with 0.22‐μm polyethersulfone [PES] membrane). hPSCs were washed once with hypertonic citrate buffer and then incubated with hypertonic citrate buffer at 37°C for 6 minutes (volumes 0.1 mL/cm2). Citrate buffer was carefully aspirated to leave colonies undisturbed on the surface, and colonies were detached into Essential 6 Medium Medium (Thermo Fisher Scientific A1516401) to create a 2X concentrate suspension according to split ratio. This suspension was added to vitronectin‐coated tissue culture plastic containing Essential 8 Medium to maintain a 1:8 to 1:12 split ratio on average.
2.2. Accelerated differentiation of hPSCs into early ectoderm population
Day 3 to 4 undifferentiated cultures were passaged as described. Within 24 hours postpassage, Essential 8 Medium was exchanged for 0.2 mL/cm2 Essential 6 Medium supplemented with 500 nM LDN‐193189 HCl (Selleckchem S7507) and 100 nM BGJ398 (Selleckchem S2183).
2.3. Differentiation of early ectoderm into rostral ectodermal progenitors
Twenty‐four hours following treatment with LDN/BGJ, medium was exchanged for Essential 6 Medium supplemented with 500 nM wntC59 (Tocris 5148), 20 ng/mL FGF2 (Peprotech 100‐18B), and 500 nM A8301 (Tocris 2939) for 24 hours.
2.4. Differentiation of early ectoderm into neuromesoderm progenitors
Twenty‐four hours following treatment with LDN/BGJ, medium was exchanged for Essential 6 Medium supplemented with 4 μM CHIR99021 (Tocris 4423), 20 ng/mL FGF2, and 500 nM A8301 for 24 hours.
2.5. Differentiation of putative ectoderm into rostral floorplate neuroepithelium
Twenty‐four hours following LDN/BGJ treatment, the medium was exchanged daily with Essential 6 Medium supplemented with 500 nM CHIR99021, 500 nM A8301, 100 nM smoothened agonist (SAG; CaymanChem 11 914), and 100 ng/mL FGF2 for 48 hours. This day 3 population was further differentiated with Essential 6 Medium supplemented with 250 nM SAG, 100 nM BGJ, and 100 nM wntc59 (Tocris 5148) for 24 hours.
2.6. Differentiation of neuromesoderm progenitors into neural crest
Following production of neuromesodermal progenitors after day 2, medium was exchanged for Essential 6 Medium Medium supplemented with 100 nM BGJ398, 30 ng/mL bone morphogenic protein 4 (BMP4, Peprotech AF‐120‐05ET), and cyclopamine (500 nM, Tocris 1623) for 24 hours. Medium was then exchanged for Essential 6 Medium supplemented with 30 ng/mL BMP4 and 20 ng/mL FGF2 for 24 hours.
2.7. Immunocytochemistry
Cultures were fixed in buffered formalin (10 wt%/vol%, Fisher Scientific 23‐305‐510) for 10 minutes, permeabilized in DPBS plus Triton X‐100 (0.2 vol%/vol%, MilliporeSigma, T8787‐100ML), blocked in DPBS plus bovine serum albumin (1 wt%/vol%, MilliporeSigma A3059‐100G) and Tween‐20 (0.1 vol%/vol%, MilliporeSigma P1379) for 2 hours, and incubated with primary antibodies overnight. Cultures were washed twice with blocking solution and incubated 1 hour with secondary antibodies. 4′,6‐diamidino‐2‐phenylindole dilactate (DAPI, Thermo Fisher D3571) was added for 10 minutes before washing three times in DPBS. Antibodies used include: SOX2 (1:500, MilliporeSigma MAB4343), NANOG (1:100, Bio‐Techne AF1997), PAX6 (1:25, Developmental Studies Hybridoma Bank PAX6), SOX1 (1:250; Bio‐Techne AF3369), OTX1/2 (1:250, abcam ab21990), Brachyury (T, 1:100, Bio‐Techne AF2085), SIX3 (1:250, abcam ab221750), HOXB4 (1:250, abcam ab133521), FOXA2 (1:500, Bio‐Techne AF2400), PAX3 (1:250, Developmental Studies Hybridoma Bank PAX3), SOX10 (1:100, Bio‐Techne AF2864), Alexa Fluor 488 Donkey anti‐Mouse (1:500, Thermo Fisher Scientific A21202), Alexa Fluor 555 Donkey anti‐Goat (1:500, Thermo Fisher Scientific A21432), and Alexa Fluor 647 Donkey anti‐Rabbit (1:500, Thermo Fisher Scientific A31573). Negative controls included unstained cultures, cultures stained with secondary‐only antibodies, and positively stained cultures known not to express the antigens of interest.
