Abstract
The pollen grains produced by flowering plants are vital for sexual reproduction. Previous studies have shown that two CCCH-type zinc-finger protein genes in Brassica campestris, BcMF30a and BcMF30c, are involved in pollen development. Due to their possible functional redundancy, gain-of-function analysis is helpful to reveal their respective biological functions. Here, we found that the phenotypes of BcMF30a and BcMF30c overexpression transgenic plants driven by their native promoters were similar, suggesting their functional redundancy. The results showed that the vegetative growth was not affected in both transgenic plants, but male fertility was reduced. Further analysis found that the abortion of transgenic pollen was caused by the degradation of pollen contents from the late uninucleate microspore stage. Subcellular localization analysis demonstrated that BcMF30a and BcMF30c could localize in cytoplasmic foci. Combined with the studies of other CCCH-type genes, we speculated that the overexpression of these genes can induce the continuous assembly of abnormal cytoplasmic foci, thus resulting in defective plant growth and development, which, in this study, led to pollen abortion. Both the overexpression and knockout of BcMF30a and BcMF30c lead to abnormal pollen development, indicating that the appropriate expression levels of these two genes are critical for the maintenance of normal pollen development.
Keywords: Brassica campestris, CCCH-type zinc-finger protein, BcMF30a, BcMF30c, pollen development, overexpression, cytoplasmic foci
1. Introduction
In flowering plants, each microspore mother cell in the anthers undergoes meiosis to produce four haploid microspores, which are initially associated in a tetrad. Then, the microspores separate from each other and undergo vacuolation and expansion, and the nuclei within them migrate toward the cell wall. Subsequently, pollen mitosis Ⅰ (PMⅠ) yields a bicellular pollen (BCP) composed of a larger vegetative cell (VC) and a smaller generative cell (GC) that is engulfed by VC. The GC further yields two sperm cells after a symmetric division (PMⅡ), and results in a tricellular pollen (TCP) grain. In Arabidopsis thaliana, nearly 14,000 genes were identified to be expressed in the male gametophyte, and their expression levels change as development progresses [1]. This clearly illustrates that this series of pollen development events is regulated by dynamic and complex changes in gene expression.
Loss-of-function mutagenesis by fast neutrons, chemicals, or gene editing that create random or targeted mutations or deletions in the genome is a conventional means to uncover the functions of a particular gene [2]. Mutant genetic study has been an effective approach to understand the molecular and cellular aspects of all phases of pollen development. A survey conducted a decade ago showed that 37 genes can affect the post-meiotic male gametophyte development by mutant analysis [3]. After the last ten years of research, gametophytic mutants have been continuously characterized, and genes required for pollen development have been identified exponentially. For instance, LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 10 co-acts with SIDECAR POLLEN/LBD27 to regulate the early stage of pollen development prior to PMⅠ [4]. An myb81-1 mutant analysis demonstrated that MYB81 is a non-redundant GAMYB family member required for polarized microspores’ progress into PMⅠ [5]. Loss of function of VAC14 led to pollen abortion due to the defective vacuolar fission after PMⅠ [6]. Four genes that encode closely related RING-type E3 ligases, APD1–4, are redundantly involved in PMⅡ regulation during male gametogenesis [7]. In addition, mutant studies also characterized a large number of genes involved in pollen development by affecting tapetum development and pollen wall formation [8,9].
However, since there are a large number of genes in Arabidopsis, rice, and many other plants that emerge as gene families [10,11], it is not always possible to characterize gene functions only by single-gene mutagenesis. There are many examples showing that mutants with single-gene disruption do not show obvious phenotypes due to genetic redundancy. For instance, Arabidopsis bHLH010, bHLH089, and bHLH090 are redundantly required for anther and pollen development meaning that single mutants can be indistinguishable from wild-type plants [12]. What is even more challenging is that it is sometimes difficult to accurately predict how many genes are functionally redundant in the same biological process. As a complementary or alternative approach to the loss-of-function approach, gain-of-function mutagenesis allows the phenotypes of the gain-of-function mutants to be observed without disturbance from other family members, which is conducive to the functional identification of redundant genes [13]. Moreover, the advantages of the gain-of-function approach also include the abilities of (a) identifying genes that confer stress resistance to plants that arise from the introduction of transgenes, e.g., the overexpression transgenic rice plants of a lipid transfer protein, Oryza sativa Drought-Induced LTP, were more tolerant to drought stress and showed less tapetal defects during the reproductive stage [14]; and (b) uncovering the functions of genes from non-model plants by using a heterologous expression system, e.g., heterologous overexpression of the stamen-specific R2R3-MYB gene BcMF28 of Brassica campestris in Arabidopsis reveals that it plays a critical regulatory role during late stamen development [15]. In addition, for genes that play a vital regulatory role, the phenotypes of a loss-of-function mutant are often the results of the inactivation of most pathways controlled by the gene, which may conceal the biological process directly regulated by this gene. Therefore, sometimes it may be easier to decipher the exact biological functions of the genes by observing the phenotypes of the gain-of-function mutants [16,17,18]. The A. thaliana genome has undergone three paleo-polyploidy events [19]. Brassica campestris (syn. B. rapa) shares these events but with the addition of a whole-genome triplication (WGT) [20], so most genes contain more duplications, which makes it more difficult to characterize gene functions only through the loss-of-function approach.
Cysteine3Histidine (CCCH)-type zinc-finger proteins form a large family that is conserved across eukaryotes. In plants, the number of tandem CCCH-type zinc-finger (TZF) proteins containing two zinc-finger motifs is usually the largest. In Arabidopsis, the TZF proteins with an arginine-rich (RR) region ahead of the TZF motif constitute the largest subfamily (RR-TZF), with a total of 11 members (AtTZF1–11) [21]. Up to now, hundreds of genes have been determined to function in various developmental events during male gametogenesis, either by loss-of-function or gain-of-function approaches. We previously identified eleven male fertility-related CCCH-type zinc-finger protein genes in Chinese cabbage, including a pair of paralogs expressed in pollen during male gametophyte development, named Brassica campestris Male Fertility 30a (BcMF30a) and BcMF30c [22]. BcMF30a and BcMF30c are two non-TZF proteins that contain a CCCH motif and two putative RNA-binding domains (RBDs), RRM and LOTUS. Genetic analysis of the double mutants constructed using CRISPR-Cas9 gene editing technology showed that partial pollen were aborted due to the degradation of pollen inclusions during microgametogenesis. However, the impact of each gene on pollen development and whether there is functional differentiation between BcMF30a and BcMF30c still remains to be ascertained.
