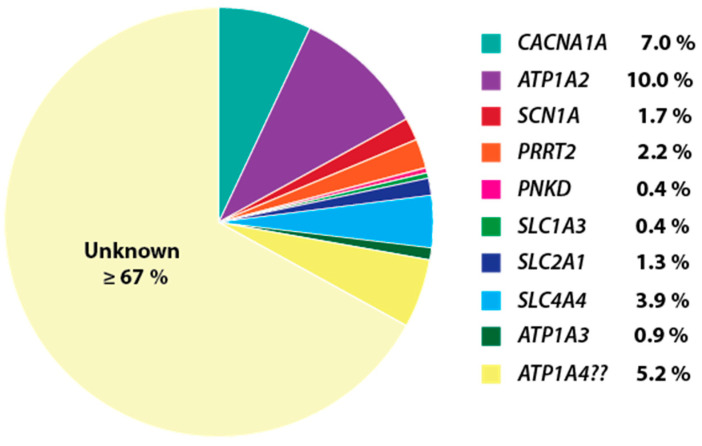
Figure 3.
The prevalence of cases referred for HM gene testing with a known or potential pathogenic variant in previously reported genes (% = estimated percentage contribution). N = 230 individuals were assessed including cases with CACNA1A, ATP1A2, and SCN1A mutations reported in Maksemous et al. [4], older cases diagnosed by Sanger sequencing of targeted exons, and additional recent samples. A total of 18.7% patients referred for HM testing had known mutations or likely pathogenic variants in the CACNA1A (7.0%), ATP1A2 (10.0%), and SCN1A (1.7%) genes. Including mutations and potentially pathogenic variants identified in genes analysed in this study increased the number of cases with a possible genetic diagnosis to 27.8% (not including variants in ATP1A4, which remains putative). For the majority of remaining cases, no rare protein-changing variants were detected in HM genes reported to date.