Abstract
Accumulating evidence has suggested that long non-coding (lnc)RNAs are widely involved in the progression of multiple diseases, including chronic obstructive pulmonary disease (COPD). The aim of the present study was to explore the function and molecule mechanism of maternally expressed gene 3 (MEG3) in cigarette smoke extract (CSE)-treated 16HBE cells. Cell viability and apoptosis were evaluated using Cell Counting Kit-8 analysis and flow cytometry, respectively. Western blot analysis was carried out to determine the protein levels of Bcl-2, Bax and cleaved caspase-3. ELISA assays were utilized to measure the protein levels of IL-1β and IL-6 and TNF-α. Cytotoxicity was assessed using a lactate dehydrogenase release assay. The expression levels of MEG3 and microRNA (miR)-181a-2-3p were detected using reverse transcription-quantitative PCR. The interaction between miR-181a-2-3p and MEG3 was predicted using DIANA tools and verified by a dual-luciferase reporter assay and RNA Immunoprecipitation assay. MEG3 expression was enhanced while miR-181a-2-3p abundance was reduced in the serum of patients with COPD and CSE-treated 16HBE cells. MEG3-knockdown or miR-181a-2-3p-overexpression inhibited CSE-induced apoptosis, inflammation and cytotoxicity in 16HBE cells. Moreover, miR-181a-2-3p directly bind to MEG3 and its knockdown reversed the inhibitory effect of MEG3 interference on apoptosis, inflammation and cytotoxicity in CSE-treated 16HBE cells. Overall, MEG3-knockdown suppressed CSE-induced apoptosis, inflammation and cytotoxicity in 16HBE cells by upregulating miR-181a-2-3p, providing a promising therapeutic target for treatment of CSE-induced COPD.
Keywords: chronic obstructive pulmonary disease, maternally expressed gene 3, microR-181a-2-3p, cell viability, apoptosis, inflammation, cytotoxicity
Introduction
Chronic obstructive pulmonary disease (COPD) is one of the leading causes of death worldwide, and its prevalence and mortality may increase further in the coming decades due to smoking and exposure to noxious agents (1). COPD is characterized by partially irreversible airflow obstruction due to inflammation of the airways and destruction of lung parenchyma (2). Tobacco smoke is the main cause of COPD development and it can induce the release of inflammatory cytokines, such as IL-1β, IL-6 and TNFα (3–5). Currently, there is no effective and specific treatment for COPD. Therefore, it is important to improve our understand of the pathogenic mechanisms at the molecular level to develop new therapeutic approaches.
Long non-coding (lnc)RNAs are a class of endogenous cellular RNAs (>200 nucleotides) that lack protein-coding capacity and serve regulatory roles in various physiopathological processes (6). In previous years, lncRNAs have been identified to have essential roles in various types of disease, including COPD (7,8). For example, lncRNA taurine upregulated 1 promotes airway remodeling by inhibiting the miR-145-5p/dual specificity protein phosphatase 6 axis in CSE-induced COPD (9). Moreover, another previous study reported that lncRNA MIR155 host gene regulated M1/M2 macrophage polarization in the progression of COPD (10). Maternally expressed gene (MEG)3, located on human chromosome 14q32.3, is a maternally imprinted gene and plays important roles in numerous diseases, such as glioma, gastric cancer and Alzheimer's disease (11,12). In addition, a previous report demonstrated that MEG3 is overexpressed in COPD tissues (13). However, the precise molecular mechanisms underlying MEG3 function in COPD progression and development remains poorly understood.
Growing evidence has shown that lncRNAs can function as competing endogenous (ce)RNAs to modulate gene expression through sponging miRNA (14,15). miRNAs are small non-coding RNAs, which negatively regulate gene expression and modulate diverse physiological processes, such as differentiation, proliferation, apoptosis and metabolism (16). An increasing number of studies have shown that dysregulation of miRNAs is associated with multiple pathological conditions, including COPD (17,18). In particular, low expression of miR-181a-2-3p has been identified in the serum of patients with COPD (19). Nevertheless, to the best of our knowledge, there is no evidence to support the interaction between MEG3 and miR-181a-2-3p in COPD. Therefore, the function of miR-181a-2-3p in the pathogenesis of COPD needs to be investigated.
The present study used cigarette smoke extract (CSE)-treated 16HBE cells as an in vitro model of COPD to investigate the biological functions of MEG3 and miR-181a-2-3p in COPD progression. In addition, the molecular mechanisms of MEG3 and miR-181a-2-3p in CSE-stimulated 16HBE cells were investigated. The results of the present study may provide a promising avenue for treatment of COPD.
Materials and methods
Serum collection and RNA isolation
Blood samples were obtained from 55 patients (median age, 58; age range: 38–81 years; male: 36, females: 19) with COPD and 47 healthy control individuals (median age: 53, age range: 36–73 years, male: 28, females; 19). These participants (no other diseases) did not receive chemotherapy, radiotherapy or other therapy prior to blood collection. All blood samples were obtained from Changning County Hospital of Traditional Chinese Medicine (Yibin, China). The participants provided written informed consent and the study was approved by The Ethics Committee of Changning County Hospital of Traditional Chinese Medicine (Yibin, China).
Blood samples from antecubital vein were collected with a special tube containing separation gel and clot activator, and then centrifuged at 1,700 × g for 10 min at room temperature. The supernatant was transferred to a new tube and centrifuged at 850 × g for 30 min at 4°C to discard the cell debris. Next, the final supernatant was transferred to labeled EP tubes and stored at −80°C until RNA extraction. The miRNeasy Serum/Plasma kit (Qiagen GmbH) was used to isolate total RNA from the serum.
Cell culture and transfection
Normal human bronchial epithelial cells (16HBE) were purchased from BeNa Culture Collection and maintained in DMEM (Hyclone; Cyvita) containing 10% (Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere with 5% CO2 at 37°C. For CSE treatment, 16HBE cells were exposed to various concentrations (0, 1, 2 and 4%) of CSE for different times (0, 12, 24 and 48 h).