Here, we analyzed the expression of BcMF30a and BcMF30c in seedlings and various tissues during reproductive growth. We also used one of the gain-of-function approaches, i.e., overexpressing coding sequences of BcMF30a and BcMF30c by their native promoters, to further investigate their biological functions during male gametogenesis. The results showed that overexpression of BcMF30a or BcMF30c could affect pollen development after microspore formation, and eventually led to pollen abortion. We also speculated that the phenotypes caused by the overexpression may be related to the variable subcellular localization properties of BcMF30a and BcMF30c.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
Brassica campestris L. ssp. chinensis var. parachinensis cv. Youqing 49 was used for the genetic transformation. Chinese cabbage and Arabidopsis were planted in a phytotron at 20 ± 2 °C under a 16/8-h light/dark cycle. Seven-day-old Arabidopsis seedlings (for GUS histochemical staining assay) were grown on synthetic Murashige and Skoog (MS) medium containing 2% Suc and 0.8% agar in a 22 °C growth chamber under a 14-h-light/10-h-dark regime. The tobacco (Nicotiana benthamiana) plants were grown in the growth room at 26 ± 2 °C under a 16/8-h light/dark cycle.
2.2. β-glucuronidase (GUS) Histochemical Staining Assay
The ProBcMF30a:GUS and ProBcMF30c:GUS transgenic Arabidopsis plants were generated earlier [22]. GUS staining of floral buds and other tissues was performed as described by Mudunkothge et al. [23]. Anther stages of Arabidopsis were referred to as described [24]. Images were obtained using a Nikon microscope (ECLIPSE 90i, Nikon, Tokyo, Japan).
2.3. Generation of BcMF30a and BcMF30c Overexpression Transgenic Chinese Cabbage
To construct the BcMF30a and BcMF30c overexpression vectors, the coding regions of BcMF30a (1608-bp) and BcMF30c (1602-bp) were amplified and subcloned into the reconstructed binary vectors, ProBcMF30a:GUS and ProBcMF30c:GUS, which have been constructed in our previous study [22]. Then, the recombinant vectors ProBcMF30a:BcMF30a and ProBcMF30c:BcMF30c were introduced into Chinese cabbage by the Agrobacterium-mediated transformation method as described previously [25]. The positive transformants were identified by the extraction of their DNA and analyzed by PCR screening and sequencing. For partial transgenic T0 line plants, the positive T1 plants were propagated and identified. The primers used are listed in Table S1.
2.4. RNA Extraction and qRT-PCR
Total RNA was extracted from inflorescences of transgenic Chinese cabbage by using RNAiso Plus (Takara, Kyoto, Japan). A PrimerScript RT reagent Kit (Takara, Kyoto, Japan) was used for preparing cDNA. A SYBR® Premix Ex Taq™ Kit (Takara, Kyoto, Japan) was used for quantitative PCR, which was conducted on a CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA). The gene encoding protein of ubiquitin conjugating enzyme (UBC10) was selected as a reference gene to normalize the quantity of total RNA. The qRT-PCR was carried in three technical replicates, and the relative gene expression levels were calculated using the 2−ΔΔCt method. All primers used are listed in Table S1.
2.5. Phenotypic Analyses, Cytological Observation, and Pollen Germination Assay
Pollen viability was investigated by staining with Alexander solution [26]. In vitro and in vivo pollen germination tests, scanning electron microscopy (SEM), and transmission electron microscopy (TEM) analyses were conducted as described by Lin et al. [27] and Xu et al. [22], with minor modifications. The images of Alexander-stained pollen grains, germinated pollens, and fertilized pistils were captured by using a fluorescent microscope (ECLIPSE 90i, Nikon, Japan).
2.6. Subcellular Localization
In our previous studies, we constructed vectors for the subcellular localization experiments, namely pFGC-eGFP-BcMF30a and pFGC-eGFP-BcMF30c [22]. The fusion vectors were transiently transformed into leaf epidermal cells of H2B-RFP transgenic tobacco plants by using an infiltrated method. The subcellular localization of eGFP-fusion proteins was analyzed 48 h after introduction under a confocal laser scanning microscope (A1, Nikon, Tokyo, Japan) with the NIS-elements AR software (Nikon) using ND acquisition with Z movement before and after heat treatment (42 °C for 90 min).
3. Results
3.1. BcMF30a and BcMF30c Share Similar Gene Expression Patterns and Both Are Highly Expressed in Bicellular Pollen
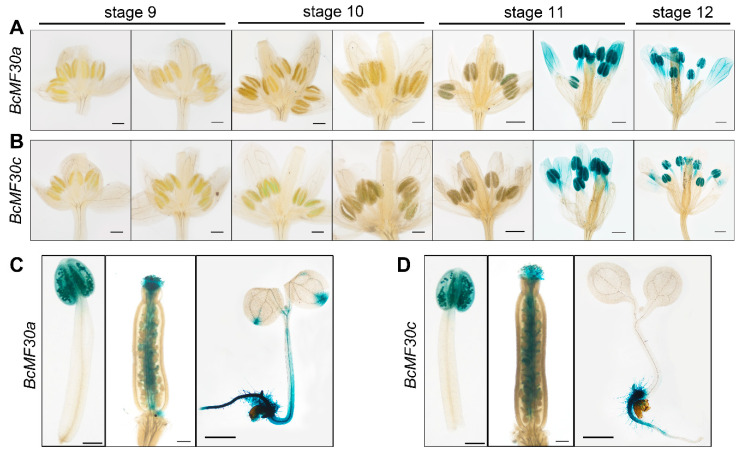
Previous studies in our lab have shown that both BcMF30a and BcMF30c are mainly expressed in pollen and pollen tubes [22]. In order to further determine at which stage of pollen development they are expressed, and whether they are expressed in other tissues, we used previously obtained transgenic plants containing ProBcMF30a:GUS or ProBcMF30c:GUS for further expression analysis. As shown in Figure 1A,B, the GUS signals in anthers reached their peak at late stage 11 and stage 12, indicating that the promoter activity of BcMF30a and BcMF30c promoters reached the highest point in anthers at the late binucleate and trinucleate stages. Since GUS histochemical staining often contaminates surrounding tissues, we performed GUS staining on each flower organ separately. For both BcMF30a and BcMF30c, GUS activity could be detected only in anthers and fertilized pistils (Figure 1C,D), but there was no GUS signal in sepals, petals, and unfertilized pistils (Figure S1). Roots of 7-day-old seedlings also showed clear GUS staining. Nevertheless, the promoter activity could be detected on the apex of cotyledons of ProBcMF30a:GUS seedlings, but not on that of ProBcMF30c:GUS seedlings (Figure 1C,D). Additionally, no GUS staining was observed in leaves, stems, and siliques (Figure S1).
Figure 1.
