Summary
Interactions between phytoplankton and bacteria play major roles in global biogeochemical cycles and oceanic nutrient fluxes. These interactions occur in the microenvironment surrounding phytoplankton cells, known as the phycosphere. Bacteria in the phycosphere use either chemotaxis or attachment to benefit from algal excretions. Both processes are regulated by quorum sensing (QS), a cell–cell signalling mechanism that uses small infochemicals to coordinate bacterial gene expression. However, the role of QS in regulating bacterial attachment in the phycosphere is not clear. Here, we isolated a Sulfitobacter pseudonitzschiae F5 and a Phaeobacter sp. F10 belonging to the marine Roseobacter group and an Alteromonas macleodii F12 belonging to Alteromonadaceae, from the microbial community of the ubiquitous diatom Asterionellopsis glacialis. We show that only the Roseobacter group isolates (diatom symbionts) can attach to diatom transparent exopolymeric particles. Despite all three bacteria possessing genes involved in motility, chemotaxis, and attachment, only S. pseudonitzschiae F5 and Phaeobacter sp. F10 possessed complete QS systems and could synthesize QS signals. Using UHPLC–MS/MS, we identified three QS molecules produced by both bacteria of which only 3‐oxo‐C16:1‐HSL strongly inhibited bacterial motility and stimulated attachment in the phycosphere. These findings suggest that QS signals enable colonization of the phycosphere by algal symbionts.
Introduction
Phytoplankton constitute the foundation of the marine food web as they are responsible for nearly half of primary production on Earth (Field et al., 1998). Through their ability to carry out photosynthesis, phytoplankton transform atmospheric carbon dioxide gas to organic matter (Simon et al., 2009) that is assimilated and remineralized by heterotrophic bacteria (Pomeroy, 1974; Burkhardt et al., 2014). In exchange, bacteria produce essential factors (e.g., vitamins) (Kazamia et al., 2012; Bertrand et al., 2015) to support the growth of phytoplankton (Amin et al., 2012). Cumulatively, phytoplankton‐bacteria symbiosis is believed to play an important role in nutrient availability and major biogeochemical cycles (Buchan et al., 2014; Durham et al., 2019).
Several bacterial lineages have been consistently observed to co‐occur with phytoplankton, such as members of the Roseobacter group, Gammaproteobacteria, and Flavobacteria (Wagner‐Döbler and Biebl, 2006; Teeling et al., 2012). Particularly, members of the Roseobacter group (hereafter roseobacters) have been shown consistently to form symbiotic relationships with phytoplankton (Amin et al., 2012). For example, roseobacters are adept at acquiring and assimilating phytoplankton metabolites (Miller et al., 2004) in exchange for a variety of cofactors. Ruegeria pomeroyi has been shown to assimilate organic sulfur compounds from the diatom Thalassiosira pseudonana in exchange for production of cobalamin, which is required for diatom growth (Durham et al., 2015). Sulfitobacter pseudonitzschiae SA11 produces the hormone indole‐3‐acetic acid (IAA) to enhance cell division of the diatom Pseudo‐nitzschia multiseries, which leads to an increase in carbon export by the diatom and the exchange of diatom‐derived organosulfur compounds and bacterial ammonia (Amin et al., 2015). Other phytoplankton such as the coccolithophore, Emiliania huxleyi, display different morphologies in response to endogenous IAA (Labeeuw et al., 2016) and exhibit enhanced cell division by roseobacters‐derived IAA (Segev et al., 2016; Bramucci et al., 2018). IAA is an endogenous hormone that regulates plant differentiation and is also produced and excreted by plant symbionts and some plant pathogens to interfere with plant root differentiation (Spaepen et al., 2007; Spaepen and Vanderleyden, 2011). Although eukaryotic phytoplankton, like diatoms and coccolithophores, are unicellular and do not undergo differentiation as defined in multicellular eukaryotes, IAA appears to have evolved the ability to manipulate the cell cycle in both groups of organisms, with related bacteria playing an important role in eukaryotic IAA perception in both marine and rhizobial environments (Seymour et al., 2017). A prerequisite for these symbiotic exchanges to occur is the intimate spatial proximity between the phytoplankton host and its symbionts.
Chemical exchanges between bacteria and phytoplankton occur in the microenvironment immediately adjacent to phytoplankton cells, known as the phycosphere (Bell and Mitchell, 1972; Seymour et al., 2017). Phytoplankton continuously exude organic matter, sometimes up to 50% of the cell's total fixed carbon (Thornton, 2014). Consequently, the phycosphere is hypothesized to harbour a significantly higher amount of phytoplankton dissolved organic matter (DOM) relative to bulk seawater due to the exudation of DOM by phytoplankton cells, and due to the negligible effects of turbulence on the diffusion of exudates within the minute phycosphere (Seymour et al., 2017). This buildup of DOM ultimately leads to bacterial attraction and colonization of the phycosphere either via chemotaxis or random encounters with phytoplankton cells (Smriga et al., 2016). Once in the phycosphere, beneficial bacteria that produce metabolites essential to phytoplankton (Mayali et al., 2011) may gain an advantage by switching their free‐living, planktonic lifestyle in bulk seawater to a surface‐attached state on phytoplankton cells. On the other hand, opportunistic bacteria benefit from phytoplankton‐derived DOM without providing apparent benefits to phytoplankton hosts (Mayali and Doucette, 2002). Compared to free‐living cells, surface‐associated cells have greater access to phytoplankton nutrients and gain protection against toxins, antibiotics, and other environmental stressors by forming a biofilm (Jefferson, 2004; Samo et al., 2018).
Successful colonization of the phycosphere can be enhanced by specific bacterial genetic traits, including chemotaxis, motility, and attachment to surfaces (Slightom and Buchan, 2009; Raina et al., 2019). Chemotaxis enables bacteria to sense changes in local concentrations of food and regulates motility accordingly. Motile bacteria generally exhibit strong chemotaxis to DOM released by phytoplankton (Miller and Belas, 2004; Miller et al., 2004; Stocker, 2012; Smriga et al., 2016), while motility and flagellar genes appear to be critical for attachment and biofilm development in many roseobacters (Miller and Belas, 2006; Bruhn et al., 2007). In the phycosphere, many bacteria may have a biphasic ‘swim‐or‐stick’ lifestyle that enables them to rapidly find food sources while minimizing energy expenditures once the food is located. During the motile phase, bacteria use chemotaxis to locate phytoplankton cells (Seymour et al., 2017). Once in the phycosphere, a ‘switch’ is turned on, causing a transition of the bacteria to a sessile lifestyle, whose phenotype includes loss of flagella and subsequent biofilm development (Geng and Belas, 2010). Despite our knowledge of bacterial behaviour, the mechanisms that regulate bacterial motility and attachment in the phycosphere have not been extensively investigated.
Quorum sensing (QS) is one of the best‐studied signalling mechanisms for bacterial cell‐to‐cell communication. Many bacteria have been shown to carry out QS by secreting small signalling molecules, known as autoinducers, to assess changes in bacterial populations and to coordinate gene expression among a whole population (Waters and Bassler, 2005). In Proteobacteria, the primary class of autoinducers is acyl‐homoserine lactones (AHLs), which are synthesized by an autoinducer synthase (LuxI) and perceived by an autoinducer regulator (LuxR) (Case et al., 2008). Bacteria use AHLs to regulate functions that are beneficial to carry out collectively, such as virulence, motility, and biofilm formation (Hammer and Bassler, 2003; Daniels et al., 2004; Antunes et al., 2010). Indeed, several Roseobacter‐group bacteria use AHLs to regulate motility, virulence, biofilm formation, and nutrient acquisition when associated with marine snow (Gram et al., 2002; Hmelo et al., 2011), red algae (Gardiner et al., 2015), and sponges (Zan et al., 2012). The addition of exogenous AHLs produced by bacterial epibionts to colonies of the cyanobacterium Trichodesmium leads to an increase in alkaline phosphatase activity and consequently phosphorus acquisition (Van Mooy et al., 2012). In contrast, the lack of complete QS systems often leads to the inability of bacteria to attach to hosts. For example, a luxR‐type gene knock‐out strain of the Roseobacter‐group member Nautella italica R11 was unable to form biofilms or attach to the multicellular red alga Delisea pulchra (Gardiner et al., 2015). Despite these examples, there is little direct evidence showing how QS regulates bacteria‐phytoplankton interactions or how QS influences inter‐ and intraspecies interactions and behaviour between bacteria within microbial consortia in the phycosphere. In this study, we examine how QS influences bacterial behaviour in the phycosphere of the ubiquitous diatom, Asterionellopsis glacialis, and whether QS provides an advantage to beneficial bacteria relative to other bacteria. Here, we hypothesize that QS regulates motility and attachment of beneficial bacteria, which may enhance their access to phytoplankton nutrients over non‐beneficial bacteria.
The diatom A. glacialis is a ubiquitous species that has been isolated from almost every major water body around the world (Korner, 1970; Kaczmarska et al., 2014) and has recently been shown to be one of several abundant and widely distributed groups of diatoms in the open ocean from the Tara Oceans expedition (Malviya et al., 2016). In addition, A. glacialis often forms blooms and dense patches worldwide (Karentz and Smayda, 1984; Franco et al., 2016) that are characterized by high DOM secretions (Abreu et al., 2003), making this diatom an ideal model system to examine interactions with bacteria. Here, we characterize AHL molecules produced by bacteria isolated from the phycosphere of A. glacialis and examine the influence of these AHLs on the ability of beneficial and opportunistic bacteria to colonize the phycosphere of A. glacialis.
Results and discussion
Bacterial attachment and influence on diatom physiology
The diatom A. glacialis strain A3 (deposited as CCMP3542) was isolated from the Persian Gulf and identified as previously described (Behringer et al., 2018). Axenic A. glacialis strain A3 cultures were generated using antibiotics as described previously (Amin et al., 2015). Under optimal growth conditions, we noticed that axenic A. glacialis strain A3 mostly existed as single cells or chains with an average of approximately three cells per chain while xenic A. glacialis strain A3 at the same cell density (~1.5 × 105 cells mL−1) formed longer chains and/or aggregated cells when the samples were observed directly without filtration (Supporting Information Fig. S1A,B). Quantifying the abundance of diatom chains spanning 1–3 cells and >3 cells per chain in axenic and xenic cultures showed that chain length did not change appreciably throughout the growth of axenic cultures with roughly half the population forming >3 cells per chain. In contrast, bacteria significantly increased the abundance of longer diatom chains, with 79.4% of the population forming >3 cells per chain relative to axenic cultures in late‐exponential to early‐stationary phases (Supporting Information Fig. S1C). In diatoms, current evidence suggests that chain length is influenced by increasing CO2 concentrations (Ramos et al., 2014) and grazing pressure (Amato et al., 2018). The finding that bacteria can influence diatom chain length is novel and consistent with observations that bacteria can influence diatom cell size and morphology (Windler et al., 2014).