The small interfering (si)RNA against MEG3 (si-MEG3) and siRNA scrambled control (si-NC; non-specific scrambled siRNA), MEG3-overexpression vector and empty vector (vector), miR-181a-2-3p mimic (miR-181a-2-3p) and its negative control (miR-NC), miR-181a-2-3p inhibitor (anti-miR-181a-2-3p) and its negative control (anti-miR-NC) were provided by Shanghai GenePharma Co., Ltd. The sequences were as follows: si-MEG3 (5′-GGGCTTCTGGAATGAGCAT-3′); si-NC (5′-UUCUCCGAACGUGUCACGUTT-3′); miR-181a-2-3p mimic (5′-ACCACUGACCGUUGACUGUACC-3′); miR-NC (5′-ACUCUAUCUGCACGCUGACUU-3′); miR-181a-2-3p inhibitor (5′-GGUACAGUCAACGGUCAGUGGU-3′); anti-miR NC (5′-CAGUACUUUUGUGUAGUACAA-3′). 16HBE cells were seeded into seeded in six-well plates (5×105 cells/well) and then transfected with oligonucleotide (50 nM miRNA mimic/inhibitor and 20 nM siRNA) or plasmid (2 µg) using the Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) when cell confluence reached 60–70%. The cells were collected for subsequent experimentation following 24 h of transfection at 37°C. Non-transfected group was used as control group.
Preparation of CSE
The preparation of CSE was performed as previously described (20). Briefly, the smoke from 10 cigarettes (China Tobacco Hunan Industrial Co, Ltd.) was bubbled through 25 ml of phosphate-buffered saline (PBS). The suspension was adjusted to pH 7.2–7.4 and filtered using a cellulose membrane (0.22 µm) to remove the bacteria. This solution was regarded as 100% CSE and diluted with PBS to obtain concentrations of 0, 1, 2 and 4%.
Cell viability assay
A Cell Counting Kit (CCK)-8 (Sangon Biotech Co., Ltd.) was utilized to evaluate cell viability. In brief, 16HBE cells (100 µl) were seeded in 96-well plates overnight. After treatment or/and transfection, CCK-8 (10 µl) reagent was added to per well and then incubated for another 3 h. Lastly, optical density (OD) value was examined under a microplate reader (Bio-Rad Laboratories, Inc.) at 450 nm.
Apoptosis assay
An apoptosis assay was conducted using an Annexin V-FITC/PI apoptosis detection kit (Sangon Biotech Co., Ltd.) to determine the apoptosis. After treatment or/and transfection, 16HBE cells were collected, centrifuged at 1,000 × g for 5 min 4°C, washed three times with PBS and resuspended in binding buffer (300 µl). Subsequently, cells were double-stained with Annexin V-FITC and PI for 20 min in the dark at room temperature. Lastly, the apoptotic rate was then analyzed using flow cytometry (Guava easyCyte HT; Luminex Corporation). The data were analyzed using GuavaSoft 3.2 software (Luminex Corporation).
Western blot assay
After treatment or/and transfection, 16HBE cells were lysed in RIPA lysis buffer (Beyotime Institute of Biotechnology) containing protease inhibitors to obtain total protein. After quantification by using bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology), an equal amount (40 µg per lane) of protein was resolved by 10–12% SDS-PAGE and then transferred onto PVDF membranes (0.2 µm; Beyotime Institute of Biotechnology). Next, membranes were blocked using 5% non-fat milk (Sangon Biotech Co., Ltd.) and then probed with specific primary antibody against Bcl-2 (1:1,000; cat. no. ab196495), Bax (1:500; cat. no. ab53154), cleaved caspase-3 (1:500; cat. no. ab49822) and GAPDH (1:3,000, cat. no. ab37168) (all Abcam) for 12 h at 4°C. Subsequently, all membranes were incubated in HRP-conjugated anti-rabbit IgG (1:4,000; cat. no. D110058; Sangon Biotech Co., Ltd.). Finally, all protein bands were observed using the ECL system (EMD Millipore). The protein levels were normalized by GAPDH, and ImageJ software version 1.50d software (National Institutes of Health) was used to evaluate the bands density.
ELISA assay
After treatment or/and transfection, the concentrations of IL-1β, IL-6 and TNF-α were detected using the following human ELISA kits: IL-1β (cat. no. E-EL-H0149c), IL-6 (cat. no. E-EL-H0102c) and TNF-α (cat. no. E-EL-H0109c (all Elabscience, Inc.). The data were presented in terms of pg per ml.
Lactate dehydrogenase (LDH) release assay
After treatment or/and transfection, the level of LDH released into cultured medium was measured using a LDH Cytotoxicity Assay kit (Beyotime Institute of Biotechnology). The results were presented as the percent of total LDH content.
RNA extraction and reverse transcription quantitative (RT-q)PCR
Referring to instruction of manufacturers, total RNA from 16HBE cells was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For detecting MEG3 expression, the first strand of cDNA was synthesized using a Prime Script RT reagent kit (Takara Bio, Inc.). For miR-181a-2-3p expression detection, cDNA was synthesized using a TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The RT conditions were conducted as per the manufacturer's protocols. Then, qPCR was performed using the SYBR-Green Master Mix on 7500 Real-time PCR system (both Applied Biosystems; Thermo Fisher Scientific, Inc.) with the following thermocycling conditions: Pre-denaturation at 95°C for 15 sec, followed by 40 cycles of denaturation at 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 40 sec. The primers used for RT-qPCR were listed as below: MEG3, Forward: 5′-CAGGATGGCAAAGGATGAAG-3′ and reverse: 5′-GCAGGTGAACACAAGCAAAGA-3′); miR-181a-2-3p, forward: 5′-GCGCGACCACTGACCGTTGAC3-3′ and reverse: 5′-ATCCAGTGCAGGGTCCGAGG-3′); GAPDH, forward: 5′-CGCTCTCTGCTCCTCCTGTTC-3′ and reverse: 5′-ATCCGTTGACTCCGACCTTCAC-3′); U6, forward: 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse: 5′-CGCTTCACGAATTTGCGTGTCAT-3′). The expression levels of MEG3 and miR-181a-2-3p were calculated using the 2−ΔΔCq method (21) and normalized to GAPDH and U6, respectively.
Dual-luciferase reporter assay
Potential binding sites of MEG3 and miR-181a-2-3p were predicted using DIANA tools (http://diana.imis.athena-innovation.gr/DianaTools/index.php). The fragment of MEG3 containing the wild-type (WT) or mutant (MUT) binding sites of miR-181a-2-3p was amplified and inserted into the pmirGLO luciferase vector (Promega Corporation), namely WT-MEG3 or MUT-MEG3. The miR-181a-2-3p or miR-NC was co-transfected with WT-MEG3 or MUT-MEG3 into 16HBE cells using the Lipofectamine 3000 reagent for 48 h as aforementioned. Lastly, the luciferase activity was determined by dual-luciferase reporter assay system (Promega Corporation), followed by normalization to Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
To further verify the relationship between MEG3 and miR-181a-2-3p, a Magna RIP kit (cat. no. 17-700; EMD Millipore) was used for RIP assay. In brief, 16HBE cells were lysed in the RIP lysis buffer, and then cell lysates were incubated in RIP buffer containing magnetic beads (50 µl; cat. no. CS203178; EMD Millipore) conjugated with anti-argonaute 2 (Anti-Ago2; cat. no. ab32381; 1:2,000; Abcam) or immunoglobulin G (Anti-IgG; cat. no. ab109489; 1:5,000, Abcam). Input and normal IgG were used as controls. After that, immunoprecipitated RNAs were isolated by proteinase K (150 µl) at 55°C for 30 min to digest the protein. Lastly, purified RNAs were determined using RTq-PCR as aforementioned.