Spatiotemporal expression patterns of BcMF30a and BcMF30c. β-glucuronidase (GUS) staining of floral buds of ProBcMF30a:GUS (A) and ProBcMF30c:GUS (B) plants at different stages of pollen development. GUS staining of anthers, fertilized pistils, and 7-day-old seedlings of ProBcMF30a:GUS (C) and ProBcMF30c:GUS (D) plants.
3.2. Overexpression of BcMF30a and BcMF30c in B. campestris Causes Reduced Male Fertility
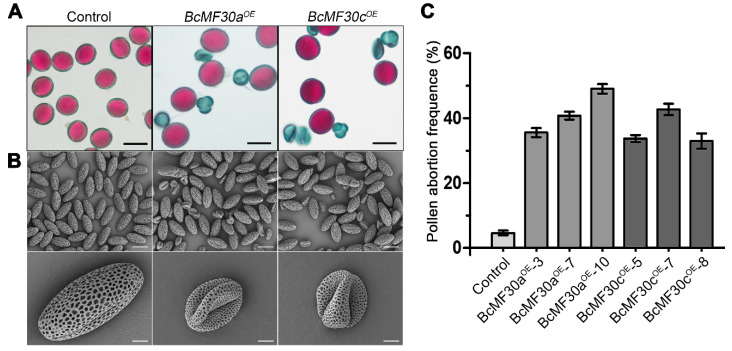
In the previous study, we constructed double-knockout mutants of BcMF30a and BcMF30c in Chinese cabbage and showed that the lack of expression of these two genes could lead to partial pollen abortion and abnormal pollen germination [22]. To further explore the biological functions of these two genes during pollen development, transgenic plants with BcMF30a and BcMF30c coding sequences driven by their native promoters were used for overexpression analysis. A total of 12 and 8 positive T0 transgenic lines (named BcMF30aOE and BcMF30cOE, respectively) were generated after introducing the constructs of ProBcMF30a:BcMF30a and ProBcMF30c:BcMF30c into Chinese cabbage calli. Since BcMF30a and BcMF30c were mainly expressed in pollen, we first paid attention to whether the pollen fertility of these transgenic plants was affected. Alexander staining revealed a reduction (14.1–46.2%) in the viability of pollen grains produced by 11 of 12 BcMF30aOE (except BcMF30aOE-9) and all BcMF30cOE T0 lines (Figure S2A). Of these, BcMF30aOE-1, 3, 7, 8, and 10 and BcMF30cOE-1, 5, 7, and 8 showed the most significant reduction (over 30%) of pollen viability when compared with the control plants. Thus, the progenies (T1 plants) of BcMF30aOE-3, 7, and 10, and BcMF30cOE-5, 7, and 8 were chosen for further analysis. As expected, all T1 plants still showed that more than 30% of pollen grains were nonviable (Figure 2A,C). SEM analysis also revealed that these aborted pollen grains shriveled and collapsed severely (Figure 2B).
Figure 2.
Cytological observation of mature pollen grains from BcMF30aOE and BcMF30cOE transgenic plants of Brassica campestris. Alexander staining (A) and scanning electron micrograph (SEM) observation (B) of BcMF30aOE and BcMF30cOE mature pollen grains. (C) Analysis of pollen abortion frequency in the T1 lines of BcMF30aOE and BcMF30cOE plants. The values are the mean ± SD (standard deviation). Bars = 25 μm in (A), and 30 and 5 μm in (B).
The BcMF30a and BcMF30c mRNA expression levels in BcMF30aOE and BcMF30cOE plants were measured. Interestingly, qRT-PCR analysis revealed that no significant increase in mRNA expression was detected, and there was even a decrease in the mRNA level of BcMF30c in BcMF30cOE-7 (Figure S2B,C). This may be due to the abortion of pollen grains, and the specificity and relatively low activation efficiency of native promoters. Since BcMF30a and BcMF30c are only expressed in developing pollen and pollen tubes during the reproductive growth stage, it is reasonable and predictable that these transgenic plants had normal vegetative growth and floral morphology (Figure S3).
3.3. The Degradation of Microspore Contents Starts from the Late Uninucleate Stage in BcMF30aOE and BcMF30cOE Transgenic Plants
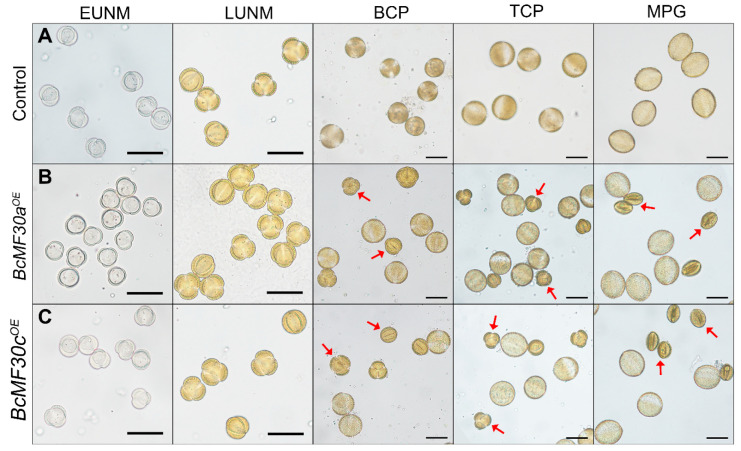
Light microscopy was used to evaluate the morphological changes of microspores during pollen development. No difference was observed between the microspores produced by the control and the transgenic plants during the microsporogenesis phase. However, at the stage of microgametogenesis, some bicellular pollen produced by the transgenic plants were obviously morphologically abnormal, and then further shriveled and aborted (Figure 3).
Figure 3.
The pollen grains produced by BcMF30aOE and BcMF30cOE transgenic plants at different developmental stages were observed by optical electron microscope. Pollen grains produced by the control plants (A), BcMF30aOE plants (B), and BcMF30cOE plants (C). Red arrows indicate the pollens with abnormal development. EUNM, early uninucleate microspore; LUNM, late uninucleate microspore; BCP, bicellular pollen; TCP, tricellular pollen; MPG, mature pollen grain. Bars = 20 μm.