2.8. Quantification of immunocytochemical counts
Cultures were imaged on a Leica DMI6000B with a DFC365FX camera running Leica Applications Suite‐Advanced Fluorescence, Version 3.1.0. Build 8587. Cell counts were automated using FIJI. 22 Briefly, four representative fields per condition were collected at ×200 total magnification. Images were subjected to local intensity thresholding, binary masks created, and segmentation performed with FIJI's built‐in watershed function. The positively stained area was measured and tabulated. Quantification represents antibody specific‐positive stained area divided by DAPI‐positive stained area. Unless otherwise indicated, statistics represent averages and SEs of mean from three biological replicates. Significance was calculated using one‐way analysis of variance (ANOVA) with Tukey's multicomparison testing.
2.9. RNA sequencing
Sequencing was performed on the NextSeq 550 platform using pair‐end reads at a read length of 76 nucleotides. An average of 12.26 million reads (11.6~13.0 million) were generated per library, with an average quality score passing quality filter above Q30. The sequencing results were analyzed using a customized pipeline (gopher‐pipelines; https://bitbucket.org/jgarbe/gopher-pipelines/overview) developed and maintained by the University of Minnesota Supercomputing Institute (MSI). The quality of Illumina sequencing data was evaluated by FastQC (v0.11.5) and the adapters and low‐quality reads were trimmed using Trimmomatic (v0.33). Trimmed sequence reads were aligned to the Homo sapiens reference genome (GRCh38) using HISAT2 (v2.0.2). Transcript abundance was then estimated using the FeatureCounts program in the Subread package (v1.4.6). The read counts generated by FeatureCounts were then filtered to include genes with at least 1 count per million and subsequently analyzed in R. Heat map was then generated using log transformed values with pheatmap package. Hierarchical clustering was performed using the Euclidean Distance and Average Link Clustering methods. Principle component analysis was performed using the prcomp function in R and visualized using ggplot2 package. Differential expression analysis was performed using edgeR package in R. Genes were retained using the criterion of at least a 2‐fold change and false discovery rate with an adjusted P value of less than .05 for gene analysis.
3. RESULTS
3.1. hPSCs exposed to combined inhibition of BMP and FGF produces an early ectodermal intermediate within 24 hours
Early embryonic fate decisions that occur between the stages of pluripotency and ectoderm formation were targeted for acceleration, which is an attractive target given its broad applicability to all neuronal differentiations. This stage is also relatively prolonged because ectodermal fate is thought to be passively acquired, which has been demonstrated in vitro most elegantly through work that showed growth factor withdrawal alone was sufficient to induce differentiation in defined hPSC monolayer culture 10 without previously necessary inhibitor‐mediated pathway modulation. 9
Rather than allow for this passive acquisition of ectodermal fate, it was hypothesized differentiation could be induced by disrupting hPSC self‐renewal. It has been shown that the undifferentiated status of hPSCs is maintained by ERK and SMAD signaling stimulated by exogenous provision of FGF2 and TGFβ1, 23 , 24 making these pathways attractive candidates to influence the undifferentiated status of hPSCs. However, previous attempts to target these pathways for directed differentiation into neuroepithelium had not substantially improved upon the historical kinetics of neuroepithelial conversion, or had only proved applicable for the differentiation of specific neural subsets. 25 , 26 , 27 , 28 Therefore, we re‐evaluated these pathways for the directed differentiation of hPSCs into neuroepithelium under defined conditions. A strategy was devised reliant upon design‐of‐experiment chemical screening with 24‐hour time point resolution using a panel of five unrelated hiPSC lines. 18 , 29 , 30 The immunocytochemical presence of NANOG was used as a primary readout for disrupted self‐renewal, given NANOG's central role in early exit from pluripotency 31 (Figure S1A). This screening strategy identified a two‐factor combination containing the activin‐like kinase 2/3 inhibitor LDN193189 (LDN, 500 nM) and the vascular endothelial growth factor receptor 2/fibroblast growth factor receptor 1 (VEGFR2/FGFR1) inhibitor SU5402 (SU, 10 μM), which reduced the number of NANOG‐positive cells in culture from 100% to approximately 43%; however, this result was not statistically significant by one‐way ANOVA and Tukey multicomparison testing (Figure S1B).
We initially selected SU for our experiments as it is often cited as an FGFR inhibitor. 28 , 32 However, SU possesses an IC50 of 20 nM to VEGFR2 and 30 nM to FGFR1. 33 The FGFR and VEGFR classes consist of four receptors each, providing eight receptors total to participate in signaling cascades, neglecting splice variants. 33 , 34 , 35 Given the possibility that the target pathways of SU could be more carefully investigated, we dissected the function of SU by repeating our experiments with the pan‐VEGFR inhibitor Axitinib and pan‐FGFR inhibitor BGJ398 (BGJ) (Figure 1A). While exposure of hPSCs to the combination of LDN and Axitinib performed similarly to controls exposed to Essential 6 Medium alone with respect to NANOG expression and morphological assessment, the combination of BGJ and LDN was capable of inducing a subtle morphological change indicated by increased flatness inferred by reduced phase‐contrast brightness around colony edges; as well as centrally located, singular nucleoli. Furthermore, cultures were shown to lose NANOG expression by greater than 60%, a difference found to be statistically significant, confirming the hypothesis that targeting FGFRs other than FGFR2 is important for the potential disruption of self‐renewal and accelerated exit from pluripotency (Figure 1B).
