Abstract
Gastric cancer (GC) is one of the most common malignancies with a high worldwide incidence rate. The association between microRNAs (miRs) and malignancy has been widely studied in recent years. The aim of the present study was to assess the clinical value of miR-4636 in patients with GC and its effect on the proliferation, migration and invasion of GC cells. Reverse transcription-quantitative PCR was used to detect the expression of miR-4636. Receiver operating characteristics curve, Kaplan-Meier survival curve and Cox regression analyses were used to evaluate the diagnostic and prognostic value of miR-4636. Transwell migration and MTT assays were used to assess the regulatory effects of miR-4636 expression on the biological function of GC. The results demonstrated that the expression of miR-4636 was significantly downregulated in GC serum and tissue samples, as well as in GC cell lines. The aberrant miR-4636 expression was closely associated with lymph node metastasis and TNM stage, and had considerable diagnostic and prognostic significance in patients with GC. Cellular experiments revealed that the overexpression of miR-4636 inhibited GC cell proliferation, migration and invasion, while the knockdown of miR-4636 led to opposite effects on the biological function of GC. In summary, decreased miR-4636 expression may serve as a biomarker for the diagnosis and prognosis of GC. Furthermore, miR-4636 overexpression significantly inhibited GC cell proliferation, migration and invasion, indicating the potential of miR-4636 as a therapeutic target for GC treatment.
Keywords: microRNA-4636, gastric cancer, diagnosis, prognosis, proliferation, migration, invasion
Introduction
Gastric cancer (GC) is the third most common malignant tumor and a leading cause of cancer-related mortality worldwide according to statistics from 2008 (1). Certain risk factors have been identified to be responsible for the initiation of GC, including excessive salt intake and Helicobacter pylori infection (2). Although the therapeutic strategies of GC are advanced, including surgical resection, radiation and chemotherapy, the overall survival rate and the prognosis of patients with GC remain poor due to a significant proportion of patients presenting with advanced tumors at initial diagnosis (3). Therefore, it is crucial to identify specific diagnostic and prognostic biomarkers to improve GC treatment.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs approximately 18–25 nucleotides in length with important regulatory effects on gene expression at the post-transcriptional level (4,5). MiRNAs bind to the 3′-untranslated region of target mRNA, leading to the inhibition of target gene expression (6). Additionally, miRNAs have been reported to be involved in various cellular processes, including proliferation, migration and invasion (7). Accumulating studies have reported the association between miRNAs and human cancers, and the results have indicated that miRNAs function as tumor suppressors or oncogenes during tumorigenesis (8,9). There are certain abnormally expressed miRNAs in GC, such as miR-216b (10) and miR-519a (11), which are closely associated with the diagnosis, prognosis and progression of GC. miR-4636 has been reported to be downregulated in cervical cancer (12) and its decreased expression has been reported in GC tissues by Zhang et al (13). However, the functional role and clinical significance of miR-4636 in GC remain unknown.
To further improve the treatment of GC, the present study analyzed the expression levels of miR-4636 in patients with GC and in GC cell lines to evaluate the diagnostic and prognostic significance of miR-4636 and its biological function in GC progression. The results may provide a potential novel biomarker and therapeutic target for the diagnosis, prognosis and treatment of GC.
Materials and methods
Patients and tissue collection
A total of 112 patients (65 males and 47 females) who underwent surgery at Zibo Central Hospital (Zibo, China) between June 2009 and May 2013 were recruited. The mean age of the patients was 64.87±11.31 years (range, 45–85 years). The inclusion criteria were as follows: i) All the patients were pathologically diagnosed with GC; ii) none of the patients had received any therapy prior to surgery; and iii) had complete clinicopathological data. The exclusion criteria were as follows: i) Patients younger than 18 years or older than 80 years; ii) pregnant or lactating patients; and iii) patients that had autoimmune diseases or other malignancies. Furthermore, 60 healthy individuals with an average age of 65.23±10.96 years (range, 46–84 years) were recruited during the same time period. The healthy volunteers included 35 males and 25 females, and had no history of malignancy. Venous blood was obtained from all the participants and serum was isolated by centrifugation at 1,500 × g for 15 min at 4°C, that was then stored at −80°C until further use. Additionally, GC tissue samples and adjacent normal tissue samples, which were located 3 cm from the edge of the tumors, were collected from the patients and frozen with liquid nitrogen at −80°C for future use. All the patients received a 5-year follow-up survey and their survival information was recorded. Prior to sampling, all the patients and healthy volunteers provided written informed consent for clinical sample use and analysis, and the experimental methods were approved by the Ethics Committee of Zibo Central Hospital (approval no. ZCHh-090618).