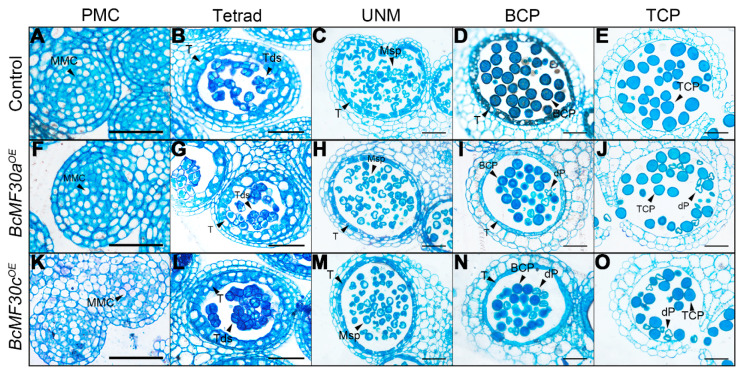
A semi-thin transverse section was conducted to investigate the precise stage at which pollen abnormality begins. No obvious defects were detected inside both BcMF30aOE and BcMF30cOE anther locules from the pollen mother cell stage to the uninucleate stage. However, during the binucleate stage, visible abnormalities appeared in these anthers, in which parts of pollen were deformed. Subsequently, the pollen contents continued to degrade until they disappeared (Figure 4).
Figure 4.
Semi-thin transverse sections of anthers from BcMF30aOE and BcMF30cOE plants of Brassica campestris. Semi-thin sections of anthers from the control (A–E), BcMF30aOE (F–J), and BcMF30cOE (K–O) plants at the pollen mother cell stage (PMC, A,F,K), tetrad stage (Tetrad, B,G,L), uninucleate microspore stage (UNM, C,H,M), bicellular pollen stage (BCP, D,I,N), and tricellular pollen stage (TCP, E,J,O). BCP, bicellular pollen; dP, degenerated pollen; MMC, microspore mother cell; Msp, microspore; T, tapetum; TCP, tricellular pollen; Tds, tetrads. Bars = 25 μm.
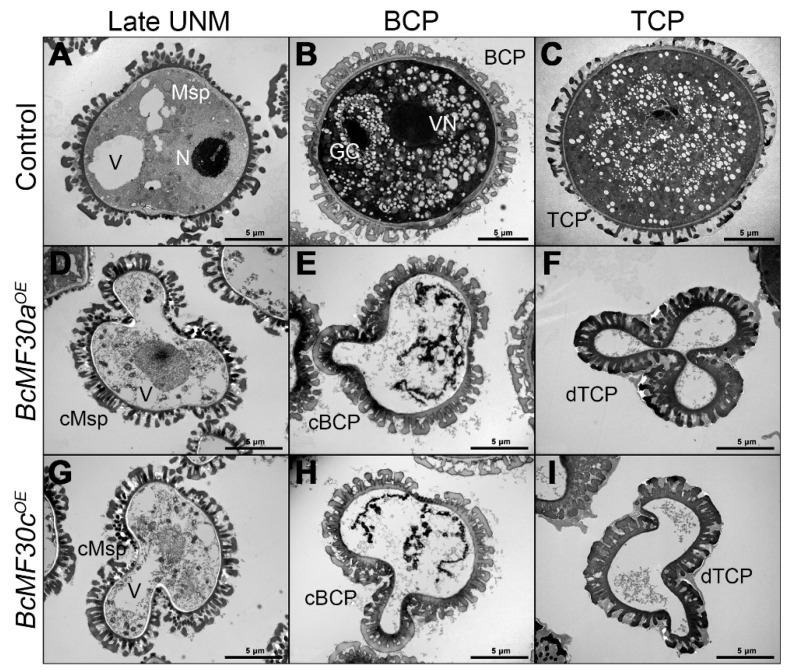
TEM was further performed to confirm the degradation process. As shown in Figure 5, the differences between the control pollen and BcMF30aOE or BcMF30cOE pollen appeared from the late uninucleate stage onwards. By this stage, the round nucleus in the control microspore was moved to the side by the large vacuole. However, the BcMF30aOE and BcMF30cOE microspores were concave in shape, with reduced cytoplasmic contents and no nuclei. At the binucleate stage, the control microspore underwent an asymmetrical mitosis and became a bicellular pollen with a generative cell located in the cytoplasm of the vegetative cell. By contrast, the cellular structures and contents of transgenic pollen further degraded. By the trinucleate stage, the mature pollen grains produced by the control plants were well developed with a dense cytoplasm, whereas the BcMF30aOE and BcMF30cOE pollen shrank severely due to the degradation of the cytoplasm, leaving only the pollen wall that seemed to be intact.
Figure 5.
Transmission electron microscopy (TEM) observation of pollen grains from BcMF30aOE and BcMF30cOE plants of Brassica campestris. Ultrastructure of pollen grains at different developmental stages from the control (A–C), BcMF30aOE (D–F), and BcMF30cOE (G–I) plants. (A,D,G), late uninucleate stage; (B,E,H), bicellular stage; (C,F,I), tricellular stage. BCP, bicellular pollen; cBCP, collapsed BCP; GC, generative cell; Msp, microspore; cMsp, collapsed Msp; N, nucleus; TCP, tricellular pollen; dTCP, degenerated TCP; V, Vacule; VN, vegetative nucleus. Bars = 5 μm.
3.4. In Vitro and In Vivo Pollen Germination Tests in BcMF30aOE and BcMF30cOE Transgenic Plants
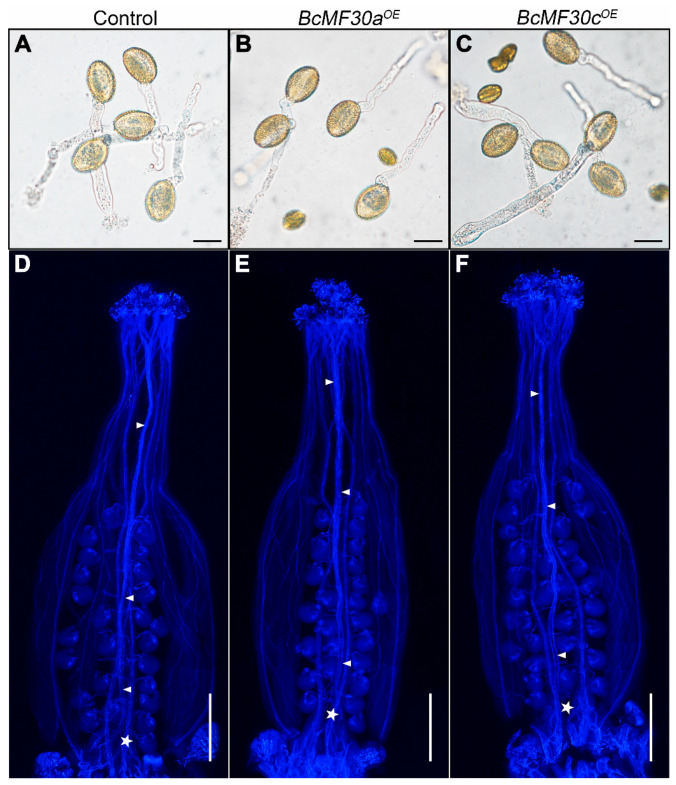
In vitro and in vivo pollen germination experiments were conducted to investigate the differences in pollen germination performance. In vitro, all shrunken pollen grains in both BcMF30aOE and BcMF30cOE plants failed to germinate as expected (Figure 6A–C). Therefore, only the plump spindle-shaped pollen grains in transgenic plants were counted for germination rate statistics. The frequencies of the pollen germination of the BcMF30aOE (89.3%) and BcMF30cOE plants (87.6%) were near to those of the control plants (91.2%). There was also no significant difference in pollen tube growth between the transgenic plants and the control plants in vivo. The results revealed that the numbers of pollen tubes that grew in BcMF30aOE and BcMF30cOE pistils were equivalent to those in the control plants after self-pollination (Figure 6D–F). These observations indicated that although the overexpression of BcMF30a and BcMF30c caused partial pollen to fail to germinate due to abortion, it did not affect the pollination and fertilization process of other viable pollen grains.