Removal of free‐living bacteria in xenic A. glacialis cultures using gravity filtration through a 3‐μm membrane filter and staining filtered diatom and bacterial cells with SYBR Green I showed large aggregates of bacteria on and/or in close proximity to diatom cells (Supporting Information Fig. S1D). Further staining of the sample with alcian blue, a dye that stains diatom transparent exopolymeric particles (TEP), showed that bacteria mostly attach to TEP (Supporting Information Fig. S1E,F), an observation consistent with previous findings (Bar‐Zeev et al., 2012). Generating biofilm on algal TEP and/or surfaces is a typical behaviour of bacteria in aquatic habitats (Kogure et al., 1981; Bagatini et al., 2014) that can enable them to persist in such environments (Geng and Belas, 2010). Forming biofilm on algal surfaces or algal TEP also protects bacteria against toxins and antibiotics and provides shelter from predation (Carvalho, 2018). For example, bacteria residing in a biofilm can tolerate antimicrobial agents at concentrations 100–1000 times those needed to kill planktonic cells (Lewis, 2001). In addition, colonizing the phycosphere by attaching to TEP may help bacteria conserve energy that would otherwise be spent on motility and chemotaxis to remain in proximity of the phycosphere (Seymour et al., 2017).
Bacterial isolation
To examine bacterial attachment and influence on diatom chain length in the phycosphere, three bacterial strains were isolated from xenic A. glacialis and classified using the 16S rRNA gene sequence identity as Sulfitobacter pseudonitzschiae F5 (Rhodobacteraceae; >99% similarity to S. pseudonitzschiae), Phaeobacter sp. F10 (Rhodobacteraceae; 97% similarity to Phaeobacter gallaeciensis) and Alteromonas macleodii F12 (Alteromonadaceae; 99% similarity to A. macleodii) (Supporting Information Fig. S2). The Roseobacter group (Rhodobacteraceae) is one of the most important groups of marine bacteria that primarily colonize both biotic (e.g., phytoplankton) and abiotic surfaces (e.g., marine snow) (Gram et al., 2002; Dang et al., 2008) and can comprise up to 25% of the marine bacterial community in some regions (Wagner‐Döbler and Biebl, 2006). They have been shown to form a substantial component of the A. glacialis microbial consortium based both on 16S rRNA gene amplicon sequencing (Behringer et al., 2018) and shotgun metagenomics (Shibl et al., 2020). Mining the 16S rRNA gene sequences of the microbial community of A. glacialis recovered after 20 days of isolation from the field (Behringer et al., 2018), we recovered reads that display 100% sequence identity to the 16S rRNA gene of Phaeobacter sp. F10 and A. macleodii F12 and 99% sequence identity to S. pseudonitzschiae F5, indicating our bacterial isolates belong to the natural population of xenic A. glacialis and that members of this population persist through time under laboratory culturing conditions.
Co‐culture of bacterial isolates with the diatom
To test whether these bacteria attach to A. glacialis, co‐cultures of each bacterium with the diatom were grown in batch cultures, and growth and attachment of both partners were monitored using microscopy. When co‐cultured with A. macleodii F12, the specific growth rate (μ) of the diatom did not exhibit significant changes relative to axenic controls (μ axenic = 1.02 ± 0.05 d−1; μ co‐culture = 1.03 ± 0.02 d−1) (Supporting Information Fig. S3A). Likewise, the growth of the diatom did not vary significantly when co‐cultured with Phaeobacter sp. F10 (μ axenic = 0.81 ± 0.02 d−1; μ co‐culture = 0.84 ± 0.04 d−1) (Supporting Information Fig. S3B). In contrast, A. glacialis co‐cultured with S. pseudonitzschiae F5 exhibited a 27.6% increase in μ relative to axenic controls (μ axenic = 0.76 ± 0.03 d−1; μ co‐culture = 0.97 ± 0.03 d−1) (Supporting Information Fig. S3C). In all co‐cultures, bacteria exhibited ~3 orders of magnitude increase in cell density, indicating uptake of diatom‐derived organic matter (Supporting Information Fig. S3D). Surprisingly, co‐cultures of S. pseudonitzschiae F5 with A. glacialis showed a significant increase in longer diatom chains than axenic cultures, similar to observations with the xenic diatom (Supporting Information Fig. S1C). This observation may be a byproduct of the enhanced growth of A. glacialis with S. pseudonitzschiae F5 or a result of a more complex mechanism of interaction.
Sulfitobacter pseudonitzschiae strains were first isolated from cultures of the toxigenic marine diatom P. multiseries obtained from the North Atlantic Ocean and the Pacific Northwest, with a model strain first coined as Sulfitobacter sp. SA11 (Amin et al., 2015). Subsequently, several additional S. pseudonitzschiae strains were isolated from the diatoms P. multiseries, Skeletonema marinoi and A. glacialis originating from the Atlantic Ocean, the Swedish coast, and the Persian Gulf, respectively (Hong et al., 2015; Töpel et al., 2019). These repetitive recoveries of nearly identical bacteria (>99% 16S rRNA gene sequence similarity) from three genera of diatoms that originated from starkly different locations with large variations in temperature, salinity and nutrients indicate that S. pseudonitzschiae is a globally distributed bacterium that may be a true symbiont of diatoms. S. pseudonitzschiae SA11 enhances the growth rate of the diatom P. multiseries by 19%–35% compared with axenic controls partially due to the activity of the hormone IAA, which S. pseudonitzschiae biosynthesizes from diatom‐derived tryptophan. Both organisms also exchange organosulfur compounds and ammonia to complement each other's metabolism (Amin et al., 2015). In this study, S. pseudonitzschiae F5 enhanced the growth rate of the diatom A. glacialis by 27.6% (Supporting Information Fig. S3C). Further genome sequencing and annotation showed that S. pseudonitzschiae F5 also possesses three complete pathways for IAA biosynthesis from tryptophan (Supporting Information Table S1), suggesting it may use the same strategy as S. pseudonitzschiae SA11 to enhance diatom growth.
Compared with Sulfitobacter, Phaeobacter species are known to colonize marine macro‐ and microalgal surfaces (e.g., Ulva australis, Thalassiosira rotula) (Rao et al., 2006; Thole et al., 2012). P. inhibens has been shown to control bacterial community assembly in the phycosphere of T. rotula (Majzoub et al., 2019). P. inhibens also produces the hormone IAA to promote the growth of the coccolithophore, E. huxleyi, similar to S. pseudonitzschiae (Segev et al., 2016). P. gallaeciensis has been shown to lyse senescent E. huxleyi by producing algicidal compounds known as roseobacticides (Seyedsayamdost et al., 2011). Although Phaeobacter sp. F10 did not enhance the growth rate of A. glacialis (Supporting Information Fig. S3B), metatranscriptomic analysis of a near‐identical metagenomically assembled genome from the A. glacialis strain A3 microbial consortium indicates that Phaeobacter sp. F10 is also a symbiont of A. glacialis (Shibl et al., 2020).
Alteromonadaceae are widespread marine opportunistic copiotrophs (López‐Pérez et al., 2012) that display algicidal activities against phytoplankton during algal blooms (Mayali and Azam, 2004). The Pseudoalteromonas and Alteromonas genera are known to effectively metabolize the organic matrix surrounding diatom frustules, exposing the silica shell to increased dissolution by the surrounding water (Bidle and Azam, 2001) and show strong algicidal activity by releasing dissolved substances (Mayali and Azam, 2004). For example, A. colwelliana shows growth inhibition and algicidal activity against the diatom Chaetoceros calcitrans (Kim et al., 1999). A. macleodii have been shown to degrade a variety of algal TEP and exopolysaccharides (Koch et al., 2019) that may enable them to benefit from phytoplankton‐derived carbon without contributing to phytoplankton metabolism. In addition, A. macleodii has been shown to compete for nitrate with the diatom Phaeodacylum tricornutum in the presence of organic carbon (Diner et al., 2016).
Bacterial attachment in the phycosphere
In order to test which bacterial strains attach to the diatom or TEP, we removed free‐living bacterial cells from co‐cultures of each strain with the diatom using gravity filtration through a 3‐μm membrane filter and stained TEP with alcian blue, and diatom and bacterial nucleic acids with SYBR Green I (Fig. 1). Both symbiotic strains, S. pseudonitzschiae F5 and Phaeobacter sp. F10, displayed strong attenuation onto filters, while no attached A. macleodii F12 were observed (Fig. 1D–F). The composite images of bright field and fluorescence showed a strong attachment preference to diatom TEP of S. pseudonitzschiae F5 and Phaeobacter sp. F10 in co‐cultures with A. glacialis, similar to observations in xenic cultures (Fig. 1G,H). Surprisingly, A. macleodii F12 did not show any attachment capacity to A. glacialis or TEP (Fig. 1) despite its ability to degrade algal polysaccharides (Koch et al., 2019). These observations suggest an inherent mechanism that enables the roseobacters but not A. macleodii to attach to TEP. To shed more light on such mechanisms, we sequenced the genomes of all three isolates.
Fig. 1.
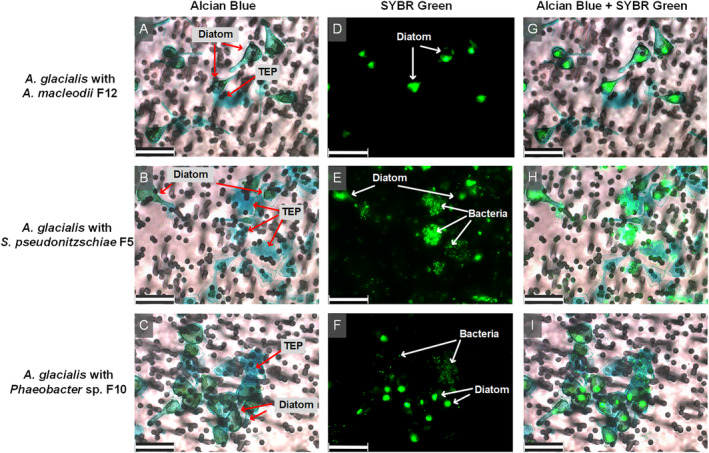
Micrographs of A. glacialis strain A3 co‐cultures with bacterial isolates. Diatom TEP were stained with alcian blue (A–C). Diatom and bacterial DNA was stained with SYBR Green I (D–F). Composite images show bacterial attachment mostly to TEP for S. pseudonitzschiae F5 and Phaeobacter sp. F10 but not A. macleodii F12 (G–I). Background in light micrographs show membrane filters. Scale bar represents 25 μm for all panels.
Genomic comparisons and AHLs identification
The genomes of the three strains were obtained by a combination of PacBio and Illumina sequencing as described in the methods. The GC contents of both Roseobacter group member genomes were more similar to each other (61.8% for S. pseudonitzschiae F5 and 60.0% for Phaeobacter sp. F10) than to A. macleodii F12 (44.9%) (Table 1), consistent with the phylogenetic similarities of S. pseudonitzschiae F5 and Phaeobacter sp. F10 compared to A. macleodii F12 (Supporting Information Fig. S2). To confirm the phylogenetic classification of the three strains based on the 16S rRNA gene, we compared the average nucleotide identity (ANI) between all three sequenced strain genomes and related publicly available genomes. Typically, an ANI value >95% between two genomes suggests they belong to the same species (Kim et al., 2014). The ANIs between S. pseudonitzschiae F5 and other S. pseudonitzschiae strain were >99% (accession numbers: GCF_000712315.1, GCF_013347125.1 and GCF_900129395.1) while the ANIs between A. macleodii F12 and other A. macleodii genomes were >96% (accession numbers: GCF_000808595.1, GCF_000172635.2 and GCF_001578515.1). On the other hand, the ANIs between Phaeobacter sp. F10 and six different Phaeobacter species were <86% (accession numbers: GCF_001888185.1, GCF_002891905.1, GCF_007923355.1, GCF_000154765.2, GCF_001251095.1 and GCF_000511385.1). These results further confirm the identity of S. pseudonitzschiae F5 and A. macleodii F12 and suggest Phaeobacter sp. F10 represents a new species.
Table 1.
