Summary
The human pathogen Vibrio cholerae serves as a model organism for many important processes ranging from pathogenesis to natural transformation, which has been extensively studied in this bacterium. Previous work has deciphered important regulatory circuits involved in natural competence induction as well as mechanistic details related to its DNA acquisition and uptake potential. However, since competence was first reported for V. cholerae in 2005, many researchers have struggled with reproducibility in certain strains. In this study, we therefore compare prominent seventh pandemic V. cholerae isolates, namely strains A1552, N16961, C6706, C6709, E7946, P27459, and the close relative MO10, for their natural transformability and decipher underlying defects that mask the high degree of competence conservation. Through a combination of experimental approaches and comparative genomics based on new whole‐genome sequences and de novo assemblies, we identify several strain‐specific defects, mostly in genes that encode key players in quorum sensing. Moreover, we provide evidence that most of these deficiencies might have recently occurred through laboratory domestication events or through the acquisition of mobile genetic elements. Lastly, we highlight that differing experimental approaches between research groups might explain more of the variations than strain‐specific alterations.
Introduction
The causative agent of cholera, Vibrio cholerae, is an interesting model organism for studying dual bacterial lifestyles due to its ability to thrive both in the human gut and in natural aquatic habitats (Colwell, 1996; Nelson et al., 2009). However, only a limited set of V. cholerae strains is responsible for cholera pandemics, while a broad variety of isolates do not cause the disease and are primarily found in the environment. Therefore, to better understand pathogen emergence, it is important to understand basic differences between pathogenic and environmental strains, as well as the capability of different species for the horizontal acquisition of genetic material.
In its environmental reservoir, V. cholerae's attachment to biotic surfaces often results in biofilm formation and the degradation of the underlying nutritious surfaces, such as the chitin polymer. Chitin is the most abundant polysaccharide in aquatic environments and the primary carbon source for many marine bacteria (Gooday, 1990), including most Vibrios (Hunt et al., 2008; Le Roux and Blokesch, 2018). Apart from its role as a major carbon source, in V. cholerae, chitin also triggers natural competence for transformation (Meibom et al., 2005), which is the state in which the bacterium is able to take up free DNA and incorporate it into its genome by double homologous recombination. As a mechanism of horizontal gene transfer (HGT), natural competence enables V. cholerae (Meibom et al., 2005) and other Vibrio species (Gulig et al., 2009; Pollack‐Berti et al., 2010; Metzger et al., 2019) to exchange DNA stretches including those that harbour genes or operons encoding new pathogenic or metabolic traits (Blokesch and Schoolnik, 2007; Miller et al., 2007).
Since the initial discovery of chitin‐induced competence in V. cholerae (Meibom et al., 2005), several studies have addressed the underlying regulatory circuits and mechanistic aspects of the DNA uptake process (Fig. 1). In addition to inducing the competence state, which includes the production of the DNA‐uptake complex (Seitz and Blokesch, 2013), chitin also induces the type VI secretion system (T6SS), a molecular killing device (Cianfanelli et al., 2016), in pandemic V. cholerae strains (Borgeaud et al., 2015). The T6SS fosters killing of adjacent bacteria in a contact‐ and kin‐discriminatory manner, thereby serving as a DNA acquisition system that enhances HGT (Borgeaud et al., 2015; Metzger and Blokesch, 2016; Veening and Blokesch, 2017). We recently showed that this link between neighbour predation and prey‐released DNA uptake allows long stretches of DNA, frequently exceeding 100 kilobases (kb) in length, to be horizontally transferred. (Matthey et al., 2019). Given the massive potential of T6SS‐mediated neighbour predation and its coupling to the concomitantly produced DNA‐uptake machinery, it is not surprising that the bacteria have evolved sophisticated regulatory circuits to control the onset of the competence state.
Fig. 1.
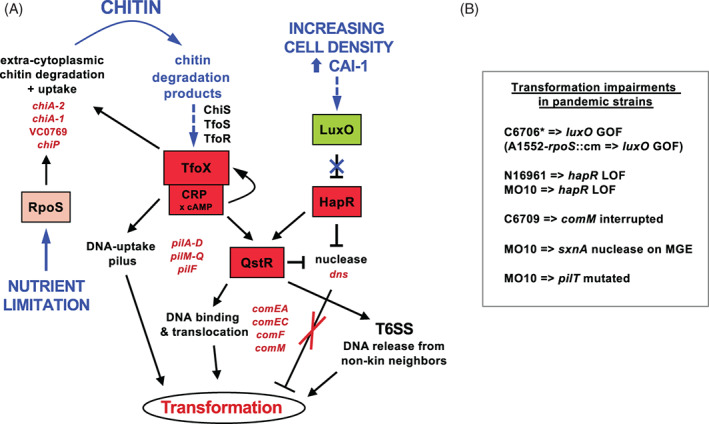
The competence regulatory circuit in V. cholerae.
A. Competence induction in V. cholerae is dependent on chitin induction and quorum sensing (mostly driven by the species‐specific autoinducer CAI‐1). The scheme is an updated version of the model proposed by Meibom et al. (2005) (see Supporting Information Fig. S1). Red boxes denote positive regulators and the most important regulated genes are shown in red writing. For details see text.
B. Causation of impaired transformation in a subset of pandemic strains. GOF, gain‐of‐function; LOF, Loss‐of‐function.
An essential inducer of competence in V. cholerae (Meibom et al., 2005), chitin is degraded by extracellular chitinases that are either produced by moulting zooplankton, by other bacteria through ‘sloppy eating’, or by starved Vibrio species themselves (Keyhani and Roseman, 1999). Chitinases predominantly release N‐acetyl‐glucosamine (GlcNAc) in the form of dimers or trimers from the chitin polymer, whereas monomeric GlcNAc is not a primary cleavage product (Gooday, 1990; Li and Roseman, 2004). Diacetylchitobiose, the GlcNAc dimer (also frequently called chitobiose), acts as a strong chemoattractant for diverse Vibrio species (Bassler et al., 1989) and is subsequently taken up via an ABC‐type transporter encoded by the genes VC0619–VC0616 in V. cholerae in conjunction with the periplasmic chitin‐binding protein Cbp (VC0620), thereby avoiding carbon catabolite repression (Li and Roseman, 2004; Blokesch, 2012b). The chitobiose is further degraded into monomeric sugars and finally shuttled into central metabolism (Gooday, 1990; Hunt et al., 2008).
Understanding of the signalling pathways that link chitin sensing to competence induction has significantly improved in recent years (Fig. 1 compared to Supporting Information Fig. S1). The chitin catabolic cascade (e.g., the chitobiose catabolic operon, VC0620–VC0611; (Li and Roseman, 2004) is mostly controlled by ChiS, a noncanonical, membrane‐embedded, one‐component sensor kinase (Klancher et al., 2020a). This pathway is important for chitin catabolism, while the sensing of chitobiose by yet another noncanonical transmembrane regulator named TfoS is, together with its regulated small RNA TfoR (Yamamoto et al., 2011; Dalia et al., 2014; Yamamoto et al., 2014), essential for the production of the major regulator of competence, TfoX (Meibom et al., 2005). Artificial TfoX production suppresses the chitin requirement and triggers competence induction even under rich medium conditions if catabolite repression does not occur (Meibom et al., 2005; Blokesch, 2012b). Because TfoX proteins are often highly conserved, their homologous or even heterologous production induces competence in diverse Vibrio species such as V. cholerae, V. parahaemolyticus, V. alginolyticus, V. fischeri, V. campbellii and V. vulnificus (Pollack‐Berti et al., 2010; Metzger et al., 2016; Metzger et al., 2019; Simpson et al., 2019).
In addition to chitin‐induced signalling through TfoX, competence induction in V. cholerae also relies on input from the bacterium's quorum sensing (QS) system (Meibom et al., 2005), which is controlled by the high cell density master QS regulator HapR (Mukherjee and Bassler, 2019). Chitin signalling and QS pathways are linked by an intermediate regulatory protein, which we named QS‐ and TfoX‐dependent regulator QstR (Lo Scrudato and Blokesch, 2013). QstR is a dual transcription factor of the LuxR‐type family (Jaskólska et al., 2018) that induces a subset of competence genes (such as comEA, comEC, comF, and comM; recently reviewed by (Dubnau and Blokesch, 2019). In addition to qstR and therefore competence gene induction, HapR plays another important role in natural transformation, namely as a repressor of the periplasmic and extracellular nuclease Dns. Previously, we showed that Dns degrades transforming material in QS‐defective strains and therefore impairs transformation (Blokesch and Schoolnik, 2008). In wild‐type cells, the encoding gene dns is, however, repressed by direct binding of HapR, a repression that is further enhanced by QstR (Blokesch and Schoolnik, 2008; Lo Scrudato and Blokesch, 2013; Jaskólska et al., 2018) (Fig. 1).
While the past 15 years since its discovery (Meibom et al., 2005) have significantly advanced our knowledge of competence regulation and DNA uptake of chitin‐induced naturally competent V. cholerae (Metzger and Blokesch, 2016; Dubnau and Blokesch, 2019), many questions remained unanswered. Moreover, selective findings or their interpretation often differ between studies from various groups, which is frequently justified by the use of different V. cholerae patient isolates. The reasons for such strain‐specific responses could be due to differences in the underlying regulatory circuits or laboratory domestication events, making the results less biologically meaningful. For instance, it is well known that the QS pathway is prone to mutations in the hapR and luxO genes (Joelsson et al., 2006; Jung et al., 2015; Stutzmann and Blokesch, 2016), with LuxO usually acting as a repressor of the hapR transcript via the Qrr sRNAs at low cell density (Mukherjee and Bassler, 2019). In this context, Kimbrough and Stabb showed that prolonged stationary phase conditions (as could occur when keeping Vibrios too long on plates) caused QS‐impairing gain‐of‐function (GOF) mutations in luxO of V. fischeri (proteins named LuxO*; Kimbrough and Stabb, 2015). A similar GOF mutation in luxO, which resulted in the G333S amino acid exchange in LuxO, led to an impaired high cell density response in the pandemic strain C6706 (Stutzmann and Blokesch, 2016). This C6706 (StrR) strain was subsequently shared among many laboratories, causing irreproducibility issues.
