Abstract
Simple Summary
Parnassius butterflies have contributed to fundamental studies in biogeography, insect–plant interactions, and other fields of conservation biology and ecology. However, the early evolutionary pattern and molecular adaptation mechanism of this alpine butterfly group to high altitudes in Qinghai–Tibet Plateau are poorly understood up to now. In this study, we report for the first time, a relatively large-scale transcriptomic dataset of eight Parnassius species and their two closely related papilionid species, a dated phylogeny based on hundreds of gene sequences, and potential genetic mechanisms underlying the high-altitude adaptation by investigating changes in evolutionary rates and positively selected genes. Overall, our findings indicate that the transcriptome data sets reported here can provide some new insights into the spatiotemporally evolutionary pattern and high altitude adaptation of Parnassius butterflies from the extrinsic and intrinsic view, and will support further expressional and functional studies that will help interested researchers to address evolution, biodiversity and conservation questions concerning Parnassius and other butterfly species.
Abstract
The early evolutionary pattern and molecular adaptation mechanism of alpine Parnassius butterflies to high altitudes in Qinghai–Tibet Plateau are poorly understood up to now, due to difficulties in sampling, limited sequence data, and time calibration issues. Here, we present large-scale transcriptomic datasets of eight representative Parnassius species to reveal the phylogenetic timescale and potential genetic basis for high-altitude adaptation with multiple analytic strategies using 476 orthologous genes. Our phylogenetic results strongly supported that the subgenus Parnassius formed a well-resolved basal clade, and the subgenera Tadumia and Kailasius were closely related in the phylogenetic trees. In addition, molecular dating analyses showed that the Parnassius began to diverge at about 13.0 to 14.3 million years ago (middle Miocene), correlated with their hostplant’s spatiotemporal distributions, as well as geological and palaeoenvironmental changes of the Qinghai–Tibet Plateau. Moreover, the accelerated evolutionary rate, candidate positively selected genes and their potentially functional changes were detected, probably contributed to the high-altitude adaptation of Parnassius species. Overall, our study provided some new insights into the spatiotemporally evolutionary pattern and high altitude adaptation of Parnassius butterflies from the extrinsic and intrinsic view, which will help to address evolution, biodiversity, and conservation questions concerning Parnassius and other butterfly species.
Keywords: accelerated diversification, conservation, divergence time, high-altitude adaptation, Parnassius butterflies, phylogeny, positive selection, Qinghai–Tibet Plateau, transcriptome
1. Introduction
The Qinghai–Tibet Plateau (QTP), as the “Third Polar” of the world, offers extreme and substantially variable environments with low level of oxygen, low temperature and strong ultraviolet radiation, making it ideal for studying the high-altitude adaptation and species radiation of organisms [1,2]. Previous studies have shown that the uplifts of QTP and its adjacent areas not only provide unique conditions for the adaptive evolution of many animal and plant groups [3,4,5], but also have greatly affected the spatiotemporally evolutionary patterns of organisms in Eurasia [6,7,8]. For example, radiations of groups of plants are shown to be triggered by extensive uplifts of the QTP [3,5], the Melitaea butterflies have been reported to diversify at significantly varying rates, contemporary with the QTP orogeny and climatic changes [6], the phylogeographic pattern of Eurasian passerines was associated with the Himalayan–Tibetan uplift [7].
Comparative analysis revealed that animals in high-altitude environments can evolve morphological, physiological and genetic adaptations to counter harsh environmental challenges, such as altered body masses, enhanced oxygen delivery, as well as metabolic and genetic changes [1,9,10]. For insects, previous research mainly focused on individual species under short-term artificial stressor treatment (e.g., hypoxia or normoxia, low, or high temperature) [1,9,11]. However, the long-term evolutionary adaptations to high altitudes among closely related species are not fully addressed. In this study, we chose to sample native butterfly species and use large scale transcriptomic data to identify potential genetic mechanisms for the high-altitude adaptations.
The genus Parnassius is a typical mountainous butterfly group, mainly distributed along a broad elevational range of 3000–5000 m on QTP and its adjacent slopes. Previous studies suggest that the Parnassius ancestors probably originated in regions of Central Asia to Western China [12,13,14], and their rapid radiations were likely related with the QTP uplift; a few species subsequently colonized at low-altitude habitats, such as Parnassius glacialis distributed in central to eastern China around 200–1000 m altitudes, suggesting a unique evolutionary history in Parnassiinae [12,13,14]. Parnassius butterflies have contributed to fundamental studies in biogeography, insect–plant interactions, and other fields of conservation biology and ecology, suitable for illustrating how modern biotas have been shaped by past geological and climatic events, and improving our ability to predict and mitigate the threats to alpine species posed by global warming and environmental disruption.
Previous studies on phylogenetic relationships of parnassian butterflies have been mainly based on a limited number of sequence markers, and the phylogenetic backbone of this group has not been resolved [12,13,14,15]. Here we use high-throughput sequencing technologies, especially RNA sequencing (RNA-Seq) [16] to massively collect the gene sequences at the genomic level to explore phylogeny, spatiotemporal diversification, and adaptive molecular evolution. We report for the first time, a relatively large-scale transcriptomic dataset of eight Parnassius species and their two closely related papilionid species, a dated phylogeny based on hundreds of gene sequences, and potential genetic mechanisms underlying the high-altitude adaptation by investigating changes in evolutionary rates and positively selected genes (PSGs).
2. Materials and Methods
2.1. Sample Collection
For de novo transcriptome sequencing, we sampled eight Parnassius species representing five subgenera and other two Papilionidae species (Sericinus montelus and Papilio polytes) as the outgroup (Supplementary Table S1). Among the eight Parnassius species, seven are high-altitude (HA) (Parnassius epaphus, Parnassius jacquemontii, Parnassius cephalus, Parnassius imperator, Parnassius acdestis, Parnassius simo, and Parnassius orleans), and one is low-altitude (LA) species (Parnassius glacialis) (Supplementary Table S1). Specimens were initially preserved in RNA stabilization solution (Sangon Biotech, Shanghai, China) in the field and transferred to −80 °C until RNA extraction. Flight muscle tissues from the thorax of one to three adult samples per species were used for purified RNA extraction.
2.2. mRNA-Seq Library Construction, Illumina Sequencing, Assembly, and Annotation
Libraries construction, Illumina sequencing and assembly were performed following the methods in previous study [17]. Total RNA was extracted using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Quality and integrity of RNA were determined using a NanoDrop spectrophotometer (Thermo Scientific, Barrington, IL, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). The sequencing library was paired-end sequenced using the Illumina NextSeq 500 platform (Shanghai Personal Biotechnology Co. Ltd., Shanghai, China). After the adaptor contamination was removed, the reads were screened from the 3′ to 5′ end to trim the bases with a quality score of Q < 20 by using 5-bp windows, and the reads with final length of less than 50 bp and ambiguous nucleotides were removed. After quality filtering, the quality reads per specimen were collected and then de novo assembled into contigs and transcripts using Trinity v2.0.2 (https://github.com/trinityrnaseq/trinityrnaseq/releases, accessed on 28 February 2019). The sequencing data has been deposited on the GenBank under BioProject number of PRJNA591246.