Figure 6.
The in vitro and in vivo pollen germination tests of BcMF30aOE and BcMF30cOE transgenic plants. (A–C) The in vitro pollen germination test. (D–F) The in vivo pollen germination and pollen tube growth. The arrows indicate the pollen tubes, and the asterisks indicate the end position of pollen tubes. Bars = 20 μm in (A–C); 100 μm in (D–F).
3.5. BcMF30a and BcMF30c Can Localize to Cytoplasmic Foci
BcMF30a and BcMF30c were reported to be located in both the nucleus and the cytoplasm under normal conditions in tobacco epidermal cells in a previous study [22]. Several studies have revealed that all AtTZFs and multiple other CCCH-type zinc-finger proteins can be localized in cytoplasmic foci similar to processing bodies (PBs) and stress granules (SGs) [28,29,30]. In order to investigate whether BcMF30a and BcMF30c were related to the cytoplasmic foci like PBs and SGs under certain conditions, the tobacco transient expression system was once again used to analyze their subcellular localization.
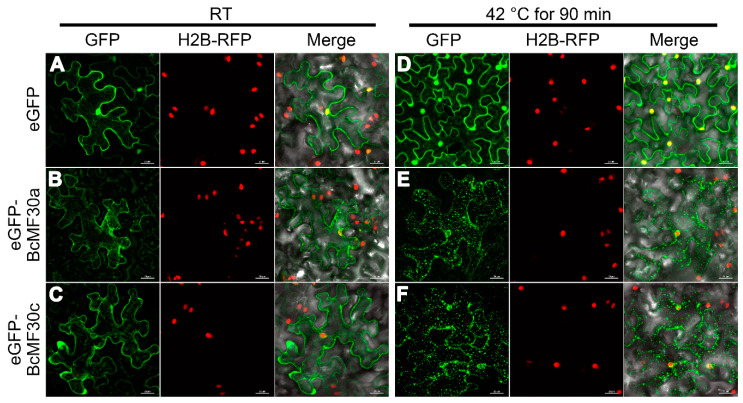
Since heat stress can usually promote or induce the formation of cytoplasmic foci like PB and SG [31], we observed and compared the localization patterns of the transiently expressed GFP fusion proteins in the epidermal cells of H2B-RFP (nuclear localization protein) transgenic tobacco before and after heat treatment. Excitingly, the heat stress changed both eGFP-BcMF30a and eGFP-BcMF30c from being mainly dispersed in the cytoplasm to being aggregated into cytoplasmic foci (Figure 7E,F). In contrast, the free-eGFP still remained as diffused localization after heat stress (Figure 7D). These results indicated that BcMF30a and BcMF30c exhibited variable subcellular localization characteristics and may play roles in mRNA regulation like PBs and SGs.
Figure 7.
BcMF30a and BcMF30c can form cytoplasmic foci in Nicotiana benthamiana leaf epidermal cells after heat treatment. eGFP, eGFP-BcMF30a, and eGFP-BcMF30c were dispersed in the cytoplasm at room temperature (RT) (A–C). After heat treatment at 42 °C for 90 min, eGFP-BcMF30a and eGFP-BcMF30c could form cytoplasmic foci (E,F), while no cytoplasmic foci were observed for eGFP (D). H2B is a marker protein of the nucleus. Pictures represent white field images (Bright), epifluorescence (GFP and RFP), and merged images (Merge). Bar =25 μm.
4. Discussion
The plant CCCH-type zinc-finger proteins play important regulator roles in multiple biological processes, such as the regulation of plant growth and development [32,33,34], adaptation to stress and hormone responses [35,36,37,38,39,40,41,42,43,44,45], and acquisition of immunity against pathogens [46,47,48]. It is worth noting that the biological functions of many CCCH-type zinc-finger protein genes were confirmed by constructing and observing the phenotypes of overexpression transgenic plants, since many single loss-of-function mutants of CCCH-type zinc-finger genes showed a subtle or no phenotype. Interestingly, although the overexpression of many CCCH-type zinc-finger genes is helpful for cells to resist certain stresses, there are some transgenic plants that show growth defects [30,36,40,41,49]. For example, constitutive overexpression of AtC3H14 and AtC3H15/CDM1 in Arabidopsis resulted in a dwarf phenotype and male sterility, respectively [33,50]. In this study, we also found that overexpressing BcMF30a or BcMF30c in Chinese cabbage plants can lead to abnormal pollen development. In order to avoid the impact of ectopic expression on vegetative growth, we used their native promoters to drive overexpression instead of the constitutive promoter CaMV35S, which is commonly used in overexpression experiments. This was also because we used native promoters to ensure that BcMF30a and BcMF30c were not expressed in tissues other than pollen in the floral buds, meaning that we have not detected a significant increase in gene expressions in the inflorescence of transgenic plants. Moreover, some plants even showed a decline in gene expressions due to a large amount of pollen abortion (Figure S2B,C). We observed that the pollen development was disrupted both in BcMF30aOE and BcMF30cOE transgenic plants. Combined with the optical electron microscope observation, semi-thin section, and ultra-thin section results, it was found that the overexpression of BcMF30a and BcMF30c led to the abnormal pollen development from late uninucleate microspores (Figure 3, Figure 4 and Figure 5). Intriguingly, the pollen abortion phenotype caused by the overexpression of these two genes is very similar to the phenotype of their double mutants [22], indicating that the appropriate expression levels of these two genes are essential for maintaining normal male gametogenesis. In addition, since there was no significant difference between the phenotypes of BcMF30aOE and BcMF30cOE plants, we believed that there was a great functional redundancy between these two genes during pollen development. However, there were small differences in the expression patterns in seedlings, in which BcMF30a could be expressed at the cotyledon apex, while BcMF30c could not (Figure 1C,D), indicating that they might have undergone functional differentiation in the morphological construction of the cotyledon apex.