Statistical analysis
Data are presented as the mean ± SD from at least three independent experiments. The statistical differences between two groups or multiple (>2) groups were assessed using unpaired Student's t-test or one-way ANOVA followed by Tukey's post hoc test. Statistical analyses were performed using Graph Prism 6.0 software (GraphPad Software Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
CSE treatment represses cell viability and promotes apoptosis, inflammation and cytotoxicity in 16HBE cells
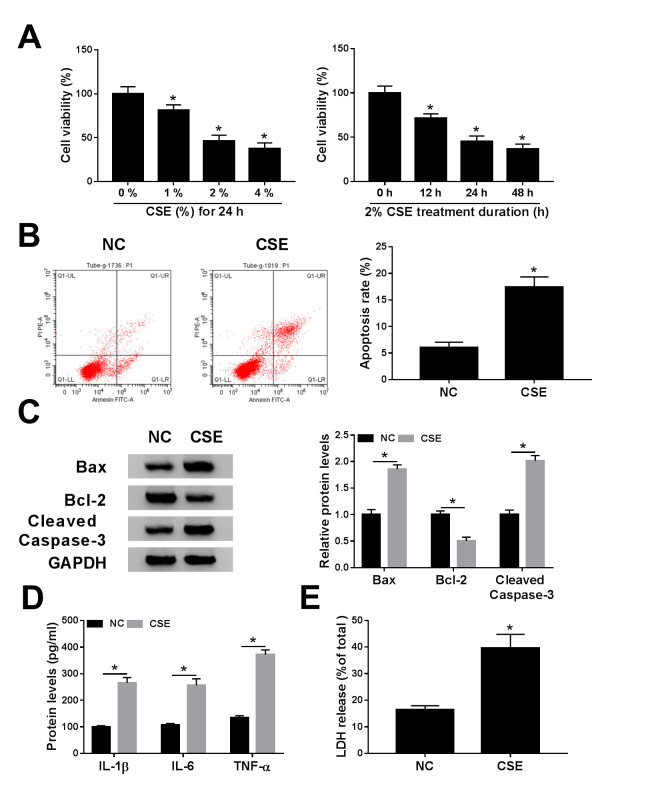
To explore the effect of CSE on COPD progression, 16HBE cells were exposed to CSE. The CCK-8 assay suggested that CSE treatment decreased viability of 16HBE cells in a dose- and time-dependent manner (Fig. 1A). The apoptosis assay revealed that apoptosis was enhanced in 16HBE cells exposed to CSE (Fig. 1B). Similarly, CSE exposure increased the protein expression of Bax (pro-apoptotic molecule) (22) and cleaved caspase-3 (a key executor in apoptotic process) (23), but decreased the protein abundance of Bcl-2 (anti-apoptotic molecule) (22) (Fig. 1C). Moreover, the levels of inflammatory cytokines, including IL-1β, IL-6 and TNF-α, were examined using ELISA analysis in CSE-induced 16HBE cells. The results reported that IL-1β, IL-6 and TNF-α levels were increased in 16HBE cells after treatment of CSE (Fig. 1D). LDH is a cytoplasmic enzyme that is released when the plasma membrane is destroyed, and can be measured in the supernatant as an indicator of cytotoxicity (24). Results showed that LDH release was increased in supernatants of 16HBE cells exposed to CSE (Fig. 1E). Overall, these data suggested that CSE might promote the progression of COPD.
Figure 1.
Effects of CSE on cell viability, apoptosis, inflammation and cytotoxicity in 16HBE cells. (A) Cell Counting Kit-8 assay was employed to determine the cell viability in 16HBE cells exposed to various concentrations (0, 1, 2 and 4%) of CSE for 0, 12, 24 or 48 h. (B-E) 16HBE cells were exposed to CSE (2% for 24 h). (B) Apoptosis rate was determined using flow cytometry analysis. (C) Western blotting was conducted to measure the protein levels of Bax, Bcl-2 and cleaved caspase-3. (D) Levels of IL-1β, IL-6 and TNF-α were examined using ELISA assays. (E) LDH release was measured by LDH release assay. *P<0.05 vs. 0% or NC. CSE, cigarette smoke extract; NC, negative control.
Increased MEG3 expression in serum of patients with COPD and CSE-treated 16HBE cells
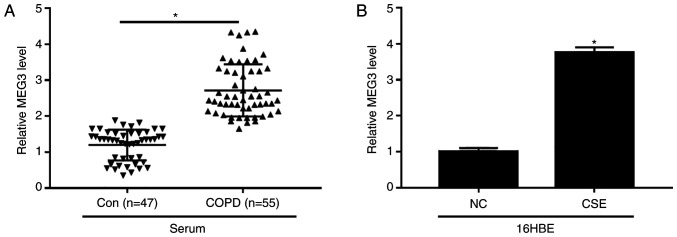
The expression of MEG3 in serum of patients with COPD and CSE-exposed 16HBE cells was examined. The data demonstrated that the level of MEG3 was evidently increased in serum of patients with COPD compared with control group (Fig. 2A). Additionally, CSE exposure also enhanced the expression of MEG3 in 16HBE cells (Fig. 2B). These results suggested that MEG3 might play an important role in COPD progression.
Figure 2.
Relative expression of MEG3 in serum of patients with COPD patients and CSE-induced 16HBE cells. (A) Expression of MEG3 was determined using RTq-PCR in the serum of patients with COPD or healthy controls. (B) MEG3 level was determined using the RT-qPCR in16HBE cells and 16HBE cells treated with CSE (2%, 24 h). *P<0.05 vs. respective control. MEG3, maternally expressed gene 3; COPD, chronic obstructive pulmonary disease; CSE, cigarette smoke extract; NC, negative control; Con, control; RT-q, reverse transcription-quantitative.