All the transcripts were searched against the non-redundant protein sequences database (NR) in the National Center for Biotechnology Information (NCBI) by using the Basic Local Alignment Search Tool (BLAST, E-value < 10−5, https://blast.ncbi.nlm.nih.gov, accessed on 28, February, 2019), and the top-hit transcripts were selected as unigenes. All unigenes were then searched against SWISS-PROT, Kyoto Encyclopedia of Genes and Genome (KEGG), and non-supervised orthologous groups (eggNOG) database (http://eggnog.embl.de/, accessed on 1 March 2019) for functional category annotation. The Blast2GO program [18] was used to obtain gene ontology (GO) annotation on the basis of GO terms with the BLASTX algorithm (Blast v2.2.28, E-value < 10−5), and the KAAS (KEGG Automatic Annotation Server, www.genome.jp/tools/kaas/, accessed on 1 March 2019) with bidirectional best-hit strategy was used to assign KEGG orthology terms (KO) and to identify the pathways involved. If a unigene could not be aligned to any of the above databases, its sequence direction and amino acid sequence prediction were conducted via the Transdecoder in the Trinity program package. Transcriptome assembly completeness was determined by Benchmarking Universal Single-Copy Orthologs (BUSCO, version 4.0.2) based on conserved insect orthologs (https://busco.ezlab.org/, accessed on 3 March 2019) with default settings.
2.3. Check for Cross-Contaminations
The CroCo 1.1 was used to check the cross-contamination level in our different sequencing runs [19], which was developed for identifying and removing cross contaminants from assembled transcriptomes. In addition, this method can also be applied to estimate relative coverage for each coding DNA sequence (CDS) of each species.
2.4. Ortholog Identification and Alignment
Single-copy orthologous genes of the ten papilionid transcriptome sequences were identified using OrthoMCL [20]. CDS regions of each species were used as queries to search against the other nine species (reciprocal best BLAST hits). The best hits with similarity > 70% and E-value < 10−6 were retrieved and only one-to-one orthologous genes existing in all ten species were retained for further analyses. All identified orthologous sequences were aligned using MAFFT v7.017 with the codon option and L-INS-i algorithm [21]. Potentially poor alignments or low quality regions were trimmed using Gblocks v.0.91b with half gaps allowed [22]. After the trimming process, alignments shorter than 100 bp were discarded.
2.5. Phylogenetic Analysis
After alignment and trimming, all amino acids and nucleotides of protein-coding genes (PCGs) were concatenated into “supergenes”. Parnassius phylogenetic trees were reconstructed using S. montelus and P. polytes as the outgroups. The best-fit nucleotide model partitioned by three codon positions was selected with PartitionFinder and ModelFinder [23,24], the best-fit amino acid model was selected with ProtTest 3 [25], both following the corrected Akaike information criterion (AICc) (Supplementary Table S2). In addition, the best-fit codon model was also used in resolving complicated phylogenies [26].
Maximum likelihood (ML) and Bayesian inference (BI) methods were used to reconstruct the species-level tree. ML analyses were performed in IQ-TREE v.1.6.1 [27], with the branch support values estimated and single branch tests using the embedded ultrafast bootstrap approach (UFBoot) and SH-like approximate likelihood ratio test (SH-aLRT), respectively [28]. Bayesian analyses were performed in MrBayes 3.2 [26], using two Markov Chain Monte Carlo (MCMC) chains running for 2 million generations with sampling each hundredth generation. Each run had four chains, one cold and three heated. When the convergence of MCMC chains was achieved (the average standard deviation of split frequencies (StdDev) < 0.01, potential scale reduction factor (PSRF) ≈ 1), the first 25% of the sampled generations were discarded as “burn-in”.
2.6. Divergence Time Estimation
Divergence dates were estimated with the program BEAST v1.8.3 [29], using both the amino acid and nucleotide data of the ten species under the uncorrelated lognormal relaxed-clock model. The amino acid data were unpartitioned with the best-fit amino acid model (Supplementary Table S2), while the nucleotide data were partitioned into codon positions with independent GTR+G model. The branching process prior was set to birth-death process.
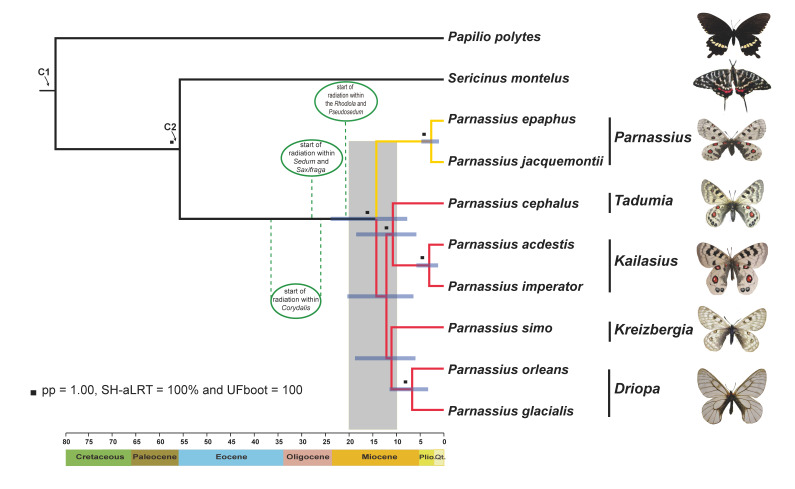
We followed a conservative approach which assumes that fossil records only provide a minimum age when setting the priors for calibration points, and used uniform prior distributions for all calibration points [30,31]. Here, we constrained the crown age of Papilionidae (Node C1, Figure 1) with a minimum age of 47.8 million years ago (Ma) based on the oldest swallowtail fossil Praepapilio colorado (early Eocene (Lutetian), the Green River formation in Colorado) [32], and that of Parnassiinae (Node C2, Figure 1) with a minimum age of 23.03 Ma (early Miocene) based on the fossil Thaites ruminiana from limestone in the Niveau du gypse d’Aix Formation of France [33]. Uniform distributions of two fossil calibrations were maximally bounded at 150 Ma, following the practice by Allio et al. [31].
Figure 1.
Phylogenetic relationships and divergence time estimates of the genus Parnassius based on amino acids of 476 orthologous genes. The tree was inferred under the maximum likelihood and Bayesian analyses. Divergence times were estimated under the birth-death prior shown with the 95% confidence intervals (blue bars); C1 and C2 show the time calibration points; the border in gray shows the time range (10–20 Ma) of the progressive extension of the uplift of the Qinghai–Tibet Plateau (QTP) and related environmental changes. Lineages in yellow and in red mostly feed on the hostplants Crassulaceae + Saxifragaceae (e.g., Sedum, Rhodiola, Pseudosedum and Saxifraga) and Papaveraceae (e.g., Corydalis), respectively.
The calibrated nodes were constrained to be monophyletic as recommended in Drummond and Bouckaert [34]. Three independent MCMC runs of 50 million generations, sampling every 5000 generations were performed. Tracer v1.6 was used to check the effective sample size values to ensure that posterior distributions were sufficiently sampled [35]. Using LogCombiner v. 1.8.3 [29], the posterior distributions of trees from the three runs were combined, discarding the first 10% trees of each run as “burn-in”. TreeAnnotator v.1.8.3 was used to extract the mean and the 95% confidence interval (CI) of the posterior distributions [29].
2.7. Evolutionary Rate and Positive Selection Analyses
The codeml program in the PAML package [36] with the free ratio model (model = 1) was used in this study to estimate the evolutionary rate following Yang et al. and Wang et al. [37,38]. After the alignment quality control, only high quality loci without alignment problems were used for analyses. The lineage-specific mean ratios of nonsynonymous (dN) to synonymous (dS) substitutions rates (ω = dN/dS) were estimated for each ortholog, and concatenated all orthologs, using the species-level tree of this study as the guide tree. Parameters, including dN, dS, dN/dS, N × dN, and S × dS values, were obtained for each branch. Genes with N × dN or S × dS < 1 or dS > 2 were filtered following the approach of the previous study [39]. Statistical significance of the differences in the dN/dS ratios along different lineages were conducted using the Wilcoxon rank sum test.