Many studies have shown that the overexpression plants of multiple CCCH-type zinc-finger protein genes (e.g., AtTZF1–6, AtC3H14, and AtC3H15/CDM1, and BcMF30a and BcMF30c in this study) exhibit abnormal growth and development. Does this indicate that there is a general molecular mechanism behind the functions of these genes? Excitingly, multiple studies have found that many TZF proteins displayed special and similar subcellular localization patterns. It has been reported that all members of RR-TZFs (AtTZF1–11) and another two TZFs (AtC3H14 and AtC3H15/CDM1) in Arabidopsis could localize to cytoplasmic foci in maize protoplasts [29]. Arabidopsis AtTZF1, 4, 5, 6, and 9, and rice OsTZF1 have also been shown to be co-localized with the markers of PBs and SGs [30,47,51]. PBs and SGs are two distinct but closely related mRNP granules which are enriched with RNA-binding proteins and translation-repressed or translationally stalled mRNAs, and are important for RNA regulation [31,52,53]. Moreover, it has been found that overexpression of TZFs, some stresses (e.g., salt stress), and hormone treatment (e.g., jasmonic acid and abscisic acid) can induce the formation of TZF-associated cytoplasmic foci [30,39,51]. In this study, we did find that the heat treatment could induce the formation of BcMF30a/BcMF30c-associated cytoplasmic foci in tobacco epidermal cells (Figure 7), indicating that the properties of these two CCCH-type zinc-finger proteins may be similar to AtC3H14, AtC3H15/CDM1, and AtTZFs in Arabidopsis. This also implies that overexpression of BcMF30a and BcMF30c may induce the formation of cytoplasmic foci. This hypothesis was also supported by the research of AtC3H18L, a novel CCCH-type zinc-finger protein identified recently [28]. Studies have shown that AtC3H18L could not only form heat stress-induced cytoplasmic foci in tobacco epidermal cells and co-localize with PB and SG markers, but it could also form cytoplasmic foci in overexpressed transgenic Arabidopsis under normal growth conditions. In mammalians, the overexpression of PB components can induce the formation of abnormally large PBs, which behave abnormally and exert aberrant function [54]. Therefore, we speculated that it might be the overexpression of BcMF30a and BcMF30c in microspores that led to the continuous formation of abnormal cytoplasmic foci, which disrupted the balance of mRNAs and ultimately resulted in pollen abortion. We believed that this hypothesis may also be one of the explanations for the growth inhibition and developmental defects of transgenic plants overexpressing TZFs. However, more evidence is needed to support this hypothesis. In addition, is the enhanced stress resistance of many overexpression transgenic plants of CCCH-type zinc-finger proteins related to the cytoplasmic foci formed by these proteins? Can overexpression of BcMF30a and BcMF30c also enhance stress resistance in a certain aspect? These issues are urgent and worthy of being revealed.
It has been proved that the TZF motif with RNA binding function in AtTZF9, rather than the putative protein–protein interaction ankyrin repeat motif, was partially responsible for its cytoplasmic foci localization [47]. Although there is just one CCCH-type motif in BcMF30a, BcMF30c, and AtC3H18L, they all contain two putative RBDs—RRM and LOTUS [22,28]—which indicates that these two RBDs may be responsible for their specific subcellular localization patterns. Further research is needed to clarify this.
5. Conclusions
The gain-of-function approach is one of the excellent methods for studying genes with redundant functions. Here, we determined that the specific overexpression of BcMF30a or BcMF30c could result in pollen abortion due to the degradation of pollen contents from the late uninucleate microspore stage. Further analysis revealed that these two proteins could form cytoplasmic foci like TZFs in Arabidopsis. Combined with the existing research of TZFs, we speculated that the high overexpression of these two proteins could induce the persistent formation of cytoplasmic foci, which would lead to pollen abortion. Together with the results that double mutants also exhibited male sterility, this suggests that appropriate expression levels of BcMF30a and BcMF30c are critical for maintaining normal pollen development. Future work should explore the functions of BcMF30a and BcMF30c in greater depth to unravel the mechanisms regulating male gametophyte development in Chinese cabbage.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/11/1287/s1, Figure S1: BcMF30a and BcMF30c were not expressed in some floral organs and tissues, Figure S2: Analysis of pollen abortion frequency and gene expression levels in BcMF30aOE and BcMF30cOE transgenic plants of Brassica campestris, Figure S3: Morphological observation of plants and floral organs of BcMF30aOE and BcMF30cOE transgenic plants of Brassica campestris, Table S1: Sequences of primers used in this study.
Author Contributions
Conceptualization, L.X. and J.C.; methodology, L.X., T.L.; validation, L.X., T.L. and X.X.; formal analysis, L.X., X.X., W.L.; investigation, L.X., T.L.; resources, J.C.; data curation, L.X.; writing—original draft preparation, L.X.; writing—review and editing, Y.Y., J.C.; visualization, L.X.; supervision, J.C.; project administration, L.X., J.C.; funding acquisition, J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grant from the National Natural Science Foundation of China (No. 31772311).