Bacterial genome statistics.
| Statistic | Sulfitobacter pseudonitzschiae F5 | Phaeobacter sp. F10 | Alteromonas macleodii F12 |
|---|---|---|---|
| Genome size (bp) | 5 087 289 | 4 008 202 | 4 727 977 |
| Number of contigs | 17 | 3 | 5 |
| Chromosome size (bp) | 3 764 450 a | 3 783 964 | 3 117 849 |
| CDs | 4991 | 3878 | 4654 |
| GC content (%) | 61.8 | 60.0 | 44.9 |
| N50 value | 179 155 | 153 588 | 3 122 272 |
| L50 value | 3 | 1 | 1 |
Sulfitobacter pseudonitzschiae F5 chromosome size estimation was not possible due to high level of repeats in the genome. The size indicated is the size of the largest contig.
The bacterium S. pseudonitzschiae F5 possesses the largest estimated genome size (5.1 Mb) compared with Phaeobacter sp. F10 (4.0 Mb) and A. macleodii F12 (4.7 Mb) (Table 1). Interestingly, A. macleodii F12 possesses a 3.1 Mb chromosome and presumably several plasmids totaling ~1.6 Mb (Table 1), consistent with previous studies showing the presence of multiple megaplasmids in A. macleodii genomes (Cusick et al., 2020). S. pseudonitzschiae F5 also had the greatest number of predicted genes (4991) compared with either Phaeobacter sp. F10 (3878) or A. macleodii F12 (4654) (Supporting Information Fig. S4A). A major difference between the three genomes was the apparent higher number of putative genes involved in membrane transport of substrates, particularly primary active transporters (e.g., amino acids, sugars) (Mishra et al., 2014). For example, the relative abundance of putative membrane transporters in the S. pseudonitzschiae F5 genome is 10.50% compared with Phaeobacter sp. F10 (8.44%) or A. macleodii F12 (7.05%) when normalized to genome size, with primary active transporters being the most abundant in the roseobacters' genomes but not in A. macleodii F12 (Supporting Information Fig. S4B). This observation suggests that both symbionts are more attuned to phycosphere metabolites than A. macleodii F12.
Chemotaxis, motility, and attachment presumably contribute to successful colonization of phytoplankton surfaces. To examine the ability of all three bacteria to interact with the diatom phycosphere, we conducted a genome‐wide comparison of genes involved in bacterial chemotaxis, motility, attachment, and quorum sensing. All three genomes contained four chemotaxis genes in a single operon‐like structure (cheA, cheR, cheW, and cheY), while a fifth gene, cheB, was present in Phaeobacter sp. F10 and A. macleodii F12, but absent in S. pseudonitzschiae F5 (Fig. 2A and Supporting Information Table S2). CheAYW together mediate a signal transduction cascade that regulates flagellar motors, while CheB and CheR together regulate the methylation state of methyl‐accepting chemotaxis proteins (Wuichet et al., 2007). Flagellar genes were present in all three genomes, although S. pseudonitzschiae F5 and Phaeobacter sp. F10 also contained a suite of flagellar structure genes absent in A. macleodii F12 (flgN, fliG, flgJ, motB, and MotA/TolQ/ExbB proton channel family protein). In addition, both S. pseudonitzschiae F5 and Phaeobacter sp. F10 contained 11 genes related to pilus formation in contrast to A. macleodii F12, which only contained a pilin flp gene, which is involved in pilus formation (Bardy et al., 2003). For attachment, genes involved in exopolysaccharide production and biofilm formation were distributed throughout the genomes of the three bacteria (Fig. 2A and Supporting Information Table S2). These lines of evidence suggest that S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 have the ability to carry out chemotaxis, construct flagella and pili structures, and form biofilm. Despite this observation, A. macleodii F12 did not attach to A. glacialis or TEP produced by A. glacialis (Fig. 1).
Fig. 2.
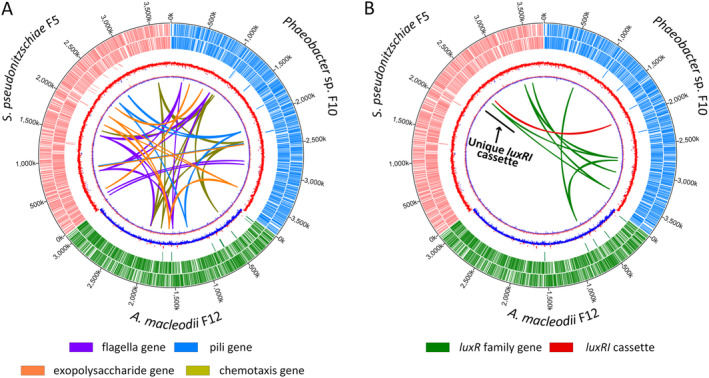
Chromosomal maps of bacterial isolate genomes showing distribution and localization of loci involved in attachment, motility, chemotaxis and quorum sensing. Tracks from the outermost to the center represent position on each chromosome, forward‐strand protein‐coding genes, reverse‐strand protein coding genes, rRNA and tRNA genes, GC content and GC skew, respectively. Chromosomes of S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 are colour‐labelled with red, blue and green, respectively.
A. Homologues related to chemotaxis, motility and attachment.
B. Homologues related to quorum sensing. Black line refers to a luxI gene homologue unique to S. pseudonitzschiae F5.
Quorum sensing (QS) autoinducers (e.g., AHLs) have been widely shown to modulate important biological functions, such as biofilm formation and motility (Bassler, 2002; Waters and Bassler, 2005), indicating QS may be able to regulate bacterial ‘swim‐or‐stick’ lifestyles in the phycosphere and suggesting that A. macleodii F12 may lack a functioning QS system. Indeed, luxI‐like genes that are responsible for biosynthesizing AHLs were only present in S. pseudonitzschiae F5 (2 homologues) and Phaeobacter sp. F10 (1 homologue), while A. macleodii F12 completely lacked apparent luxI homologues (Fig. 2B and Supporting Information Table S2). Indeed, luxI is rarely present in Alteromonas species. We found only two of 67 publicly available genomes belonging to the Alteromonadaceae contain putative AHL synthases. In addition to AHL biosynthesis, a transcriptional regulator, encoded by a luxR family gene, is required to perceive AHLs and coordinate gene expression among bacterial populations. All three genomes contained luxR family genes; while S. pseudonitzschiae F5 possessed four putative luxR‐family genes and Phaeobacter sp. F10 possessed six, A. macleodii F12 possessed only one (Fig. 2B and Supporting Information Table S2). Typically, apparent luxR genes are found adjacent to or near a luxI gene on bacterial chromosomes in a single operon‐like structure, such as in S. pseudonitzschiae F5 and Phaeobacter sp. F10 (Supporting Information Table S2). However, some bacteria also have putative ‘solo’ luxR genes without associated putative luxI genes, which is the case in A. macleodii F12 and for the additional putative luxR homologues present in S. pseudonitzschiae F5 and Phaeobacter sp. F10. It has been hypothesized that bacteria possessing ‘solo’ luxR genes do so to detect and respond to exogenous signals from other bacterial populations (Hudaiberdiev et al., 2015) or that these solo genes represent the loss of QS function (Subramoni and Venturi, 2009). Some solo LuxR proteins have low specificity for AHL binding compared with LuxR proteins coupled with LuxI proteins (Subramoni and Venturi, 2009). For example, SdiA, a solo transcriptional regulator that is present in members of Salmonella, Escherichia, and Klebsiella, could bind seven different AHL molecules (Yao et al., 2006; Janssens et al., 2007). Potentially, the solo luxR genes in A. macleodii F12 and the roseobacters may respond to a wide range of AHLs from other bacteria.
To characterize AHLs produced by S. pseudonitzschiae F5 and Phaeobacter sp. F10, we purchased a suite of commercially available AHL standards (Supporting Information Table S3) and used ultra‐high‐performance liquid chromatography–tandem mass spectrometry (UHPLC–MS/MS) to identify potential AHLs in bacterial cultures. Three AHLs were recovered from S. pseudonitzschiae F5 and Phaeobacter sp. F10 pure culture supernatants with ionized parent masses of m/z 270.1700, 352.2480, and 354.2640 and were identified as 3‐oxo‐C10‐HSL, 3‐oxo‐C16:1‐HSL and 3‐oxo‐C16‐HSL, respectively, using high‐resolution m/z values, daughter‐ion fragmentation and retention times (Fig. 3 and Supporting Information Fig. S5). No standard from the AHL library matched any metabolite in the supernatant of A. macleodii F12, consistent with its lack of luxI‐like genes (Fig. 2). We attempted to measure AHLs in co‐cultures of the roseobacters and the diatom but due to the significantly lower bacterial abundance in cocultures compared with pure bacterial cultures we were not successful in detecting AHLs. The identification of AHL production by diatom symbionts prompted examination of the influence of these signalling molecules on the ability of the symbionts to attach to diatom TEP.
Fig. 3.
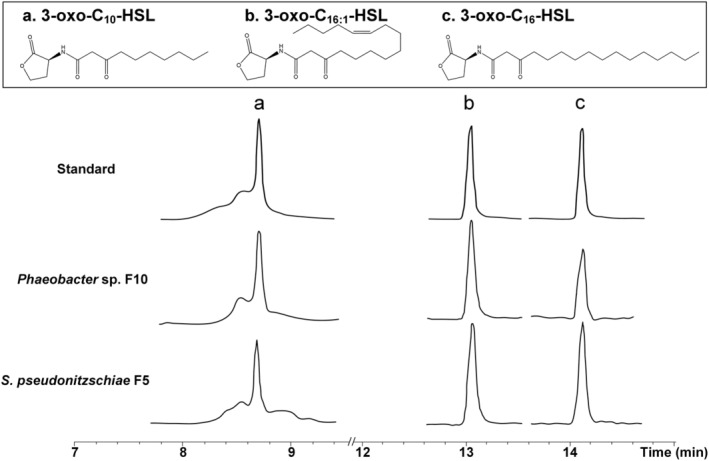
Identification of acyl‐homoserine lactones (AHLs) from roseobacters isolates. Structures and UHPLC–MS/MS chromatograms of AHLs of S. pseudonitzschiae F5 or Phaeobacter sp. F10 isolated from pure culture supernatants compared with purchased standards. For complete MS/MS spectra refer to Supporting Information Fig. S5.
Influence of AHLs on bacterial motility and biofilm formation
Cell attachment is the first step in the process of biofilm formation, which requires motility for initial attachment to surfaces (Slightom and Buchan, 2009). Consequently, we examined the influence of AHLs on motility and biofilm formation since both functions are regulated by QS in bacteria (Hammer and Bassler, 2003; Daniels et al., 2004). All three AHLs and two QS inhibitors (QSIs), 2(5H)‐furanone and furanone C‐30 (Ponnusamy et al., 2010; He et al., 2012), were used to test the motility and biofilm formation capabilities of S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12. QSIs were used to confirm that any phenotypes observed using AHLs were due to QS regulation and not byproducts of other processes. Since the A. macleodii F12 genome contained a putative solo luxR gene (Fig. 2B), we suspected A. macleodii F12 might still respond to AHLs produced by S. pseudonitzschiae F5 and Phaeobacter sp. F10 despite lacking the ability to synthesize AHLs.