To find genes that potentially experienced positive selection underlying the high-altitude adaptation, the branch-site model (model = 2) in the PAML package was used to detect positively selected genes (PSGs) [36], with the Parnassius common ancestor being specified as the foreground branch. To avoid false positive results, the following rigorous criteria were used: The dN/dS ratio (ω) greater than 1 on the foreground branch; the positively selected sites with a posterior probability greater than 0.95 [40]; and the p-value ≤ 0.05 in the likelihood ratio test [41]. Finally, the substitution specificity of positively selected sites in the candidate PSGs was recognized with more insect homologous gene sequences in NCBI nr/nt database. Candidate PSGs without identical substitutions at the positively selected sites were used for later analysis.
2.8. Protein Three-Dimensional (3D) Structure Simulation
The PSGs’ functional domains were obtained from the PROSITE database (https://prosite.expasy.org, accessed on 20 December 2019). The 3D structures of PSGs were predicated using Phyre2 [42] and the SWISS-MODEL server (https://swissmodel.expasy.org, accessed on 20 December 2019), then evaluated on the SAVES website (https://servicesn.mbi.ucla.edu/SAVES, accessed on 27 December 2019). All the high-confidence 3D structures obtained both on Phyre2 and SWISS-MODEL servers were visualized using PyMOL [43]. The electrostatic potentials at the surfaces of the predicted protein structures were obtained using PyMOL with ABPS and PDB2PQR [44].
3. Results
3.1. Sequencing, De Novo Assembly, and Annotation
For Parnassius species, about 515 million clean reads with 74.75 Gbp in length were obtained from Illumina sequencing, and assembled into more than 361 thousand unigenes with a mean of 493 bp in size (Supplementary Table S3). In total, average of 25,908 (56.87%) unigenes per Parnassius sample had significant matches to currently known proteins in databases (Supplementary Table S4). Of all databases, NR had the largest match value (24,810, 54.35%), followed by eggNOG (24,761, 54.40%), SwissProt (17,513, 37.47%) and GO (16,555, 35.43%) databases (Supplementary Table S4). The results of annotation, including NR, GO category distribution, eggNOG functional classification and the KEGG pathway of unigenes were similar (Supplementary Figures S1–S3), suggesting high consistencies of de novo assembly and annotation. Average of 1304 (78.6%) complete and fragmented BUSCOs were identified in Parnassius species, while 1449 (87.4%) and 1379 (83.2%) found in S. montelus and P. polytes, respectively (Supplementary Figure S4). Thus, our results indicate that the 10 transcriptomes were well assembled and relatively complete. The BUSCO analysis showed that our assembled unigenes were better than the majority of other 46 arthropod species in assembly and annotation completeness [45]. The cross-contamination check using CroCo recovered a low level of cross contamination with a mean of 108 out of 10,000 (1.08%) contigs contaminated by species (Supplementary Table S5).
3.2. Phylogenetic Analysis
After our alignment treatments and trimming for quality control, 476 single-copy orthologous protein-coding genes were recovered from the determined sequence data, covering 435,231 nucleotides or 145,077 amino acids (Supplementary dataset S1). Nearly all the resultant phylogenetic trees were robust when the best-fit amino acid and codon models were used (posterior probabilities (PP) > 0.95, SH-aLRT ≥ 80% and UFboot ≥ 95; Figure 1 and Supplementary Figure S5). The subgenus Parnassius, which use Crassulaceae as their hostplant, formed a basal clade, sister to other subgenera mostly specialized in feeding on the hostplant Corydalis, supporting the results of previous analyses [13,14]. The subgenus Tadumia was shown to be more closely related to Kailasius than to the Kreizbergia + Driopa clade (Figure 1), consistent with the ML analysis of previous study [13]. The nucleotide model-based ML and Bayesian analyses produced basically the same phylogenies, with some weak node supports as found in previous studies [12,13,14,15,46] (Supplementary Figure S5).
3.3. Divergence Time Estimates
Multiple molecular dating analyses using amino acid and nucleotide datasets, yielded similar divergence time estimates (Table 1). Our estimates of evolutionary timescale suggested that the diversification of Parnassius initiated about 13.0 to 14.3 Ma, followed by successive splittings during 9.9 to 12.1 Ma (Figure 1 and Table 1, birth-death process), supporting the previous hypotheses of their rapidly adaptive radiations [13,14]. These diversifications are largely coeval with the progressive QTP orogenic events and related environmental changes that occurred around 10–20 Ma [47,48,49], and their hostplants’ rapid diversification from the QTP region to Central Asia (e.g., Corydalis, Rhodiola, Pseudosedum, Sedum, and Saxifraga) [50,51,52]. Though somewhat different from those of previous studies [12,13,53], our dating estimates are congruent with other studies in recent years [14,31,46].
Table 1.
Comparison of crown ages with the 95% confidence interval (CI) of major lineages and the most recent common ancestor (MRCA) of major splits in genus Parnassius across different datasets.
| Major Lineage/Split | Amino Acid Dataset | Nucleotide Dataset | ||
|---|---|---|---|---|
| Mean Age | 95% CI | Mean Age | 95% CI | |
| Genus Parnassius | 14.3 | 7.7–24.0 | 13.0 | 6.8–22.3 |
| Tadumia + Kailasius/Kreizbergia + Driopa | 12.1 | 6.4–20.4 | 11.2 | 5.9–19.3 |
| Tadumia/Kailasius | 10.8 | 5.8–18.6 | 9.9 | 5.2–17.1 |
| Kreizbergia/Driopa | 11.1 | 6.0–18.8 | 10.2 | 5.3–17.5 |
3.4. Accelerated Evolution of the Subgeneric Diversification
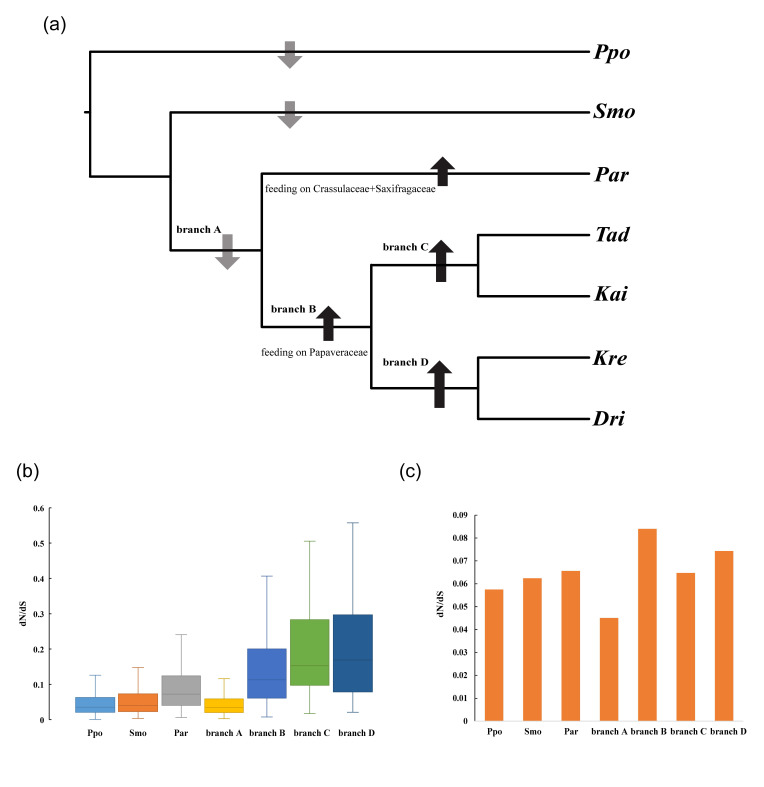
To better understand the evolutionary dynamics of Parnassius butterflies, we compared the overall difference in selective constraints in different branches. Each putative single copy orthologous gene was evaluated for evolutionary parameters including dN, dS, and dN/dS, using the free-ratio model in PAML. Across all 476 orthologous genes, the branch of subgenus Parnassius (mostly feed on the hostplants Crassulaceae and Saxifragaceae) had a significantly higher ratio of dN/dS than branch Ppo (Papilio polytes, Wilcoxon rank sum test, p < 6.4 × 10−19), branch Smo (Sericinus montelus, Wilcoxon rank sum test, p < 1.1 × 10−11) and branch A (ancestral branch of genus Parnassius, Wilcoxon rank sum test, p < 7.5 × 10−24), while the branch B, C and D (ancestral branches of other subgenera which mostly feed on the hostplant Papaveraceae) all had higher ratios than other branches mentioned above, including the branch of subgenus Parnassius (Wilcoxon rank sum test, p < 3.0 × 10−3, Figure 2a,b). In addition, analysis of concatenated gene sequence was also conducted and showed the same result in general: Ancestral branches of subgenera in Parnassius harbored higher dN/dS ratios, suggesting accelerated evolutionary rate for Crassulaceae+Saxifragaceae feeders (subgenus Parnassius) and Papaveraceae feeders (all Parnassius except subgenus Parnassius) (Figure 2c). All above indicated that accelerated evolution occurred after the early splitting event in the genus Parnassius and in the process of subgeneric origin and diversification (Figure 2a), which is likely to be caused by their hostplant shifts [14].