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Honys D., Twell D. Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol. 2004;5:R85. doi: 10.1186/gb-2004-5-11-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Østergaard L., Yanofsky M.F. Establishing gene function by mutagenesis in Arabidopsis thaliana. Plant J. 2004;39:682–696. doi: 10.1111/j.1365-313X.2004.02149.x. [DOI] [PubMed] [Google Scholar]
- 3.Twell D. Male gametophyte development. In: Pua E.C., Davey M.R., editors. Plant Developmental Biology-Biotechnological Pespectives. 1st ed. Volume 1. Springer; Berlin/Heidelberg, Germany: 2010. pp. 225–244. [Google Scholar]
- 4.Kim M.-J., Kim M., Lee M.R., Park S.K., Kim J. Lateral Organ Boundaries Domain (LBD) 10 interacts with Sidecar Pollen/Lbd27 to control pollen development in Arabidopsis. Plant J. 2015;81:794–809. doi: 10.1111/tpj.12767. [DOI] [PubMed] [Google Scholar]
- 5.Oh S.-A., Hoai T.N.T., Park H.-J., Zhao M., Twell D., Honys D., Park S.-K. MYB81, a microspore-specific GAMYB transcription factor, promotes pollen mitosis I and cell lineage formation in Arabidopsis. Plant J. 2020;101:590–603. doi: 10.1111/tpj.14564. [DOI] [PubMed] [Google Scholar]
- 6.Zhang W.-T., Li E., Guo Y.-K., Yu S.-X., Wan Z.-Y., Ma T., Li S., Hirano T., Sato M.H., Zhang Y. Arabidopsis VAC14 is Critical for Pollen Development through Mediating Vacuolar Organization. Plant Physiol. 2018;177:1529–1538. doi: 10.1104/pp.18.00495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo G., Gu H., Liu J., Qu L.-J. Four closely-related RING-type E3 ligases, APD1-4, are involved in pollen mitosis II regulation in Arabidopsis. J. Integr. Plant Biol. 2012;54:814–827. doi: 10.1111/j.1744-7909.2012.01152.x. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J., Lou Y., Xu X., Yang Z.-N. A genetic pathway for tapetum development and function in Arabidopsis. J. Integr. Plant Biol. 2011;53:892–900. doi: 10.1111/j.1744-7909.2011.01078.x. [DOI] [PubMed] [Google Scholar]
- 9.Quilichini T.D., Samuels A.L., Douglas C.J. ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell. 2014;26:4483–4498. doi: 10.1105/tpc.114.130484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaul S., Koo H.L., Jenkins J., Rizzo M., Rooney T., Tallon L.J. The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 11.Goff S.A., Ricke D., Lan T.-H., Presting G., Wang R., Dunn M., Glazebrook J., Sessions A., Oeller P., Varma H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 12.Zhu E., You C., Wang S., Cui J., Niu B., Wang Y., Qi J., Ma H., Chang F. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 2015;83:976–990. doi: 10.1111/tpj.12942. [DOI] [PubMed] [Google Scholar]
- 13.Kondou Y., Higuchi M., Matsui M. High-throughput characterization of plant gene functions by using gain-of-function technology. Annu. Rev. Plant Biol. 2010;61:373–393. doi: 10.1146/annurev-arplant-042809-112143. [DOI] [PubMed] [Google Scholar]
- 14.Guo C., Ge X., Ma H. The rice OsDIL gene plays a role in drought tolerance at vegetative and reproductive stages. Plant Mol. Biol. 2013;82:239–253. doi: 10.1007/s11103-013-0057-9. [DOI] [PubMed] [Google Scholar]
- 15.Shen X., Hu Z., Xiang X., Xu L., Cao J. Overexpression of a stamen-specific R2R3-MYB gene BcMF28 causes aberrant stamen development in transgenic Arabidopsis. Biochem. Biophys. Res. Commun. 2019;518:726–731. doi: 10.1016/j.bbrc.2019.08.119. [DOI] [PubMed] [Google Scholar]
- 16.Yu Y., Li Y., Li L., Lin J., Zheng C., Zhang L. Overexpression of PwTUA1, a pollen-specific tubulin gene, increases pollen tube elongation by altering the distribution of alpha-tubulin and promoting vesicle transport. J. Exp. Bot. 2009;60:2737–2749. doi: 10.1093/jxb/erp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Li S., Zhou L.-Z., Fox E., Pao J., Sun W., Zhou C., McCormick S. Overexpression of Arabidopsis thaliana PTEN caused accumulation of autophagic bodies in pollen tubes by disrupting phosphatidylinositol 3-phosphate dynamics. Plant J. 2011;68:1081–1092. doi: 10.1111/j.1365-313X.2011.04761.x. [DOI] [PubMed] [Google Scholar]
- 18.Gui C.-P., Dong X., Liu H.-K., Huang W.-J., Zhang D., Wang S.-J., Barberini M.L., Gao X.-Y., Muschietti J., McCormick S., et al. Overexpression of the tomato pollen receptor kinase LePRK1 rewires pollen tube growth to a blebbing mode. Plant Cell. 2014;26:3538–3555. doi: 10.1105/tpc.114.127381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowers J.E., Chapman B.A., Rong J., Paterson A.H. Ecological and immunological determinants of influenza evolution. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., Bai Y., Mun J.-H., Bancroft I., Cheng F., et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011;43:1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 21.Wang D., Guo Y., Wu C., Yang G., Li Y., Zheng C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008;9:44. doi: 10.1186/1471-2164-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu L., Xiong X., Liu W., Liu T., Yu Y., Cao J. BcMF30a and BcMF30c, Two Novel Non-Tandem CCCH Zinc-Finger Proteins, Function in Pollen Development and Pollen Germination in Brassica campestris ssp. chinensis. Int. J. Mol. Sci. 2020;21:6428. doi: 10.3390/ijms21176428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mudunkothge J.M., Mudunkothge B.A. The GUS reporter system in flower development studies. In: Riechmann J.L., Wellmer F., editors. Flower Development: Methods and Protocols. 1st ed. Volume 1110. Springer; New York, NY, USA: 2014. pp. 295–304. [DOI] [PubMed] [Google Scholar]
- 24.Sanders P.M., Bui A.Q., Weterings K., McIntire K.N., Hsu Y.-C., Lee P.Y., Truong M.T., Beals T.P., Goldberg R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999;11:297–322. doi: 10.1007/s004970050158. [DOI] [Google Scholar]
- 25.Yu X., Cao J., Ye W., Wang Y. Construction of an antisense CYP86MF gene plasmid vector and production of a male-sterile Chinese cabbage transformant by the pollen-tube method. J. Hortic. Sci. Biotechnol. 2004;79:833–839. doi: 10.1080/14620316.2004.11511851. [DOI] [Google Scholar]
- 26.Alexander M.P. Differential staining of aborted and non-aborted pollen. Stain Technol. 1969;44:117–122. doi: 10.3109/10520296909063335. [DOI] [PubMed] [Google Scholar]
- 27.Lin S., Dong H., Zhang F., Qiu L., Wang F., Cao J., Huang L. BcMF8, a putative arabinogalactan protein-encoding gene, contributes to pollen wall development, aperture formation and pollen tube growth in Brassica campestris. Ann. Bot. 2014;113:777–788. doi: 10.1093/aob/mct315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L., Liu T., Xiong X., Liu W., Yu Y., Cao J. AtC3H18L is a stop-codon read-through gene and encodes a novel non-tandem CCCH zinc-finger protein that can form cytoplasmic foci similar to mRNP granules. Biochem. Biophys. Res. Commun. 2020;528:140–145. doi: 10.1016/j.bbrc.2020.05.081. [DOI] [PubMed] [Google Scholar]