In this study, we compared the chitin‐induced competence and transformation behaviour in several of the most prominent pandemic O1 El Tor V. cholerae isolates and the close relative O139 strain MO10. Through comparative analysis of de novo assembled genomes of strains A1552, N16961, C6706, C6709, E7946, and P27459 based on long‐read, whole‐genome, PacBio sequencing data combined with experimental validation, we show that the competence regulon is completely conserved. We also provide evidence that all strains with a fully functional quorum sensing circuit behave in a highly comparable manner with respect to competence induction and natural transformability, which suggests that previous studies differed due to experimental differences and not due to extensive strain variations. Moreover, we found that comM of strain C6709 is interrupted by a mobile genetic element and show that this interruption impairs the strain's natural transformability.
Results and discussion
The primary role of CAI‐1 in competence induction is conserved among pandemic strains
Competence regulation in V. cholerae has been extensively studied, and strain‐specific responses have frequently been reported, even for patient isolates representing the current ongoing seventh pandemic cholera. In contrast to environmental isolates, these pandemic strains are thought to be highly similar if not clonal, at least with respect to the core genome (Mutreja et al., 2011). We therefore aimed to first compare the impact of QS on competence induction and transformation, using two prominent V. cholerae isolates, namely strains A1552 and C6706, both of which belong to the Latin American clade of seventh pandemic strains (Blokesch, 2012c; Domman et al., 2017) (Supporting Information Table S1). We previously showed that the competence and transformation of strain A1552 was primarily driven by the species‐specific cholera autoinducer 1 (CAI‐1), while the effect of the more universal autoinducer AI‐2 (Ng and Bassler, 2009) seemed negligible in this context (Suckow et al., 2011). In the absence of the CAI‐1 synthase CqsA, natural transformation was mostly abrogated or very close to the detection limit, while the absence of the AI‐2 synthase LuxS did not significantly impact the bacteria's transformability. The absence of both autoinducer synthases reflected the hapR‐deficient phenotype that lacked transformability (Suckow et al., 2011; Lo Scrudato and Blokesch, 2012). Using strain C6706, Antonova and Hammer came to slightly different conclusions (Antonova and Hammer, 2011). They found that while a hapR mutant was likewise non‐transformable, both cqsA‐minus and surprisingly cqsA/luxS‐double mutants were still significantly transformable (Antonova and Hammer, 2011). To address this conflicting result, we repeated these transformation experiments with the two strains A1552 and C6706 and their derivatives (C6706 mutants kindly provided by B. Hammer) in parallel using an optimized transformation protocol based on chitin flakes as the competence inducer (Marvig and Blokesch, 2010 and Method section). As shown in Fig. 2, the direct side‐by‐side comparison resulted in almost identical transformation phenotypes for both pandemic strains and their autoinducer synthase single or double mutants.
Fig. 2.
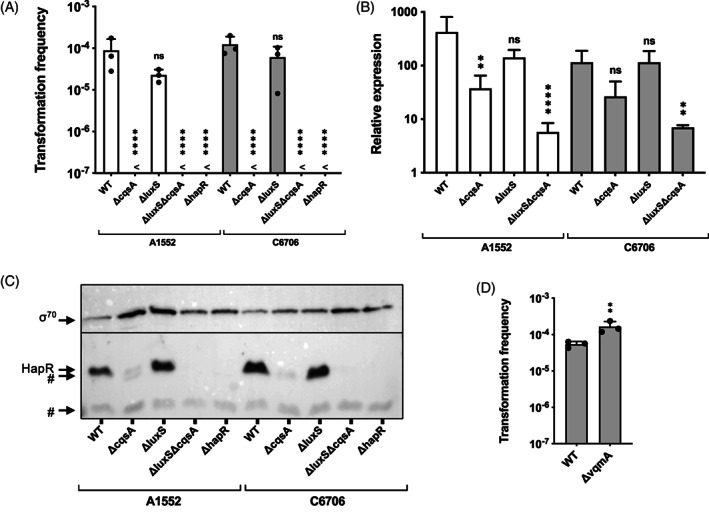
Absence of the species‐specific CAI‐1 severely impacts transformation in monocultures. V. cholerae strains lacking the autoinducer synthases CqsA (for CAI‐1) and/or LuxS (for AI‐2) were tested for their natural transformability on chitin (A), their hapR transcripts (B), and their HapR protein levels (C). Wild‐type (WT) and hapR‐minus (∆hapR) strains served as controls. The experiments were performed with mutants in two different strain backgrounds (A1552 and C6706). Panels (A) and (D): Transformation was tested on chitin surfaces and transformation frequencies are given on the Y‐axis. Panel (D) shows that the VqmA‐DPO QS system is dispensable for natural transformation. Panel (C): Sigma70 detections served as a loading control. The known cross‐reacting bands of the Anti‐HapR antibodies are indicated by # in the Western blot analysis. All experiments were performed three independent times and the mean values are shown for all graphs (±SD). <, below detection limit. Statistical analysis compared each mutant to their parental WT strains. Statistical analyses on the log‐transformed data: (A, B) one‐way ANOVA with a Sidak's multiple comparisons test; (D) two‐tailed t‐test. The detection limit was used for statistical analysis for conditions in which no transformants were recovered. ** P < 0.01; ****P < 0.0001; ns, not significant.
To better understand the impact of the autoinducer synthases on QS, and therefore natural competence, we next tested the strains' hapR transcript levels using quantitative reverse transcription PCR (qRT‐PCR). We saw reduced hapR transcript levels for those strains that lacked the CAI‐1 synthase and a highly significant hapR transcription reduction for the strains that lacked both autoinducer synthases (∆luxS∆cqsA; Fig. 2B). As hapR is known to be post‐transcriptionally regulated by several Qrr sRNAs, we also checked its protein production by western blotting. These data showed a highly significant reduction in HapR protein levels in CAI‐synthase mutants of strain A1552 and C6706 (∆cqsA), while the AI‐2 synthase mutants (∆luxS) retained high HapR levels (Fig. 2C). Importantly, no HapR was detectable in the absence of cqsA and luxS, no matter which pandemic strain background was tested (Fig. 2C). We therefore conclude that CAI‐1 plays a primary role in competence regulation in V. cholerae. This result supports our previous work showing the strong dominance of the species‐specific autoinducer CAI‐1 over the universal autoinducer AI‐2 with respect to natural competence and transformation of V. cholerae in monoculture (Suckow et al., 2011; Lo Scrudato and Blokesch, 2012). Furthermore, this result is in line with a more recent study on a similar species‐specific repression of the chitobiose utilization locus (Klancher et al., 2020b).
Since competence induction is linked to a high cell density state, we wondered if the newly identified VqmA‐DPO QS system of V. cholerae was also involved in natural transformation. Like HapR, the transcription factor VqmA (Liu et al., 2006), upon binding to the cognate autoinducer 3,5‐dimethylpyrazin‐2‐ol (DPO), indirectly represses biofilm formation (Papenfort et al., 2017), suggesting that both QS pathways might share common gene targets. However, testing a vqmA mutant for its natural transformability showed that the system is not essential for transformation (Fig. 2D).
The alternative sigma factor RpoS does not foster HapR protein production
The data shown above confirmed the primary role of HapR in competence induction and nuclease repression, so we therefore sought to investigate the production of HapR itself. In the first publication on chitin‐induced natural competence by the Schoolnik laboratory, the authors showed that a rpoS mutant of V. cholerae was non‐transformable and suggested that this phenotype might be based on RpoS's effect on hapR expression (Meibom et al., 2005). Previous work had suggested that RpoS was required for hapR transcription, since an rpoS mutant showed reduced hapR transcript levels (Yildiz et al., 2004) and decreased HA/protease (HapA) activity (Yildiz and Schoolnik, 1998) (the encoding hapA gene is positively regulated by HapR). However, recent work challenged this interpretation by showing that RpoS did not affect QS‐dependent competence regulation and instead exerted its effect via the production of extracellular chitinases, therefore using the chitin‐induction branch of the competence network (Dalia, 2016) (Fig. 1). We therefore repeated the initial experiment that was reported from the Schoolnik laboratory (Meibom et al., 2005; co‐authored by the senior author of this study), and compared the WT, hapR‐, and rpoS‐minus strains for their natural transformability on chitin, which confirmed the absence of transformants for the two mutants (Fig. 3A). Moreover, both mutants remained non‐transformable even upon chitin‐independent TfoX production (Fig. 3C; rpoS::cm). Given these conflicting results compared to the convincing data provided by Dalia (Dalia, 2016), we genetically reengineered the rpoS mutant in two ways: first, by transferring the inactivated rpoS allele containing a chloramphenicol‐resistance cassette from the original rpoS mutant strain (rpoS::cm corresponding to FY1; Yildiz and Schoolnik, 1998; Meibom et al., 2005) into the WT strain via natural transformation (mutant rpoS::cm‐new); and second, by designing a completely new in‐frame rpoS deletion strain (mutant ∆rpoS) using natural transformation coupled to flip recombination (TransFLP; Marvig and Blokesch, 2010; Silva and Blokesch, 2010; Blokesch, 2012a). Surprisingly, both of these new mutants were naturally transformable on chitin at levels that were statistically insignificant from the parental WT strain (Fig. 3A). Consistently, we also showed that the in‐frame deleted rpoS mutant was fully transformable under TfoX‐producing conditions in rich medium (Fig. 3C).
Fig. 3.
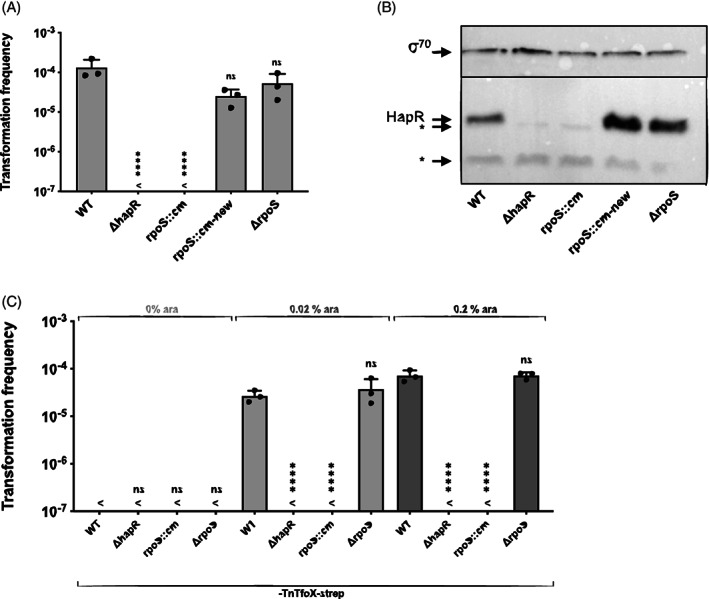
RpoS‐deficient V. cholerae are producing HapR and are naturally transformable.