The addition of 2 μM of each AHL to each bacterium showed statistically significant inhibition of motility in S. pseudonitzschiae F5 and Phaeobacter sp. F10 (P < 0.01) by all three AHLs, while both QSIs enhanced motility in S. pseudonitzschiae F5, as expected, and furanone C‐30 enhanced motility in Phaeobacter sp. F10 (Fig. 4A,B). Specifically, 3‐oxo‐C16:1‐HSL exhibited the strongest inhibition of motility in S. pseudonitzschiae F5 by 78.2% (P < 0.001) compared with the two other AHLs (28.6% and 31.8%). Despite two AHLs, 3‐oxo‐C16‐HSL and 3‐oxo‐C10‐HSL, weakly inhibiting motility in A. macleodii F12, both QSIs also weakly inhibited its motility, suggesting A. macleodii F12 does not respond or weakly responds to roseobacters' QS molecules (Fig. 4A,B). Bacterial motility is characterized by a circular swimming zone in motility assays or by dendritic morphology, which is typical for swarming motility caused by a locally restricted movement on agar plates (Michael et al., 2016). Phaeobacter sp. F10 and A. macleodii F12 displayed a typical circular swimming zone while S. pseudonitzschiae F5 displayed a dendritic motility phenotype (Fig. 4A).
Fig. 4.
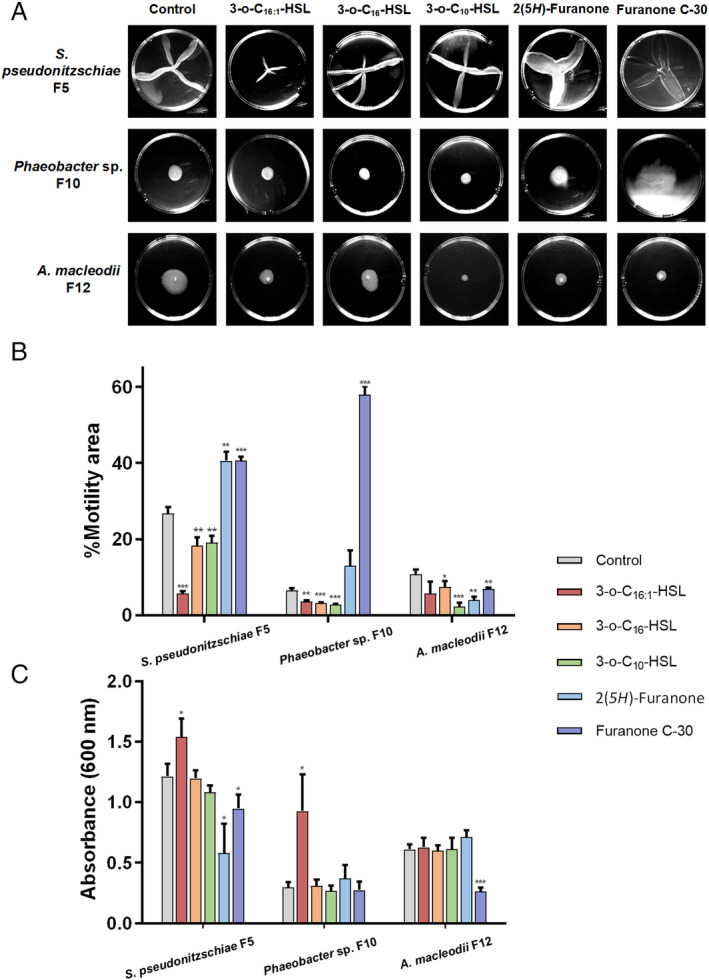
AHLs control bacterial motility and attachment in Roseobacter group bacteria.
A. Motility assays of S. pseudonitzschiae F5, Phaeobacter sp. F10 and A. macleodii F12 with three AHL molecules: 3‐oxo‐C10‐HSL, 3‐oxo‐C16‐HSL and 3‐oxo‐C16:1‐HSL, and two quorum sensing inhibitors (QSIs): 2(5H)‐furanone and furanone C‐30. Assays were conducted by inoculating 0.25% soft agar plates with each bacterium and incubating for 3 days.
B. Percent motility of S. pseudonitzschiae F5, Phaeobacter sp. F10 and A. macleodii F12 in the presence of 2 μM AHLs or QSIs. All error bars represent standard deviation (SD) of triplicate experiments. *P < 0.05, **P < 0.01, ***P < 0.001).
C. Biofilm quantification in the presence of 10 μg ml−1AHLs or QSIs. Assays were conducted by incubating 1 × 106 bacterial cells with each molecule in 96‐well plates for 24 h. Attachment was quantified using absorbance of solubilized crystal violet at 600 nm. All error bars represent standard deviation (SD) of triplicate experiments. *P < 0.05, **P < 0.01, ***P < 0.001).
To assess biofilm formation, a standardized crystal violet assay was conducted as described previously (O'Toole and Kolter, 1998). Interestingly, 10 μg ml−1of 3‐oxo‐C16:1‐HSL significantly enhanced biofilm formation in S. pseudonitzschiae F5 and Phaeobacter sp. F10 by 26.7% and 211%, respectively (P < 0.05), while 3‐oxo‐C10‐HSL and 3‐oxo‐C16‐HSL did not influence biofilm formation in either bacteria (Fig. 4C). These results suggest each AHL molecule regulates different sets of functions. Similar observations have been shown previously (Su et al., 2018; Hou et al., 2019). In contrast, both QSIs inhibited biofilm formation of S. pseudonitzschiae F5 by 52.2% and 21.9%, respectively, relative to the control. None of the AHLs influenced biofilm formation of A. macleodii F12 (Fig. 4C).
These findings suggest that specific AHLs produced by diatom symbionts promote bacterial colonization of the phycosphere by enhancing the bacterial capacity to form biofilms and reduce motility, functions that are essential to successfully colonize the phycosphere. In contrast, A. macleodii F12 is unable to respond to these molecules or to effectively attach to TEP. Other mechanisms must also be functioning to further prevent opportunists, like A. macleodii F12, from benefitting from diatom exudates. One such mechanism is the regulation of microbial consortia by the eukaryotic host, A. glacialis. A. glacialis has recently been shown to release the unusual metabolites, rosmarinic acid and azelaic acid, in the phycosphere in response to bacteria (Shibl et al., 2020). Azelaic acid promoted the growth of both symbionts and concomitantly inhibited the growth of A. macleodii F12. Similarly, rosmarinic acid inhibited the motility of S. pseudonitzschiae F5 and Phaeobacter sp. F10 in an identical way to 3‐oxo‐C16:1‐HSL, while upregulating motility in A. macleodii F12 (Shibl et al., 2020). Rosmarinic acid has been shown to interfere with QS in a plant pathogen (Hammer and Bassler, 2003), suggesting this molecule acts as a QSI. A similar mechanism of interfering with QS, a process known as quorum quenching, is used by the marine macroalga D. pulchra to inhibit swarming motility of Serratia liquefaciens by producing halogenated furanones (Rasmussen et al., 2000). Cumulatively, our observations suggest that S. pseudonitzschiae F5 and Phaeobacter sp. F10 switch their free‐living mode to a surface‐attached mode to remain in the phycosphere by releasing AHLs. In addition, the adaptation of both strains to diatom metabolites (Shibl et al., 2020) ensures the success of these bacteria in the phycosphere relative to other bacteria that cannot colonize the phycosphere.
QS gene homology and organization in the Roseobacter group
To shed light on the prevalence of QS systems in the Roseobacter group, we constructed a phylogenetic tree of 52 sets of the LuxR‐ITS‐LuxI sequences (luxRI cassettes) in 32 Roseobacter group representative genomes. The tree revealed multiple conserved QS gene topologies that were distributed across the Roseobacter group (Fig. 5).
Fig. 5.

Maximum‐likelihood phylogenetic tree of 52 LuxR‐ITS‐LuxI loci and neighbouring genes in 32 roseobacterial genomes. Group B is highlighted by red branches at the top of the tree. Colour shades indicate the different groups of luxR/I cassettes. Bacteria isolated in this study are bold‐faced. Length shows the sequence lengths of LuxR‐ITS‐LuxI loci. D = dendritic motility, M = motility, A = attachment. Y and N indicate whether a bacterium is able or unable to carry out each function. Isolation origin of bacteria highlighted in red indicates a biological host origin. The number of AHL molecules reported from each bacterium is provided, if known. Note that information on which cassette makes which AHL molecule is scarce. Some bacteria are present twice in the tree because they possess more than one Lux‐like operon. Gene neighbourhood abbreviations: HK, histidine kinase; CHP, conserved hypothetical protein; TctA, TctA family transmembrane transporter; RND, RND multidrug efflux pump; AL, adenylosuccinate lyase; TF, trigger factor; Tran, transposase; TetR, TetR family transcriptional regulator; TR, transcriptional regulator. Bootstrap values >50% calculated from 100 iterations are shown at branch nodes. *Full references are listed in the Supporting Information.
In Fig. 5, half of the roseobacters genomes contained more than one pair of luxRI (18/31) and more than two AHL molecules were identified in nine strains. It has been shown that >80% of bacteria in the Roseobacter group possess at least one luxRI gene cassette (Buchan et al., 2016). Phaeobacter species consistently appear to produce more than three AHLs using 2–3 luxI homologues (Fig. 5 and Supporting Information Table S4). For example, Phaeobacter gallaeciensis DSM 26640 produces up to eight different AHLs via only three luxI homologues (Ziesche et al., 2015). Cude and Buchan (2013) provide a classification of the genetic makeup and organization of the luxRI gene cassettes and neighbouring genes, with the B topology (B group) being the most common cassette structure (Fig. 5). In our work, the B group contained 26 luxRI cassettes distributed in 26 genomes (Fig. 5). Both S. pseudonitzschiae F5 and Phaeobacter sp. F10 possess one cassette that belongs to this group, while a unique luxRI cassette only occurs in S. pseudonitzschiae F5. Interestingly, both S. pseudonitzschiae F5 and Phaeobacter sp. F10 strains produced the same three AHLs molecules (Fig. 3 and Supporting Information Fig. S5), suggesting these molecules must be a byproduct of a homologous luxI, common to both S. pseudonitzschiae F5 and Phaeobacter sp. F10. This finding is consistent with previous studies showing that a single luxI‐type gene is responsible for biosynthesizing more than one AHL molecule (Ortori et al., 2007; Hansen et al., 2015). LuxI proteins use S‐adenosylmethionine (SAM) and acylated acyl‐carrier protein (ACP) to catalyse the acylation and lactonization of AHL molecules (Churchill and Chen, 2011). Potentially, the luxI homologue common to S. pseudonitzschiae F5 and Phaeobacter sp. F10 does not discriminate well between different substrates and can accept acyl chains of varying lengths, enabling it to biosynthesize three AHLs. This lack of specificity could allow different AHL molecules produced by a single protein to bind different transcriptional regulators and thus regulate different functions. By regulating the availability of substrates needed to make each AHL, different AHLs may be produced depending on the environmental condition while continuously expressing the same AHL synthase (Yates et al., 2002). For example, different temperatures have been shown to influence the types and concentrations of seven different AHLs produced by one luxI in the fish pathogen Aliivibrio salmonicida (Hansen et al., 2015). The diversity of substrates and selectivity levels of different luxI homologues also influence the proportions of various AHLs produced by roseobacters (Ziesche et al., 2018). S. pseudonitzschiae F5 and Phaeobacter sp. F10 were isolated from the same diatom, indicating horizontal gene transfer may be responsible for their identical production of AHLs. The second luxI gene in S. pseudonitzschiae F5 likely produces one or more other AHL molecules that we were not able to characterize due to the limitation of AHL mass spectrometry standards. As mentioned before, S. pseudonitzschiae F5 displayed a dendritic motility phenotype, which is absent in most roseobacters (Bartling et al., 2018). It is not clear what ecological advantage dendritic motility have on bacteria like S. pseudonitzschiae. Interestingly, two additional S. pseudonitzschiae strains displayed this phenotype previously (Bartling et al., 2018), indicating it is common in this species for unknown reasons.