Cell culture and transfection
The GC cell lines AGS, HGC27, HS746T and MKN45 and the gastric epithelial cell line GES-1 were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. The cells were cultured in RPMI-1640 medium (BioTek Corporation) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5% CO2.
GC cell lines AGS and MGN45 were subjected to cell transfection. Sequences for 50 nM miR-4636 mimics (5′-AACUCGUGUUCAAAGCCUUUAG-3′), 100 nM miR-4636 inhibitors (5′-CUAAAGGCUUUGAACACGAGUU-3′) or their respective negative controls (50 nM mimic NC, 5′-UUCUCCGAACGUGUCACGU-3′ and 100 nM inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′) were transfected into AGS and MGN45 cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C according to the manufacturer's protocol. After 48 h of cell transfection, the cells were used for subsequent experiments. The cells transfected with only transfection reagents were used as a mock group.
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from GC tissues and cell lines using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized from RNA using a PrimeScript RT reagent kit (Takara Bio, Inc.), according to the manufacturer's protocol. The expression levels of miR-4636 were detected by qPCR, which was conducted using a SYBR-Green I Master Mix kit (Invitrogen; Thermo Fisher Scientific, Inc.) on a 7500 Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling conditions were as follows: 95°C, 10 min for initial denaturation, 40 cycles of 95°C, 30 sec for denaturation, 58°C, 20 sec for annealing, 72°C, 30 sec for elongation, and 72°C, 10 min for final extension. U6 was used as the internal control and the final expression values of miR-4636 were quantified using the 2−ΔΔCq method (14). The sequences of primers used were as follows: miR-4636 forward, 5′-GCCGAGAACTCGTGTTCAA-3′ and reverse, 5′-CTCAACTGGTGTCGTGGA-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
Cell proliferation assay
At 48-h post-transfection, AGS and MKN45 cells (3×103 cells/well) were seeded into 96-well plates and the cell proliferation was analyzed using a MTT assay. The cell plates were retained in an incubator at 37°C for 3 days. A total of 10 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA) was added to each well every 24 h followed by incubation for a further 4 h. Following this, 150 µl DMSO was added in each well to dissolve the violet formazan crystals. Following incubation, the absorbance of the cultures was determined at a wavelength of 570 nm using a microplate reader.
Cell migration and invasion assay
The invasion and migration abilities of AGS and MKN45 cells were analyzed using Transwell chambers with 8 µm pore membranes (Corning, Inc.) precoated with Matrigel (for invasion assays) or without Matrigel (for migration assays). GC cells (3×105 cells/well) were seeded into the upper chamber with serum-free RPMI-1640 medium. The lower chambers contained medium supplemented with 10% FBS. Following 24 h of incubation at 37°C, the cells in the lower chambers were stained using 0.1% crystal violet for 10 min at room temperature, and were calculated under an inverted light microscope (Olympus Corporation).
Statistical analysis
The statistical analyses were performed using SPSS (version no. 21.0; IBM Corp.) and GraphPad Prism (version no. 7.0; GraphPad Software, Inc.) software. Differences between groups were analyzed using Student's t-test (paired t-test for tissue miR-4636 expression examination; unpaired t-test for serum miR-4636 expression between healthy controls and cancer patients) or one-way ANOVA followed by Tukey's post-hoc test. A χ2 test was used to assess the association between miR-4636 expression and the clinicopathological features of patients with GC. A receiver operating characteristics (ROC) curve was performed to evaluate the diagnostic value of serum miR-4636 expression. The survival curves for patients with GC were constructed using the Kaplan-Meier (KM) method and log-rank test. A multivariate Cox regression analysis was applied to evaluate the prognostic value of miR-4636. P<0.05 was considered to indicate a statistically significant difference.