Figure 2.
Phylogenetic tree used in this study (a) and branch specific ratios of nonsynonymous (dN) to synonymous (dS) substitutions rates (dN/dS ratios) obtained from different data sets (b,c). Arrows in (a) indicate decreased or increased dN/dS ratios. The dN/dS ratios for branches were estimated from each ortholog (b) and concatenated all orthologs (c). Ppo, Papilio polytes; Smo, Sericinus montelus; Par, subgenus Parnassius; Tad, subgenus Tadumia; Kai, subgenus Kailasius; Kre, subgenus Kreizbergia; Dri, subgenus Driopa.
3.5. Positively Selected Genes
After applying rigorous criteria to identify genes that are likely to be important in high-altitude adaptation for Parnassius species, five candidate PSGs with high quality alignments remained on the list (Table 2 and Supplementary Table S6). Moreover, when the substitutions for these five genes were examined with more available insect sequences, no identical substitutions were found in those insects (Supplementary Table S7). Three-dimensional (3D) structure simulations were also conducted to examine the possible effects of these mutations on the protein structure, and fortunately, three candidate PSGs (ILF2, PRPF17, and CDASE) can be aligned to the suitable templates in the PDB data bank to build homology 3D structures.
Table 2.
Results of positive selection analysis for the branch of the genus Parnassius.
| Gene | Model with Positive Selection (Model A) |
Null Model (Model A null) |
LRT | p-Value | Biological Category | |||
|---|---|---|---|---|---|---|---|---|
| L1 | Parameters | Positively Selected Sites | L0 | Parameters | 2ΔL | |||
| ILF2 | −1335.05 | k = 1.73 | Ser149Gln 0.98 * | −1337.71 | k = 1.71 | 5.32 | 0.02 | RNA splicing and DNA damage response |
| p0 = 0.97 | p0 = 0.95 | |||||||
| ω0 = 0.02 | ω0 = 0.02 | |||||||
| ps = 0.02 | ||||||||
| ωs = 25.86 | ||||||||
| PRPF17 | −4662.37 | k = 1.65 | Ala289Cys 0.98 * | −4664.23 | k = 1.64 | 3.72 | 0.05 | pre-mRNA splicing and mRNA surveillance |
| p0 = 0.97 | p0 = 0.95 | |||||||
| ω0 = 0.03 | ω0 = 0.03 | |||||||
| ps = 0.01 | ||||||||
| ωs = 14.87 | ||||||||
| SHF | −3105.63 | k = 2.04 | Val300Arg 1.00 ** | −3108.38 | k = 2.02 | 5.50 | 0.02 | morphogenesis and patterning |
| p0 = 0.94 | Lys304Arg 0.98 * | p0 = 0.91 | ||||||
| ω0 = 0.02 | Asp314Arg 0.98 * | ω0 = 0.02 | ||||||
| ps = 0.03 | ||||||||
| ωs = 5.34 | ||||||||
| CDASE | −11227.05 | k = 1.95 | Arg307Val 0.97 * | −11229.24 | k = 1.94 | 4.38 | 0.03 | development and stress responses |
| p0 = 0.92 | His605Ala 0.98 * | p0 = 0.91 | ||||||
| ω0 = 0.05 | ω0 = 0.05 | |||||||
| ps = 0.01 | ||||||||
| ωs = 9.96 | ||||||||
| PGM2 | −5402.44 | k = 1.79 | Cys504Thr 1.00 ** | −5406.12 | k = 1.77 | 7.36 | 0.01 | glycogen metabolism and environmental adaptation |
| p0 = 0.88 | p0 = 0.86 | |||||||
| ω0 = 0.06 | ω0 = 0.06 | |||||||
| ps = 0.01 | ||||||||
| ωs = 15.05 | ||||||||
* if the posterior probability is >0.95, and ** if the probability is >0.99 in the Bayes empirical Bayes (BEB) calculation; the numerical order of positively selected sites referred to the corresponding genes of Bombyx mori; L1, Log-likelihood value in Model A; L0, Log-likelihood value in Model A null; LRT, likelihood ratio test; 2△L, equal to 2 × (L1-L0).
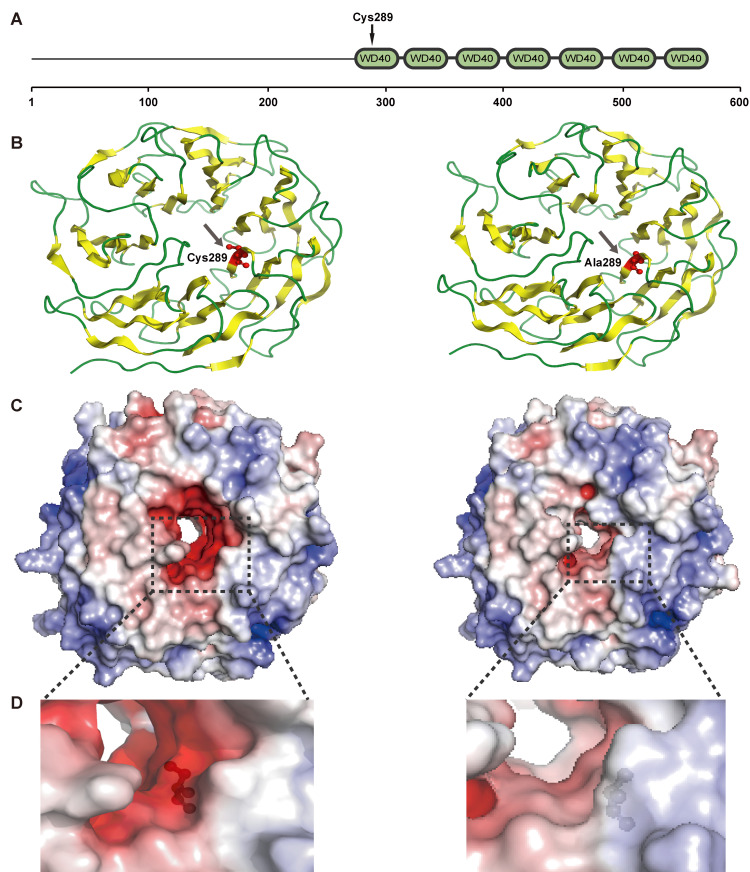
Two of the candidate PSGs, ILF2 (interleukin enhancer-binding factor 2) and PRPF17 (pre-mRNA processing factor 17), are both involved in genetic information processing. ILF2 (also known as NF45) is a regulator of RNA splicing and DNA damage response that promotes mRNA processing and transcripts stabilization involved in homologous recombination [54], and dimerizes with ILF3 (also known as NF90) to form a protein complex involved in a variety of cellular processes [55]. The candidate positively selected amino acid mutation (G149Q) of ILF2 in swallowtail butterflies is located within the DZF domain (Supplementary Table S7 and Figure S6), likely altering its affinity to ILF3 and the protein complex activity in RNA splicing and DNA damage response. PRPF17 encodes a structurally and functionally conserved protein for pre-mRNA splicing and is related to the cell cycle progression [56], required for splicing-dependent NMD (nonsense mediated mRNA decay), serving to increase fidelity and efficiency of gene expression and to promote organism’s viability and development [57]. The A289C mutation of PRPF17 is found to be located at the first one of seven repeated WD40 motifs, which are known to serve as platforms for the assembly of protein complexes or mediators of transient interplay among other proteins [58]. Due to the mutation of the hydrophobic alanine to a hydrophilic cysteine, it could change the regional electrostatic potential as shown by comparison of the electrostatic potential maps (Figure 3), and is likely to form the disulfide bonds. It is probable that the A289C substitution is contributed to the protein-protein interaction involved in diverse functions.