MEG3-knockdown attenuates CSE-induced apoptosis, inflammation and cytotoxicity in 16HBE cells
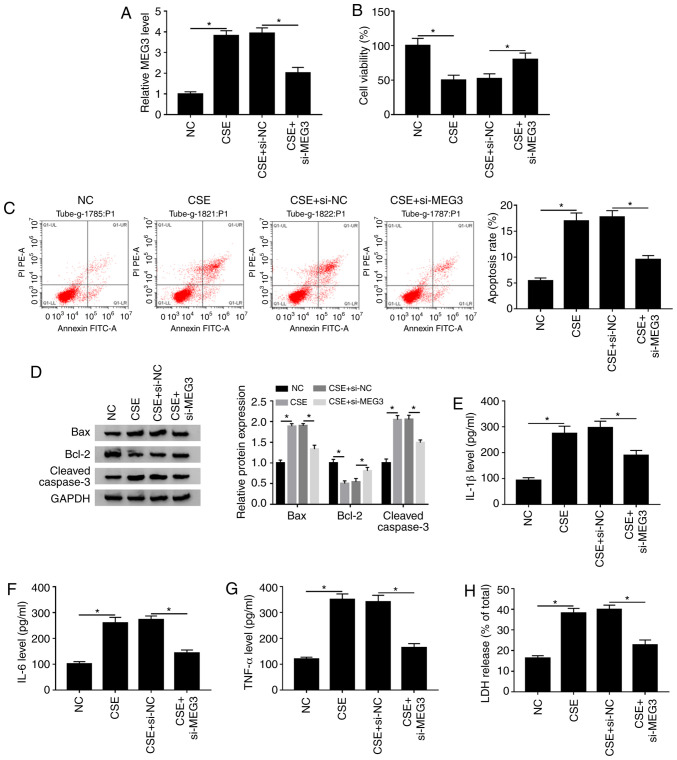
To investigate whether MEG3 was involved in the CSE-mediated the progression of COPD, si-NC or si-MEG3 was transfected into CSE-treated 16HBE cells. It was reported that the expression of MEG3 was decreased after transfection with si-MEG in 16HBE cells compared with the si-NC (Fig. S1A), suggesting that MEG3 was successfully knocked down in 16HBE cells. The RT-qPCR assay showed that knockdown of MEG3 obviously reversed increase of MEG3 expression caused by CSE in 16HBE cells (Fig. 3A). The CCK-8 assay indicated that MEG3 interference markedly abrogated the inhibitory effect of CSE treatment on the viability of 16HBE cells (Fig. 3B). Next, the impact of MEG3 on apoptosis of CSE-treated 16HBE cells was further explored. As shown in Fig. 3C and D, silencing MEG3 weakened the effect of CSE treatment on promotion of apoptotic rate, Bax and cleaved caspase-3 expression decreased and the level of Bcl-2 increased compared with the NC group. Moreover, knockdown of MEG3 also abated the promoting effects of CSE treatment on IL-1β, IL-6 and TNF-α levels as well as LDH release compared with the NC (Fig. 3E-H). These findings suggested that MEG3-downregulation overcame CSE-mediated decreased cell viability and decreased CSE-induced apoptosis, inflammation and cytotoxicity in 16HBE cells.
Figure 3.
MEG3-downregulation inhibits CSE-mediated apoptosis, inflammation and cytotoxicity in 16HBE cells. 16HBE cells were transfected with si-NC or si-MEG3 and then stimulated with CSE. (A) MEG3 expression was assessed using reverse transcription-quantitative PCR. (B) Cell viability was evaluated using Cell Counting Kit-8 analysis. (C) Flow cytometry was used to detect apoptosis. (D) Western blotting was used to measure the protein expression of Bax, Bcl-2 and cleaved caspase-3. Levels of (E) IL-1β, (F) IL-6 and (G) TNF-α were analyzed using ELISA kita. (H) LDH release assay was utilized to assess the cytotoxicity. *P<0.05 vs. respective control. MEG3, maternally expressed gene 3; CSE, cigarette smoke extract; NC, negative control; si, small interfering.
miR-181a-2-3p is a direct target of MEG3
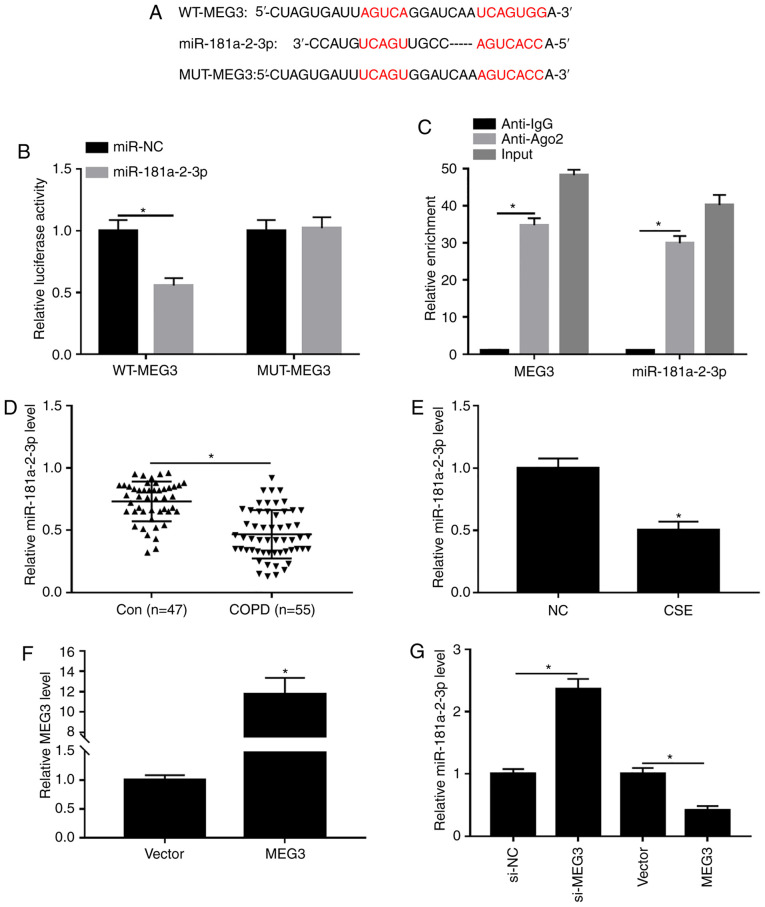
It is widely reported that lncRNAs can exert their functions through binding with their downstream miRNAs (25). Thus, the predicted target miRNAs of MEG3 were identified using DIANA tools. As presented in Fig. 4A, MEG3 contained potential binding sites of miR-181a-2-3p. Moreover, the transfection efficiency of miR-181a-2-3p and anti-miR-181a-2-3p was examined using RTq-PCR. The results showed that transfection of miR-181a-2-3p increased the expression of miR-181a-2-3p compared with miR-NC group, and transfection of anti-miR-181a-2-3p decreased the expression of miR-181a-2-3p (Fig. S1B), indicating that miR-181a-2-3p and anti-miR-181a-2-3p were successfully transfected into 16HBE cells. Subsequently, the interaction between MEG3 and miR-181a-2-3p was validated by dual-luciferase reporter and RIP assays. Results showed that miR-181a-2-3p-overexpression significantly decreased the luciferase activity of WT-MEG3 in16HBE cells, whereas miR-181a-2-3p upregulation had no significant impact on the luciferase activity of MUT-MEG3 (Fig. 4B). RIP analysis demonstrated that the enrichment of MEG3 and miR-181a-2-3p was greatly enhanced in Anti-Ago2 group compared with the Anti-IgG group (Fig. 4C). Next, miR-181a-2-3p expression in serum of patients with COPD and CSE-induced 16HBE cells was measured. As exhibited in Fig. 4D and E, miR-181a-2-3p abundance was significantly decreased in serum of patients with COPD and CSE-exposed 16HBE cells compared with respective controls. In addition, the effect of MEG3 on expression of miR-181a-2-3p in 16HBE cells was further explored. The transfection efficiency of MEG3 was assessed using RTq-PCR. As a result, MEG3 expression was significantly increased in 16HBE cells transfected with MEG3 compared with vector group, suggesting MEG3 had been successfully transfected into 16HBE cells (Fig. 4F). Furthermore, it was reported that knockdown of MEG3 enhanced miR-181a-2-3p abundance, whereas MEG3-overexpression decreased the levels of miR-181a-2-3p (Fig. 4G). Taken together, these data demonstrated that miR-181a-2-3p could directly bind to MEG3 and was negatively modulated by MEG3.