(A and C) WT and rpoS‐or hapR‐minus strains were tested for natural transformation under chitin‐dependent (A) or chitin‐independent (C) conditions. Details for (A) as in Fig. 2A.
B. Strains for HapR detection by Western blotting were grown in plain LB medium and processed as those samples in Fig. 2C. For panel (C), the strains carried inducible TfoX on a transposon and were grown in LB medium in the absence (0%), or presence (0.02% or 0.2%) of arabinose as an inducer as indicated above the graph. All experiments were done three independent times and their average is shown in the bar graphs (±SD). <, below detection limit. Statistical analyses: (A) one‐way ANOVA with Tukey's multiple comparisons test. The detection limit was used for statistical analysis for conditions in which no transformants were recovered. ****P < 0.0001; ns, not significant.
These results generated two new questions, namely (i) why the original rpoS mutant was non‐transformable while the newly engineered mutants, which were fully verified for the absence of rpoS by PCR and Sanger sequencing, were fully transformable; and (ii) why the results under chitin‐inducing conditions differed compared to Dalia's work (Dalia, 2016). To address the first question, and given that the original rpoS mutant phenocopied several HapR‐deficient phenotypes (Yildiz and Schoolnik, 1998), we first sequenced the strain's hapR and luxO genes. While the hapR gene was identical to the gene in the parental A1552 strain, luxO had acquired a point mutation at base pair 359 (base pair 317 according to the initial luxO ORF annotation; (Heidelberg et al., 2000). This mutation resulted in a valine‐to‐alanine exchange of amino acid 120 (V120A; V106A according to the old luxO annotation), which is identical to the LuxO*[V106A] GOF mutation in V. fischeri (Kimbrough and Stabb, 2015). Given this finding, we next tested all rpoS mutants for their HapR production and showed the absence of this transcription regulator in the original rpoS::cm mutant, consistent with a LuxO* GOF phenotype (Fig. 3B). Importantly, both newly engineered rpoS mutants produced copious amounts of HapR at high cell densities (Fig. 3C), challenging the general notion that RpoS positively regulates HapR production (Yildiz et al., 2004; Conner et al., 2016). At this point, we cannot distinguish whether the GOF luxO mutation was introduced into the rpoS::cm mutant strain when it was initially generated (Yildiz and Schoolnik, 1998) or whether it appeared through extended lab domestication while the strain was transferred between group members in the Schoolnik laboratory (we confirmed the luxO mutation in three independent aliquots that were transferred from the Schoolnik laboratory to the Blokesch laboratory in 2009). However, this luxO mutation could explain the initial observation by Yildiz and Schoolnik, who showed that the rpoS::cm mutant could be fully complemented for RpoS protein production, but only poorly compensated for the lack of HA/protease production and/or its secretion (Yildiz and Schoolnik, 1998).
Contribution of RpoS to the expression of the chitin utilization gene
While the LuxO* GOF mutation explained why previous work concluded that RpoS was required for natural competence and transformation (Meibom et al., 2005), the question remained of why the newly generated rpoS mutants were highly transformable when grown on chitin flakes, contradicting recent results (Dalia, 2016), (Fig. 3A). Previously, Dalia showed that an rpoS mutant was barely transformable, with transformation frequencies being reduced by more than six orders of magnitude compared with the WT when grown on insoluble chitin (Dalia, 2016). This transformation deficiency was caused by the inability of the RpoS mutant to produce the extracellular chitinases ChiA‐1 and ChiA‐2 (Dalia, 2016), both of which are known to contribute to growth on chitin and to be induced by chitin oligosaccharides (Meibom et al., 2004). To understand why the rpoS mutants were fully transformable in our study, we first checked the transcript levels of chiA‐1 and chiA‐2 upon growth of WT or the rpoS‐minus strain on chitin. As shown in Fig. 4A, both chiA‐1 and chiA‐2 transcript levels were indeed significantly reduced in the rpoS mutant. Notably, the hapR transcripts were also slightly reduced (2.4‐fold; Fig. 4A), which most likely reflects the reduced chitin degradation capacity and growth of the rpoS‐minus strain. The rpoS mutant not only contained lower chiA‐1 and chiA‐2 transcript levels but also had reduced expression levels of other chitin catabolism pathway genes, such as the chitoporin‐encoding gene chiP, cbp, as well as the uncharacterized putative chitinase‐encoding gene VC0769 (Supporting Information Fig. S2A).
Fig. 4.
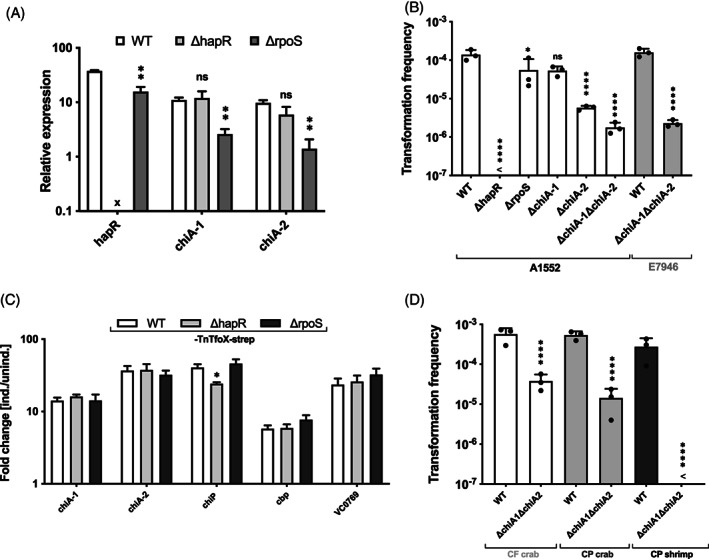
RpoS and its regulated chitinases are conditionally dispensable for transformation.
A. RpoS influences chitinase transcript levels when grown on chitin. Relative expression levels of hapR, chiA‐1 and chiA‐2 for chitin‐grown WT, ∆hapR, or ∆rpoS V. cholerae strains. Details as in Fig. 2B.
B. Chitinases enhance natural transformation but are not essential. The indicated WT and knock‐out strains (in A1552 or E7946 background) were tested for their natural transformability on chitin flakes (as in Fig. 2A).
C. RpoS is dispensable after TfoX induction. Expression levels of representative chitin utilization genes were tested under TfoX‐non‐inducing and TfoX‐inducing conditions (see also Supporting Information Fig. S2) and the fold change upon TfoX induction is shown on the Y‐axis. (D) Commercially available chitin source support transformation in a chitinases‐dependent or ‐independent manner. WT and chitinase‐negative (∆chiA‐1∆chiA‐2) V. cholerae cells were scored for their natural transformability after their inoculation on different chitin sources: chitin flakes (CF) from crab shells, chitin powder (CP) from crab shells, and CP from shrimp. <, below detection limit. All data are based on three independent biological experiments and depict average values (±SD). Statistical analyses were performed to compare mutants to the WT using these tests: (A, C) multiple t tests corrected for multiple comparisons using the Holm‐Sidak method; (B, D) one‐way ANOVA with Sidak's multiple comparisons test. The detection limit was used for statistical analysis for conditions in which no transformants were recovered. *P < 0.05; **P < 0.01; ****P < 0.0001; ns, not significant. For panel C, only the statistically significant difference is shown. x, not applicable as hapR‐minus strain.
Given that we confirmed reduced chiA‐1 and chiA‐2 expression/transcript levels but that the rpoS mutant was still transformable, we wondered whether our experimental conditions did not require chitinases for competence induction. We therefore tested a ∆chiA‐1, a ∆chiA‐2, and a ∆chiA‐1∆chiA‐2 strain for natural transformation on chitin flakes and the expression of the chitin utilization genes. As shown in Fig. 4B, we observed that a chiA‐1‐minus strain was transformable at levels close to the WT and maintained high expression levels of chiP, cbp, and VC0769 (Supporting Information Fig. S2A). In contrast, the chiA‐2‐ and chiA‐1/chiA‐2‐deficient mutants showed reduced transcript levels of the chitin utilization genes and therefore depicted transformation rates that were reduced by approximately two orders of magnitude (Fig. 4B). This latter result was confirmed for a double chiA knockout in a different strain background (E7946; kindly provided by A. Dalia). These results indicate that these two primary chitinases are not essential for chitin‐induced transformation on chitin flakes but do enhance its efficiency, while the RpoS mutant apparently still produced sufficient chitinases to maintain WT transformation levels.
Finally, we tested the transcript levels of chitin utilization genes in bacteria grown to high cell density in LB medium with or without artificial TfoX induction. These experiments showed lower chiA‐1 and chiA‐2 levels for the rpoS mutant under TfoX‐inducing conditions (Supporting Information Fig. S2B). However, the fold induction values upon TfoX production, which were observed to be between 10 and 100 for the chitin utilization genes, did not differ between a WT or rpoS‐minus strain (Fig. 4C). We therefore suggest that V. cholerae produces low levels of chitinase under starvation conditions and in the absence of carbon catabolite repression (Blokesch, 2012b). Once chitin degradation products are released from the polysaccharide and sensed by V. cholerae, TfoX production is triggered, making RpoS dispensable for the high‐level expression of chiA‐1 and chiA‐2 as well as other chitin utilization genes. Notably, except for chiP, none of the tested chitin utilization genes showed highly significant expression variations in a hapR‐minus strain under chitin‐grown or TfoX‐inducing conditions (Supporting Information Fig. S2). Therefore, the previous claim of a QS‐dependent regulation of chitin utilization genes is not supported (Sun et al., 2015).