Figure 3.
Structure of PRPF17 protein sequence of the P. acdestis (left) and S. montelus (right), respectively. (A): The PRPF17 protein domain with seven repeated WD40 motifs; the candidate positively selected site is indicated by black arrow. (B): The structures of sheet and loop are shown in yellow and green, respectively, with the positively selected residues shown with the red ball and stick representation. (C): Electrostatic potential map of PRPF17 domain; the range of electrostatic surface potential is shown from −7 kT/e (red color) to +7 kT/e (blue color). (D): The zoomed-in picture of electrostatic potential around the positively selected residues. Template in the Protein Data Bank: 5mqf.1.
The other two candidate PSGs, SHF (protein shifted or WIF-1-like protein) and CDASE (neutral ceramidase), are both involved in the signal transduction associated with environmental information processing. Serving as a component in the Drosophila Hedgehog signaling pathway, SHF is required for both stability and diffusion of cholesterol-modified Hedgehog, crucial to the morphogenesis and patterning (e.g., the wing development) [59]. All three positively selected sites of SHF here are located in the fourth epidermal growth factor (EGF)-like domain, different from the missense mutations between two known Shf alleles in the third EGF-like domain (see Figure 1B,C in Glise et al. [60]), implying that the positive selection of these selected sites and alleles resulted from different environmental conditions. CDASE plays important roles in insect development and stress responses (e.g., starvation, abnormal temperature, ultraviolet radiation and insecticides) through regulating the biosynthesis of sphingolipids [61], and in maintaining the photoreceptor homeostasis associated with UV damage response for Drosophila [62]. Two candidate positively selected mutations (the probable R307V and H605A) of CDASE are located in the N-terminal of the ZnF_CHCC domain and the highly conserved C-terminal tail of the enzyme, respectively (Supplementary Table S7 and Figure S7). According to the changes of hydrophobic properties and electrostatic pattern after ionization, these two mutations of CDASE may alter the affinity for zinc ion binding, and play important roles in stabilization and interaction of the enzyme with the plasma membranes [63].
The last one candidate PSG, PGM2 (phosphoglucomutase 2), plays an important role in the regulation of glycogen biosynthesis associated with the energy metabolism and stress resistance for environmental adaptation [64]. The positively selected site of PGM2 herein is located adjacent to the phosphorylation site of the domain 4 compared to the PGM1, and is probably associated with efficient catalysis [65]. Interestingly, the similar induced physicochemical change has been found in other proteins of the α-D-phosphohexomutase superfamily [66], and the positive selection of this gene was also detected in high-altitude dwelling Phrynocephalus and Rana lineages [2,4].
4. Discussion
The Parnassius internal phylogeny were not fully resolved, mainly due to the usage of only a limited number of molecular markers in previous studies, regardless of potentially homoplastic morphological characters used or not [12,13,14]. The main unresolved issues are as follows: (1) The equivocal position of its subgenus Parnassius, an important lineage for ancestral hostplant inference and biogeographic reconstruction; (2) unstable relationships of other subgenera due to the short branches on the trees, with similar mitochondrial and nuclear sequence datasets used in previous analyses [12,13,14,46]. Therefore, further efforts should focus on more inclusive sampling of genes as well as more Parnassius species [12]. However, previous studies indicated that the phylogenetic tree topology of Parnassius remained unresolved, even by sampling as many Parnassius species as possible (14,15,46). Rapid advances of RNA-Seq technologies have significantly accelerated the speed of gene gathering and genomics studies at reasonable cost. In this work, the Parnassius phylogenetic trees were reconstructed based on a large-scale transcriptomic datasets for the first time, using multiple analytic methods. Our results showed that all the topologies were basically the same with strong supports on nodes, regardless of the selection of sequences (nucleotides or amino acids), models, and tree-constructing methods (Figure 1, Supplementary Figure S5). These results indicated that high-throughput transcriptomic sequencing could represent a powerful approach to phylogenomics in a group that has previously been hardly to resolve.
Independent evidence indicated that the progressive extension of the QTP uplifts occurred during the Miocene period, responsible for the origin and radiation of the current biota in the region [48,49,67]. For the lineage of Parnassius, the significant up-shift in speciation rates has been detected, coincident with the colonization to the QTP [14], which can partially be explained by the QTP uplifts of providing novel, high-altitude habitats for the colonization of Parnassius common ancestor. Nevertheless, one of the premises of the QTP colonization for Parnassius species should be that their hostplants survived and diversified in the QTP region no later than its herbivores. This kind of temporal correlation between butterflies and their host plants has indirectly been corroborated by a survey, which indicated that butterflies along the Andean slope between 2400 and 5000 m acclimation to novel habitats depends on their larval hostplant use [68]. Previous phylogeographic studies indicated the Corydalis (Papaveraceae) colonized the QTP and Central Asia at about 37–26 Ma in Oligocene, the Sedum and Rhodiola-Pseudosedum clade (Crassulaceae) as well as Saxifraga (Saxifragaceae) originated in the QTP region at about 28.0–21.0 Ma [50,51,52]. Our dating estimates using transcriptomic data showed that the subgenus Parnassius (feeding on Crassulaceae+Saxifragaceae) diverged from other subgenera (feeding on Papaveraceae) probably in the Miocene (95% CI 7.7–24.0 Ma, Figure 1 and Table 1). Therefore, based on multiple independent studies above, the diversification pattern of Parnassius lineages are compatible with the spatiotemporal distribution pattern of their hostplants, supporting the underlying coevolutionary scenario between Parnassius and hostplants [69,70,71], mainly under the coupled effect of common extrinsic factors (e.g., mountain building, climate change and the resultant aridification event) in the Miocene [14]. Similar diversification patterns have also been found in alpine butterflies of the genus Erebia [72], the arcesia and fergana species groups in the genus Melitaea [6], and other animal groups [7,73]. Interestingly, the time range for diversification of Parnassius lineages is nearly contemporary with that of recent angiosperms’ radiation in western China (15.29–18.86 Ma) [5]. The coevolutionary scenario between Parnassius and hostplants can serve as a theoretical basis of conservation strategy for Parnassius and other alpine butterflies.