Figure 4.
Interaction between miR-181a-2-3p and MEG3. (A) Putative binding sites between miR-181a-2-3p and MEG3 were predicted using DIANA tools. (B) Effect of miR-181a-2-3p-overexpression on luciferase activities of WT-MEG3 and MUT-MEG3 was evaluated using a dual-luciferase luciferase reporter assay. (C) Enrichment of MEG3 or miR-181a-2-3p was measured using an RNA immunoprecipitation assay in 16HBE cells incubated with Anti-Ago2 or Anti-IgG. (D) Relative abundance of miR-181a-2-3p was detected using RT-qPCR in serum of patients with COPD or healthy controls. (E) Relative miR-181a-2-3p expression was analyzed using RT-qPCR in16HBE cells and 16HBE cells treated with CSE (2%, 24 h). (F) Expression of MEG3 was assessed by RT-qPCR assay in 16HBE cells transfected with vector or MEG3. (G) RT-qPCR was carried out to evaluate the level of miR-181a-2-3p in 16HBE cells transfected with si-NC, si-MEG3, vector or MEG3. *P<0.05 vs. respective control. MEG3, maternally expressed gene 3; CSE, cigarette smoke extract; NC, negative control; si, small interfering; WT, wild-type; MUT, mutant; RT-q, reverse transcription-quantitative; COPD, chronic obstructive pulmonary disease; vector, MEG3-overexpression vector negative control.
miR-181a-2-3p inhibits CSE-induced apoptosis, inflammation and cytotoxicity in 16HBE cells
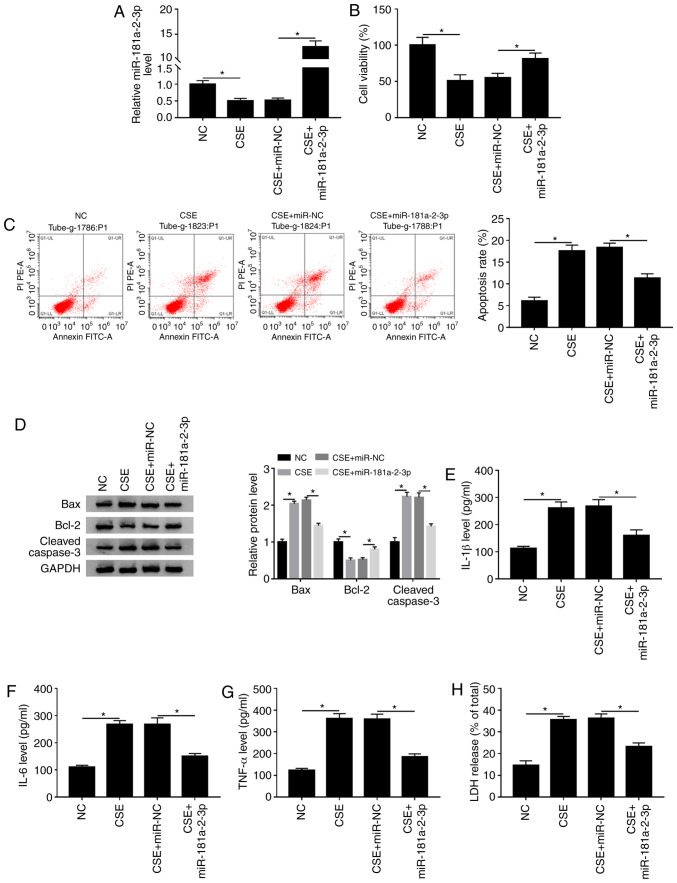
To determine the biological function of miR-181a-2-3p in CSE-treated 16HBE cells, the overexpression plasmid of miR-181a-2-3p was constructed. The data demonstrated that treatment with CSE led to a significant decrease in miR-181a-2-3p level compared with the NC, while this effect was abated by addition of miR-181a-2-3p (Fig. 5A). Subsequently, it was examined whether miR-181a-2-3p upregulation could play biological roles in cell viability, apoptosis, inflammation and cytotoxicity in CSE-exposed 16HBE cells. It was reported that miR-181a-2-3p upregulation reversed the effect of CSE on the inhibition of cell viability in 16HBE cells (Fig. 5B). Moreover, it was shown that miR-181a-2-3p upregulation decreased CSE-induced apoptosis (Fig. 5C). In addition, western blotting demonstrated that miR-181a-2-3p-overexpression decreased CSE-mediated promotion of Bax and cleaved caspase-3 expression and increased Bcl-2 expression (Fig. 5D). Furthermore, miR-181a-2-3p-overexpression also abated the promotive effects of CSE on inflammatory cytokine (IL-1β, IL-6 and TNF-α) levels and LDH release (Fig. 5E-H). Collectively, these findings demonstrated that miR-181a-2-3p repressed CSE-induced apoptosis, inflammation and cytotoxicity in16HBE cells.
Figure 5.