Different chitin sources lead to experimental variations
In contrast to a previous study (Dalia, 2016), the rpoS‐minus strain and the chitinase double mutant were still highly transformable when V. cholerae is grown on chitin. Therefore, we next tested if the chitin source could influence experimental outcomes. To our surprise, we found that the commercially available chitin flakes (#C9213) and chitin powder (#C7170) from Sigma had changed composition while maintaining the same product numbers within the past years (i.e. initially isolated from crab shells but now isolated from shrimp). While the chitin itself should not differ between these two sources, the preparation procedure might not be the same (no information was provided by the manufacturer). We therefore tested the crab‐derived chitin flakes and powder, following the standard chitin induction protocol in our laboratory (Marvig and Blokesch, 2010) with minor modifications (see Methods section), and compared those chitin sources to the new chitin powder derived from shrimp. As shown in Fig. 4D, chitin flakes and chitin powder derived from crab shells resulted in high levels of transformation of V. cholerae WT cells and reduced but still relatively high‐level transformation of the chiA‐1/chiA‐2 mutant. When shrimp‐derived chitin was used as sole carbon source, the WT maintained full transformability, whereas no transformants were detectable for the double mutant strain (Fig. 4D). We therefore suggest that either the input material or different preparation methods used by Sigma for crab‐ or shrimp‐derived chitin result in varying chitin mixtures, which most likely contain different ratios of the insoluble polysaccharide chitin and soluble, short‐length, chitin‐derived oligosaccharide. As the latter do not require chitinase activity for competence induction (Dalia, 2016), this could explain the different transformation outcomes.
Natural competence and transformation are strongly conserved in seventh pandemic V. cholerae isolates
While we established above that experimental conditions could explain different results between research groups (e.g. RpoS‐dependent transformation phenotype), we still wanted to address whether the well‐studied seventh pandemic strains had comparable natural transformation capabilities, given that some studies reported frequencies that were more than 100‐fold above those commonly observed in our group. We therefore tested six well‐studied O1 El Tor strains for chitin‐induced natural transformation: three isolates from the Latin American cholera outbreak in Peru in the 1990s (strains A1552, C6706, and C6709), two Bangladeshi isolates from the 1970s (N16961 and P27459), and strain E7946, which was isolated in Bahrain in 1978 (Supporting Information Table S1). It should be noted that several laboratories are currently working with a QS‐impaired C6706 variant that carries a luxO* GOF mutation (luxO[G333S]; Stutzmann and Blokesch, 2016). We received a copy of this mutated C6706 variant from several laboratories, and it was transformable at levels that were one to two orders of magnitude below the level of strain A1552 (Stutzmann and Blokesch, 2016). Here, to confirm the causality of this luxO[G333S] GOF mutation for the impaired transformability, we introduced this single point mutation into strain A1552 (Supporting Information Figs. S3A and B). Given the widespread usage of the mutated C6706 variant, we decided to include an old stock of the C6706 isolate in our study (kindly provided by J. Mekalanos), which reflects a less laboratory‐domesticated version of the strain before it was rendered streptomycin resistant [named C6706 (StrS)]. As shown in Fig. 5, none of these other O1 El Tor pandemic strains showed lower transformation frequencies under chitin‐ (Fig. 5A) or tfoX‐ (Supporting Information Fig. S3C) inducing conditions when compared with the WT strain used in our laboratory (strain A1552), except for C6709 (see below) and N16961. The non‐transformability of strain N16961 was consistent with a previous report and was caused by a frameshift mutation in hapR that caused its QS deficiency (Meibom et al., 2005). Hence, repairing the single‐base‐pair mutation (strain N16961‐hapR Rep) was sufficient to fully restore HapR production (Fig. 5B) and therefore the strain's natural transformability (Fig. 5A).
Fig. 5.
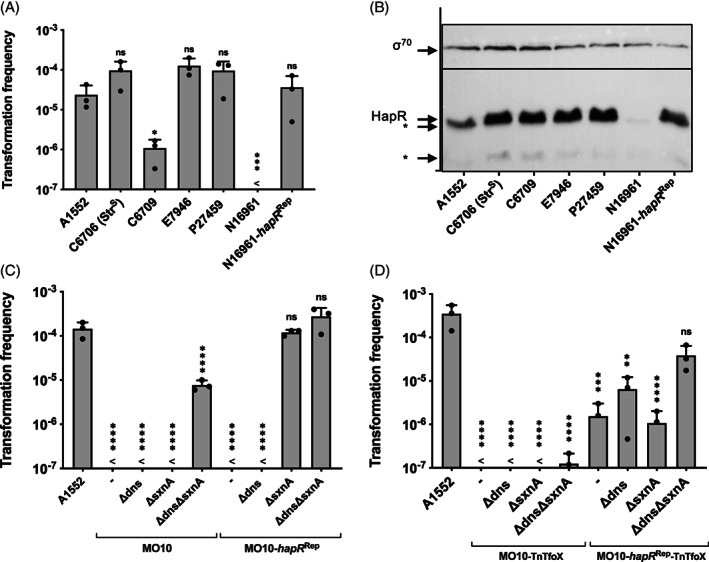
Pandemic V. cholerae strains are transformation variable.
A, B. Different V. cholerae O1 El Tor pandemic isolates were tested for their natural transformability (A) or their HapR production (B). Strain N16961‐hapR Rep is a genetically engineered N16961 variant with a repaired hapR gene.
C, D. The O139 serogroup strain MO10 and its hapR‐repaired variant (MO10‐hapR Rep) were tested for their natural transformability on chitin (C; together with the indicated knock‐out mutants of these two strain backgrounds) or under TfoX‐inducible conditions (D). Details as described in Fig. 2. Statistical analyses were performed to compare diverse strains to the reference strain of this study, A1552, using a one‐way ANOVA on log‐transformed data with Sidak's multiple comparisons test. <, below detection limit. The detection limit was used for statistical analysis for conditions in which no transformants were recovered. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
The O139 serogroup strain MO10 is non‐transformable due to multiple factors
It has long been known that hapR‐mutated strains are impaired in their natural transformability, which is also true for the well‐studied strain MO10 (Blokesch and Schoolnik, 2007). This O139 serogroup strain is closely related to the O1 El Tor pandemic lineage and had acquired a novel O‐antigen‐encoding gene cluster by horizontal gene transfer (Johnson et al., 1994; Blokesch and Schoolnik, 2007). We therefore repaired the hapR copy of strain MO10 (known to carry a deleterious mutation; Joelsson et al., 2006) resulting in strain MO10‐hapR Rep. Surprisingly, this strain maintained its transformation‐negative phenotype upon chitin induction (Fig. 5C) or artificial TfoX production (Fig. 5D), despite the fact that selected transcript scoring suggested a full restoration of HapR's regulatory capacity (such as hapA induction or vpsA repression; Supporting Information Fig. S4). Furthermore, the nuclease gene dns was repressed by the repaired HapR protein (Supporting Information Fig. S4), consistent with the fact that not even deleting dns restored transformation (Fig. 5C); this is in direct contrast to a transformation rescue in strain N16961 through dns deletion (Blokesch and Schoolnik, 2008).
We previously reported that the ATPase PilT, which is involved in DNA uptake and therefore transformation via its role in retracting the DNA‐uptake pilus, was mutated in strain MO10 (pilT[R206S]; Adams et al., 2019b). However, we excluded that this was the primary reason for the strains' non‐transformability due to the action of the secondary retraction ATPase PilU. Indeed, we showed that PilU compensates for PilT[R206S]'s impaired functionality when reconstituted in strain A1552 (Adams et al., 2019b). Hence, we considered that other genes related to competence or quorum sensing might be mutated in this strain and therefore whole‐genome sequenced our laboratory stock of strain MO10 and de novo assembled its genome (Supporting Information Table S2). Upon closer inspection of its genome, we realized that apart from the mutations in hapR and pilT, all of the genes encoding other competence‐related proteins were 100% identical to their homologues in A1552 (Supporting Information Table S3).
Intriguingly, Dalia and colleagues had previously shown that clinical V. cholerae isolates from Haiti were poorly transformable due to a Dns homologous nuclease IdeA, whose gene was not repressed by HapR and QstR upon competence induction (Dalia et al., 2015), in contrast to dns (Blokesch and Schoolnik, 2008). Moreover, the ideA gene was located on a specific integrative and conjugative element (ICE) known as the VchInd5 (Dalia et al., 2015). The first ICE of any V. cholerae strains was described by Waldor and colleagues in 1996 in strain MO10 and named SXT (Waldor et al., 1996). When we inspected the SXT‐contained genes for putative nuclease‐encoding genes, this search yielded locus tag H6M50_12490 in the large chromosome 1 of MO10. The gene product of H6M50_12490, which equals ORF s62 in the original SXT description (though predicted to start 33 bp downstream of the annotated start of s62; Genbank accession number AY055428.1; Beaber et al., 2002) shows 93%/97% and 51%/64% identity/similarity to IdeA and Dns, respectively, strongly suggesting its role as yet another extracellular nuclease (Blokesch, 2017). We therefore named this gene sxnA for SXT‐encoded nuclease A, and deleted the gene from the wild‐type MO10 strain; its dns‐minus mutant; MO10‐hapR Rep; or MO10‐hapR Rep∆dns. By deleting sxnA, the last three strains became naturally transformable on chitin surfaces, with the HapR‐repaired variants reaching frequencies in the range of those observed for strain A1552 (Fig. 5C). Similar results were obtained under chitin‐independent transformation conditions, where we observed increasing transformation frequencies for strains lacking both nuclease in the presence of a repaired hapR allele (Fig. 5D); the latter condition significantly induced qstR and comEA while repressing dns upon artificial TfoX induction (Supporting Information Fig. S4). On the basis of these results, we can conclude that the O139 serogroup strain carries, in principle, a functional competence induction system. However, competence induction is non‐functional in this strain due its QS impairment. In addition, through the horizontal acquisition of the SXT element, the strain became non‐transformable, as surrounding DNA can be degraded by the SxnA nuclease.