Multiple extrinsic factors can drive diversification of Parnassius lineages; however, currently little is known about the intrinsic genetic basis of the high-altitude adaptation for this butterfly genus. In this work, analyses of evolutionary rate estimation suggested that accelerated evolution occurred in the process of subgeneric origin and diversification in this alpine Parnassius butterfly group. Similar results of genome-wide rapid evolution were also found in Tibetan fishes, highland-dwelling lizard and ranid frog lineages [4,37,38]. In addition, two of the candidate PSGs here along the Parnassius lineages, PRPF17 and PGM2, seem to be homologous with those (PRPF4B, PGM2, PGM2L1, and PGM5, respectively) detected along the highland-dwelling lizard and frog lineages [2,4]. Therefore, both the accelerated evolution and the partial overlap of PSGs as well as related functions of all five PSGs herein, especially DNA damage response and energy metabolism pathways, probably exhibit numerous convergent evolutionary changes among different animal groups relating to high altitude adaptation. Nevertheless, three PSGs herein (ILF2, SHF, and CDASE), involved in insect development, morphogenesis, and stress responses (e.g., starvation, abnormal temperature, ultraviolet radiation, and DNA damage), are not shared with previously reported genes in other animals. This observation suggests that Parnassius butterflies may have employed different genetic toolkits to adapt to extreme and substantially variable environments of the QTP, and can serve as a foundation for future studies aiming to mitigate the threats to alpine butterflies posed by climate changes and environmental disruptions. However, this hypothesis needs to be further confirmed by population genomics studies in the future.
Overall, at least several intrinsic traits may contribute to the Parnassius adaptive evolution: The relatively faster evolutionary rates for subgeneric origin than those of other low-altitude papilionids revealed by genome-wide elevated dN/dS ratios; the five detected PSGs with functions as mentioned above in promoting stabilization of genetic information processing for viability and development, stress responses (e.g., DNA damage, starvation, abnormal temperature, and ultraviolet radiation), and energy metabolism, et al., suggesting their multifaceted strategies for long-lasting adaptation to high-altitude environments of low level of oxygen, low temperature and strong ultraviolet radiation. In the future, samplings of additional taxa and more high-quality genomic data are needed to clarify the evolutionary spatiotemporal pattern of Parnassius butterflies and the underlying extrinsic and intrinsic factors more effectively.
5. Conclusions
Our large-scale transcriptomic data of eight representative Parnassius species and their two closely related papilionid species are valuable in establishing well-resolved phylogeny and a more reasonable timescale for the divergences of genus Parnassius, which initiated in the middle Miocene, followed by successive subgeneric diversifications that may be historically correlated with their hostplant spatiotemporal distributions as well as the geological and environmental changes in the QTP region. Additionally, our study provides the first report about the genetic mechanism of QTP high-altitude adaptation of the main Parnassius butterfly lineages at the whole genome scale, that is, the accelerated evolution for subgeneric origin and diversification correlated with hostplant shifts, candidate PSGs and their potentially functional changes may attribute to both the convergent evolution and the unique strategies for long-lasting adaptation to high-altitude environments, which will help interested researchers to address evolution, biodiversity and conservation questions concerning Parnassius and other butterfly species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/11/754/s1, Figure S1: Results of annotation in the NR database for Parnassius species, Figure S2: Characteristics of GO term of biological process, cellular component and molecular function for Parnassius species, Figure S3: Characteristics of eggNOG functional category and KEGG pathway classification of unigenes for Parnassius species, Figure S4: BUSCO analysis of transcriptome completeness, Figure S5: The species-level Parnassius phylogenetic relationships using maximum likelihood and Bayesian inference methods, Figure S6: Structure of ILF2 protein sequence of the P. acdestis (left) and S. montelus (right), respectively, Figure S7: Structure of CDASE protein sequence of the P. acdestis (left) and S. montelus (right), respectively, Table S1: Information of samples used for transcriptome sequencing, Table S2: Substitution model of each partition with different partitioning schemes, Table S3: Summary of assembly results of transcriptome data for species, Table S4: Summary of functional annotation of Unigenes, Table S5: Results of cross-contamination check using the CroCo program, Table S6: Overview of the positively selected genes and their quality assessment, Table S7: Amino acid substitution specificity information for positively selected sites along the lineage leading to the genus Parnassius, Dataset S1: Phylogenomic dataset including 476 orthologous genes in nucleotides format.
Author Contributions
Conceptualization, J.H. and Q.Y.; methodology, C.S. (Chengyong Su) and J.M.; software, C.S. (Chengyong Su); validation, J.H. and Q.Y.; formal analysis, C.S. (Chengyong Su); investigation, T.X.; resources, Y.W.; data curation, C.S. (Chengyong Su) and X.S.; writing—original draft preparation, C.S. (Chengyong Su); writing—review and editing, C.S. (Chengyong Su), C.L., and J.H.; visualization, T.X., C.S. (Chengcai Si), L.L., J.M., and C.L.; supervision, J.H. and Q.Y.; project administration, J.H.; funding acquisition, J.H. and Q.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Strategic Priority Research Program of Chinese Academy of Sciences to Q.Y. [Grant No. XDB26000000], National Science Foundation of China to J.H. [Grants Nos. 41472028, 41972029]; and the State Key Laboratory of Palaeobiology and Stratigraphy (Nanjing Institute of Geology and Palaeontology, CAS) [Grant No. Y626040108].