Overexpression of miR-181a-2-3p has similar effects to knockdown of MEG3 in CSE-exposed 16HBE cells. 16HBE cells were exposed to CSE after transfection with miR-NC or miR-181a-2-3p. (A) Relative miR-181a-2-3p level was examined using reverse transcription-quantitative-PCR. (B) Cell Counting Kit-8 analysis was utilized to assess cell viability. (C) Apoptosis was analyzed using flow cytometry analysis. (D) Western blotting was used to examine the protein expression of Bax, Bcl-2 and cleaved caspase-3. Levels of (E) IL-1β, (F) IL-6 and (G) TNF-α were analyzed using ELISA kits. (H) Cytotoxicity was evaluated by LDH release assay. *P<0.05. MEG3, maternally expressed gene 3; CSE, cigarette smoke extract; NC, negative control.
miR-181a-2-3p-knockdown partly abates the inhibitory effect of MEG3 interference on apoptosis, inflammation and cytotoxicity in CSE-treated 16HBE cells
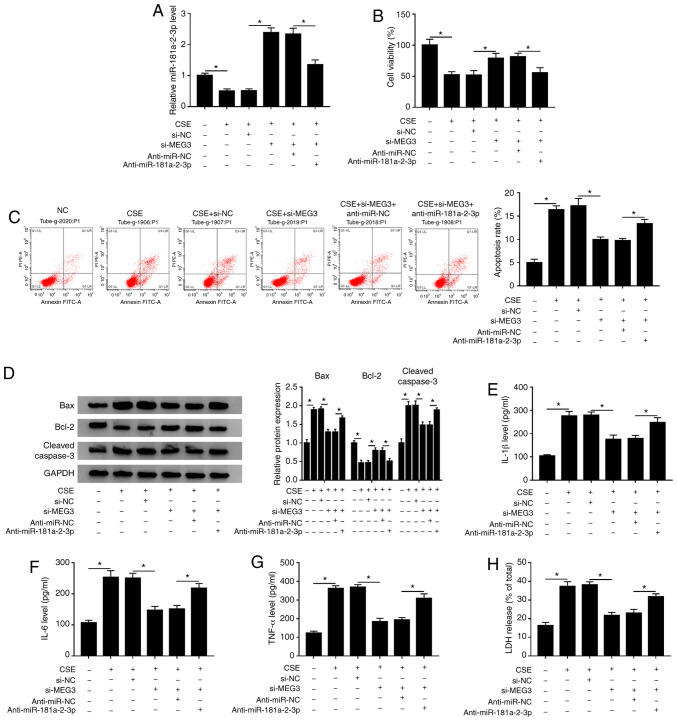
Based on the aforementioned findings, it was speculated that MEG3-knockdown inhibited CSE- induced apoptosis, inflammation and cytotoxicity via regulating miR-181a-2-3p. To validate this hypothesis, rescue experiments were performed in CSE-exposed 16HBE cells. The data showed that transfection of si-MEG3 reversed the inhibitory effect of CSE on miR-181a-2-3p expression, whereas co-transfection of anti-miR-181a-2-3p again weakened the promotive effect of MEG3-knockdown on miR-181a-2-3p level (Fig. 6A). Moreover, miR-181a-2-3p interference inhibited the promotion of cell viability mediated si-MEG3 in CSE-treated 16HBE cells (Fig. 6B). Furthermore, miR-181a-2-3p interference effectively abated the antiapoptotic effect induced by silencing MEG3 in CSE-treated 16HBE cells (Fig. 6C). In addition, miR-181a-2-3p downregulation reversed the impact of si-MEG3 on inhibition of Bax and cleaved caspase-3 protein expression, and promotion of Bcl-2 protein level in CSE-treated 16HBE cells (Fig. 6D). Besides, the suppressive effects of MEG3-knockdown on inflammatory cytokine (IL-1β, IL-6 and TNF-α) levels and LDH release were also abolished by miR-181a-2-3p-downregulation in CSE-exposed 16HBE cells (Fig. 6E-H). To sum up, these data indicated that MEG3-knockdown inhibited CSE-mediated apoptosis, inflammation and cytotoxicity by upregulating miR-181a-2-3p.
Figure 6.
miR-181a-2-3p downregulation reverses the suppressive effects of MEG3-knockdown on apoptosis, inflammation and cytotoxicity in CSE-treated 16HBE cells. 16HBE cells were transfected with si-NC, si-MEG3, si-MEG3 + anti-miR-NC, or si-MEG3 + anti-miR-181a-2-3p before treatment of CSE. (A) Relative miR-181a-2-3p abundance was analyzed using RT-qPCR. (B) Cell Counting Kit-8 analysis was applied to assess the cell viability. (C) Flow cytometry analysis was performed to measure the apoptotic rate. (D) Western blotting was conducted to examine the protein levels of Bax, Bcl-2 and cleaved caspase-3. Levels of (E) IL-1β, (F) IL-6 and (G) TNF-α were analyzed using ELISA kits. (H) LDH release assay was used to evaluate cytotoxicity. *P<0.05 vs. respective control. MEG3, maternally expressed gene 3; CSE, cigarette smoke extract; NC, negative control; si, small interfering; RT-q, reverse transcription-quantitative; COPD, chronic obstructive pulmonary disease.
Discussion
COPD is a progressive lung disease that is primarily caused by cigarette smoke-induced chronic inflammation (26). Increasing evidence has suggested that lncRNAs are commonly dysregulated in a range of diseases and serve as critical regulators of pathological processes, such as differentiation, immunity and inflammation (27,28). Therefore, research on lncRNA may help improve the diagnosis and treatment of COPD.
As a relatively well-studied lncRNA, MEG3 has been confirmed to widely participate in the progression of multiple human diseases. For instance, Lu et al (29) demonstrated that MEG3 represses non-small cell lung cancer cell proliferation and promotes apoptosis through increased p53 expression. Qiu et al (30) highlighted that MEG3-downregulation aggravated retinal vessel dysfunction in vivo through activating PI3K/Akt signaling pathway. Besides, several studies have demonstrated that the expression of MEG3 is enhanced in tissues of patients with COPD and might participate in the development of the disease (13,31,32). Consistent with these results, the present study reported that MEG3 expression was increased in serum of patients with COPD and CSE-stimulated 16HBE cells. Furthermore, it was observed that si-MEG3 reversed CSE-induced apoptosis, inflammation and cytotoxicity. These results combined with previous studies demonstrated that MEG3 serves a role in the progression of COPD.