Comparative genomics of de‐novo‐assembled pandemic V. cholerae genomes
To follow up on previous claims of major strain differences within the seventh pandemic clade for several tested phenotypes, we next decided to whole‐genome sequence the above‐mentioned pandemic strains C6706(StrS), C6709, E7946, and P27459 using a long‐read PacBio sequencing approach and to de novo assemble their respective genomes. The de novo assembly was important, due to previous assembly artefacts, which we know from personal communications to have caused extensive experimental failures and frustration related to recombineering approaches in diverse research groups, missing rRNA clusters in old genome data, as well as a known bona fide genomic inversion in strain A1552 (Val et al., 2016; Matthey et al., 2018; Kemter et al., 2018; Matthey et al., 2019). Pairwise comparisons between these strains' de‐novo‐assembled genomes and those that we recently reported for strain A1552 and N16961 (using the same sequencing approach; Matthey et al., 2018; Matthey et al., 2019), showed overall identity levels above 95% for both chromosomes (Fig. 6A). Moreover, when we scored gene products related to chitin utilization, competence regulation, DNA‐uptake and translocation, as well as competence‐related recombination, we saw that almost all of these proteins were 100% identical (Supporting Information Table S3). From this analysis, we conclude that pandemic patient isolates are indeed highly similar, supporting previous large‐scale comparative genomic studies based on the strains' core genomes (Mutreja et al., 2011; Domman et al., 2017; Weill et al., 2017; Weill et al., 2019). Importantly for this study, all six pandemic isolates maintained their chitin‐utilization genes and their chitin‐dependent competence inducibility, suggesting that under certain circumstances, these phenotypes might be positively selected.
Fig. 6.
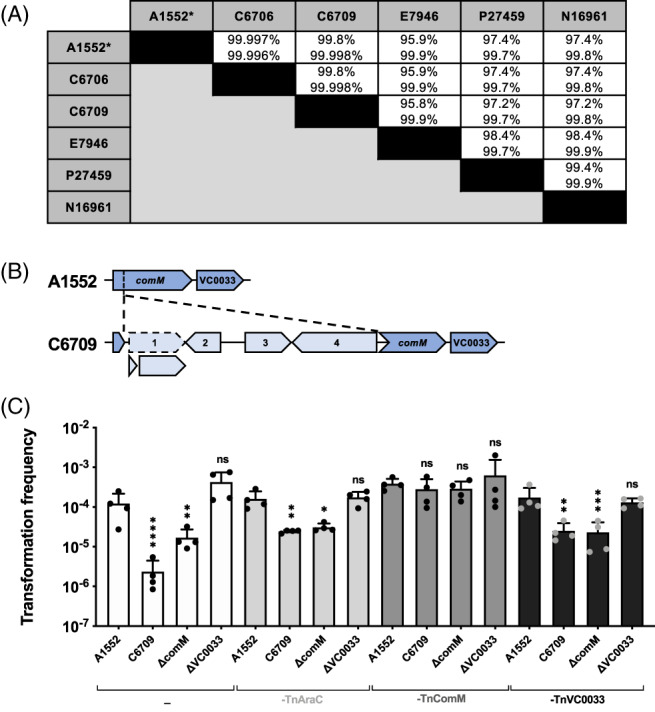
Whole genome comparisons revealed a comM interruption in strain C6709.
A. Pairwise identity values for genome comparisons of the indicated strains (upper and lower value for chr1 and chr2, respectively). *, the intrachromosomal inversion of strain A1552 was in silico reverted before comparison.
B. A mobile genetic elements interrupts comM in strain C6709. Scheme of the genomic region of comM in strain A1552 (dark blue) and its interruption by an MGE (light blue) in C6709. The NCBI Prokaryotic Genome Annotation Pipeline (PGAP) predicted one ORF for the region 1 (dashed arrow), despite a frameshift‐causing mutation that results in a premature stop codon (shown by the two split ORFs; mutation confirmed by Sanger sequencing).
C. The interrupted comM is causative for the decreased transformability of strain C6709. V. cholerae strains A1552, C6709, and the knockout strains A1552∆comM and A1552∆VC0033 were tested for their natural transformability on chitin (+0.2% ara). The strains were either transposon‐deficient or carried a TnAraC (control), TnComM, and TnVC0033 transposon. The experiment was performed four independent times and the mean values are shown (±SD). Statistical analyses were performed to compare each strain to A1552 within the same category (e.g., −, TnAraC, TnComM, TnVC0033) using a one‐way ANOVA on log‐transformed data with Sidak's multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Transformation is impaired in strain C6709 due to an interruption of comM
While the data above supported the notion that the underlying causes of non‐transformability of selected patient isolates of V. cholerae are often a result of an impaired QS circuit (potentially caused by sampling biases (Blokesch, 2012c), sample storage, or laboratory domestication, we wondered why transformation of strain C6709 was reduced compared to other isolates (Fig. 5A). Since we recently showed that the TfoX‐induced T6SS, which is exquisitely co‐regulated with competence induction in pandemic V. cholerae (Borgeaud et al., 2015), was fully functional in strain C6709 (Metzger et al., 2019), we excluded a regulatory defect. Instead, the comparative genomic analysis revealed that the comM competence gene in strain C6709 is interrupted by a 5′161 bp mobile genetic element (MGE) (Fig. 6B and Supporting Information Table S3). This MGE interrupted comM after base pair 197, causing an aborted gene product of 79 amino acids. The MGE carried four putative genes. The first gene (locus tag GTF73_00105) encoded an acyltransferase‐encoding gene, which was, however, separated into two ORFs due to a frameshift mutation. We confirmed the authenticity of this frameshift mutation by Sanger sequencing. The second gene coded for a protein of unknown function, while the third and fourth genes encoded a plasmid‐recombination enzyme and an unknown protein belonging to the recombinase family respectively.
As we and others have shown that comM‐deficient V. cholerae mutants showed reduced transformation levels (~5‐ to 10‐fold in Jaskólska et al., 2018 and >100‐fold in Nero et al., 2018; note the different transforming material used in both studies), we reasoned that comM's interruption or a potential polar effect of the integrated MGE onto the comM‐successive gene VC0033 might be causing the lowered transformability of C6709. We therefore constructed inducible copies of comM and VC0033 inside a site‐specific and araC‐ plus PBAD‐carrying transposon. We transferred those constructs (as well as the parental transposon as a control) into the chromosomes of A1552, C6709, as well as ∆comM‐ and ∆VC0033 knockout strains. We then tested the strains' natural transformability on chitin (under arabinose‐induced conditions). As shown in Fig. 6C, expression of a full‐length comM gene restored the transformability of strain C6709 while provision of VC0033 had no effect. The latter result is consistent with the VC0033‐minus mutant that has no transformation defect (Fig. 6C and Jaskólska et al., 2018). We also observed that even the provision of the araC‐carrying transposon without any inducible gene increased the transformation frequency of strain C6709 to the level of the ∆comM mutant of strain A1552. This result was highly reproducible (even in an added fourth biologically independent replicate; Fig. 6C), suggesting a positive impact of either the transposon or the AraC repressor on transformation in this strain. Based on these comM complementation data, we can conclude that the impairment of natural transformation in strain C6709 was indeed caused by the comM interruption. In support of this assumption, previous work has shown that ComM and its homolog RadA in S. pneumoniae (Marie et al., 2017; Nero et al., 2018) act as helicases/branch migration factors and assist in the recombination of incoming ssDNA into the double‐stranded bacterial genome through their interaction with RecA. The inactivation of comM in the Latin American isolate C6709, which otherwise shows more than 99.8% pairwise identity with the two other Peruvian isolates C6706 and A1552, suggests a recent acquisition of the MGE before or after sampling. Interestingly, Croucher and colleagues reported that comM is frequently interrupted by MGEs in other naturally transformable bacteria such as Aggregatibacter actinomycetemcomitans, Acinetobacter baumannii, and Mannheimia succiniciproducens (Croucher et al., 2016). By searching for orthologs of the apparently site‐specific integrase within the integrated MGEs, these authors identified several additional examples of comM interruption across diverse bacterial species including pathogens such as Mannheimia haemolytica, Francisella philomiragia, and Pseudomonas syringae (Croucher et al., 2016). Based on this finding and the assumption that comM interruption would abrogate transformation similarly to interruption or mutation of other core competence genes, the authors concluded that selfish MGEs tend to interrupt competence genes to limit their curing from the genome (Croucher et al., 2016). Natural transformation might therefore serve as a general genome maintenance tool in competent bacteria (Croucher et al., 2016). This is a very interesting hypothesis, and our data of the comM interruption in strain C6709 support the idea in principle. However, in contrast to interruptions of the essential DNA uptake/translocation genes comEA, comEC, and comF (Seitz and Blokesch, 2013), which are all co‐regulated with comM (Jaskólska et al., 2018), the abrogation of ComM production had only minor effects on V. cholerae's transformability under the tested conditions (Fig. 6C). In our experiments, the transforming DNA was fully homologous to the competent acceptor strain A1552 and the closely related C6709 strain apart from a resistance marker, consistent with the idea that DNA is acquired from closely related, non‐kin neighbour strains through type VI secretion‐mediated predation (Borgeaud et al., 2015). However, ComM might contribute to a more efficient insertion of heterologous incoming DNA, as previously suggested (Nero et al., 2018), and interruption of its gene might indeed protect the integrated MGE from its excision. However, further mechanistic studies are needed to fully address this idea.
Conclusions
The current study provides in‐depth insight into the conservation of chitin utilization and natural competence in the seventh pandemic clade of V. cholerae. This finding therefore raises new questions on the evolution of pandemic strains. For instance, V. cholerae O139 isolates such as strain MO10 are mostly considered as serogroup‐converted O1 El Tor strains. Such serogroup conversion events have occurred frequently in nature in the past (Chun et al., 2009) and can easily be accomplished by natural transformation, as we have experimentally demonstrated (Blokesch and Schoolnik, 2007). However, given the frequent possession of SXT/R391‐type ICE elements in recent isolates such as those isolated from Yemen (Weill et al., 2019), it is tempting to speculate that the transfer of large genomic regions such as the one that encodes the O‐antigen biosynthesis might be significantly impaired through ICE‐encoded nucleases, as previously shown for IdeA (Dalia et al., 2015).