FIGURE 1.
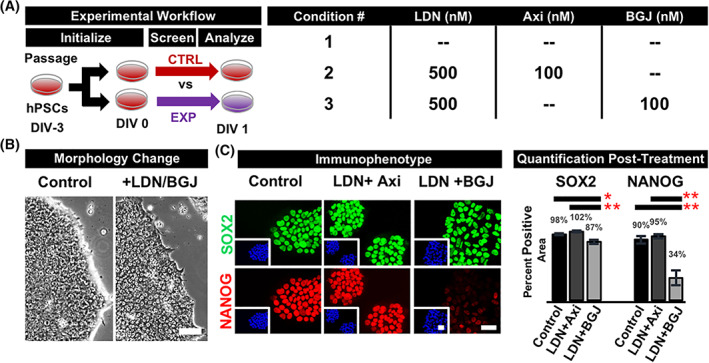
Combined antagonism of FGF and BMP disrupts hPSC self‐renewal. hPSCs were screened with inhibitor combinations and compared 24 hours later to untreated cultures demonstrate FGF to be the predominant self‐renewal pathway. A, Treatment with LDN193189 (LDN) and the pan‐FGFR inhibitor BGJ398 (BGJ) but not the pan‐VEGFR inhibitor Axitinib (Axi) promotes a subtle morphological change under phase‐contrast microscopy (B), with regularly‐spaced cells, lower nuclear‐to‐cytoplasmic ratio, flatter colonies evidenced by reduced phase‐bright edges, and centrally localized nucleoli. C, The novel combination of LDN/BGJ‐treatment reduces NANOG expression by 70% while maintaining SOX2 positivity. Quantification indicates averages and SE for five unrelated hiPSCs. One‐way ANOVA with Tukey multiple comparison testing showed significance at *P < .01 and **P < .001. Nuclei stained blue with DAPI. Scale bars = 50 μm. ANOVA, analysis of variance; BGJ, fibroblast growth factor pathway inhibitor BGJ398; BMP, bone morphogenic protein; DAPI, 4′,6‐diamidino‐2‐phenylindole dilactate; DIV, days in vitro; E8, Essential 8; FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; hiPSCs, human induced pluripotent stem cells; hPSC, human pluripotent stem cell; LDN, bone morphogenic protein pathway inhibitor LDN193189; VEGFR, vascular endothelial growth factor receptor
To contextualize the identity of the induced population, it was compared to other possible early fate decisions by differentiating hPSCs using standard methods (Figure 2A). RNA was collected from three unrelated hPSC lines cultured using Essential 8 Medium, 23 cultured into ectoderm over 6 days using “dual SMAD inhibition”, 9 cultured into primitive streak over 2 days, 36 or following 24‐hour treatment with LDN/BGJ (Figure 2B). Heatmap analysis demonstrated biological replicates to cluster according to treatment, with each treatment group containing unique upregulated gene expression clusters (Figure 2C). The top 500 upregulated genes from each cluster were extracted and assessed for molecular function (PANTHER gene ontology) to identify sequence‐specific DNA‐binding proteins (ie, transcription factors). The top 15 upregulated transcription factors were listed for each cluster and compared across treatments. This analysis validated the identity of the three standard treatment groups, as cluster 1 genes upregulated with E8 treatment contained canonical pluripotency factors POU5F1 (OCT4), 37 ZFP42 (REX1), 38 and MYC 39 ; cluster 2 genes upregulated with LDN/SB treatment contained the ectoderm‐associated genes SALL2, 40 PAX6, 41 and SOX2 42 ; and cluster 4 genes upregulated with CHIR treatment contained primitive streak‐associated genes T, 43 CDX1, 44 CDX2, 43 and several HOX genes. 45 Application of this validated methodology to cluster 3 genes upregulated with LDN/BGJ treatment identified ZIC2, 46 ZIC3, 46 TRIM24, 47 and LHX5, 48 known to be involved in early gastrulation events related to lateral‐rostral ectoderm formation. Furthermore, cultures exposed to LDN/BGJ treatment expressed factors associated with pluripotency (OCT4) and standard ectoderm (SOX2 and SOX11 49 ), but at reduced levels (Figure 2D). These results are interpreted to represent the formation of an early intermediate ectoderm population from hPSCs within 24 hours by treatment with LDN/BGJ.
FIGURE 2.