Several studies have suggested that lncRNAs exert their functions via interacting with miRNA (33,34). For example, lncRNA X-inactive specific transcript promotes glioma tumorigenicity and angiogenesis through serving as a molecular sponge of miR-29 (35). lncRNA metallothionein 1J, pseudogene functions as a ceRNA to regulate F-box/WD repeat-containing protein 7 in gastric cancer by competitively binding to miR-92a-3p (36). MEG3 is also involved in a variety of diseases by serving as a sponge for miRNA. For example, MEG3-knockdown inhibits osteosarcoma cell viability, migration and invasion and promotes apoptosis through sponging miR-127 (37). Moreover, MEG3 suppresses the proliferation of chronic myeloid leukemia cells by acting as a sponge of miRNA-21 (38). Understanding the exact molecular mechanism underlying the biological effects of lncRNAs may contribute to the development of lncRNA-directed diagnosis and therapy for COPD. Using DIANA tools, the present study observed that MEG3 contained binding sites for miR-181a-2-3p. Subsequently, the prediction was confirmed using luciferase reporter and RIP assays. miR-181a-2-3p, a member of miR-181 family, has is abnormally expressed in numerous diseases, such as cervical cancer (39), child acute lymphoblastic leukemia (40) and gastric cancer (41). Some studies have shown that miR-181a serves as a novel marker for the inflammatory response (42,43). Besides, Kim et al (19) showed that miR-181a-2-3p expression is decreased in lung tissues and serum of patients with COPD, and its knockdown promotes inflammatory responses in cadmium-treated bronchial epithelial cells. The present study reported that miR-181a-2-3p abundance was decreased in the serum of patients with COPD and CSE-stimulated 16HBE cells. Some recent studies have shown that hemolysis, which occurs during blood collection or sample processing, can have significant impact on the levels of certain miRNAs in plasma and serum (44–46). However, hemolysis miRNA controls were not considered in the plasma analysis. In future studies, hemolysis miRNA controls (miR-23a and −451a) should be included in the plasma analysis. The function experiments demonstrated that overexpression of miR-181a-2-3p blocked the pro-apoptosis, pro-inflammation and pro-cytotoxicity effects induced by CSE in 16HBE cells, suggesting that miR-181a-2-3p might relieve these CSE-induced effects in COPD. Rescue experiments were performed to determine whether the function of MEG3 was regulated by miR-181a-2-3p. As expected, miR-181a-2-3p interference reversed the inhibitory effects of MEG3-knockdown on apoptosis, inflammation and cytotoxicity in CSE-stimulated 16HBE cells. As miRNAs exert their biological functions through modulating their downstream targets (47), future research should continue to investigate the downstream targets of miR-181a-2-3p to understand the underlying mechanism of its function in COPD.
In conclusion, the present study demonstrated that MEG3-knockdown inhibited CSE-induced apoptosis, inflammation and cytotoxicity in 16HBE cells by upregulating miR-181a-2-3p. The study reported a novel molecular mechanism of COPD progression and might help us to improve our understanding the pathogenesis of COPD. Furthermore, understanding this mechanism might accelerate the development of lncRNA-targeted diagnostic and therapeutic agents for COPD.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
SF and YR conceived and designed the study. SF, WZ and HZ performed the experiments. CW performed the statistical analysis and interpreted the data. YR drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by The Ethics Committee of Changning County Hospital of Traditional Chinese Medicine (Yibin, China). Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pauwels RA, Rabe KF. Burden and clinical features of chronic obstructive pulmonary disease (COPD) Lancet. 2004;364:613–620. doi: 10.1016/S0140-6736(04)16855-4. [DOI] [PubMed] [Google Scholar]
- 2.Nussbaumer-Ochsner Y, Rabe KF. Systemic manifestations of COPD. Chest. 2011;139:165–173. doi: 10.1378/chest.10-1252. [DOI] [PubMed] [Google Scholar]
- 3.Tanni SE, Pelegrino NR, Angeleli AY, Correa C, Godoy I. Smoking and tumor necrosis factor-alpha mediated systemic inflammation in COPD patients. J Inflamm (Lond) 2010;7:29. doi: 10.1186/1476-9255-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rusznak C, Mills PR, Devalia JL, Sapsford RJ, Davies RJ, Lozewicz S. Effect of cigarette smoke on the permeability and IL-1 β and sICAM-1 release from cultured human bronchial epithelial cells of never-smokers, smokers, and patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2000;23:530–536. doi: 10.1165/ajrcmb.23.4.3959. [DOI] [PubMed] [Google Scholar]
- 5.Wu H, Yang S, Wu X, Zhao J, Zhao J, Ning Q, Xu Y, Xie J. Interleukin-33/ST2 signaling promotes production of interleukin-6 and interleukin-8 in systemic inflammation in cigarette smoke-induced chronic obstructive pulmonary disease mice. Biochem Biophys Res Commun. 2014;450:110–116. doi: 10.1016/j.bbrc.2014.05.073. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz JE, Hongjae S, Spector DL. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Lalevée S, Feil R. Long noncoding RNAs in human disease: Emerging mechanisms and therapeutic strategies. Epigenomics. 2015;7:877–879. doi: 10.2217/epi.15.55. [DOI] [PubMed] [Google Scholar]
- 9.Gu W, Yuan Y, Wang L, Yang H, Li S, Tang Z, Li Q. Long non-coding RNA TUG1 promotes airway remodelling by suppressing the miR-145-5p/DUSP6 axis in cigarette smoke-induced COPD. J Cell Mol Med. 2019;23:7200–7209. doi: 10.1111/jcmm.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li N, Liu Y, Cai J. LncRNA MIR155HG regulates M1/M2 macrophage polarization in chronic obstructive pulmonary disease. Biomed Pharmacother. 2019;117:109015. doi: 10.1016/j.biopha.2019.109015. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Bian EB, He XJ, Ma CC, Zong G, Wang HL, Zhao B. Epigenetic repression of long non-coding RNA MEG3 mediated by DNMT1 represses the p53 pathway in gliomas. Int J Oncol. 2016;48:723–733. doi: 10.3892/ijo.2015.3285. [DOI] [PubMed] [Google Scholar]
- 12.Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song B, Ye L, Wu S, Jing Z. Long non-coding RNA MEG3 regulates CSE-induced apoptosis and inflammation via regulating miR-218 in 16HBE cells. Biochem Biophys Res Commun. 2020;521:368–374. doi: 10.1016/j.bbrc.2019.10.135. [DOI] [PubMed] [Google Scholar]