We also deciphered strain‐specific competence deficiencies and showed that they are frequently based on laboratory domestication events leading to QS impairment. Moreover, we tested different chitin sources from the same commercial supplier and found that the source from which the chitin is derived or the preparation method could impact experimental outcomes. Finally, we showed that rpoS‐deficient V. cholerae are transformable in the presence of certain chitin sources and that these mutants are not impaired in QS‐related competence regulation, consistent with a previous report (Dalia, 2016). We extended this analysis to show that RpoS does not influence HapR protein production, even under chitin‐independent conditions, which supports the notion that previous connections between RpoS and HapR should be reconsidered. Furthermore, while the domestication effect in luxO in the rpoS‐deficient strain cannot be tracked back to its origin, the absence of HapR in this strain could potentially explain other observations reported in the literature (e.g., the absence of its fully complementability; Yildiz and Schoolnik, 1998). This also challenges the role of RpoS in controlling the mucosal escape response of V. cholerae (Nielsen et al., 2006) instead of mirroring the QS impairment. In light of this finding, we suggest that a proper re‐evaluation of previously described RpoS‐dependent phenotypes would provide some clarification about its role.
Experimental procedures
Bacterial strains, plasmids and culture conditions
Bacterial strains (V. cholerae and E. coli) and plasmids used in this study are listed in the Supporting Information Table S1. The bacteria were cultured at 30°C in Lysogeny broth medium (LB; 10 g l−1 of tryptone, 5 g l−1 of yeast extract, 10 g l−1 of sodium chloride; Carl Roth) medium under aerobic conditions. LB agar plates contained 1.5% w/v agar and were supplemented with antibiotics where required. The final concentrations of antibiotics were 75, 100, 100, 2.5, and 50 μg ml−1 for kanamycin, streptomycin, ampicillin, chloramphenicol, and gentamicin, respectively. Thiosulfate citrate bile salts sucrose (TCBS) agar was prepared following the manufacturer's instructions (Fluka). Half‐concentrated and HEPES‐buffered defined artificial seawater medium (DASW) (Meibom et al., 2005) was used for the natural transformation experiments on chitinous surfaces.
Vibrio cholerae strain constructions
Vibrio cholerae mutant strains were genetically engineered using chitin‐induced transformation methods combined with flip recombination (TransFLP; Marvig and Blokesch, 2010; De Souza Silva and Blokesch, 2010; Blokesch, 2012a; Borgeaud and Blokesch, 2013). Briefly, to construct the rpoS and vqmA knockout strains, the genes' flanking regions as well as the aph (KanR) resistance marker were PCR amplified (with Pwo polymerase; Roche) and joined by splicing using overlap extension PCR. The PCR fragments were provided to chitin‐induced competent V. cholerae, and natural transformants were selected using selective agar plates. Transformants were confirmed for the correctness by colony PCR (with GoTaq polymerase; Promega) followed by Sanger sequencing of the genetically engineered loci (Microsynth, Switzerland). The resistance marker was subsequently removed by flip recombination (De Souza Silva and Blokesch, 2010; Blokesch, 2012a).
The TnAraC (Adams et al., 2019a) miniTn7 transposon was cloned by synthesizing the fragment SYN‐araC‐NotI (IDT via LubioScience, Switzerland) containing araC, PBAD, and a NdeI restriction site at an optimal distance from the promoter. The synthesized fragment was PCR‐amplified, NotI‐digested, and cloned into an equally digested miniTn7 carried on plasmid pGP704 (Müller et al., 2007). To render them arabinose‐inducible, the genes comM and VC0033 were subsequently cloned inside the NdeI cloning site of TnAraC and the resulting miniTn7 derivatives (TnComM and TnVC0033) were transferred into the respective V. cholerae strains by triparental mating (Bao et al., 1991).
Natural transformation on chitin surfaces
To measure the natural transformation efficiency, V. cholerae bacteria were grown on chitin flakes as previously described with minor modifications (De Souza Silva and Blokesch, 2010; Marvig and Blokesch, 2010). Briefly, overnight cultures were back‐diluted 1:20 in 0.5x DASW + HEPES + vitamins and added to pre‐autoclaved chitin flakes (crab‐shell‐derived; #C9213, Sigma) in Eppendorf tubes. After incubation at 30°C for ~24 h, genomic DNA (gDNA) from strain A1552‐LacZ‐Kan (Marvig and Blokesch, 2010) was added as the transforming material and the mixture was incubated for 6 h. Transformants were identified by selective plating, and transformation frequencies were determined by dividing the number of resistant transformants by the total number of colony‐forming units (CFU) on plain LB agar plates. Averages of at least three biologically independent experiments are provided. Transformation frequencies were log‐transformed (Keene, 1995) and subjected to statistical analysis as indicated in the figure legends. When no transformants were recovered, the value was set to the detection limit to allow for statistical analysis (using the Prism software from GraphPad; version 8.4.3).
To test the transformation with different chitin sources (chitin flakes; crab‐shell‐ or shrimp‐shell‐derived; #C9213, Sigma; or chitin powder; #C7170, Sigma), the protocol was slightly changed. Briefly, bacteria from overnight cultures were harvested by centrifugation and resuspended in 0.5x DASW + HEPES + vitamins medium to reach an optical density at 600 nm of 0.1. An amount of 1 ml of this suspension was added to each chitin source and incubated at 30°C for ~24 h. The next day, the supernatant was replaced by fresh medium before the transforming gDNA of strain A1552‐lacZ‐Cat (Supporting Information Table S1) was added. The bacteria were detached by vortexing after another 8 h of incubation and spotted on selective and plain LB agar plates after serial dilution.
Natural transformation in the absence of chitin
Natural transformation assays of inducible tfoX‐carrying strains were performed following a previously established protocol (Lo Scrudato and Blokesch, 2012; Metzger and Blokesch, 2016). Briefly, V. cholerae strains were grown at 30°C for 3 h in LB medium (with ot with arabinose) to an optical density at 600 nm (OD600) of ~1.0. At this point, 0.5 ml of the cultures were transferred to an Eppendorf tube and supplemented with 1 μg of A1552‐lacZ‐Kan gDNA. Cells were further incubated for 4 h under shaking conditions. Serial dilutions were spotted on selective and plain LB agar plates to enumerate the number of transformants and the number of total cells, respectively. Transformation frequencies were calculated as described above.
Gene expression profiling by quantitative reverse transcription PCR
Quantitative reverse transcription PCR (qRT‐PCR)‐based transcript scoring was done for cultures grown at 30°C in LB medium for 6 h if not indicated otherwise. For chitin‐dependent expression analysis, the bacteria were inoculated onto chitin flakes submerged in 0.5x DASW + HEPES + vitamins, as noted above for the transformation assay. After an incubation period at 30°C for 24 h, the samples (four parallel tubes of each) were centrifuged for 3 min at maximum speed in a tabletop centrifuge before the supernatant was discarded. The bacteria‐ and chitin‐flakes‐containing pellet was resuspended in 1 ml of TRI reagent (Sigma) and vortexed. The chitin material was sedimented by a quick spin for 30 s at maximum speed. The supernatants was transferred to a new sterile Eppendorf tube and flash frozen on dry ice before their storage at −80°C. RNA extraction, DNase treatment, reverse transcription, and qPCR were performed as previously described (Lo Scrudato and Blokesch, 2012). Expression is shown relative to the housekeeping gene gyrA. Averages of at least three biologically independent experiments (±standard deviation) are provided. Statistical analyses using GraphPad's Prism (version 8.4.3) were performed on the log‐transformed data (Keene, 1995), with details provided in the figure legends.
SDS‐PAGE and western blotting
Cell lysates were prepared as described previously (Metzger et al., 2016). Briefly, after growing the bacterial cultures in LB medium at 30°C for 6 h, bacterial cell pellets were resuspended in Laemmli buffer, adjusting for the total number of bacteria according to the OD600 values. Proteins were separated on sodium dodecyl sulfate (SDS)‐polyacrylamide gels and transferred onto PVDF membranes by western blotting. Primary antibodies against HapR (#A000542; Lo Scrudato and Blokesch, 2012) were used at 1:3000–1:5000 dilutions and goat anti‐rabbit horseradish peroxidase (HRP) (diluted 1:10 000–1:20 000; Sigma‐Aldrich, Switzerland) was used as the secondary antibody. Sigma70 detection served as a loading control using anti‐E. coli Sigma70 coupled with HRP (BioLegend, USA distributed via Brunschwig, Switzerland; used at a 1:10 000 dilution). Lumi‐LightPLUS western blotting substrate (Roche, Switzerland) was used as an HRP substrate, and the signals were detected using a ChemiDoc XRS+ station (BioRad).
Long‐read whole‐genome sequencing (PacBio)
Genomic DNA was isolated from the pandemic V. cholerae O1 El Tor strains C6706, C6709, E7946, and P27459 cultured in LB medium using Qiagen's Genomic DNA Buffer set combined with 100/G Genomic‐tips. Sequencing was performed by the Genomic Technology Facility of the University of Lausanne. For sample preparation, the DNA samples were sheared in Covaris g‐TUBEs to obtain fragments with a mean length of 20 kb.
The sheared DNA was used to prepare each library with the PacBio SMRTbell Template Prep Kit 1 according to the manufacturer's recommendations (Pacific Biosciences). The resulting libraries were size‐selected on a BluePippin system (Sage Science) for molecules larger than 15 kb. Each library was sequenced on one SMRT cell with P6/C4 chemistry and MagBeads on a PacBio RSII system at 360 min movie length. Genome assembly was performed using the protocol ‘RS_HGAP_Assembly.3' in SMRT Pipe v2.3.0, and circularization of the genomes was achieved with the Minimus assembler of the AMOS software package 3.1.0 using the default parameters (Sommer et al., 2007). The assembled genomes were annotated using Prokka‐1.12 (Seemann, 2014) but were re‐annotated upon submission to the NCBI database using with their own pipeline (PGAP annotation). The genomic data and NCBI accession numbers are summarized in the Supporting Information Table S2.
For the whole‐genome sequencing of strain MO10, slight variations from this protocol were applied. Briefly, the isolated DNA was sheared with a Megaruptor (Diagenode, Denville, NJ) to obtain 10–15 kb fragments on average and the size distribution was verified on a Fragment Analyser (Advanced Analytical Technologies, Ames, IA). Library preparation was done with the PacBio SMRTbell Express Template Prep Kit 2.0 (Pacific Biosciences, Menlo Park, CA, USA) according to the manufacturer's recommendations. The library was pooled with other libraries processed at the same time and the pool was size‐selected with Ampure PacBio beads to eliminate fragments <3 kb. It was sequenced with v3.0/v3.0 chemistry and diffusion loading on a PacBio Sequel instrument (Pacific Biosciences, Menlo Park, CA, USA) at 600 min movie length, pre‐extension time of 120 min using one SMRT cell 1 M v3. Genome assembly was performed using the protocol Microbial Assembly in SMRT Link Version 8.