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ding D., Liu G.J., Hou L., Gui W.Y., Chen B., Kang L. Genetic variation in PTPN1 contributes to metabolic adaptation to high-altitude hypoxia in Tibetan migratory locusts. Nat. Commun. 2018;9:4991. doi: 10.1038/s41467-018-07529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun Y.B., Fu T.T., Jin J.Q., Murphy R.W., Hillis D.M., Zhang Y.P., Che J. Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations. Proc. Natl. Acad. Sci. USA. 2018;115:E10634–E10641. doi: 10.1073/pnas.1813593115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen J., Zhang J.Q., Nie Z.L., Zhong Y., Sun H. Evolutionary diversifications of plants on the Qinghai-Tibetan Plateau. Front. Genet. 2014;5:4. doi: 10.3389/fgene.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y.Z., Wang L.Z., Han J., Tang X.L., Ma M., Wang K., Zhang X., Ren Q., Chen Q., Qiu Q. Comparative transcriptomic analysis revealed adaptation mechanism of Phrynocephalus erythrurus, the highest altitude Lizard living in the Qinghai-Tibet Plateau. BMC Evol. Biol. 2015;15:101. doi: 10.1186/s12862-015-0371-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu L.M., Mao L.F., Yang T., Ye J.F., Liu B., Li H.L., Sun M., Miller J.T., Mathews S., Hu H.H., et al. Evolutionary history of the angiosperm flora of China. Nature. 2018;554:234–238. doi: 10.1038/nature25485. [DOI] [PubMed] [Google Scholar]
- 6.Leneveu J., Chichvarkhin A., Wahlberg N. Varying rates of diversification in the genus Melitaea (Lepidoptera: Nymphalidae) during the past 20 million years. Biol. J. Linn. Soc. 2009;97:346–361. doi: 10.1111/j.1095-8312.2009.01208.x. [DOI] [Google Scholar]
- 7.Päckert M., Martens J., Sun Y.H., Tietze D.T. Evolutionary history of passerine birds (Aves: Passeriformes) from the Qinghai-Tibetan plateau: From a pre-Quarternary perspective to an integrative biodiversity assessment. J. Ornithol. 2015;156:S355–S365. doi: 10.1007/s10336-015-1185-6. [DOI] [Google Scholar]
- 8.Zhao Z., Li S. Extinction vs. Rapid radiation: The juxtaposed evolutionary histories of coelotine spiders support the Eocene–Oligocene orogenesis of the Tibetan Plateau. Syst. Biol. 2017;66:988–1006. doi: 10.1093/sysbio/syx042. [DOI] [PubMed] [Google Scholar]
- 9.Zhao D.J., Zhang Z.Y., Cease A., Harrison J., Kang L. Efficient utilization of aerobic metabolism helps Tibetan locusts conquer hypoxia. BMC Genom. 2013;14:631. doi: 10.1186/1471-2164-14-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou D., Haddad G.G. Genetic analysis of hypoxia tolerance and susceptibility in Drosophila and humans. Annu. Rev. Genom. Hum. Genet. 2013;14:25–43. doi: 10.1146/annurev-genom-091212-153439. [DOI] [PubMed] [Google Scholar]
- 11.Cui M.M., Hu P., Wang T., Tao J., Zong S. Differential transcriptome analysis reveals genes related to cold tolerance in seabuckthorn carpenter moth, Eogystia hippophaecolus. PLoS ONE. 2017;12:e0187105. doi: 10.1371/journal.pone.0187105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nazari V., Zakharov E.V., Sperling F.A. Phylogeny, historical biogeography, and taxonomic ranking of Parnassiinae (Lepidoptera, Papilionidae) based on morphology and seven genes. Mol. Phylogenet. Evol. 2007;42:131–156. doi: 10.1016/j.ympev.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 13.Michel F., Rebourg C., Cosson E., Descimon H. Molecular phylogeny of Parnassiinae butterflies (Lepidoptera: Papilionidae) based on the sequences of four mitochondrial DNA segments. Ann. Soc. Entomol. Fr. 2008;44:1–36. doi: 10.1080/00379271.2008.10697541. [DOI] [Google Scholar]
- 14.Condamine F.L., Rolland J., Höhna S., Sperling F.A., Sanmartín I. Testing the role of the Red Queen and Court Jester as drivers of the macroevolution of Apollo butterflies. Syst. Biol. 2018;67:940–964. doi: 10.1093/sysbio/syy009. [DOI] [PubMed] [Google Scholar]
- 15.Zheng B., Wang Y., Xia C., Huang D., Cao Y., Hao J., Zhu C. The complete mitochondrial genome of Parnassius actius (Lepidoptera: Papilionidae: Parnassinae) with the related phylogenetic analysis. Zool. Syst. 2018;43:1–17. doi: 10.11865/zs.201801. [DOI] [Google Scholar]
- 16.Wilhelm B.T., Landry J.R. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Hu J.W., You F., Wang Q., Weng S.D., Liu H., Wang L.J., Zhang P.J., Tan X.G. Transcriptional responses of olive flounder (Paralichthys olivaceus) to low temperature. PLoS ONE. 2014;9:e108582. doi: 10.1371/journal.pone.0108582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conesa A., Götz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008;2008:619832. doi: 10.1155/2008/619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simion P., Belkhir K., Francois C., Veyssier J., Rink J.C., Manuel M., Philippe H., Telford M.J. A software tool ‘CroCo’ detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 2018;16:28. doi: 10.1186/s12915-018-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L., Stoeckert C.J., Jr., Roos D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 23.Lanfear R., Calcott B., Ho S.Y.W., Guindon S. PartitionFinder: Combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 24.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darriba D., Taboada G.L., Doallo R., Posada D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoang D.T., Chernomor O., von Haeseler A., Minh B.Q., Vinh L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond A.J., Suchard M.A., Xie D., Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chazot N., Wahlberg N., Freitas A.V.L., Mitter C., Labandeira C., Sohn J.C., Sahoo R.K., Seraphim N., de Jong R., Heikkila M. Priors and posteriors in Bayesian timing of divergence analyses: The age of butterflies revisited. Syst. Biol. 2019;68:797–813. doi: 10.1093/sysbio/syz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Allio R., Scornavacca C., Nabholz B., Clamens A.L., Sperling F.A., Condamine F.L. Whole genome shotgun phylogenomics resolves the pattern and timing of swallowtail butterfly evolution. Syst. Biol. 2020;69:38–60. doi: 10.1093/sysbio/syz030. [DOI] [PubMed] [Google Scholar]
- 32.Durden C.J., Rose H. Butterflies from the Middle Eocene: The Earliest Occurrence of Fossil Papilionoidea (Lepidoptera) Texas Memorial Museum, The University of Texas at Austin; Austin, TX, USA: 1978. [Google Scholar]
- 33.Scudder S.H. Fossil butterflies. Mem. Am. Assoc. Adv. Sci. 1875;1:1–99. [Google Scholar]
- 34.Drummond A.J., Bouckaert R.R. Bayesian Evolutionary Analysis with BEAST. Cambridge University Press; London, UK: 2015. pp. 98–125. [Google Scholar]
- 35.Rambaut A., Suchard M.A., Xie D., Drummond A.J. Tracer v1.6. [(accessed on 20 December 2019)]; Available online: http://beast.bio.ed.ac.uk/Tracer.
- 36.Yang Z.H. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 37.Yang L., Wang Y., Zhang Z., He S. Comprehensive transcriptome analysis reveals accelerated genic evolution in a Tibet fish, Gymnodiptychus pachycheilus. Genome Biol. Evol. 2014;7:251–261. doi: 10.1093/gbe/evu279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Yang L., Zhou K., Zhang Y., Song Z., He S. Evidence for adaptation to the Tibetan Plateau inferred from Tibetan loach transcriptomes. Genome Biol. Evol. 2015;7:2970–2982. doi: 10.1093/gbe/evv192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman M., Sterner K.N., Islam M., Uddin M., Sherwood C.C., Hof P.R., Hou Z.C., Lipovich L., Jia H., Grossman L.I. Phylogenomic analyses reveal convergent patterns of adaptive evolution in elephant and human ancestries. Proc. Natl. Acad. Sci. USA. 2009;106:20824–20829. doi: 10.1073/pnas.0911239106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Z.H., Wong W.S.W., Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J.Z., Nielsen R., Yang Z.H. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 42.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeLano W.L. Pymol: An open-source molecular graphics tool. Ccp4 Newslett. Protein Crystallogr. 2002;40:82–92. [Google Scholar]