Conclusions
Here, we have shown that two diatom symbionts use AHL molecules to inhibit their motility and enhance biofilm formation, processes that likely control their ability to attach to diatom TEP and thus colonize the phycosphere. In contrast, an opportunist bacterium was incapable of attaching to diatom TEP and was found to lack the ability to synthesize AHL molecules. The ability of symbionts to switch their lifestyle from motile bacteria entering the phycosphere to permanent residents of this microenvironment is essential to marine bacteria that rely on phycosphere exudates to survive. This ‘swim‐or‐stick’ switch appears also to be partially controlled by diatoms that can make quorum sensing mimics, such as rosmarinic acid. Significant work is needed to delineate the importance of bacterial intraspecies signalling vs eukaryotic host interference in bacterial communication.
Experimental procedures
Diatom growth
Asterionellopsis glacialis strain A3 was deposited for this work in the National Center for Marine Algae and Microbiota (NCMA) under the accession number CCMP3542. Axenic A. glacialis strain A3 (CCMP3542) was generated as described previously (Behringer et al., 2018). All diatom cultures were grown at 22°C in a 12:12 h light/dark diurnal cycle (125 μE m−2 s−1) in semi‐continuous batch cultures (Brand et al., 1981). Since in vivo fluorescence linearly correlates with cell numbers during exponential growth in batch cultures (Wood et al., 2005), diatom growth was monitored by measuring in vivo fluorescence of chlorophyll a (relative fluorescence units, RFU) using a 10‐AU fluorimeter (Turner Designs, San Jose, CA, USA). Specific growth rates (μ) of diatoms were calculated from the linear regression of the natural log of RFU versus time during the exponential growth phase of cultures. The standard deviation of μ was calculated using values from biological replicates (n = 5 unless otherwise indicated) over the exponential growth period. The relationship between cell density and RFU of A. glacialis strain A3 can be formulated with the following equation: y = 14.958 × − 0.519, where y is cells/μL and x is the RFU value, calculated from the regression line of the linear portion of the growth curve. The adjusted R 2 value for this curve is 0.98 (P < 0.001).
Bacterial isolation, identification and phylogenetic analysis
To assess whether specific bacteria attach to A. glacialis strain A3, we cultivated bacteria from the xenic A. glacialis strain A3 culture as described previously (Shibl et al., 2020). Briefly, bacteria were isolated from the diatom at early stationary phase by serially diluting 0.5 ml 1000 times in sterile f/2 media (Guillard, 1975) and subsequently spreading 200 μl onto (i) marine agar plates, (ii) plates containing per litre of seawater: 15 g agar and 2 g carbon source (sodium succinate or glucose) and (iii) sterilized A. glacialis strain A3 culture suspension with 1.5% agar. Plates were incubated at 25°C in the dark for 3–7 days and single colonies with unique morphologies were re‐streaked three times to eliminate cross‐contamination before storage at −80°C in 15% glycerol stocks.
Bacterial isolates were identified using direct PCR (Hofmann and Brian, 1991). Isolates were incubated in 3 ml marine broth at 25°C in the dark until optical density (600 nm) reached ~1.0; subsequently, 2 μl of each bacterium was used as a DNA template in PCR. The 16S rRNA gene from all bacteria was amplified by universal primers (27F, 1492R) as previously described (Amin et al., 2015). PCR products were purified by the Wizard PCR purification kit (Promega) and sequenced using Sanger sequencing (Apical Scientific, Malaysia). Sequences were aligned with 16S rRNA gene sequences from GenBank using ClustalW Multiple Alignment in BioEdit 7.0.9.0. A neighbour‐joining (NJ) phylogenetic tree was constructed with BIONJ (Gascuel, 1997) using Kimura's two‐parameter model. Another tree was constructed using the maximum likelihood (ML) methods using PhyML 3.0 (Guindon et al., 2010) with GTR + R + I substitution model as determined by SMS (Lefort et al., 2017). The final 16S rRNA gene phylogenetic consensus tree was generated and edited using FigTree 1.4.2 (Rambaut, 2014). 16S rRNA genes of S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 were compared with the microbial community of A. glacialis recovered after 20 days of isolation from the field (Behringer et al., 2018) using the BLASTn tool in BLAST+ (Camacho et al., 2009). Average nucleotide identities (ANI) shared between the three isolates and their closely related species were calculated using the enveomics toolbox (http://enve-omics.ce.gatech.edu/ani/).
Co‐culture generation and growth
For co‐cultures, axenic A. glacialis strain A3 was inoculated from an acclimated, mid‐exponential phase growing culture into 25 ml sterile f/2 media to achieve an initial diatom cell density of ~4000 cells mL−1. Bacteria were grown in marine broth (ZoBell, 1941) in the dark overnight from a single colony at 26°C and shaking at 180 rpm. Cultures were centrifuged at 4000 rpm for 10 min followed by washing twice with sterile f/2 medium. Subsequently, cultures were used to inoculate axenic A. glacialis cultures at an initial bacterial density of ~1 × 104 cells mL−1. Bacterial counts in co‐cultures were quantified by staining 1 ml fresh bacterial cells with 1× SYBR Safe stain (Edvotek Corp. USA), incubating stained samples in the dark at room temperature for 1 hour and using a CyFlow Space flow cytometer (Partec, Münster, Germany). Specific growth rates of diatoms were calculated as described above. Specific growth rates (μ) of bacteria were calculated from the linear regression of the natural log of cell counts from 2 to 6 days of cultures. The standard deviation of μ was calculated using values from biological replicates (n = 3 unless otherwise indicated).
Bright‐field and fluorescence microscopy
To observe transparent exopolymeric particles (TEP) distribution and their attached bacteria, two staining steps were applied. Alcian blue was first used to stain TEP in diatom cultures. A 1‐ml culture in mid‐exponential phase was gently filtered by gravity onto 3‐μm 25 mm polycarbonate membrane filters (Whatman) to remove free‐living bacteria (~1 μm). Subsequently, 1 ml alcian blue in 0.06% glacial acetic acid (pH 2.5) was allowed to gently run down the tube wall onto the filter to stain samples for 10 min at room temperature (Long and Azam, 1996). Excess alcian blue was removed using a gentle wash with PBS buffer and gently filtered by gravity. Subsequently, filters that retained diatoms (~25 μm for a single cell), TEP particles and attached bacteria were fixed with Moviol‐Sybr Green I as described previously (Lunau et al., 2005). Filters were visualized using a DMI6000B epifluorescence microscope (Leica) with a DMC2900 colour brightfield camera (Leica). Bright‐field microscopy was used for TEP observation. Fluorescence microscopy was used for bacteria and diatom nucleic acid observations. L5 fluorescence filter was used for SYBR Green I staining of both bacterial and algal nucleic acids with an excitation wavelength range of 480–656 nm and an emission wavelength at 590 nm. Y5 fluorescence filter was used for algal chlorophyll autofluorescence with an excitation wavelength of 620 nm and an emission wavelength of 700 nm. Both bright‐field and fluorescence images were merged using the LAS X software (Leica Microsystems, Germany). To observe diatom chain length, aliquots from A. glacialis cultures were directly placed onto microscopy slides and were visualized using the DMI6000B microscope described before. The chain length of axenic, xenic A. glacialis strain A3 and co‐culture of the diatom with S. pseudonitzschiae F5 were quantified using an inverted microscope (Eclipse Ti‐U, Nikon, Japan).
Bacterial genome sequencing and assembly
Genomic DNA of S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 were extracted from pure bacterial cultures using an E.Z.N.A. Bacterial DNA Kit (Omega BIO‐TEK) according to the manufacturer's instructions. DNA yield and quality were measured and checked using a Qubit 3.0 Fluorometer (Invitrogen; Life Technologies) and gel electrophoresis.
Genome sequencing of all three bacterial genomes was conducted using the Illumina and PacBio platforms at Apical Scientific Sdn. Bhd (Malaysia) for S. pseudonitzschiae F5 and Phaeobacter sp. F10 and at Novogene (China) for A. macleodii F12. In brief, 100–1000 ng DNA was fragmented by acoustic disruption using a Covaris S220 system (Covaris, Woburn, MA) for Illumina sequencing. Subsequently, DNA libraries were built using a NEBNext® Ultra™ II DNA Library Prep Kit (NEB E7645S/L, New England BioLabs®), and quantification and quality control of generated libraries were performed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The libraries were then sequenced with TruSeq SBS Kit v4‐HS reagents on an Illumina HiSeq 2500 platform (Illumina, San Diego, CA). For PacBio sequencing, DNA was sheared to ~15–20 kb using a Megarupter (Diagenode) and sequenced on a PacBio RS II system (Pacific Biosciences, Menlo Park, CA).
High‐quality PacBio sub‐reads were assembled by aligning the Illumina paired‐end 150 bp reads with the Basic Local Alignment via Successive Refinement (BLASR) aligner, a tool that combines data from short read alignments with optimization methods from whole genome alignment (Chaisson and Tesler, 2012) and were trimmed using bbduk, aligned with bbmap, and the resulting sorted bam file and the Pacbio consensus reference were used as input to pilon (−genome genome.fasta, −bam input. bam) (Walker et al., 2014) for further error correction and polishing. The assemblies were further polished with arrow in Canu 1.7 with parameter corOutCoverage = 60 (Koren et al., 2017). The largest circular contig in each assembly represented the chromosome size for each bacterium. In addition, plasmid contigs were BLASTed against closely related genomes to confirm these were indeed plasmids. For S. pseudonitzschiae F5, we were not able to close the chromosome due to high number of repeats in the genome and thus exact chromosome size could not be determined (Table 1). The consensus reference was annotated by Prokka (Seemann, 2014) and RAST and checked for completeness by BUSCO (Simão et al., 2015). Circa 1.2.1 (http://omgenomics.com/circa/) was used to draw chromosomal circos plots and chord diagrams to compare the same gene locations in different genomes. All genome sequences and their annotations can be accessed at GenBank under the accession numbers WKFG01000000 and CP046140‐CP046144.
Mining publicly available Alteromonas genomes for autoinducer synthase genes was performed on MicroScope (https://mage.genoscope.cns.fr/microscope/home/index.php). Identification and classification of transporter proteins in the three bacterial genomes was done using 6097 membrane transport protein sequences downloaded from the Transporter Classification Database (TCDB) (Saier Jr et al., 2016). To this end, Gblast2 program (http://www.tcdb.org/labsoftware.php) with a cutoff e‐value < 1e−20 was used and sequences with alignment scores <100 were removed. The chemotaxis, flagellar protein, pili, exopolysaccharides and quorum sensing related gene identities were confirmed by BLASTp against the model Roseobacter group strain R. pomeroyi DSS‐3.