Pairwise comparisons between the de‐novo‐assembled genomes were performed using the Geneious software (version 10.2.6) and a progressive Mauve algorithm assuming collinear genomes and performing full alignments. For these analyses, the large inversion between the two rRNA clusters of strain A1552 (Matthey et al., 2018; Matthey et al., 2019) was in silico reverted for a simplified alignment of all genomes.
While comparing the assembled genomes, we observed a frameshift mutation in the gene VC0860 (part of the minor pilin gene cluster; Seitz and Blokesch, 2013) for strain E7946. Compared to the sequence of the homologous gene in N16961 (Heidelberg et al., 2000) and A1552 (Matthey et al., 2018), the E7946 genome missed an ‘A’ base from a stretch of six consecutive As. Since we suspected that this might represent a sequencing artefact, the corresponding region was PCR amplified and Sanger sequenced. The data showed that the strain indeed carries all 6 A bases without the frameshift mutation (see also Supporting Information Table S3).
Data Availability Statement
All sequencing data have been deposited on NCBI under BioProject accession number PRJNA598947 (BioSamples SAMN13734987, SAMN13734988, SAMN13734999, SAMN13734989, and SAMN15756141). The whole genome sequences are available in GenBank under accession numbers CP047295/CP047296 (C6706), CP047297/CP047298 (C6709), CP047303/CP047304 (E7946), CP047299/CP047300 (P27459), and CP060094/CP060095 (MO10). The raw reads are available from the Sequence Read Archive (SRA) under submission numbers SRR10828860, SRR10828861, SRR10828863, SRR10828864, and SRR12407298).
Supporting information
Appendix S1: Supplementary Information
Acknowledgements
The authors thank Noémie Matthey and Natália Drebes Dörr for the preparation of genomic DNA of strains C6706, C6709, E7946, P27459, and MO10 and help with Sanger sequencing, Candice Stoudmann and Tiziana Scrignari for technical assistance, the staff of the Lausanne Genomic Technologies Facility at the University of Lausanne for sample processing and bioinformatic analysis, and members of the Blokesch laboratory for discussion. The authors also thank J. Mekalanos for providing the original V. cholerae C6706 strain (StrS) and A. Camilli, J. Reidl, A. Dalia, and B. Hammer for sharing previously published strains. This work was supported by the European Research Council (Consolidator grant 724630) and the Swiss National Science Foundation (grant 310030_185022). M.B. is a Howard Hughes Medical Institute (HHMI) International Research Scholar (grant 55008726). The purchase of the Pacific Biosciences RS II instrument at the University of Lausanne was partially supported by the Loterie Romande through the Fondation pour la Recherche en Médecine Génétique.
References
- Adams, D.W. , Stutzmann, S. , Stoudmann, C. , and Blokesch, M. (2019a) DNA‐uptake pili of Vibrio cholerae are required for chitin colonization and capable of kin recognition via sequence‐specific self‐interaction. Nat Microbiol 4: 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, D.W. , Pereira, J.M. , Stoudmann, C. , Stutzmann, S. , and Blokesch, M. (2019b) The type IV pilus protein PilU functions as a PilT‐dependent retraction ATPase. PLoS Genet 15: e1008393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonova, E.S. , and Hammer, B.K. (2011) Quorum‐sensing autoinducer molecules produced by members of a multispecies biofilm promote horizontal gene transfer to Vibrio cholerae . FEMS Microbiol Lett 322: 68–76. [DOI] [PubMed] [Google Scholar]
- Bao, Y. , Lies, D.P. , Fu, H. , and Roberts, G.P. (1991) An improved Tn7‐based system for the single‐copy insertion of cloned genes into chromosomes of Gram‐negative bacteria. Gene 109: 167–168. [DOI] [PubMed] [Google Scholar]
- Bassler, B. , Gibbons, P. , and Roseman, S. (1989) Chemotaxis to chitin oligosaccharides by Vibrio furnissii, a chitinivorous marine bacterium. Biochem Biophys Res Commun 161: 1172–1176. [DOI] [PubMed] [Google Scholar]
- Beaber, J.W. , Hochhut, B. , and Waldor, M.K. (2002) Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae . J Bacteriol 184: 4259–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch, M. (2012a) TransFLP – a method to genetically modify V. cholerae based on natural transformation and FLP‐recombination. J Vis Exp 68: e3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch, M. (2012b) Chitin colonization, chitin degradation and chitin‐induced natural competence of Vibrio cholerae are subject to catabolite repression. Environ Microbiol 14: 1898–1912. [DOI] [PubMed] [Google Scholar]
- Blokesch, M. (2012c) A quorum sensing‐mediated switch contributes to natural transformation of Vibrio cholerae . Mob Genet Elements 2: 224–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch, M. (2017) In and out‐contribution of natural transformation to the shuffling of large genomic regions. Curr Opin Microbiol 38: 22–29. [DOI] [PubMed] [Google Scholar]
- Blokesch, M. , and Schoolnik, G.K. (2007) Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog 3: e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch, M. , and Schoolnik, G.K. (2008) The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae . J Bacteriol 190: 7232–7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgeaud, S. , and Blokesch, M. (2013) Overexpression of the tcp gene cluster using the T7 RNA polymerase/promoter system and natural transformation‐mediated genetic engineering of Vibrio cholerae. PLoS One 8 8: e53952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgeaud, S. , Metzger, L.C. , Scrignari, T. , and Blokesch, M. (2015) The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science 347: 63–67. [DOI] [PubMed] [Google Scholar]
- Chun, J. , Grim, C.J. , Hasan, N.A. , Lee, J.H. , Choi, S.Y. , Haley, B.J. , et al (2009) Comparative genomics reveals mechanism for short‐term and long‐term clonal transitions in pandemic Vibrio cholerae . Proc Natl Acad Sci U S A 106: 15442–15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfanelli, F.R. , Monlezun, L. , and Coulthurst, S.J. (2016) Aim, load, fire: the type VI secretion system, a bacterial Nanoweapon. Trends Microbiol 24: 51–62. [DOI] [PubMed] [Google Scholar]
- Colwell, R.R. (1996) Global climate and infectious disease: the cholera paradigm. Science 274: 2025–2031. [DOI] [PubMed] [Google Scholar]
- Conner, J.G. , Teschler, J.K. , Jones, C.J. , and Yildiz, F.H. (2016) Staying alive: Vibrio cholerae's cycle of environmental survival, transmission, and dissemination. Microbiol Spectr 4 [Epub ahead of print]. 10.1128/microbiolspec.VMBF-0015-2015 [DOI] [Google Scholar]
- Croucher, N.J. , Mostowy, R. , Wymant, C. , Turner, P. , Bentley, S.D. , and Fraser, C. (2016) Horizontal DNA transfer mechanisms of bacteria as weapons of Intragenomic conflict. PLoS Biol 14: e1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalia, A.B. (2016) RpoS is required for natural transformation of Vibrio cholerae through regulation of chitinases. Environ Microbiol 18: 3758–3767. [DOI] [PubMed] [Google Scholar]
- Dalia, A.B. , Lazinski, D.W. , and Camilli, A. (2014) Identification of a membrane‐bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae . MBio 5: e01028–e01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalia, A.B. , Seed, K.D. , Calderwood, S.B. , and Camilli, A. (2015) A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae . Proc Natl Acad Sci U S A 112: 10485–10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza Silva, O. , and Blokesch, M. (2010) Genetic manipulation of Vibrio cholerae by combining natural transformation with FLP recombination. Plasmid 64: 186–195. [DOI] [PubMed] [Google Scholar]
- Domman, D. , Quilici, M.L. , Dorman, M.J. , Njamkepo, E. , Mutreja, A. , Mather, A.E. , et al (2017) Integrated view of Vibrio cholerae in the Americas. Science 358: 789–793. [DOI] [PubMed] [Google Scholar]
- Dubnau, D. , and Blokesch, M. (2019) Mechanisms of DNA uptake by naturally competent bacteria. Annu Rev Genet 53: 217–237. [DOI] [PubMed] [Google Scholar]
- Gooday, G.W. (1990) Physiology of microbial degradation of chitin and chitosan. Biodegradation 1: 177–190. [Google Scholar]
- Gulig, P.A. , Tucker, M.S. , Thiaville, P.C. , Joseph, J.L. , and Brown, R.N. (2009) USER friendly cloning coupled with chitin‐based natural transformation enables rapid mutagenesis of Vibrio vulnificus . Appl Environ Microbiol 75: 4936–4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg, J.F. , Eisen, J.A. , Nelson, W.C. , Clayton, R.A. , Gwinn, M.L. , Dodson, R.J. , et al (2000) DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae . Nature 406: 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt, D.E. , Gevers, D. , Vahora, N.M. , and Polz, M.F. (2008) Conservation of the chitin utilization pathway in the Vibrionaceae . Appl Environ Microbiol 74: 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskólska, M. , Stutzmann, S. , Stoudmann, C. , and Blokesch, M. (2018) QstR‐dependent regulation of natural competence and type VI secretion in Vibrio cholerae . Nucleic Acids Res 46: 10619–10634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joelsson, A. , Liu, Z. , and Zhu, J. (2006) Genetic and phenotypic diversity of quorum‐sensing systems in clinical and environmental isolates of Vibrio cholerae . Infect Immun 74: 1141–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.A. , Salles, C.A. , Panigrahi, P. , Albert, M.J. , Wright, A.C. , Johnson, R.J. , and Morris, J.G., Jr. (1994) Vibrio cholerae O139 synonym bengal is closely related to Vibrio cholerae El tor but has important differences. Infect Immun 62: 2108–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, S.A. , Chapman, C.A. , and Ng, W.L. (2015) Quadruple quorum‐sensing inputs control Vibrio cholerae virulence and maintain system robustness. PLoS Pathog 11: e1004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, O.N. (1995) The log transformation is special. Stat Med 14: 811–819. [DOI] [PubMed] [Google Scholar]