Results
Downregulation of miR-4636 in patients with GC and GC cell lines
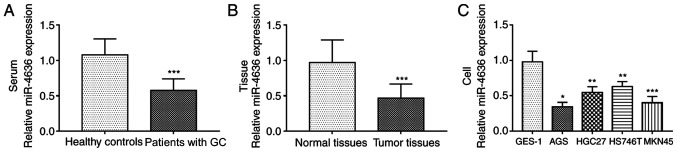
The serum expression levels of miR-4636 in patients with GC were significantly lower compared with healthy controls (P<0.001; Fig. 1A). Additionally, the expression of miR-4636 in GC tissues was significantly downregulated compared with normal tissues (P<0.001; Fig. 1B). The expression levels of miR-4636 in GC cell lines were significantly lower compared with GES-1 cells (all, P<0.05; Fig. 1C).
Figure 1.
Expression of miR-4636 in patients with GC and GC cell lines. (A) Expression of miR-4636 in serum samples collected from healthy individuals and patients with GC. ***P<0.001 vs. healthy controls. (B) Expression of miR-4636 in GC tissues and adjacent normal tissues. ***P<0.001 vs. normal tissues. (C) Expression of miR-4636 in the GC cell lines. *P<0.05, **P<0.01 and ***P<0.001 vs. GES-1 cells. miR, microRNA; GC, gastric cancer.
Association of miR-4636 with the clinicopathological characteristics of patients with GC
As presented in Table I, GC patients were divided into low and high miR-4636 expression groups based on the mean expression value (serum, 0.576; tissue, 0.470). The χ2 test results revealed that the expression of miR-4636 in both serum and tissue samples was associated with lymph node metastasis and TNM stage (both, P<0.05). No significant association was found between miR-4636 and age, sex, tumor size or differentiation (all, P>0.05).
Table I.
Association between miR-4636 and the clinical characteristics of patients with gastric cancer.
| miR-4636 in serum | miR-4636 in tissues | ||||||
|---|---|---|---|---|---|---|---|
| Characteristic | Total (n=112) | Low (n=60) | High (n=52) | P-value | Low (n=62) | High (n=50) | P-value |
| Age (years) | 0.821 | 0.734 | |||||
| <60 | 40 | 22 | 18 | 23 | 17 | ||
| ≥60 | 72 | 38 | 34 | 39 | 33 | ||
| Sex | 0.945 | 0.572 | |||||
| Female | 47 | 25 | 22 | 24 | 22 | ||
| Male | 65 | 35 | 30 | 38 | 28 | ||
| Tumor size (cm) | 0.143 | 0.068 | |||||
| <5 | 52 | 24 | 28 | 24 | 28 | ||
| ≥5 | 60 | 36 | 24 | 38 | 22 | ||
| Differentiation | 0.054 | 0.052 | |||||
| Well/moderate | 58 | 26 | 32 | 27 | 31 | ||
| Poor | 54 | 34 | 20 | 35 | 19 | ||
| Lymph node metastasis | 0.026 | 0.010a | |||||
| Negative | 52 | 22 | 30 | 22 | 30 | ||
| Positive | 60 | 38 | 22 | 40 | 20 | ||
| TNM stage | 0.017 | 0.019a | |||||
| I–II | 49 | 20 | 29 | 21 | 28 | ||
| III–IV | 63 | 40 | 23 | 42 | 22 | ||
P<0.05. miR, microRNA.