QS genes analysis
The luxR‐like and luxI‐like genes of S. pseudonitzschiae F5 and Phaeobacter sp. F10 were predicted by RAST and Prokka. Maximum likelihood phylogenetic trees of multiple copies of LuxI‐like, genetically linked LuxR‐like and their internal transcribed spacer (ITS) sequences from S. pseudonitzschiae F5, Phaeobacter sp. F10, and 30 publicly accessible Roseobacter group genomes were constructed using PhyML 3.0 (Guindon et al., 2010) with HKY85 substitution model, which was selected according to SMS (Smart Model Selection in PhyML) (Lefort et al., 2017). These 30 strains were chosen based on several criteria: (i) Strains must have published information about at least two of the following: AHLs molecules, motility or attachment lifestyles, (ii) all strains have whole genomes publicly available, and one of the following two criteria: (iii) Strains are phylogenetically related to S. pseudonitzschiae F5 or Phaeobacter sp. F10, or (iv) Strains are well studied as model roseobacters. The luxRI and ITS sequences of Pseudomonas aeruginosa PAO1 (RefSeq accession number NC_002516.2) were used as an outgroup. Phylogeny was tested with a fast likelihood‐based method aBayes (Anisimova et al., 2011) with 100 bootstraps.
AHLs extraction and identification
To extract AHL molecules, S. pseudonitzschiae F5 and Phaeobacter sp. F10 strains were re‐plated from glycerol stocks and subsequently single picked colonies were inoculated overnight in 25‐ml marine broth at 26°C in the dark with shaking at 180 rpm. Overnight cultures were subsequently transferred to 1 l sterile marine broth in triplicates and grown under the same conditions for 20 h. Finally, bacterial cells were removed by centrifugation (15 min, 7000 rpm) when the optical density (600 nm) reached ~1.0. To further remove residual bacteria, the supernatant was further filtered through 0.2‐μm polycarbonate membrane filters (Whatman, NJ, USA). Oasis HLB solid‐phase extraction (SPE) cartridges (3 cc, 540 mg) were activated according to the manufacturer's instructions and were subsequently used to remove organic molecules from the filtrates as previously described (Wang et al., 2017). Finally, extracts were eluted with 5 ml 0.1% (v/v) formic acid in methanol and dried using an evaporator (SpeedVac SC210A; Thermo Savant, Holbrook, NY, USA) and were stored at −20°C for subsequent UHPLC–MS/MS analysis. AHL standards (Supporting Information Table S3) were acquired from Cayman Chemicals and Sigma‐Aldrich to help identify AHLs from S. pseudonitzschiae F5 and Phaeobacter sp. F10.
AHLs were analysed using an Agilent 1290 HPLC system coupled to a Bruker Impact II Q‐ToF‐MS (Bruker, Germany). Metabolites were separated using a reversed‐phase separation method. In RP mode, medium‐polarity and non‐polar metabolites were separated using an Eclipse Plus C18 column (50 mm × 2.1 mm ID) (Agilent, USA). Chromatographic separation consisted of MilliQ‐H2O + 0.2% formic acid (buffer A), and acetonitrile + 0.2% formic acid (buffer C) at a flowrate of 0.4 ml. The initial mobile phase composition was 90% A and 5% C followed by a gradient to 100% C over 18 min. The column was maintained at 100% C for 2 min followed by cleaning with isopropanol for 3 min. The column was allowed to equilibrate for 4 min using the initial mobile phase composition. Detection was carried out in the positive ionization mode with the following parameters: mass range = 50–1300 m/z measured at 6 Hz, ESI source parameters: dry gas temperature = 220°C, dry gas flow = 10.0 l min−1, Nebulizer pressure = 2.2 bar, Capillary V = 3000 V, end plate Offset: 500 V; MS‐ToF tuning parameters: Funnel 1 RF = 150 Vpp, Funnel 2 RF = 200 Vpp, isCID Energy = 0 eV, Hexapole RF = 50 Vpp, Ion Energy = 4.0 eV, Low Mass = 90 m/z, Collision Energy = 7.0 eV, pre Pulse storage = 5 μs.
Data were processed and analysed with Metaboscape 3.0 (Bruker, Bremen). Processing was conducted with the T‐Rex3D algorithm with an intensity threshold of 500 and a minimum peak length of 10 spectra. Spectra were lock‐mass calibrated, and features were only created if detected in a minimum of three samples. The presence of a specific AHL was determined by comparing the high‐resolution parent ion mass, daughter ion masses and retention times of the purchased standards relative to bacterial extracts.
Bacterial motility and biofilm formation assays
Bacterial motility assay was performed using semisolid (0.25% w/v) marine agar plates supplemented with a final concentration of 2 μM of each AHL or QS inhibitor (QSI) as described previously (Zan et al., 2012). The QSIs 2(5H)‐furanone and (Z‐)‐4‐Bromo‐5‐(bromomethylene)‐2(5H)‐furanone (furanone C‐30) were purchased from Sigma‐Aldrich. S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 strains were grown in marine broth overnight and then were gently inoculated using a sterilized toothpick into the centre of the agar surface. Triplicate plates were incubated at 26°C for 3 days after which motility plates were observed using the Uvitec Cambridge Fire‐reader imaging system. The proportion of motile area (percent motility) was measured using ImageJ software (http://rsb.info.nih.gov/ij/). Bacterial motility is characterized by a circular swimming zone in the motility assay or by dendritic morphology, which is typical of swarming motility caused by a locally restricted movement on agar plates (Michael et al., 2016).
Bacterial biofilm formation was quantified for S. pseudonitzschiae F5, Phaeobacter sp. F10, and A. macleodii F12 strains using the crystal violet assay (O'Toole and Kolter, 1998). Briefly, bacteria were cultured in marine broth overnight and then diluted to ~1 × 105 cells mL−1. 100 μl aliquots were transferred to a 96‐well suspension culture plate (Greiner bio‐one, CELLSTAR, Monroe, NC, USA). A previously optimized final concentration of 10 μg ml−1 AHL or QSI were added to each well in triplicates (Ren et al., 2001; Zhu et al., 2019) while controls received no AHL or QSI addition. The plate was incubated at 26°C for 24 h and subsequently 25 μl of 0.1% crystal violet was added to each well and incubated at room temperature for 15 min. Wells were rinsed twice with 150 μl Milli‐Q water to remove free‐living bacteria and then dried at 60°C for 10 min. Finally, stained biomass was solubilized in 95% ethanol for 1 h and absorbance readings were measured using a plate reader (BioTek, Winooski, VT) at 600 nm. Unpaired t‐test was used to compare the significant differences of control and other variables in bacterial swarming and biofilm formation.
Supporting information
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.
Acknowledgements
The authors would like to thank the China Scholarship Council for the financial support of C.F. This research was partially carried out using the Core Technology Platform resources at New York University Abu Dhabi. This work was funded by an NYU Abu Dhabi grant (AD179) and a NOAA grant (NA19NOS4780183) to S.A.A.
References
- Abreu, P.C. , Rörig, L.R. , Garcia, V. , Odebrecht, C. , and Biddanda, B. (2003) Decoupling between bacteria and the surf‐zone diatom Asterionellopsis glacialis at Cassino Beach, Brazil. Aquat Microb Ecol 32: 219–228. [Google Scholar]
- Amato, A. , Sabatino, V. , Nylund, G.M. , Bergkvist, J. , Basu, S. , Andersson, M.X. , et al (2018) Grazer‐induced transcriptomic and metabolomic response of the chain‐forming diatom Skeletonema marinoi . ISME J 12: 1594–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin, S. , Hmelo, L. , Van Tol, H. , Durham, B. , Carlson, L. , Heal, K. , et al (2015) Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522: 98–101. [DOI] [PubMed] [Google Scholar]
- Amin, S.A. , Parker, M.S. , and Armbrust, E.V. (2012) Interactions between diatoms and bacteria. Microbiol Mol Biol Rev 76: 667–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova, M. , Gil, M. , Dufayard, J.‐F. , Dessimoz, C. , and Gascuel, O. (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood‐based approximation schemes. Syst Biol 60: 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes, L.C.M. , Ferreira, R.B. , Buckner, M.M. , and Finlay, B.B. (2010) Quorum sensing in bacterial virulence. Microbiology 156: 2271–2282. [DOI] [PubMed] [Google Scholar]
- Bagatini, I.L. , Eiler, A. , Bertilsson, S. , Klaveness, D. , Tessarolli, L.P. , and Vieira, A.A.H. (2014). Host‐specificity and dynamics in bacterial communities associated with bloom‐forming freshwater phytoplankton. PLoS ONE, 9(1): e85950 10.1371/journal.pone.0085950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar‐Zeev, E. , Berman‐Frank, I. , Girshevitz, O. , and Berman, T. (2012) Revised paradigm of aquatic biofilm formation facilitated by microgel transparent exopolymer particles. Proc Natl Acad Sci USA. 109: 9119–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardy, S.L. , Ng, S.Y. , and Jarrell, K.F. (2003) Prokaryotic motility structures. Microbiology 149: 295–304. [DOI] [PubMed] [Google Scholar]
- Bartling, P. , Vollmers, J. , and Petersen, J. (2018) The first world swimming championships of roseobacters‐phylogenomic insights into an exceptional motility phenotype. Syst Appl Microbiol 41: 544–554. [DOI] [PubMed] [Google Scholar]
- Bassler, B.L. (2002) Small talk: cell‐to‐cell communication in bacteria. Cell 109: 421–424. [DOI] [PubMed] [Google Scholar]
- Behringer, G. , Ochsenkühn, M.A. , Fei, C. , Fanning, J. , Koester, J.A. , and Amin, S.A. (2018) Bacterial communities of diatoms display strong conservation across strains and time. Front Microbiol 9: 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, W. , and Mitchell, R. (1972) Chemotactic and growth responses of marine bacteria to algal extracellular products. Biol Bull 143: 265–277. [Google Scholar]
- Bertrand, E.M. , McCrow, J.P. , Moustafa, A. , Zheng, H. , McQuaid, J.B. , Delmont, T.O. , et al (2015) Phytoplankton–bacterial interactions mediate micronutrient colimitation at the coastal Antarctic Sea ice edge. Proc Natl Acad Sci U S A 112: 9938–9943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidle, K.D. , and Azam, F. (2001) Bacterial control of silicon regeneration from diatom detritus: significance of bacterial ectohydrolases and species identity. Limnol Oceanogr 46: 1606–1623. [Google Scholar]
- Bramucci, A.R. , Labeeuw, L. , Orata, F.D. , Ryan, E.M. , Malmstrom, R.R. , and Case, R.J. (2018) The bacterial symbiont Phaeobacter inhibens shapes the life history of its algal host Emiliania huxleyi . Front Mar Sci 5: 188. [Google Scholar]
- Brand, L.E. , Guillard, R.R. , and Murphy, L.S. (1981) A method for the rapid and precise determination of acclimated phytoplankton reproduction rates. J Plankton Res 3: 193–201. [Google Scholar]
- Bruhn, J.B. , Gram, L. , and Belas, R. (2007) Production of antibacterial compounds and biofilm formation by Roseobacter species are influenced by culture conditions. Appl Environ Microbiol 73: 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan, A. , LeCleir, G.R. , Gulvik, C.A. , and González, J.M. (2014) Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12: 686–698. [DOI] [PubMed] [Google Scholar]