- Kemter, F.S. , Messerschmidt, S.J. , Schallopp, N. , Sobetzko, P. , Lang, E. , Bunk, B. , et al (2018) Synchronous termination of replication of the two chromosomes is an evolutionary selected feature in Vibrionaceae. PLoS Genet 14: e1007251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyhani, N.O. , and Roseman, S. (1999) Physiological aspects of chitin catabolism in marine bacteria. Biochim Biophys Acta 1473: 108–122. [DOI] [PubMed] [Google Scholar]
- Kimbrough, J.H. , and Stabb, E.V. (2015) Antisocial luxO mutants provide a stationary‐phase survival advantage in Vibrio fischeri ES114. J Bacteriol 198: 673–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klancher, C.A. , Yamamoto, S. , Dalia, T.N. , and Dalia, A.B. (2020a) ChiS is a noncanonical DNA‐binding hybrid sensor kinase that directly regulates the chitin utilization program in Vibrio cholerae . Proc Natl Acad Sci U S A 117: 20180–20189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klancher, C.A. , Newman, J.D. , Ball, A.S. , van Kessel, J.C. , and Dalia, A.B. (2020b) Species‐specific quorum sensing represses the chitobiose utilization locus in Vibrio cholerae . Appl Environ Microbiol [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux, F. , and Blokesch, M. (2018) Eco‐evolutionary dynamics linked to horizontal gene transfer in Vibrios. Annu Rev Microbiol 72: 89–110. [DOI] [PubMed] [Google Scholar]
- Li, X. , and Roseman, S. (2004) The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two‐component chitin catabolic sensor/kinase. Proc Natl Acad Sci U S A 101: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. , Hsiao, A. , Joelsson, A. , and Zhu, J. (2006) The transcriptional regulator VqmA increases expression of the quorum‐sensing activator HapR in Vibrio cholerae . J Bacteriol 188: 2446–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Scrudato, M. , and Blokesch, M. (2012) The regulatory network of natural competence and transformation of Vibrio cholerae . PLoS Genet 8: e1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Scrudato, M. , and Blokesch, M. (2013) A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res 41: 3644–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie, L. , Rapisarda, C. , Morales, V. , Berge, M. , Perry, T. , Soulet, A.L. , et al (2017) Bacterial RadA is a DnaB‐type helicase interacting with RecA to promote bidirectional D‐loop extension. Nat Commun 8: 15638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvig, R.L. , and Blokesch, M. (2010) Natural transformation of Vibrio cholerae as a tool‐optimizing the procedure. BMC Microbiol 10: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthey, N. , Drebes Dörr, N.C. , and Blokesch, M. (2018) Long‐read‐based genome sequences of pandemic and environmental Vibrio cholerae strains. Microbiol Resour Announc 7: e01574–e01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthey, N. , Stutzmann, S. , Stoudmann, C. , Guex, N. , Iseli, C. , and Blokesch, M. (2019) Neighbor predation linked to natural competence fosters the transfer of large genomic regions in Vibrio cholerae . Elife 8: e48212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom, K.L. , Blokesch, M. , Dolganov, N.A. , Wu, C.‐Y. , and Schoolnik, G.K. (2005) Chitin induces natural competence in Vibrio cholerae . Science 310: 1824–1827. [DOI] [PubMed] [Google Scholar]
- Meibom, K.L. , Li, X.B. , Nielsen, A.T. , Wu, C.Y. , Roseman, S. , and Schoolnik, G.K. (2004) The Vibrio cholerae chitin utilization program. Proc Natl Acad Sci U S A 101: 2524–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger, L.C. , and Blokesch, M. (2016) Regulation of competence‐mediated horizontal gene transfer in the natural habitat of Vibrio cholerae . Curr Opin Microbiol 30: 1–7. [DOI] [PubMed] [Google Scholar]
- Metzger, L.C. , Matthey, N. , Stoudmann, C. , Collas, E.J. , and Blokesch, M. (2019) Ecological implications of gene regulation by TfoX and TfoY among diverse Vibrio species. Environ Microbiol 21: 2231–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger, L.C. , Stutzmann, S. , Scrignari, T. , Van der Henst, C. , Matthey, N. , and Blokesch, M. (2016) Independent regulation of type VI secretion in Vibrio cholerae by TfoX and TfoY. Cell Rep 15: 951–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.C. , Keymer, D.P. , Avelar, A. , Boehm, A.B. , and Schoolnik, G.K. (2007) Detection and transformation of genome segments that differ within a coastal population of Vibrio cholerae strains. Appl Environ Microbiol 73: 3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S. , and Bassler, B.L. (2019) Bacterial quorum sensing in complex and dynamically changing environments. Nat Rev Microbiol 17: 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, J. , Miller, M.C. , Nielsen, A.T. , Schoolnik, G.K. , and Spormann, A.M. (2007) vpsA‐ and luxO‐independent biofilms of Vibrio cholerae . FEMS Microbiol Lett 275: 199–206. [DOI] [PubMed] [Google Scholar]
- Mutreja, A. , Kim, D.W. , Thomson, N.R. , Connor, T.R. , Lee, J.H. , Kariuki, S. , et al (2011) Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477: 462–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, E.J. , Harris, J.B. , Morris, J.G.J. , Calderwood, S.B. , and Camilli, A. (2009) Cholera transmission: the host pathogen and bacteriophage dynami. Nat Rev Microbiol 7: 693–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nero, T.M. , Dalia, T.N. , Wang, J.C. , Kysela, D.T. , Bochman, M.L. , and Dalia, A.B. (2018) ComM is a hexameric helicase that promotes branch migration during natural transformation in diverse gram‐negative species. Nucleic Acids Res 46: 6099–6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.L. , and Bassler, B.L. (2009) Bacterial quorum‐sensing network architectures. Annu Rev Genet 43: 197–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, A.T. , Dolganov, N.A. , Otto, G. , Miller, M.C. , Wu, C.Y. , and Schoolnik, G.K. (2006) RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog 2: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort, K. , Silpe, J.E. , Schramma, K.R. , Cong, J.P. , Seyedsayamdost, M.R. , and Bassler, B.L. (2017) A Vibrio cholerae autoinducer‐receptor pair that controls biofilm formation. Nat Chem Biol 13: 551–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack‐Berti, A. , Wollenberg, M.S. , and Ruby, E.G. (2010) Natural transformation of Vibrio fischeri requires tfoX and tfoY . Environ Microbiol 12: 2302–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Seitz, P. , and Blokesch, M. (2013) DNA‐uptake machinery of naturally competent Vibrio cholerae . Proc Natl Acad Sci U S A 110: 17987–17992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, C.A. , Podicheti, R. , Rusch, D.B. , Dalia, A.B. , and van Kessel, J.C. (2019) Diversity in natural transformation frequencies and regulation across Vibrio species. MBio 10: e02788–e02819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, D.D. , Delcher, A.L. , Salzberg, S.L. , and Pop, M. (2007) Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzmann, S. , and Blokesch, M. (2016) Circulation of a quorum‐sensing‐impaired variant of Vibrio cholerae strain C6706 masks important phenotypes. mSphere 1: e00098–e00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckow, G. , Seitz, P. , and Blokesch, M. (2011) Quorum sensing contributes to natural transformation of Vibrio cholerae in a species‐specific manner. J Bacteriol 193: 4914–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, S. , Tay, Q.X. , Kjelleberg, S. , Rice, S.A. , and McDougald, D. (2015) Quorum sensing‐regulated chitin metabolism provides grazing resistance to Vibrio cholerae biofilms. ISME J 9: 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Val, M.E. , Marbouty, M. , de Lemos Martins, F. , Kennedy, S.P. , Kemble, H. , Bland, M.J. , et al (2016) A checkpoint control orchestrates the replication of the two chromosomes of Vibrio cholerae . Sci Adv 2: e1501914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening, J.W. , and Blokesch, M. (2017) Interbacterial predation as a strategy for DNA acquisition in naturally competent bacteria. Nat Rev Microbiol 15: 621–629. [DOI] [PubMed] [Google Scholar]
- Waldor, M.K. , Tschäpe, H. , and Mekalanos, J.J. (1996) A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol 178: 4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill, F.X. , Domman, D. , Njamkepo, E. , Almesbahi, A.A. , Naji, M. , Nasher, S.S. , et al (2019) Genomic insights into the 2016‐2017 cholera epidemic in Yemen. Nature 565: 230–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill, F.X. , Domman, D. , Njamkepo, E. , Tarr, C. , Rauzier, J. , Fawal, N. , et al (2017) Genomic history of the seventh pandemic of cholera in Africa. Science 358: 785–789. [DOI] [PubMed] [Google Scholar]
- Yamamoto, S. , Izumiya, H. , Mitobe, J. , Morita, M. , Arakawa, E. , Ohnishi, M. , and Watanabe, H. (2011) Identification of a chitin‐induced small RNA that regulates translation of the tfoX gene, encoding a positive regulator of natural competence in Vibrio cholerae . J Bacteriol 193: 1953–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S. , Mitobe, J. , Ishikawa, T. , Wai, S.N. , Ohnishi, M. , Watanabe, H. , and Izumiya, H. (2014) Regulation of natural competence by the orphan two‐component system sensor kinase ChiS involves a non‐canonical transmembrane regulator in Vibrio cholerae . Mol Microbiol 91: 326–347. [DOI] [PubMed] [Google Scholar]
- Yildiz, F.H. , and Schoolnik, G.K. (1998) Role of rpoS in stress survival and virulence of Vibrio cholerae . J Bacteriol 180: 773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz, F.H. , Liu, X.S. , Heydorn, A. , and Schoolnik, G.K. (2004) Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol 53: 497–515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Information
Data Availability Statement
All sequencing data have been deposited on NCBI under BioProject accession number PRJNA598947 (BioSamples SAMN13734987, SAMN13734988, SAMN13734999, SAMN13734989, and SAMN15756141). The whole genome sequences are available in GenBank under accession numbers CP047295/CP047296 (C6706), CP047297/CP047298 (C6709), CP047303/CP047304 (E7946), CP047299/CP047300 (P27459), and CP060094/CP060095 (MO10). The raw reads are available from the Sequence Read Archive (SRA) under submission numbers SRR10828860, SRR10828861, SRR10828863, SRR10828864, and SRR12407298).