Clinical significance of miR-4636 in the diagnosis and prognosis of GC
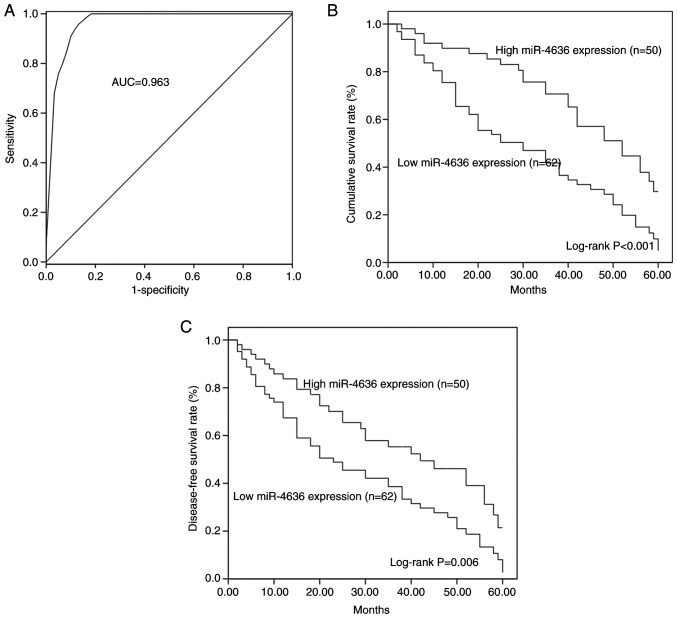
A ROC curve based on serum miR-4636 levels was constructed, which demonstrated that the area under the curve (AUC) was 0.963, indicating the diagnostic accuracy of serum miR-4636 in patients with GC (Fig. 2A). At a cut-off value of 0.855, the values for diagnosis sensitivity and specificity were 97.3 and 86.7%, respectively.
Figure 2.
Clinical significance of miR-4636 in the diagnosis and prognosis of GC. (A) A receiver operating characteristics curve based on serum miR-4636 levels of patients with GC (AUC, 0.963). (B) The Kaplan-Meier overall survival curves for patients with GC and different expression patterns of miR-4636 (log-rank, P<0.001). (C) The disease-free survival curves for patients with GC with high or low miR-4636 expression (log-rank, P=0.006). miR, microRNA; GC, gastric cancer; AUC, area under the curve.
By evaluating the 5-year follow-up data, the KM method was used to construct the overall survival and disease-free survival curves of patients with GC. Patients with GC and low miR-4636 expression had shorter survival times compared with patients with a high miR-4636 expression (log-rank, P<0.001; Fig. 2B). Furthermore, a low expression of miR-4636 was associated with poor disease-free survival in patients with GC (log-rank P=0.006; Fig. 2C). The multivariate Cox regression analysis revealed that aberrant miR-4636 expression (HR=3.522; 95% CI=2.016–6.154; P<0.001; Table II) and TNM stage (HR=1.800; 95% CI=1.093–2.966; P=0.021) were two independent prognostic factors for the overall survival rate in patients with GC.
Table II.
Cox regression analysis in patients with gastric cancer.
| Parameter | HR value | 95% CI | P-value |
|---|---|---|---|
| miR-4636 | 3.522 | 2.016–6.154 | <0.001 |
| Age | 1.681 | 0.997–2.829 | 0.053 |
| Sex | 1.397 | 0.880–2.217 | 0.156 |
| Tumor size | 1.377 | 0.855–2.217 | 0.189 |
| Differentiation | 1.149 | 0.717–1.842 | 0.564 |
| Lymph node metastasis | 1.527 | 0.927–2.516 | 0.097 |
| TNM stage | 1.800 | 1.093–2.966 | 0.021 |
miR, microRNA.