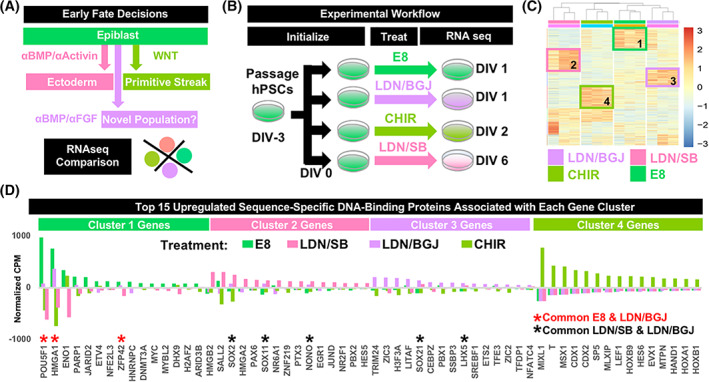
RNA sequencing of early embryonic populations identifies LDN/BGJ‐treated cultures to be a novel, early ectodermal population. A, Possible early embryonic fate decisions were identified as controls to contextualize cultures treated with 500 nM LDN and 100 nM BGJ for 24 hours. B, Three unrelated, independent hiPSC lines were maintained undifferentiated, or differentiated into either ectoderm or primitive streak using standard protocols containing 250 nM LDN and 10 μM SB435142 for 6 days or 6 μM CHIR99021 for 2 days, respectively. These controls allowed for the contextualization of cultures treated with 500 nM LDN and 100 nM BGJ for 24 hours. C, Total RNA was extracted, libraries created, and 20 M reads sequenced for each sample. Counts‐per‐million (CPM) values for each gene were normalized to generate heatmaps, demonstrating biological replicates for each treatment to cluster appropriately, and identified unique clusters of upregulated genes for each treatment group. D, CPMs for each treatment group were averaged, and GO analysis for the top 500 upregulated genes for each cluster was performed to identify sequence‐specific DNA‐binding proteins (transcription factors). The top 15 upregulated transcription factors for each group were plotted, and transcription factor overlaps indicated with asterisks. Each standard treatment group expressed germ‐layer specific transcription factors, with LDN/BGJ‐treated cultures demonstrating residual OCT4 expression, but also transcripts associated with ectoderm, such as SOX2, SOX11, LHX5, ZIC2, ZIC3, and TRIM24. BGJ, fibroblast growth factor pathway inhibitor BGJ398; CHIR, wnt pathway agonist CHIR99021; DIV, days in vitro; E8, Essential 8; hiPSCs, human induced pluripotent stem cells; GO, gene ontology; LDN, bone morphogenic protein pathway inhibitor LDN193189; SB, SB435142
3.2. Putative early intermediate ectoderm responds immediately to morphogens to produce rostro‐caudal and medial‐lateral neurodevelopmental populations
Practical applications of this population for manufacturing specific neuronal subtypes would require its appropriate response to patterning morphogens. To demonstrate this critical property, three unrelated putative early ectoderm cultures produced following 24‐hour treatment with LDN/BGJ were exposed for an additional 24 hours to either suppressed (500 nM wntC59) or enhanced (4 μM CHIR99021) wnt signaling, conditions shown to produce either rostral or caudal neurodevelopmental populations, 50 , 51 , 52 , 53 , 54 , 55 respectively (Figure 3A). Following either treatment, appropriate rostral or caudal landmarks were shown to emerge as indicated by the acquisition of SIX3 and PAX6‐positive telencephalic neuroepithelium; or HOXB4, SOX2, and T‐positive neuromesoderm 56 , 57 (Figure 3B). To demonstrate the medial‐lateral patterning competency of the putative ectoderm, directed differentiation protocols were used following developmentally guided principles targeting the midbrain floorplate and neural crest. After 24 hours of treatment with LDN/BGJ, cultures further exposed to wnt and hedgehog signaling demonstrated rapid acquisition of floorplate marker FOXA2 by day 4 (Figure S2A). Similarly, LDN/BGJ‐treated cultures produced a PAX3/SOX10‐positive neural crest phenotype by day 4 when further exposed to increased BMP signaling and decreased FGF and hedgehog signaling (Figure S2B). In both cases, these populations were produced with greater than 70% efficiency. This provides evidence that the early intermediate ectodermal population produced with LDN/BGJ possesses maximal rostro‐caudal/medial‐lateral patterning flexibility demonstrated in other systems, useful for generating full‐spectrum neuronal diversity from hPSCs in a drastically accelerated fashion.
FIGURE 3.