- 14.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, Downward J. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature. 2014;505:212–217. doi: 10.1038/nature12785. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Ardekani AM, Naeini MM. The role of microRNAs in human diseases. Avicenna J Med Biotechnol. 2010;2:161–179. [PMC free article] [PubMed] [Google Scholar]
- 17.Hobbs BD, Tantisira KG. MicroRNAs in COPD: Small molecules with big potential. Eur Respir J. 2019;53:1900515. doi: 10.1183/13993003.00515-2019. [DOI] [PubMed] [Google Scholar]
- 18.Diao X, Zhou J, Wang S, Ma X. Upregulation of miR-132 contributes to the pathophysiology of COPD via targeting SOCS5. Exp Mol Pathol. 2018;105:285–292. doi: 10.1016/j.yexmp.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Kim DY, Heo HR, Choi SS, Hong SH, Kim WJ. Role of miRNA-181a-2-3p in cadmium-induced inflammatory responses of human bronchial epithelial cells. J Thorac Dis. 2019;11:3055–3069. doi: 10.21037/jtd.2019.07.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter A, O'Donnell RA, Powell RM, Sanders MW, Holgate ST, Djukanovic R, Davies DE. Autocrine ligands for the epidermal growth factor receptor mediate interleukin-8 release from bronchial epithelial cells in response to cigarette smoke. Am J Respir Cell Mol Biol. 2002;27:85–90. doi: 10.1165/ajrcmb.27.1.4789. [DOI] [PubMed] [Google Scholar]
- 21.Danpure C. Lactate dehydrogenase and cell injury. Cell Biochem Funct. 1984;2:144–148. doi: 10.1002/cbf.290020305. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Edlich F. BCL-2 proteins and apoptosis: Recent insights and unknowns. Biochem Biophys Res Commun. 2018;500:26–34. doi: 10.1016/j.bbrc.2017.06.190. [DOI] [PubMed] [Google Scholar]
- 24.Porter AG, Jänicke RU. Emerging roles of caspase-3 in apoptosis. Cell Death Differ. 1999;6:99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 25.Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non-coding RNA SNHG15 promotes CDK14 expression via miR-486 to accelerate non-small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233:7164–7172. doi: 10.1002/jcp.26543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Zielinski J. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 27.Manel E. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 28.Yan B, Wang ZH, Liu JY, Tao ZF, Li XM, Qin J. Long noncoding RNAs: Versatile players in biologcial processes and human disorders. Epigenomics. 2014;6:375–379. doi: 10.2217/epi.14.29. [DOI] [PubMed] [Google Scholar]
- 29.Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP, Hou YY. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer. 2013;13:461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiu GZ, Tian W, Fu HT, Li CP, Liu B. Long noncoding RNA-MEG3 is involved in diabetes mellitus-related microvascular dysfunction. Biochem Biophys Res Commun. 2016;471:135–141. doi: 10.1016/j.bbrc.2016.01.164. [DOI] [PubMed] [Google Scholar]
- 31.Tang W, Shen Z, Guo J, Sun S. Screening of long non-coding RNA and TUG1 inhibits proliferation with TGF-β induction in patients with COPD. Int J Chron Obstruct Pulmon Dis. 2016;11:2951–2964. doi: 10.2147/COPD.S109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Zheng M, Pu J, Zhou Y, Hong W, Fu X, Peng Y, Zhou W, Pan H, Li B, Ran P. Identification of abnormally expressed lncRNAs induced by PM2.5 in human bronchial epithelial cells. Biosci Rep. 2018;38:BSR20171577. doi: 10.1042/BSR20171577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18:780–788. doi: 10.1093/bib/bbw053. [DOI] [PubMed] [Google Scholar]
- 34.Momen-Heravi F, Bala S. Emerging role of non-coding RNA in oral cancer. Cell Signal. 2018;42:134–143. doi: 10.1016/j.cellsig.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Cheng Z, Li Z, Ma K, Li X, Tian N, Duan J, Xiao X, Wang Y. Long non-coding RNA XIST promotes glioma tumorigenicity and angiogenesis by acting as a molecular sponge of miR-429. J Cancer. 2017;8:4106–4116. doi: 10.7150/jca.21024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, Zhao Q, Wu D, Gong W, Du M, et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17:87. doi: 10.1186/s12943-018-0829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Kong D. Knockdown of lncRNA MEG3 inhibits viability, migration, and invasion and promotes apoptosis by sponging miR-127 in osteosarcoma cell. J Cell Biochem. 2018;119:669–679. doi: 10.1002/jcb.26230. [DOI] [PubMed] [Google Scholar]
- 38.Li Z, Yang L, Liu X, Nie Z, Luo J. Long noncoding RNA MEG3 inhibits proliferation of chronic myeloid leukemia cells by sponging microRNA21. Biomed Pharmacother. 2018;104:181–192. doi: 10.1016/j.biopha.2018.05.047. [DOI] [PubMed] [Google Scholar]
- 39.Ravindresh C. let-7i-5p, miR-181a-2-3p and EGF/PI3K/SOX2 axis coordinate to maintain cancer stem cell population in cervical cancer. Sci Rep. 2018;8:7840. doi: 10.1038/s41598-018-26292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nabhan M, Louka ML, Khairy E, Tash F, Ali-Labib R, El-Habashy S. MicroRNA-181a and its target Smad 7 as potential biomarkers for tracking child acute lymphoblastic leukemia. Gene. 2017;628:253–258. doi: 10.1016/j.gene.2017.07.052. [DOI] [PubMed] [Google Scholar]
- 41.Gorur A, Balci Fidanci S, Dogruer Unal N, Ayaz L, Akbayir S, Yildirim Yaroglu H, Dirlik M, Serin MS, Tamer L. Determination of plasma microRNA for early detection of gastric cancer. Mol Biol Rep. 2013;40:2091–2096. doi: 10.1007/s11033-012-2267-7. [DOI] [PubMed] [Google Scholar]
- 42.Xie W, Li Z, Li M, Xu N, Zhang Y. miR-181a and inflammation: miRNA homeostasis response to inflammatory stimuli in vivo. Biochem Biophys Res Commun. 2013;430:647–652. doi: 10.1016/j.bbrc.2012.11.097. [DOI] [PubMed] [Google Scholar]
- 43.Weidong X, Mengnan L, Naihan X, Qing L, Nunu H, Jie H, Yaou Z, Tobias E. miR-181a regulates inflammation responses in monocytes and macrophages. PLoS One. 2013;8:e58639. doi: 10.1371/journal.pone.0058639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirschner MB, Edelman JJ, Kao SC, Vallely MP, van Zandwijk N, Reid G. The impact of hemolysis on cell-free microRNA biomarkers. Front Genet. 2013;4:94. doi: 10.3389/fgene.2013.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myklebust MP, Rosenlund B, Gjengstø P, Bercea BS, Karlsdottir Á, Brydøy M, Dahl O. Quantitative PCR measurement of miR-371a-3p and miR-372-p is influenced by hemolysis. Front Genet. 2019;10:463. doi: 10.3389/fgene.2019.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;13:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.