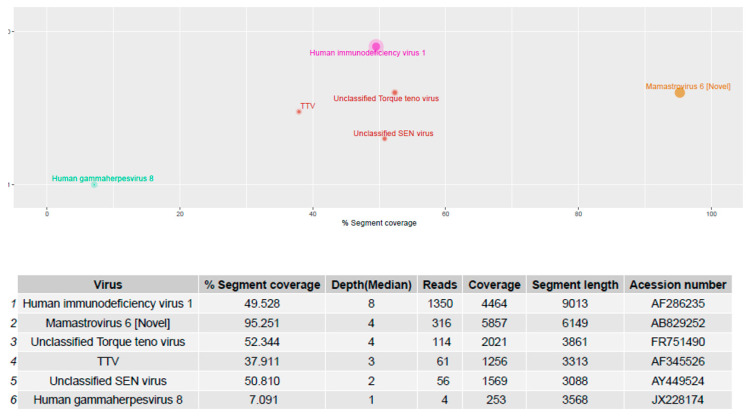
Figure 6.
Human blood samples were sequenced and reads generated using the RNA protocol [9] were aligned to the Virosaurus database. The result is easy to interpret and confirms that the patient was positive for a novel human astrovirus, HIV-1 and HHV-8 sequences, as previously reported [23]. Top panel: 2D representation of detected sequences with %segment coverage in the X-axis, and depth (median) in Y-axis; bottom panel: raw data. Size of dots is relative to number of reads. Anellovirus (TTV) sequences were also detected. The Virosaurus hierarchy allows allocating reads to viral entities: at the level of virus (HIV-1, HHV-8, MastV-6) or higher (TTV). Unclassified SEN viruses are TTV-like genomes. Mamastrovirus (Novel) is a subtyping, allowing differentiating between novel (i.e., MLB and VA/HMO) and classical human astroviruses.