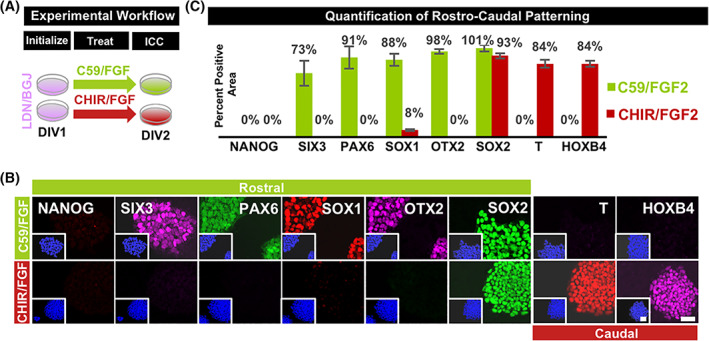
Treatment with LDN/BGJ unlocks immediate access to rostro‐caudal extremes of ectodermal patterning. A, Ectoderm derived using LDN/BGJ was exposed to either wntC59 (500 nM) and FGF2 (10 ng/mL) or CHIR99021 (4 μM) and FGF2 (10 ng/mL) to produce either forebrain or neuromesoderm phenotypes by 48 hours in vitro. B, Rostral landmarks upregulated following treatment with wntc59 include SIX3, PAX6, and OTX2; but not caudal neuromesodermal landmarks T and HOXB4. Conversely, caudal neuromesoderm landmarks SOX2, T, and HOXB4 were upregulated; but not rostral forebrain landmarks SIX3, PAX6, and OTX2. C, Differentiation was seen to be greater than 70% efficient in either rostro‐caudal direction. Quantification indicates averages and SE for three unrelated hiPSC lines. Nuclei stained blue with DAPI. Scale bars = 50 μm. BGJ, fibroblast growth factor pathway inhibitor BGJ398; C59, wnt pathway inhibitor WNTC59; CHIR, wnt pathway agonist CHIR99021; DAPI, 4′,6‐diamidino‐2‐phenylindole dilactate; DIV, days in vitro; E8, Essential 8; FGF2, fibroblast growth factor 2; hiPSCs, human induced pluripotent stem cells; hPSC, human pluripotent stem cell; LDN, bone morphogenic protein pathway inhibitor LDN193189
4. DISCUSSION
Traditional methods for producing ectoderm describe the direct, but gradual and asynchronous generation of a PAX6/SOX1 expressing obligate anterior population 58 from hPSCs over the course of 7 to 12 days. This contrasts with the methods disclosed in this work which produce within 24 hours a previously uncharacterized population we have termed “primal ectoderm” before later acquiring a PAX6/SOX1 phenotype by 48 hours. This new method for producing PAX6/SOX1‐positive cultures proceeds rapidly and discretely in a defined and controllable fashion. Rapid and step‐wise differentiation processes for neural lineages are currently lacking, and this report is likely the first of a new generation of carefully designed and efficient protocols for the manufacture of hPSC‐derived neuronal subpopulations. However, additional work is now required to use the populations described in this report for terminal maturation into electrophysiologically functional neurons.
The identity of the “primal” ectoderm described in this study now requires further investigation. Recent studies describe that both epiblast cells in early embryonic differentiation and human embryonic stem cells (hESCs) undergoing neural differentiation in vitro, acquire axial identity prior to overt neural induction and suggest that the in vitro patterned, “pre‐ectodermal” populations generated in this study may indeed have an in vivo counterpart. 59 , 60 A “pre‐ectodermal” designation is used because early ectoderm has been classically defined as a PAX6‐expressing primitive neuroepithelium.
It is possible that neural induction described in previous studies also occurs via the primal ectoderm now described in this study. In their recent publication generating neural tissue from hESCs following dual SMAD inhibition, Rifes et al define a “pre‐ectodermal” population as an LHX5‐positive intermediate, 60 a marker seen in the populations produced in our current study. Additional evidence for a previously undefined, primal ectoderm may also be seen in the original dual SMAD inhibition protocol. 9 In this seminal study, hPSCs differentiating with LDN/SB through a 6‐day time course maintained an OCT4‐positive/PAX6‐negative phenotype for 4 days, before transitioning into an undefined OCT4‐negative/PAX6‐negative phenotype on day 5, just prior to adopting the classically defined OCT4‐negative/PAX6‐positive primitive ectodermal phenotype. 9 This differentiation trajectory suggests the presence of a primal ectoderm without PAX6 expression. A direct comparison of this OCT4‐negative/PAX6‐negative population produced in 5 days using LDN/SB with the population described in this study produced after 24 hours incubation with LDN/BGJ would provide additional evidence that the first committed step toward ectoderm requires reexamination.
5. CONCLUSION
The protocols provided here represent a new entry point for generating ectoderm from hPSCs and provide an accelerated differentiation trajectory that can be rapidly expanded upon for further protocol development using developmentally guided principles. Continued refinement of protocols based on this technology platform will result in additional gains in differentiation speed and efficiency. This will accelerate the adoption of hPSC‐derived neural cell types for use in basic research, drug discovery, and cell therapy applications due to ease‐of‐use considerations, experimental iteration speed, abridged manufacturing timelines, and cost advantages.
CONFLICT OF INTEREST
P.W. declared employment/leadership position, patent holder, and stock ownership with Anatomic Incorporated. V.T. declared employment/leadership position and stock ownership with Anatomic Incorporated. W.C.L. declared employment/leadership position and stock ownership with Recombinetics. All the other authors declared no potential conflicts of interest. The University of Minnesota has filed patent application WO/2018/208836 related to the methods, compositions, and uses described in the article.
AUTHOR CONTRIBUTIONS
P.W.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; V.T., S.N., M.S.M.: collection and/or assembly of data, data analysis and interpretation, manuscript writing; W.C.L.: manuscript preparation, administrative support; A.M.P.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript, administrative support; J.R.D.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript, financial support, administrative support.