miR-4636 overexpression inhibits GC cell proliferation
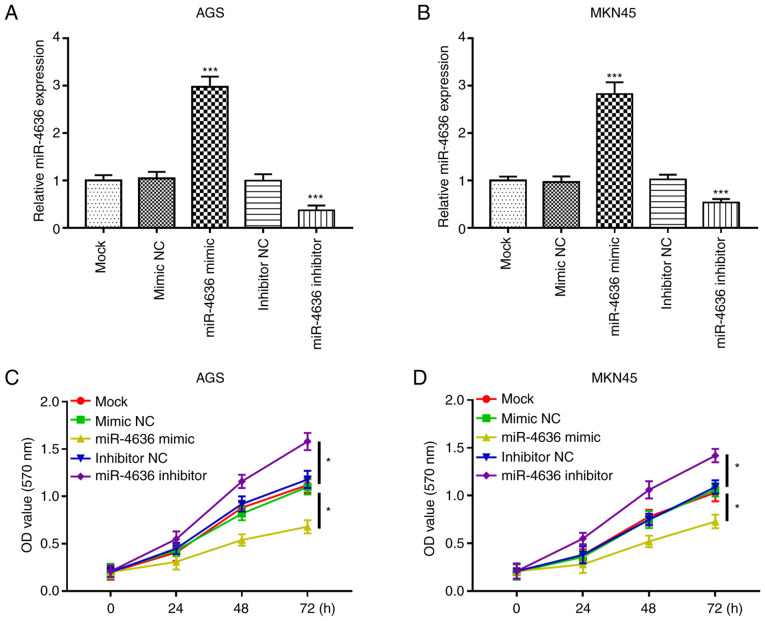
The functional role of miR-4636 in GC progression was further investigated. GC cell lines AGS and MKN45 were selected for the cell experiments due to their significantly lower expression of miR-4636 expression compared with the normal cell line GES-1. The expression of mi-4636 was upregulated by miR-4636 mimics and downregulated by miR-4636 inhibitors compared with the mock group in the AGS and MKN45 cell lines (all, P<0.001; Fig. 3A and B). Furthermore, the MTT assay results demonstrated that cell proliferation in the AGS and MKN45 cell lines was significantly inhibited by the overexpression of miR-4636 and promoted by the knockdown of miR-4636 compared with the mock group (all, P<0.05; Fig. 3C and D).
Figure 3.
Dysregulation of miR-4636 regulates cell proliferation of AGS and MKN45 cells. The expression of miR-4636 in (A) AGS and (B) MKN45 cells was upregulated by miR-4636 mimics and downregulated by miR-4636 inhibitors. Proliferation in (C) AGS and (D) MKN45 cells was inhibited by the overexpression of miR-4636 and promoted by the knockdown of miR-4636. *P<0.05 and ***P<0.001 vs. the mock group. miR, microRNA; NC, negative control.
Effects of miR-4636 on GC cell migration and invasion
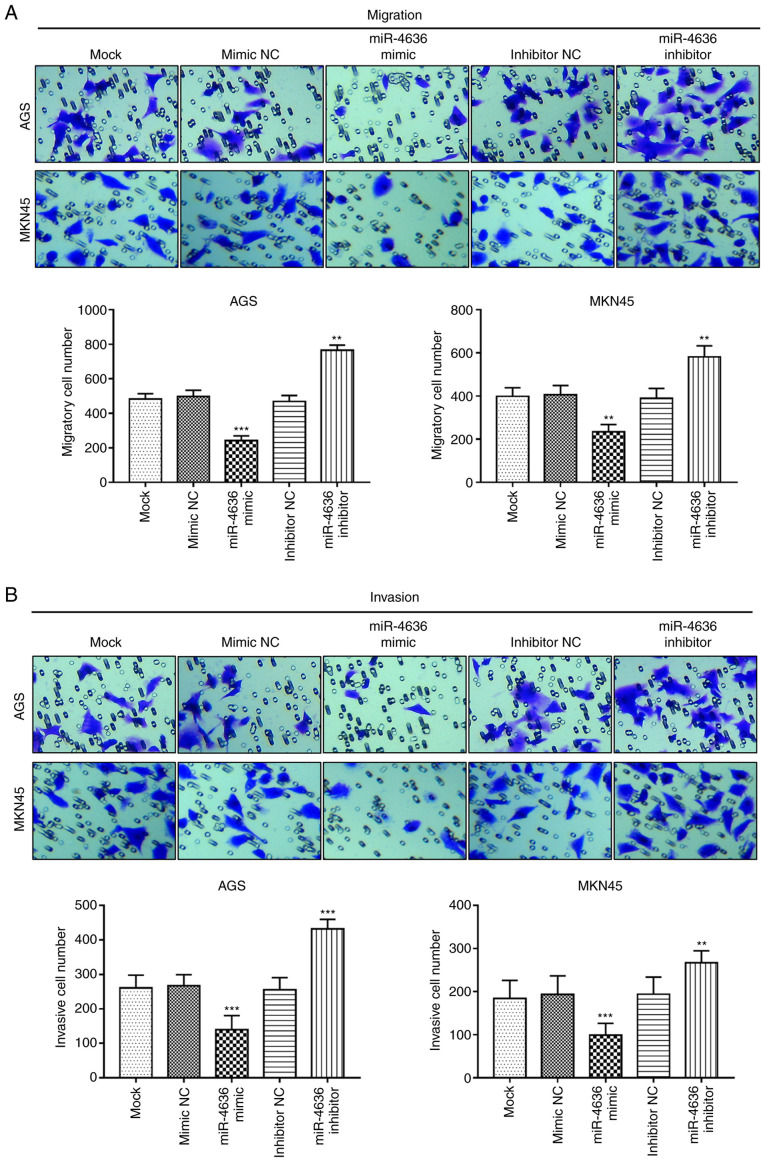
The Transwell assay results demonstrated that the overexpression of miR-4636 inhibited GC cell migration, while the miR-4636 knockdown promoted cell migration in the AGS and MKN45 cell lines compared with the mock group (P<0.01 or P<0.001; Fig. 4A). Furthermore, the cell invasion ability of the AGS and MKN5 cells was suppressed by the overexpression of miR-4636 and enhanced by the knockdown of miR-4636 compared with the mock group (P<0.01 or P<0.001; Fig. 4B).