- 44.Unni S., Huang Y., Hanson R.M., Tobias M., Krishnan S., Li W.W., Nielsen J.E., Baker N.A. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 2011;32:1488–1491. doi: 10.1002/jcc.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Theissinger K., Falckenhayn C., Blande D., Toljamo A., Gutekunst J., Makkonen J., Jussila J., Lyko F., Schrimpf A., Schulz R., et al. De Novo assembly and annotation of the freshwater crayfish Astacus astacus transcriptome. Mar. Genom. 2016;28:7–10. doi: 10.1016/j.margen.2016.02.006. [DOI] [PubMed] [Google Scholar]
- 46.Condamine F.L., Sperling F.A.H., Kergoat G.J. Global biogeographical pattern of swallowtail diversification demonstrates alternative colonization routes in the Northern and Southern hemispheres. J. Biogeogr. 2013;40:9–23. doi: 10.1111/j.1365-2699.2012.02787.x. [DOI] [Google Scholar]
- 47.Guo Z.T., Ruddiman W.F., Hao Q.Z., Wu H.B., Qiao Y.S., Zhu R.X., Peng S.Z., Wei J.J., Yuan B.Y., Liu T.S. Onset of Asian desertification by 22 Myr ago inferred from loess deposits in China. Nature. 2002;416:159–163. doi: 10.1038/416159a. [DOI] [PubMed] [Google Scholar]
- 48.Wang C.S., Zhao X.X., Liu Z.F., Lippert P.C., Graham S.A., Coe R.S., Yi H., Zhu L., Liu S., Li Y. Constraints on the early uplift history of the Tibetan Plateau. Proc. Natl. Acad. Sci. USA. 2008;105:4987–4992. doi: 10.1073/pnas.0703595105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Favre A., Packert M., Pauls S.U., Jahnig S.C., Uhl D., Michalak I., Muellner-Riehl A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 2015;90:236–253. doi: 10.1111/brv.12107. [DOI] [PubMed] [Google Scholar]
- 50.Zhang J.Q., Meng S.Y., Allen G.A., Wen J., Rao G.Y. Rapid radiation and dispersal out of the Qinghai-Tibetan Plateau of an alpine plant lineage Rhodiola (Crassulaceae) Mol. Phylogenet. Evol. 2014;77:147–158. doi: 10.1016/j.ympev.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 51.Pérez-Gutiérrez M.A., Romero-García A.T., Fernández M.C., Blanca G., Salinas-Bonillo M.J., Suárez-Santiago V.N. Evolutionary history of fumitories (subfamily Fumarioideae, Papaveraceae): An old story shaped by the main geological and climatic events in the Northern Hemisphere. Mol. Phylogenet. Evol. 2015;88:75–92. doi: 10.1016/j.ympev.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 52.Ebersbach J., Muellner-Riehl A.N., Michalak I., Tkach N., Hoffmann M.H., Roser M., Sun H., Favre A. In and out of the Qinghai-Tibet Plateau: Divergence time estimation and historical biogeography of the large arctic-alpine genus Saxifraga L. J. Biogeogr. 2016;44:900–910. doi: 10.1111/jbi.12899. [DOI] [Google Scholar]
- 53.Omoto K., Yonezawa T., Shinkawa T. Molecular systematics and evolution of the recently discovered “Parnassian” butterfly (Parnassius davydovi Churkin, 2006) and its allied species (Lepidoptera, Papilionidae) Gene. 2009;441:80–88. doi: 10.1016/j.gene.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 54.Marchesini M., Ogoti Y., Fiorini E., Samur A.A., Nezi L., D’Anca M., Storti P., Samur M.K., Ganan-Gomez I., Fulciniti M.T., et al. ILF2 is a regulator of RNA splicing and DNA damage response in 1q21-amplified multiple myeloma. Cancer Cell. 2017;32:88–100. doi: 10.1016/j.ccell.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolkowicz U.M., Cook A.G. NF45 dimerizes with NF90, Zfr and SPNR via a conserved domain that has a nucleotidyltransferase fold. Nucleic Acids Res. 2012;40:9356–9368. doi: 10.1093/nar/gks696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dahan O., Kupiec M. The Saccharomyces cerevisiae gene CDC40/PRP17 controls cell cycle progression through splicing of the ANC1 gene. Nucleic Acids Res. 2004;32:2529–2540. doi: 10.1093/nar/gkh574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: An intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 2015;16:665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 58.Li D., Roberts R. WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorfinkiel N., Sierra J., Callejo A., Ibanez C., Guerrero I. The Drosophila ortholog of the human Wnt inhibitor factor Shifted controls the diffusion of lipid-modified Hedgehog. Dev. Cell. 2005;8:241–253. doi: 10.1016/j.devcel.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 60.Glise B., Miller C.A., Crozatier M., Halbisen M.A., Wise S., Olson D.J., Vincent A., Blair S.S. Shifted, the Drosophila ortholog of Wnt inhibitory factor-1, controls the distribution and movement of Hedgehog. Dev. Cell. 2005;8:255–266. doi: 10.1016/j.devcel.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Shi X.X., Huang Y.J., Begum M.A., Zhu M.F., Li F.Q., Zhang M.J., Zhou W.W., Mao C.G., Zhu Z.R. A neutral ceramidase, NlnCDase, is involved in the stress responses of brown planthopper, Nilaparvata lugens (Stål) Sci. Rep. 2018;8 doi: 10.1038/s41598-018-19219-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Acharya J.K., Dasgupta U., Rawat S.S., Yuan C.Q., Sanxaridis P.D., Yonamine I., Karim P., Nagashima K., Brodsky M.H., Tsunoda S., et al. Cell-nonautonomous function of ceramidase in photoreceptor homeostasis. Neuron. 2008;57:69–79. doi: 10.1016/j.neuron.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Inoue T., Okino N., Kakuta Y., Hijikata A., Okano H., Goda H.M., Tani M., Sueyoshi N., Kambayashi K., Matsumura H., et al. Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase. J. Biol. Chem. 2009;284:9566–9577. doi: 10.1074/jbc.M808232200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maliekal P., Sokolova T., Vertommen D., Veiga-Da-Cunha M., Van Schaftingen E. Molecular identification of mammalian phosphopentomutase and glucose-1,6-bisphosphate synthase, two members of the alpha-D-phosphohexomutase family. J. Biol. Chem. 2007;282:31844–31851. doi: 10.1074/jbc.M706818200. [DOI] [PubMed] [Google Scholar]
- 65.Beamer L.J. Mutations in hereditary phosphoglucomutase 1 deficiency map to key regions of enzyme structure and function. J. Inherit. Metab. Dis. 2015;38:243–256. doi: 10.1007/s10545-014-9757-9. [DOI] [PubMed] [Google Scholar]
- 66.Muenks A.G., Stiers K.M., Beamer L.J. Sequence-structure relationships, expression profiles, and disease-associated mutations in the paralogs of phosphoglucomutase 1. PLoS ONE. 2017;12:e0183563. doi: 10.1371/journal.pone.0183563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ding W.N., Ree R.H., Spicer R.A., Xing Y.W. Ancient orogenic and monsoon-driven assembly of the world’s richest temperate alpine flora. Science. 2020;369:578–581. doi: 10.1126/science.abb4484. [DOI] [PubMed] [Google Scholar]
- 68.Despland E. Butterflies of the high-altitude Atacama Desert: Habitat use and conservation. Front. Genet. 2014;5:334. doi: 10.3389/fgene.2014.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wheat C.W., Vogel H., Wittstock U., Braby M.F., Underwood D., Mitchell-Olds T. The genetic basis of a plant-insect coevolutionary key innovation. Proc. Natl. Acad. Sci. USA. 2007;104:20427–20431. doi: 10.1073/pnas.0706229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fordyce J.A. Host shifts and evolutionary radiations of butterflies. Proc. R. Soc. B Biol. Sci. 2010;277:3735–3743. doi: 10.1098/rspb.2010.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Su C.Y., Shi Q.H., Sun X.Y., Ma J.Y., Li C.X., Hao J.S., Yang Q. Dated phylogeny and dispersal history of the butterfly subfamily Nymphalinae (Lepidoptera: Nymphalidae) Sci. Rep. 2017;7:8799. doi: 10.1038/s41598-017-08993-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peña C., Witthauer H., Klečková I., Fric Z., Wahlberg N. Adaptive radiations in butterflies: Evolutionary history of the genus Erebia (Nymphalidae: Satyrinae) Biol. J. Linn. Soc. 2015;116:449–467. doi: 10.1111/bij.12597. [DOI] [Google Scholar]
- 73.Drovetski S.V., Semenov G., Drovetskaya S.S., Fadeev I.V., Red’kin Y.A., Voelker G. Geographic mode of speciation in a mountain specialist Avian family endemic to the Palearctic. Ecol. Evol. 2013;3:1518–1528. doi: 10.1002/ece3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.