Supporting information
Supplemental Figure 1 Identification of a novel two‐factor condition that reduces NANOG expression.
hPSCs were screened with inhibitors to candidate self‐renewal pathways and analyzed 24 hours later in comparison to undifferentiated cultures. A) A small‐molecule combination of the activin‐like kinase 2/3 inhibitor LDN193189 (LDN) and vascular endothelial growth factor receptor 2 (VEGFR2)/fibroblast growth factor receptor 1 (FGFR1) inhibitor SU5402 (SU) was identified and a design‐of‐experiment performed. B) LDN/SU were necessary and sufficient to reduce the number of NANOG‐positive cells in culture by 60% compared to undifferentiated controls. Quantification indicates averages and SE for five unrelated hiPSC lines. One‐way ANOVA with Tukey multiple comparison testing showed no statistical significance., E8: Essential 8, DIV: days in vitro, LDN: bone morphogenic protein pathway inhibitor LDN193189, SU: VEGFR2/FGFR2 inhibitor SU5402. Nuclei stained blue with DAPI. Scale bars 50 μM.
Supplemental Figure 2 Rostro‐caudalized ectoderm can be further patterned medially‐laterally to efficiently produce numerous neurodevelopmental populations in drastically accelerated fashion.
A) A rostral‐medial ectodermal phenotype of the midbrain floorplate expressing OTX2 and FOXA2 was produced with 70% efficiency within four days in vitro. B) Caudal‐lateral neural crest expressing PAX3 and SOX10 can be produced with greater than 70% efficiency from hPSCs within four days in vitro. Quantification indicates averages and SE for three unrelated hiPSCs. hPSC: human pluripotent stem cell, E8: Essential 8, LDN: bone morphogenic protein pathway inhibitor LDN193189, BGJ: fibroblast growth factor pathway inhibitor BGJ398, FGF2: fibroblast growth factor 2, A8: TGFβ pathway inhibitor A8301, CHIR: wnt pathway agonist CHIR99021, BMP4: bone morphogenic protein 4, SAG: sonic hedgehog pathway agonist Smoothened Agonist. Nuclei stained blue with DAPI. Scale bars 50 μM.
ACKNOWLEDGMENTS
The authors thank the laboratories of Jakub Tolar and Deborah Ferrington for providing a panel of hPSCs for validating various aspects of differentiation; and Seunghyun Lim for his analysis of RNA‐sequencing data. J.R.D. and A.M.P. acknowledge support from the Minnesota Spinal Cord Injury and Traumatic Brain Injury Research Grant Program and a generous donation from an anonymous philanthropic donor that initiated this study.
Walsh P, Truong V, Nayak S, et al. Accelerated differentiation of human pluripotent stem cells into neural lineages via an early intermediate ectoderm population. Stem Cells. 2020;38:1400–1408. 10.1002/stem.3260
Ann M. Parr and James R. Dutton contributed equally to this work.
Funding information Minnesota Spinal Cord Injury and Traumatic Brain Injury Research Grant Program
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
REFERENCES
- 1. Thomson JA, Itskovitz‐Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145‐1147. [DOI] [PubMed] [Google Scholar]
- 2. Yu J, Vodyanik MA, Smuga‐Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917‐1920. [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors [in Eng]. Cell. 2007;131(5):861‐872. [DOI] [PubMed] [Google Scholar]
- 4. Munoz‐Sanjuan I, Brivanlou AH. Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci. 2002;3(4):271‐280. [DOI] [PubMed] [Google Scholar]
- 5. Itskovitz‐Eldor J, Schuldiner M, Karsenti D, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6(2):88‐95. [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19(12):1129‐1133. [DOI] [PubMed] [Google Scholar]
- 7. Perrier AL, Tabar V, Barberi T, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101(34):12543‐12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yao S, Chen S, Clark J, et al. Long‐term self‐renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci USA. 2006;103(18):6907‐6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lippmann ES, Estevez‐Silva MC, Ashton RS. Defined human pluripotent stem cell culture enables highly efficient neuroepithelium derivation without small molecule inhibitors. Stem Cells. 2014;32(4):1032‐1042. [DOI] [PubMed] [Google Scholar]
- 11. Li XJ, Zhang X, Johnson MA, Wang ZB, LaVaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136(23):4055‐4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan Y, Yang D, Zarnowska ED, et al. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23(6):781‐790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Liu H, Sauvey C, Yao L, Zarnowska ED, Zhang SC. Directed differentiation of forebrain GABA interneurons from human pluripotent stem cells. Nat Protoc. 2013;8(9):1670‐1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu J, Zhong X, Liu H, et al. Generation of serotonin neurons from human pluripotent stem cells. Nat Biotechnol. 2016;34(1):89‐94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoon K, Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8(6):709‐715. [DOI] [PubMed] [Google Scholar]
- 16. Mason I. Initiation to end point: the multiple roles of fibroblast growth factors in neural development. Nat Rev Neurosci. 2007;8(8):583‐596. [DOI] [PubMed] [Google Scholar]