Figure 4.
Effects of miR-4636 on cell migration and invasion in AGS and MKN45 cells. (A) Cell migration was inhibited by the upregulated expression of miR-4636 and enhanced by decreased miR-4636 expression in both cell lines. (B) Cell invasion in AGS and MKN45 cells was significantly suppressed by the upregulation of miR-4636 and promoted by the downregulation of miR-4636. Magnification, ×200. **P<0.01 and ***P<0.001 vs. the mock group. miR, microRNA; NC, negative control.
Discussion
GC is a heterogeneous malignant disease (15). Although the global incidence and mortality of GC have declined significantly over the past decades, GC remains a leading cause of cancer-associated death worldwide based on statistics from 2010 (16). Despite the development of therapeutic strategies for GC, prognosis remains poor, with the median survival time ranging from 4–12 months (17,18). Previous studies have reported that certain aberrantly expressed miRNAs were associated with disease development and served as biomarkers to improve the diagnosis and prognosis of various cancers, such as lung cancer and hepatocellular carcinoma (19–21). Moreover, aberrant miRNA expression has been documented to be involved in the regulation of tumor progression. For instance, the downregulated expression of miR-145 was detected in colorectal cancer and correlated with tumor size, grade of differentiation, invasion, metastasis and clinical stage (22). In GC, certain aberrant miRNAs participate in tumorigenesis and tumor progression. Cai et al (23) revealed that the downregulated expression of miR-519a predicted the poor prognosis of GC and was involved in the regulation of GC progression. Additionally, Li and Zou (24) reported that miR-652 was significantly elevated in GC tissues and cell lines compared with normal tissues and cells and functioned as an oncogene in GC by promoting tumor progression through targeting RAR-related orphan receptor α (24). These previous studies demonstrated that miRNAs serve roles in tumor progression. Therefore, the present study aimed to investigate the biological function and clinical value of miR-4636 in GC.
miR-4636 has been reported to be deregulated in certain human cancers. For instance, Yin et al (12) revealed that miR-4636 expression levels were correlated with gross tumor volume and the depth of invasion in cervical cancer. Notably, Zhang et al (13) reported that miR-4636 expression was reduced in GC tissues. The present study confirmed downregulated expression of miR-4636 in GC tissues compared with normal tissues. Additionally, the expression of miR-4636 was decreased in GC serum samples and cell lines compared with controls. The expression of miR-4636 was significantly associated with lymph node metastasis and TNM stage in patients with GC. However, there was no significant association between miR-4636 expression and tumor size, which may be due to the limited sample size. Therefore, further investigation with larger study populations are required. These findings indicated that miR-4636 acted as a potential tumor suppressor and may be involved in GC development.