- Buchan, A. , Mitchell, A. , Cude, W.N. , and Campagna, S. (2016) Acyl‐homoserine lactone‐based quorum sensing in members of the marine bacterial Roseobacter clade: complex cell‐to‐cell communication controls multiple physiologies In Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. Hoboken, NJ: Wiley, pp. 225–233. [Google Scholar]
- Burkhardt, B.G. , Watkins‐Brandt, K.S. , Defforey, D. , Paytan, A. , and White, A.E. (2014) Remineralization of phytoplankton‐derived organic matter by natural populations of heterotrophic bacteria. Mar Chem 163: 1–9. [Google Scholar]
- Camacho, C. , Coulouris, G. , Avagyan, V. , Ma, N. , Papadopoulos, J. , Bealer, K. , and Madden, T.L. (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, C.C. (2018) Marine biofilms: a successful microbial strategy with economic implications. Front Mar Sci 5: 126. [Google Scholar]
- Case, R.J. , Labbate, M. , and Kjelleberg, S. (2008) AHL‐driven quorum‐sensing circuits: their frequency and function among the proteobacteria. ISME J 2: 345–349. [DOI] [PubMed] [Google Scholar]
- Chaisson, M.J. , and Tesler, G. (2012) Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, M.E. , and Chen, L. (2011) Structural basis of acyl‐homoserine lactone‐dependent signaling. Chem Rev 111: 68–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cude, W.N. , and Buchan, A. (2013) Acyl‐homoserine lactone‐based quorum sensing in the Roseobacter clade: complex cell‐to‐cell communication controls multiple physiologies. Front Microbiol 4: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusick, K.D. , Polson, S.W. , Duran, G. , and Hill, R.T. (2020) Multiple megaplasmids confer extremely high levels of metal tolerance in Alteromonas strains. Appl Environ Microbiol 86: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, H. , Li, T. , Chen, M. , and Huang, G. (2008) Cross‐ocean distribution of Rhodobacterales bacteria as primary surface colonizers in temperate coastal marine waters. Appl Environ Microbiol 74: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels, R. , Vanderleyden, J. , and Michiels, J. (2004) Quorum sensing and swarming migration in bacteria. FEMS Microbiol Rev 28: 261–289. [DOI] [PubMed] [Google Scholar]
- Diner, R.E. , Schwenck, S.M. , McCrow, J.P. , Zheng, H. , and Allen, A.E. (2016) Genetic manipulation of competition for nitrate between heterotrophic bacteria and diatoms. Front Microbiol 7: 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham, B.P. , Sharma, S. , Luo, H. , Smith, C.B. , Amin, S.A. , Bender, S.J. , et al (2015) Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci U S A 112: 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durham, B.P. , Boysen, A.K. , Carlson, L.T. , Groussman, R.D. , Heal, K.R. , Cain, K.R. , et al (2019) Sulfonate‐based networks between eukaryotic phytoplankton and heterotrophic bacteria in the surface ocean. Nat Microbiol 4: 1706–1715. [DOI] [PubMed] [Google Scholar]
- Field, C.B. , Behrenfeld, M.J. , Randerson, J.T. , and Falkowski, P. (1998) Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281: 237–240. [DOI] [PubMed] [Google Scholar]
- Franco, A.D.O.D.R. , They, N.H. , Canani, L.G.D.C. , Maggioni, R. , and Odebrecht, C. (2016) Asterionellopsis tropicalis (Bacillariophyceae): a new tropical species found in diatom accumulations. J Phycol 52: 888–895. [DOI] [PubMed] [Google Scholar]
- Gardiner, M. , Fernandes, N.D. , Nowakowski, D. , Raftery, M. , Kjelleberg, S. , Zhong, L. , et al (2015) VarR controls colonization and virulence in the marine macroalgal pathogen Nautella italica R11. Front Microbiol 6: 1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel, O. (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14: 685–695. [DOI] [PubMed] [Google Scholar]
- Geng, H. , and Belas, R. (2010) Molecular mechanisms underlying Roseobacter–phytoplankton symbioses. Curr Opin Biotechnol 21: 332–338. [DOI] [PubMed] [Google Scholar]
- Gram, L. , Grossart, H.‐P. , Schlingloff, A. , and Kiørboe, T. (2002) Possible quorum sensing in marine snow bacteria: production of acylated homoserine lactones by Roseobacter strains isolated from marine snow. Appl Environ Microbiol 68: 4111–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard, R.R. (1975) Culture of phytoplankton for feeding marine invertebrates In Culture of Marine Invertebrate Animals. New York, NY: Springer, pp. 29–60. [Google Scholar]
- Guindon, S. , Dufayard, J.‐F. , Lefort, V. , Anisimova, M. , Hordijk, W. , and Gascuel, O. (2010) New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hammer, B.K. , and Bassler, B.L. (2003) Quorum sensing controls biofilm formation in Vibrio cholerae . Mol Microbiol 50: 101–104. [DOI] [PubMed] [Google Scholar]
- Hansen, H. , Purohit, A.A. , Leiros, H.‐K.S. , Johansen, J.A. , Kellermann, S.J. , Bjelland, A.M. , and Willassen, N.P. (2015) The autoinducer synthases LuxI and AinS are responsible for temperature‐dependent AHL production in the fish pathogen Aliivibrio salmonicida . BMC Microbiol 15: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Z. , Wang, Q. , Hu, Y. , Liang, J. , Jiang, Y. , Ma, R. , et al (2012) Use of the quorum sensing inhibitor furanone C‐30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. Int J Antimicrob Agents 40: 30–35. [DOI] [PubMed] [Google Scholar]
- Hmelo, L.R. , Mincer, T.J. , and Van Mooy, B.A. (2011) Possible influence of bacterial quorum sensing on the hydrolysis of sinking particulate organic carbon in marine environments. Environ Microbiol Rep 3: 682–688. [DOI] [PubMed] [Google Scholar]
- Hofmann, M. , and Brian, D. (1991) Sequencing PCR DNA amplified directly from a bacterial colony. Biotechniques 11: 30–31. [PubMed] [Google Scholar]
- Hong, Z. , Lai, Q. , Luo, Q. , Jiang, S. , Zhu, R. , Liang, J. , and Gao, Y. (2015) Sulfitobacter pseudonitzschiae sp. nov., isolated from the toxic marine diatom Pseudo‐nitzschia multiseries . Int J Syst Evol Microbiol 65: 95–100. [DOI] [PubMed] [Google Scholar]
- Hou, H. , Zhu, Y. , Wang, Y. , Zhang, G. , and Hao, H. (2019) AHLs regulate biofilm formation and swimming motility of Hafnia alvei H4. Front Microbiol 10: 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudaiberdiev, S. , Choudhary, K.S. , Vera Alvarez, R. , Gelencsér, Z. , Ligeti, B. , Lamba, D. , and Pongor, S. (2015) Census of solo LuxR genes in prokaryotic genomes. Front Cell Infect Microbiol 5: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens, J.C. , Metzger, K. , Daniels, R. , Ptacek, D. , Verhoeven, T. , Habel, L.W. , et al (2007) Synthesis of N‐acyl homoserine lactone analogues reveals strong activators of SdiA, the Salmonella enterica serovar Typhimurium LuxR homologue. Appl Environ Microbiol 73: 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, K.K. (2004) What drives bacteria to produce a biofilm? FEMS Microbiol Lett 236: 163–173. [DOI] [PubMed] [Google Scholar]
- Kaczmarska, I. , Mather, L. , Luddington, I.A. , Muise, F. , and Ehrman, J.M. (2014) Cryptic diversity in a cosmopolitan diatom known as Asterionellopsis glacialis (Fragilariaceae): implications for ecology, biogeography, and taxonomy. Am J Bot 101: 267–286. [DOI] [PubMed] [Google Scholar]
- Karentz, D. , and Smayda, T.J. (1984) Temperature and seasonal occurrence patterns of 30 dominant phytoplankton species in Narragansett Bay over a 22‐year period (1959–1980). Mar Ecol Prog Ser 18: 277–293. [Google Scholar]
- Kazamia, E. , Czesnick, H. , Nguyen, T.T.V. , Croft, M.T. , Sherwood, E. , Sasso, S. , et al (2012) Mutualistic interactions between vitamin B12‐dependent algae and heterotrophic bacteria exhibit regulation. Environ Microbiol 14: 1466–1476. [DOI] [PubMed] [Google Scholar]
- Kim, J.H. , Park, J.H. , Song, Y.H. , and Chang, D.S. (1999) Isolation and characterization of the marine bacterium, Alteromonas sp. SR‐14 inhibiting the growth of diatom, Chaetoceros species. Korean J Fish Aquat Sci 32: 155–159. [Google Scholar]
- Kim, M. , Oh, H.‐S. , Park, S.‐C. , and Chun, J. (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64: 346–351. [DOI] [PubMed] [Google Scholar]
- Koch, H. , Dürwald, A. , Schweder, T. , Noriega‐Ortega, B. , Vidal‐Melgosa, S. , Hehemann, J.‐H. , et al (2019) Biphasic cellular adaptations and ecological implications of Alteromonas macleodii degrading a mixture of algal polysaccharides. ISME J 13: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogure, K. , Simidu, U. , and Taga, N. (1981) Bacterial attachment to phytoplankton in sea water. J Exp Mar Biol Ecol 56: 197–204. [Google Scholar]
- Koren, S. , Walenz, B.P. , Berlin, K. , Miller, J.R. , Bergman, N.H. , and Phillippy, A.M. (2017) Canu: scalable and accurate long‐read assembly via adaptive k‐mer weighting and repeat separation. Genome Res 27: 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner, H. (1970) Morphologie und Taxonomie der Diatomeen‐gattung Asterionella . Nova Hedwigia 20: 557–724. [Google Scholar]
- Labeeuw, L. , Khey, J. , Bramucci, A.R. , Atwal, H. , de la Mata, A.P. , Harynuk, J. , and Case, R.J. (2016) Indole‐3‐acetic acid is produced by Emiliania huxleyi coccolith‐bearing cells and triggers a physiological response in bald cells. Front Microbiol 7: 828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort, V. , Longueville, J.‐E. , and Gascuel, O. (2017) SMS: smart model selection in PhyML. Mol Biol Evol 34: 2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, K. (2001) Riddle of biofilm resistance. Antimicrob Agents Chemother 45: 999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, R.A. , and Azam, F. (1996) Abundant protein‐containing particles in the sea. Aquat Microb Ecol 10: 213–221. [Google Scholar]
- López‐Pérez, M. , Gonzaga, A. , Martin‐Cuadrado, A.‐B. , Onyshchenko, O. , Ghavidel, A. , Ghai, R. , and Rodriguez‐Valera, F. (2012) Genomes of surface isolates of Alteromonas macleodii: the life of a widespread marine opportunistic copiotroph. Sci Rep 2: 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunau, M. , Lemke, A. , Walther, K. , Martens‐Habbena, W. , and Simon, M. (2005) An improved method for counting bacteria from sediments and turbid environments by epifluorescence microscopy. Environ Microbiol 7: 961–968. [DOI] [PubMed] [Google Scholar]
- Majzoub, M.E. , Beyersmann, P.G. , Simon, M. , Thomas, T. , Brinkhoff, T. , and Egan, S. (2019) Phaeobacter inhibens controls bacterial community assembly on a marine diatom. FEMS Microbiol Ecol 95: fiz060. [DOI] [PubMed] [Google Scholar]
- Malviya, S. , Scalco, E. , Audic, S. , Vincent, F. , Veluchamy, A. , Poulain, J. , et al (2016) Insights into global diatom distribution and diversity in the world's ocean. Proc Natl Acad Sci U S A 113: E1516–E1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayali, X. , and Doucette, G.J. (2002) Microbial community interactions and population dynamics of an algicidal bacterium active against Karenia brevis (Dinophyceae). Harmful Algae 1: 277–293. [Google Scholar]