- 17. Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene‐free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85(8):348‐362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geng Z, Walsh PJ, Truong V, et al. Generation of retinal pigmented epithelium from iPSCs derived from the conjunctiva of donors with and without age related macular degeneration. PLoS One. 2017;12(3):e0173575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lindborg BA, Brekke JH, Vegoe AL, et al. Rapid induction of cerebral organoids from human induced pluripotent stem cells using a chemically defined hydrogel and defined cell culture medium. Stem Cells Translational Medicine. 2016;5(7):970‐979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye L, Zhang S, Greder L, et al. Effective cardiac myocyte differentiation of human induced pluripotent stem cells requires VEGF. PLoS One. 2013;8(1):e53764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsh P, Truong V, Hill C, et al. Defined culture conditions accelerate small‐molecule‐assisted neural induction for the production of neural progenitors from human‐induced pluripotent stem cells. Cell Transplant. 2017;26(12):1890‐1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schindelin J, Arganda‐Carreras I, Frise E, et al. Fiji: an open‐source platform for biological‐image analysis. Nat Methods. 2012;9(7):676‐682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen G, Gulbranson DR, Hou Z, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beers J, Gulbranson DR, George N, et al. Passaging and colony expansion of human pluripotent stem cells by enzyme‐free dissociation in chemically defined culture conditions. Nat Protoc. 2012;7(11):2029‐2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Greber B, Coulon P, Zhang M, et al. FGF signalling inhibits neural induction in human embryonic stem cells. EMBO J. 2011;30(24):4874‐4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qi Y, Zhang XJ, Renier N, et al. Combined small‐molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nat Biotechnol. 2017;35(2):154‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yoo YD, Huang CT, Zhang X, Lavaute TM, Zhang SC. Fibroblast growth factor regulates human neuroectoderm specification through ERK1/2‐PARP‐1 pathway. Stem Cells. 2011;29(12):1975‐1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chambers SM, Qi Y, Mica Y, et al. Combined small‐molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30(7):715‐720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferrington DA, Ebeling MC, Kapphahn RJ, et al. Altered bioenergetics and enhanced resistance to oxidative stress in human retinal pigment epithelial cells from donors with age‐related macular degeneration. Redox Biol. 2017;13:255‐265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kudva YC, Ohmine S, Greder LV, et al. Transgene‐free disease‐specific induced pluripotent stem cells from patients with type 1 and type 2 diabetes. Stem Cells Translational Medicine. 2012;1(6):451‐461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17(1):42‐49. [DOI] [PubMed] [Google Scholar]
- 32. Buchholz DE, Pennington BO, Croze RH, Hinman CR, Coffey PJ, Clegg DO. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Translational Medicine. 2013;2(5):384‐393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun L, Tran N, Liang C, et al. Design, synthesis, and evaluations of substituted 3‐[(3‐ or 4‐carboxyethylpyrrol‐2‐yl)methylidenyl]indolin‐2‐ones as inhibitors of VEGF, FGF, and PDGF receptor tyrosine kinases. J Med Chem. 1999;42(25):5120‐5130. [DOI] [PubMed] [Google Scholar]
- 34. Zhang X, Ibrahimi OA, Olsen SK, Umemori H, Mohammadi M, Ornitz DM. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281(23):15694‐15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cross MJ, Dixelius J, Matsumoto T, Claesson‐Welsh L. VEGF‐receptor signal transduction. Trends Biochem Sci. 2003;28(9):488‐494. [DOI] [PubMed] [Google Scholar]
- 36. Lian X, Bao X, Zilberter M, et al. Chemically defined, albumin‐free human cardiomyocyte generation. Nat Methods. 2015;12(7):595‐596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu G, Scholer HR. Role of Oct4 in the early embryo development. Cell Regen. 2014;3(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Son MY, Choi H, Han YM, Sook Cho Y. Unveiling the critical role of REX1 in the regulation of human stem cell pluripotency. Stem Cells. 2013;31(11):2374‐2387. [DOI] [PubMed] [Google Scholar]
- 39. Chappell J, Dalton S. Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb Perspect Med. 2013;3(12):a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kelberman D, Islam L, Lakowski J, et al. Mutation of SALL2 causes recessive ocular coloboma in humans and mice. Hum Mol Genet. 2014;23(10):2511‐2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pera EM, Kessel M. Patterning of the chick forebrain anlage by the prechordal plate. Development. 1997;124(20):4153‐4162. [DOI] [PubMed] [Google Scholar]
- 42. Thomson M, Liu SJ, Zou LN, Smith Z, Meissner A, Ramanathan S. Pluripotency factors in embryonic stem cells regulate differentiation into germ layers. Cell. 2011;145(6):875‐889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mendjan S, Mascetti VL, Ortmann D, et al. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15(3):310‐325. [DOI] [PubMed] [Google Scholar]