Given the results by Yin et al (12), which indicated that miR-4636 may serve as a clinical biomarker in cervical cancer, the diagnostic and prognostic value of miR-4636 in patients with GC was evaluated. The ROC curve of serum miR-4636 levels indicated a relatively high diagnostic value of miR-4636 in patients with GC. Furthermore, by analyzing the recorded survival data from a 5-year follow-up survey, the association between miR-4636 and the survival rate of patients with GC was investigated. The KM curves demonstrated that patients with lower miR-4636 expressions had shorter overall survivals time and poor disease-free survival compared with patients with higher miR-4636 expression. The Cox regression analysis revealed that the expression of miR-4636 was an independent factor for the prognosis of GC. Therefore, miR-4636 may serve as a biomarker for the diagnosis and prognosis of GC.
Numerous downregulated miRNAs in human malignancies have been reported to be involved in the regulation of tumor progression (25). These functional miRNAs have been proposed as candidate therapeutic targets for the treatment of cancer (26). In the pathogenesis of GC, previous studies have reported that miR-625-3p inhibited tumor cell migration (27) and that miR-26a-5p overexpression suppressed GC cell proliferation, migration and invasion (12). The current study demonstrated that the overexpression of miR-4636 significantly inhibited GC cell proliferation, migration and invasion, while downregulated expression resulted in the opposite effects on these biological processes. Therefore, we hypothesized that miR-4636 may be a tumor suppressor in GC progression. However, to the best of our knowledge, miR-4636-related signaling pathways or target genes have not been reported until now. Thus, the underlying molecular mechanisms involved in the functional role of miR-4636 in GC require further investigation. Furthermore, the current study was a preliminary examination of the biological function of miR-4636 in vitro. Therefore, the role of miR-4636 in GC tumorigenesis requires corroboration by further investigations, including colony formation analysis, wound healing assays, flow cytometry and in vivo experiments.
In conclusion, the present study demonstrated that miR-4636 expression was decreased in the serum and tissues of patients with GC and that miR-4636 dysregulation may serve as a biomarker for the diagnosis and prognosis of GC. Additionally, the results revealed a significant inhibitory effect of miR-4636 on GC cell proliferation, migration and invasion, indicating that may miR-4636 be a potential therapeutic target for the treatment of GC. Methods to increase miR-4636 expression may help in the development of novel therapeutic approaches for GC therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Authors' contributions
JT and CG conducted the clinical study, analyzed clinical data and wrote the manuscript. YH and CZ performed the cell experiments. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Prior to sampling, all the patients and healthy volunteers provided written informed consent for clinical sample use and analysis, and the experimental methods were approved by the Ethics Committee of Zibo Central Hospital, Zibo, China (approval no. ZCHh-090618).
Patient consent for publication
Written informed consent for publication was obtained.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Piazuelo MB, Correa P. Gastric cancer: Overview. Colomb Med (Cali) 2013;44:192–201. [PMC free article] [PubMed] [Google Scholar]
- 2.Correa P. Human gastric carcinogenesis: A multistep and multifactorial process-First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 3.Stock M, Otto F. Gene deregulation in gastric cancer. Gene. 2005;360:1–19. doi: 10.1016/j.gene.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Li M, Zang W, Ma Y, Wang N, Li P, Wang T, Zhao G. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr) 2013;36:385–394. doi: 10.1007/s13402-013-0144-6. [DOI] [PubMed] [Google Scholar]
- 6.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Liu F, Fu Y, Chen X, Zhang D. MiR-520d-5p functions as a tumor-suppressor gene in cervical cancer through targeting PTK2. Life Sci. 2020;254:117558. doi: 10.1016/j.lfs.2020.117558. [DOI] [PubMed] [Google Scholar]