- Mayali, X. , and Azam, F. (2004) Algicidal bacteria in the sea and their impact on algal blooms 1. J Eukaryot Microbiol 51: 139–144. [DOI] [PubMed] [Google Scholar]
- Mayali, X. , Franks, P.J. , and Burton, R.S. (2011) Temporal attachment dynamics by distinct bacterial taxa during a dinoflagellate bloom. Aquat Microb Ecol 63: 111–122. [Google Scholar]
- Michael, V. , Frank, O. , Bartling, P. , Scheuner, C. , Goker, M. , Brinkmann, H. , and Petersen, J. (2016) Biofilm plasmids with a rhamnose operon are widely distributed determinants of the 'swim‐or‐stick’ lifestyle in roseobacters. ISME J 10: 2498–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, T.R. , and Belas, R. (2004) Dimethylsulfoniopropionate metabolism by Pfiesteria‐associated Roseobacter spp. Appl Environ Microbiol 70: 3383–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, T.R. , and Belas, R. (2006) Motility is involved in Silicibacter sp. TM1040 interaction with dinoflagellates. Environ Microbiol 8: 1648–1659. [DOI] [PubMed] [Google Scholar]
- Miller, T.R. , Hnilicka, K. , Dziedzic, A. , Desplats, P. , and Belas, R. (2004) Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol 70: 4692–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, N.K. , Chang, J. , and Zhao, P.X. (2014) Prediction of membrane transport proteins and their substrate specificities using primary sequence information. PLoS One 9: e100278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole, G.A. , and Kolter, R. (1998) Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28: 449–461. [DOI] [PubMed] [Google Scholar]
- Ortori, C.A. , Atkinson, S. , Chhabra, S.R. , Cámara, M. , Williams, P. , and Barrett, D.A. (2007) Comprehensive profiling of N‐acylhomoserine lactones produced by Yersinia pseudotuberculosis using liquid chromatography coupled to hybrid quadrupole–linear ion trap mass spectrometry. Anal Bioanal Chem 387: 497–511. [DOI] [PubMed] [Google Scholar]
- Pomeroy, L.R. (1974) The ocean's food web, a changing paradigm. Bioscience 24: 499–504. [Google Scholar]
- Ponnusamy, K. , Paul, D. , Kim, Y.S. , and Kweon, J.H. (2010) 2 (5H)‐Furanone: a prospective strategy for biofouling‐control in membrane biofilm bacteria by quorum sensing inhibition. Braz J Microbiol 41: 227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina, J.‐B. , Fernandez, V. , Lambert, B. , Stocker, R. , and Seymour, J.R. (2019) The role of microbial motility and chemotaxis in symbiosis. Nat Rev Microbiol 17: 284–294. [DOI] [PubMed] [Google Scholar]
- Rambaut, A. (2014) FigTree 1.4. 2 software. Institute of Evolutionary Biology, Univ Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/
- Ramos, J.B. , Schulz, K.G. , Brownlee, C. , Sett, S. , and Azevedo, E.B. (2014) Effects of increasing seawater carbon dioxide concentrations on chain formation of the diatom Asterionellopsis glacialis . PLoS One 9: e90749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, D. , Webb, J.S. , and Kjelleberg, S. (2006) Microbial colonization and competition on the marine alga Ulva australis . Appl Environ Microbiol 72: 5547–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen, T.B. , Manefield, M. , Andersen, J.B. , Eberl, L. , Anthoni, U. , Christophersen, C. , et al (2000) How Delisea pulchra furanones affect quorum sensing and swarming motility in Serratia liquefaciens MG1. Microbiology 146: 3237–3244. [DOI] [PubMed] [Google Scholar]
- Ren, D. , Sims, J.J. , and Wood, T.K. (2001) Inhibition of biofilm formation and swarming of Escherichia coli by (5Z)‐4‐bromo‐5‐(bromomethylene)‐3‐butyl‐2 (5H)‐furanone. Environ Microbiol 3: 731–736. [DOI] [PubMed] [Google Scholar]
- Saier, M.H., Jr. , Reddy, V.S. , Tsu, B.V. , Ahmed, M.S. , Li, C. , and Moreno‐Hagelsieb, G. (2016) The transporter classification database (TCDB): recent advances. Nucleic Acids Res 44: D372–D379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samo, T.J. , Kimbrel, J.A. , Nilson, D.J. , Pett‐Ridge, J. , Weber, P.K. , and Mayali, X. (2018) Attachment between heterotrophic bacteria and microalgae influences symbiotic microscale interactions. Environ Microbiol 20: 4385–4400. [DOI] [PubMed] [Google Scholar]
- Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30: 2068–2069. [DOI] [PubMed] [Google Scholar]
- Segev, E. , Wyche, T.P. , Kim, K.H. , Petersen, J. , Ellebrandt, C. , Vlamakis, H. , et al (2016) Dynamic metabolic exchange governs a marine algal‐bacterial interaction. Elife 5: e17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost, M.R. , Carr, G. , Kolter, R. , and Clardy, J. (2011) Roseobacticides: small molecule modulators of an algal‐bacterial symbiosis. J Am Chem Soc 133: 18343–18349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour, J.R. , Amin, S.A. , Raina, J.‐B. , and Stocker, R. (2017) Zooming in on the phycosphere: the ecological interface for phytoplankton–bacteria relationships. Nature Microbiol 2: 17065. [DOI] [PubMed] [Google Scholar]
- Shibl, A.A. , Isaac, A. , Ochsenkühn, M.A. , Cardenas, A. , Fei, C. , Behringer, G. et al. (2020) Diatom modulation of microbial consortia through use of two unique secondary metabolites. ( 10.1101/2020.06.11.144840). [DOI] [PMC free article] [PubMed]
- Simão, F.A. , Waterhouse, R.M. , Ioannidis, P. , Kriventseva, E.V. , and Zdobnov, E.M. (2015) BUSCO: assessing genome assembly and annotation completeness with single‐copy orthologs. Bioinformatics 31: 3210–3212. [DOI] [PubMed] [Google Scholar]
- Simon, N. , Cras, A.‐L. , Foulon, E. , and Lemée, R. (2009) Diversity and evolution of marine phytoplankton. C R Biol 332: 159–170. [DOI] [PubMed] [Google Scholar]
- Slightom, R.N. , and Buchan, A. (2009) Surface colonization by marine roseobacters: integrating genotype and phenotype. Appl Environ Microbiol 75: 6027–6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smriga, S. , Fernandez, V.I. , Mitchell, J.G. , and Stocker, R. (2016) Chemotaxis toward phytoplankton drives organic matter partitioning among marine bacteria. Proc Natl Acad Sci U S A 113: 1576–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen, S. , and Vanderleyden, J. (2011) Auxin and plant‐microbe interactions. Cold Spring Har Perspect Biol 3: a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen, S. , Vanderleyden, J. , and Remans, R. (2007) Indole‐3‐acetic acid in microbial and microorganism‐plant signaling. FEMS Microbiol Rev 31: 425–448. [DOI] [PubMed] [Google Scholar]
- Stocker, R. (2012) Marine microbes see a sea of gradients. Science 338: 628–633. [DOI] [PubMed] [Google Scholar]
- Su, Y. , Tang, K. , Liu, J. , Wang, Y. , Zheng, Y. , and Zhang, X.‐H. (2018) Quorum sensing system of Ruegeria mobilis Rm01 controls lipase and biofilm formation. Front Microbiol 9: 3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramoni, S. , and Venturi, V. (2009) LuxR‐family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155: 1377–1385. [DOI] [PubMed] [Google Scholar]
- Teeling, H. , Fuchs, B.M. , Becher, D. , Klockow, C. , Gardebrecht, A. , Bennke, C.M. , et al (2012) Substrate‐controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611. [DOI] [PubMed] [Google Scholar]
- Thole, S. , Kalhoefer, D. , Voget, S. , Berger, M. , Engelhardt, T. , Liesegang, H. , et al (2012) Phaeobacter gallaeciensis genomes from globally opposite locations reveal high similarity of adaptation to surface life. ISME J 6: 2229–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, D.C. (2014) Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Euro J Phycol 49: 20–46. [Google Scholar]
- Töpel, M. , Pinder, M.I. , Johansson, O.N. , Kourtchenko, O. , Clarke, A.K. , and Godhe, A. (2019) Complete genome sequence of novel Sulfitobacter pseudonitzschiae strain SMR1, isolated from a culture of the marine diatom Skeletonema marinoi . J Genom 7: 7–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mooy, B.A. , Hmelo, L.R. , Sofen, L.E. , Campagna, S.R. , May, A.L. , Dyhrman, S.T. , et al (2012) Quorum sensing control of phosphorus acquisition in Trichodesmium consortia . ISME J 6: 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner‐Döbler, I. , and Biebl, H. (2006) Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol 60: 255–280. [DOI] [PubMed] [Google Scholar]
- Walker, B.J. , Abeel, T. , Shea, T. , Priest, M. , Abouelliel, A. , Sakthikumar, S. , et al (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Ding, L. , Li, K. , Schmieder, W. , Geng, J. , Xu, K. , et al (2017) Development of an extraction method and LC–MS analysis for N‐acylated‐l‐homoserine lactones (AHLs) in wastewater treatment biofilms. J Chromatogr B 1041: 37–44. [DOI] [PubMed] [Google Scholar]
- Waters, C.M. , and Bassler, B.L. (2005) Quorum sensing: cell‐to‐cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346. [DOI] [PubMed] [Google Scholar]
- Windler, M. , Bova, D. , Kryvenda, A. , Straile, D. , Gruber, A. , and Kroth, P.G. (2014) Influence of bacteria on cell size development and morphology of cultivated diatoms. Phycol Res 62: 269–281. [Google Scholar]
- Wood, A.M. , Everroad, R. , and Wingard, L. (2005) Measuring growth rates in microalgal cultures. Algal Cult Tech 18: 269–288. [Google Scholar]
- Wuichet, K. , Alexander, R.P. , and Zhulin, I.B. (2007) Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis In Methods in Enzymology. Amsterdam, Netherlands: Elsevier, pp. 3–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y. , Martinez‐Yamout, M.A. , Dickerson, T.J. , Brogan, A.P. , Wright, P.E. , and Dyson, H.J. (2006) Structure of the Escherichia coli quorum sensing protein SdiA: activation of the folding switch by acyl homoserine lactones. J Mol Biol 355: 262–273. [DOI] [PubMed] [Google Scholar]
- Yates, E.A. , Philipp, B. , Buckley, C. , Atkinson, S. , Chhabra, S.R. , Sockett, R.E. , et al (2002) N‐acylhomoserine lactones undergo lactonolysis in a pH‐, temperature‐, and acyl chain length‐dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa . Infect Immun 70: 5635–5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan, J. , Cicirelli, E.M. , Mohamed, N.M. , Sibhatu, H. , Kroll, S. , Choi, O. , et al (2012) A complex LuxR‐LuxI type quorum sensing network in a roseobacterial marine sponge symbiont activates flagellar motility and inhibits biofilm formation. Mol Microbiol 85: 916–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziesche, L. , Rinkel, J. , Dickschat, J.S. , and Schulz, S. (2018) Acyl‐group specificity of AHL synthases involved in quorum‐sensing in Roseobacter group bacteria. Beilstein J Org Chem 14: 1309–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziesche, L. , Bruns, H. , Dogs, M. , Wolter, L. , Mann, F. , Wagner‐Döbler, I. , et al (2015) Homoserine lactones, methyl oligohydroxybutyrates, and other extracellular metabolites of macroalgae‐associated bacteria of the Roseobacter clade: identification and functions. Chembiochem 16: 2094–2107. [DOI] [PubMed] [Google Scholar]
- ZoBell, C.E. (1941) Studies on marine bacteria. I the cultural requirements of heterotrophic aerobes. J Mar Res 4: 42–75. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information.
Appendix S2. Supporting Information.