- 44. Meyer BI, Gruss P. Mouse Cdx‐1 expression during gastrulation. Development. 1993;117(1):191‐203. [DOI] [PubMed] [Google Scholar]
- 45. Forlani S, Lawson KA, Deschamps J. Acquisition of Hox codes during gastrulation and axial elongation in the mouse embryo. Development. 2003;130(16):3807‐3819. [DOI] [PubMed] [Google Scholar]
- 46. McMahon AR, Merzdorf CS. Expression of the zic1, zic2, zic3, and zic4 genes in early chick embryos. BMC Res Notes. 2010;3:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hintze M, Prajapati RS, Tambalo M, et al. Cell interactions, signals and transcriptional hierarchy governing placode progenitor induction. Development. 2017;144(15):2810‐2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng G, Westerfield M. Lhx5 promotes forebrain development and activates transcription of secreted Wnt antagonists. Development. 2006;133(16):3191‐3200. [DOI] [PubMed] [Google Scholar]
- 49. Yan B, Neilson KM, Moody SA. foxD5 plays a critical upstream role in regulating neural ectodermal fate and the onset of neural differentiation. Dev Biol. 2009;329(1):80‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Arenas E, Denham M, Villaescusa JC. How to make a midbrain dopaminergic neuron. Development. 2015;142(11):1918‐1936. [DOI] [PubMed] [Google Scholar]
- 51. Sances S, Bruijn LI, Chandran S, et al. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat Neurosci. 2016;19(4):542‐553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yuan F, Fang KH, Cao SY, et al. Efficient generation of region‐specific forebrain neurons from human pluripotent stem cells under highly defined condition. Sci Rep. 2015;5:18550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Arber C, Precious SV, Cambray S, et al. Activin A directs striatal projection neuron differentiation of human pluripotent stem cells. Development. 2015;142(7):1375‐1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vadodaria KC, Marchetto MC, Mertens J, Gage FH. Generating human serotonergic neurons in vitro: methodological advances. Bioessays. 2016;38(11):1123‐1129. [DOI] [PubMed] [Google Scholar]
- 55. Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012;7(10):1836‐1846. [DOI] [PubMed] [Google Scholar]
- 56. Gouti M, Tsakiridis A, Wymeersch FJ, et al. In vitro generation of neuromesodermal progenitors reveals distinct roles for wnt signalling in the specification of spinal cord and paraxial mesoderm identity. PLoS Biol. 2014;12(8):e1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lippmann ES, Williams CE, Ruhl DA, et al. Deterministic HOX patterning in human pluripotent stem cell‐derived neuroectoderm. Stem Cell Reports. 2015;4(4):632‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pankratz MT, Li XJ, Lavaute TM, et al. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells. 2007;25(6):1511‐1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Metzis V, Steinhauser S, Pakanavicius E, et al. Nervous system regionalization entails axial allocation before neural differentiation. Cell. 2018;175(4):1105‐1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rifes P, Isaksson M, Rathore GS, et al. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat Biotechnol. 2020. https://doi:10.1038/s41587-020-0525-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 Identification of a novel two‐factor condition that reduces NANOG expression.
hPSCs were screened with inhibitors to candidate self‐renewal pathways and analyzed 24 hours later in comparison to undifferentiated cultures. A) A small‐molecule combination of the activin‐like kinase 2/3 inhibitor LDN193189 (LDN) and vascular endothelial growth factor receptor 2 (VEGFR2)/fibroblast growth factor receptor 1 (FGFR1) inhibitor SU5402 (SU) was identified and a design‐of‐experiment performed. B) LDN/SU were necessary and sufficient to reduce the number of NANOG‐positive cells in culture by 60% compared to undifferentiated controls. Quantification indicates averages and SE for five unrelated hiPSC lines. One‐way ANOVA with Tukey multiple comparison testing showed no statistical significance., E8: Essential 8, DIV: days in vitro, LDN: bone morphogenic protein pathway inhibitor LDN193189, SU: VEGFR2/FGFR2 inhibitor SU5402. Nuclei stained blue with DAPI. Scale bars 50 μM.
Supplemental Figure 2 Rostro‐caudalized ectoderm can be further patterned medially‐laterally to efficiently produce numerous neurodevelopmental populations in drastically accelerated fashion.
A) A rostral‐medial ectodermal phenotype of the midbrain floorplate expressing OTX2 and FOXA2 was produced with 70% efficiency within four days in vitro. B) Caudal‐lateral neural crest expressing PAX3 and SOX10 can be produced with greater than 70% efficiency from hPSCs within four days in vitro. Quantification indicates averages and SE for three unrelated hiPSCs. hPSC: human pluripotent stem cell, E8: Essential 8, LDN: bone morphogenic protein pathway inhibitor LDN193189, BGJ: fibroblast growth factor pathway inhibitor BGJ398, FGF2: fibroblast growth factor 2, A8: TGFβ pathway inhibitor A8301, CHIR: wnt pathway agonist CHIR99021, BMP4: bone morphogenic protein 4, SAG: sonic hedgehog pathway agonist Smoothened Agonist. Nuclei stained blue with DAPI. Scale bars 50 μM.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author.