- 9.Yu HY, Pan SS. MiR-202-5p suppressed cell proliferation, migration and invasion in ovarian cancer via regulating HOXB2. Eur Rev Med Pharmacol Sci. 2020;24:2256–2263. doi: 10.26355/eurrev_202003_20491. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Zhang L, Song Q, Chen Z. MicroRNA-216b regulates cell proliferation, invasion and cycle progression via interaction with cyclin T2 in gastric cancer. Anticancer Drugs. 2020;31:623–631. doi: 10.1097/CAD.0000000000000915. [DOI] [PubMed] [Google Scholar]
- 11.Cai H, Lin H, Cao W, Sun J, Huang Y, Fang Y. The downregulation of miR-519a predicts poor prognosis and contributes to tumor progression in gastric cancer. Int J Clin Exp Pathol. 2019;12:2496–2505. [PMC free article] [PubMed] [Google Scholar]
- 12.Yin S, Yang M, Li X, Zhang K, Tian J, Luo C, Bai R, Lu Y, Wang M. Peripheral blood circulating microRNA-4636/-143 for the prognosis of cervical cancer. J Cell Biochem. 2020;121:596–608. doi: 10.1002/jcb.29305. [DOI] [PubMed] [Google Scholar]
- 13.Zhang C, Zhang CD, Ma MH, Dai DQ. Three-microRNA signature identified by bioinformatics analysis predicts prognosis of gastric cancer patients. World J Gastroenterol. 2018;24:1206–1215. doi: 10.3748/wjg.v24.i11.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Strong VE. Progress in gastric cancer. Updates Surg. 2018;70:157–159. doi: 10.1007/s13304-018-0543-3. [DOI] [PubMed] [Google Scholar]
- 16.Cheng XJ, Lin JC, Tu SP. Etiology and prevention of gastric cancer. Gastrointest Tumors. 2016;3:25–36. doi: 10.1159/000443995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wagner AD, Unverzagt S, Grothe W, Kleber G, Grothey A, Haerting J, Fleig WE. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2010:CD004064. doi: 10.1002/14651858.CD004064.pub3. [DOI] [PubMed] [Google Scholar]
- 18.Pyrhonen S, Kuitunen T, Nyandoto P, Kouri M. Randomised comparison of fluorouracil, epidoxorubicin and methotrexate (FEMTX) plus supportive care with supportive care alone in patients with non-resectable gastric cancer. Br J Cancer. 1995;71:587–591. doi: 10.1038/bjc.1995.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Felden J, Villanueva A. Role of molecular biomarkers in liver transplantation for hepatocellular carcinoma. Liver Transpl. 2020;26:823–831. doi: 10.1002/lt.25731. [DOI] [PubMed] [Google Scholar]
- 20.Pan QZ, Liu Q, Zhou YQ, Zhao JJ, Wang QJ, Li YQ, Tang Y, Gu JM, He J, Chen SP, et al. CIK cell cytotoxicity is a predictive biomarker for CIK cell immunotherapy in postoperative patients with hepatocellular carcinoma. Cancer Immunol Immunother. 2020;69:825–834. doi: 10.1007/s00262-020-02486-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backes C, Meese E, Keller A. Specific miRNA disease biomarkers in blood, serum and plasma: Challenges and prospects. Mol Diagn Ther. 2016;20:509–518. doi: 10.1007/s40291-016-0221-4. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q, Yang W, Luo Y, Hu S, Zhu L. Correlation between miR-21 and miR-145 and the incidence and prognosis of colorectal cancer. J Buon. 2018;23:29–35. [PubMed] [Google Scholar]
- 23.Cai H, Lin H, Cao W, Sun J, Huang Y, Fang Y. Downregulation of miR-519a predicts poor prognosis and contributes to tumor progression in gastric cancer. Oncol Res Treat. 2020;43:19–26. doi: 10.1159/000504054. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Zou X. MiR-652 serves as a prognostic biomarker in gastric cancer and promotes tumor proliferation, migration, and invasion via targeting RORA. Cancer Biomark. 2019;26:323–331. doi: 10.3233/CBM-190361. [DOI] [PubMed] [Google Scholar]
- 25.Hu ML, Xiong SW, Zhu SX, Xue XX, Zhou XD. MicroRNAs in gastric cancer: From bench to bedside. Neoplasma. 2019;66:176–186. doi: 10.4149/neo_2018_180703N439. [DOI] [PubMed] [Google Scholar]
- 26.Jin W, Han H, Liu D. Downregulation miR-539 is associated with poor prognosis of gastric cancer patients and aggressive progression of gastric cancer cells. Cancer Biomark. 2019;26:183–191. doi: 10.3233/CBM-190384. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Zhou HC, Zhang Y, Huang H. MicroRNA-625-3p inhibits gastric cancer metastasis through modulating EZH2. Eur Rev Med Pharmacol Sci. 2020;24:1177–1185. doi: 10.26355/eurrev_202002_20169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.