- 29.Pomeranz M., Lin P.-C., Finer J., Jang J.-C. AtTZF gene family localizes to cytoplasmic foci. Plant Signal. Behav. 2010;5:190–192. doi: 10.4161/psb.5.2.10988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bogamuwa S., Jang J.-C. The Arabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid- and gibberellic acid-mediated regulation of seed germination. Plant Cell Environ. 2013;36:1507–1519. doi: 10.1111/pce.12084. [DOI] [PubMed] [Google Scholar]
- 31.Guzikowski A.R., Chen Y.S., Zid B.M. Stress-induced mRNP granules: Form and function of processing bodies and stress granules. Wiley Interdiscip. Rev. RNA. 2019;10:e1524. doi: 10.1002/wrna.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu P., Chai M., Yang J., Ning G., Wang G., Ma H. The Arabidopsis CALLOSE DEFECTIVE MICROSPORE1 gene is required for male fertility through regulating callose metabolism during microsporogenesis. Plant Physiol. 2014;164:1893–1904. doi: 10.1104/pp.113.233387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi Z.-H., Zhang C., Xu X.-F., Zhu J., Zhou Q., Ma L.-J., Niu J., Yang Z.-N. Overexpression of AtTTP affects ARF17 expression and leads to male sterility in Arabidopsis. PLoS ONE. 2015;10:e0117317. doi: 10.1371/journal.pone.0117317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chai G., Kong Y., Zhu M., Yu L., Qi G., Tang X., Wang Z., Cao Y., Yu C., Zhou G. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J. Exp. Bot. 2015;66:2595–2609. doi: 10.1093/jxb/erv060. [DOI] [PubMed] [Google Scholar]
- 35.Wang W., Liu B., Xu M., Jamil M., Wang G. ABA-induced CCCH tandem zinc finger protein OsC3H47 decreases ABA sensitivity and promotes drought tolerance in Oryza sativa. Biochem. Biophys. Res. Commun. 2015;464:33–37. doi: 10.1016/j.bbrc.2015.05.087. [DOI] [PubMed] [Google Scholar]
- 36.Lin P.-C., Pomeranz M.C., Jikumaru Y., Kang S.G., Hah C., Fujioka S., Kamiya Y., Jang J.-C. The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 2011;65:253–268. doi: 10.1111/j.1365-313X.2010.04419.x. [DOI] [PubMed] [Google Scholar]
- 37.Kim D.H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., Kamiya Y., Choi G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20:1260–1277. doi: 10.1105/tpc.108.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou T., Yang X., Wang L., Xu J., Zhang X. GhTZF1 regulates drought stress responses and delays leaf senescence by inhibiting reactive oxygen species accumulation in transgenic Arabidopsis. Plant Mol. Biol. 2014;85:163–177. doi: 10.1007/s11103-014-0175-z. [DOI] [PubMed] [Google Scholar]
- 39.Jan A., Maruyama K., Todaka D., Kidokoro S., Abo M., Yoshimura E., Shinozaki K., Nakashima K., Yamaguchi-Shinozaki K. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol. 2013;161:1202–1216. doi: 10.1104/pp.112.205385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee S.-j., Jung H.J., Kang H., Kim S.Y. Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 2012;53:673–686. doi: 10.1093/pcp/pcs023. [DOI] [PubMed] [Google Scholar]
- 41.Huang P., Ju H.-W., Min J.-H., Zhang X., Chung J.-S., Cheong H.-S., Kim C.S. Molecular and physiological characterization of the Arabidopsis thaliana Oxidation-related Zinc Finger 2, a plasma membrane protein involved in ABA and salt stress response through the ABI2-mediated signaling pathway. Plant Cell Physiol. 2012;53:193–203. doi: 10.1093/pcp/pcr162. [DOI] [PubMed] [Google Scholar]
- 42.Kong Z., Li M., Yang W., Xu W., Xue Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006;141:1376–1388. doi: 10.1104/pp.106.082941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seok H.-Y., Nguyen L.V., Park H.-Y., Tarte V.N., Ha J., Lee S.-Y., Moon Y.-H. Arabidopsis non-TZF gene AtC3H17 functions as a positive regulator in salt stress response. Biochem. Biophys. Res. Commun. 2018;498:954–959. doi: 10.1016/j.bbrc.2018.03.088. [DOI] [PubMed] [Google Scholar]
- 44.Wang W., Zheng H., Wang Y., Han G., Sui N. Overexpression of CCCH zinc finger protein gene delays flowering time and enhances salt tolerance in Arabidopsis by increasing fatty acid unsaturation. Acta Physiol. Plant. 2018;40:1842. doi: 10.1007/s11738-018-2775-8. [DOI] [Google Scholar]
- 45.Sun J., Jiang H., Xu Y., Li H., Wu X., Xie Q., Li C. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007;48:1148–1158. doi: 10.1093/pcp/pcm088. [DOI] [PubMed] [Google Scholar]
- 46.Tabassum N., Eschen-Lippold L., Athmer B., Baruah M., Brode M., Maldonado-Bonilla L.D., Hoehenwarter W., Hause G., Scheel D., Lee J. Phosphorylation-dependent control of an RNA granule-localized protein that fine-tunes defence gene expression at a post-transcriptional level. Plant J. 2020;101:1023–1039. doi: 10.1111/tpj.14573. [DOI] [PubMed] [Google Scholar]
- 47.Maldonado-Bonilla L.D., Eschen-Lippold L., Gago-Zachert S., Tabassum N., Bauer N., Scheel D., Lee J. The Arabidopsis tandem zinc finger 9 protein binds RNA and mediates pathogen-associated molecular pattern-triggered immune responses. Plant Cell Physiol. 2014;55:412–425. doi: 10.1093/pcp/pct175. [DOI] [PubMed] [Google Scholar]
- 48.Guo Y.-H., Yu Y.-P., Wang D., Wu C.-A., Yang G.-D., Huang J.-G., Zheng C.-C. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009;183:62–75. doi: 10.1111/j.1469-8137.2009.02838.x. [DOI] [PubMed] [Google Scholar]
- 49.Huang P., Chung M.-S., Ju H.-W., Na H.-S., Lee D.J., Cheong H.-S., Kim C.S. Physiological characterization of the Arabidopsis thaliana oxidation-related zinc finger 1, a plasma membrane protein involved in oxidative stress. J. Plant Res. 2011;124:699–705. doi: 10.1007/s10265-010-0397-3. [DOI] [PubMed] [Google Scholar]
- 50.Kim W.-C., Kim J.-Y., Ko J.-H., Kang H., Kim J., Han K.-H. AtC3H14, a plant-specific tandem CCCH zinc-finger protein, binds to its target mRNAs in a sequence-specific manner and affects cell elongation in Arabidopsis thaliana. Plant J. 2014;80:772–784. doi: 10.1111/tpj.12667. [DOI] [PubMed] [Google Scholar]
- 51.Pomeranz M.C., Hah C., Lin P.-C., Kang S.G., Finer J.J., Blackshear P.J., Jang J.-C. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010;152:151–165. doi: 10.1104/pp.109.145656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kedersha N., Ivanov P., Anderson P. Stress granules and cell signaling: More than just a passing phase? Trends Biochem. Sci. 2013;38:494–506. doi: 10.1016/j.tibs.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parker R., Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 54.Kedersha N., Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.