Abstract
Orofacial cleft (OFC) is one of the most prevalent birth defects, leading to substantial and long-term burdens in a newborn’s quality of life. Although studies revealed several genetic variants associated with the birth defect, novel approaches may provide additional clues about its etiology. Using the Center for Craniofacial and Dental Genetics project data (n = 10,542), we performed linear mixed-model analyses to study the genetic compositions of OFC and investigated the dependence among identified loci using conditional analyses. To identify genes associated with OFC, we conducted a transcriptome-wide association study (TWAS) based on predicted expression levels. In addition to confirming the previous findings at four loci, 1q32.2, 8q24, 2p24.2 and 17p13.1, we untwined two independent loci at 1q32.2, TRAF3IP3 and IRF6. The sentinel SNP in TRAF3IP3 (rs2235370, p-value = 5.15 × 10−9) was independent of the sentinel SNP at IRF6 (rs2235373, r2 < 0.3). We found that the IRF6 effect became nonsignificant once the 8q24 effect was conditioned, while the TRAF3IP3 effect remained significant. Furthermore, we identified nine genes associated with OFC in TWAS, implicating a glutathione synthesis and drug detoxification pathway. We identified some meaningful additions to the OFC etiology using novel statistical methods in the existing data.
Keywords: orofacial cleft, mixed model genome-wide association study, expression imputation, transcriptome-wide association study, PrediXcan, EPACTS
1. Introduction
Orofacial cleft (OFC) is a group of congenital deformities showing a gap or breakage in oral features. The gap most commonly appears in the palate (CP; cleft palate), which is the “roof” of the mouth, upper lip (CL; cleft lip) or both (CLP; cleft lip and cleft palate). The majority (~70%) of OFC is nonsyndromic, without additional features [1]. Although it varies among populations, the prevalence of all OFC makes it one of the most common birth defects (approximately 1.2 out of 1000 live births worldwide) [2]. OFC is a significant public health challenge since newborns with these anomalies encounter substantial surgical, nutritional, dental, speech, medical and behavioral challenges in the later phases of their lifetimes, leading to nominal economic and societal burdens [3]. The formation of the head and the face in an embryo is a biological event elaborately coordinated by signaling and pathways [1]. The etiology of OFC is considered a complex disorder that involves not only genetic influences but also interactions with environmental factors.
Studies have shown that genetic components play a significant role in OFC; however, the specific genetic architecture behind the etiology is still unclear. OFC showed a strong familial aggregation in that relatives of cleft cases showed higher risks of being affected than the general population, and the risk decreased as the genetic distance increased [4,5]. Twin studies also confirmed the role of genetics in the OFC status in monozygotic twins that showed higher concordance rates than dizygotic twins [6]. While segregation analyses rarely found definitive results [7], linkage studies mapped several loci for OFC [8,9,10]. Recently, genome-wide association studies (GWAS) in OFC have successfully identified common and rare variants, and their interactions with environmental factors [11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26]. Besides, there have been active investigations on candidate genes such as RUNX2, GREM1, MTHFR, folate-related genes, ESRRG and SNAI1 [27,28,29,30,31]. However, the functional implications of these loci are still obscure, except for IRF6 [32,33]. In addition to association studies with human genotypes and phenotypes, the functional mechanisms of OFC genes were investigated using animal models. Earlier animal studies have demonstrated the role of Irf6 gene in epithelial proliferation and differentiation, which are crucial for craniofacial morphogenesis and ectodermal formation [34]. An understanding of the Irf6 mechanism has been furthered by a recent finding that showed the role of Irf6 in the osteoblast differentiation of craniofacial bones [35]. Although animal models can help us understand the functional mechanisms involved in human OFC etiology, there are certainly limitations such as the limited number of homologs of human OFC genes and the inability to evaluate the roles of common human genetic variations in animal models.
We performed secondary analyses on Center for Craniofacial and Dental Genetics (CCDG) data on dbGaP (accession: phs000774.v2.p1) in order to understand the contributions of different genetic components to OFC using novel statistical strategies. Our study is aimed at improving the current understanding in OFC genetics for the following reasons. First, we performed a mega-analysis of samples from multiple ethnic groups together by an applying mixed model-based genetic association analysis with an estimated genetic relatedness matrix. The previous study [19] analyzed case-parent trios and the unrelated case-control set separately, and then meta-analyzed them. Furthermore, to adjust for the population structure in the case-control analysis, the principal components (PCs) of ancestry were estimated using the maximal subset of unrelated subjects and a pruned set of SNPs. Our study design reduces the complexity of the analysis while taking the full set of genetic information. Second, we performed a conditional analysis to examine independent genetic influences among multiple OFC loci. Last, we explored the functional implications of human genetic variations on OFC etiology utilizing a computational model that predicted gene expression levels based on the genetic variation in various types of human tissues.
2. Materials and Methods
2.1. Data Acquisition and Subjects
Genotype and phenotype data were accessed from the dbGaP repository with the accession ID phs000774.v2.p1. The repository includes nonsyndromic cleft cases and their noncleft relatives, as well as controls (n = 11,925), mostly from studies led by the University of Pittsburgh Center for Craniofacial and Dental Genetics (CCDG) and also from genetic studies by the University of Iowa. The detailed specifics of the cohort were previously described [19,36], but to summarize briefly, samples were genotyped for 589,945 SNPs on the Illumina Human610-Quadv.1_BBeadChip (Illumina Inc., San Diego, CA, USA), and genotypes were phased using SHAPEIT (Olivier Delaneau, Paris, France and Oxford, United Kingdom). Phased genotypes were imputed with IMPUTE2 software (Bryan Howie and Jonathan Marchini, Oxford, UK) using the 1000 Genomes Phase 1 reference panel (The 1000 Genomes Project Consortium). QC procedures of genotype data are publicly available online at the CCDG study website (http://www.ccdg.pitt.edu/docs/Marazita_ofc_QC_report_feb2015.pdf). The types of nonsyndromic OFC were originally categorized as either cleft lip, cleft palate, or cleft lip and palate. In this study, any cleft types (cleft lip and/or palate) were treated as a case (“OFC”), and subjects without any OFC phenotype as a “control”. Subjects with both the genotypes and phenotypes (n = 10,542) were included in this study.
2.2. Conditional Genome-Wide Association Study Based on Efficient Mixed-Model Association Expedited (EMMAX) Model
We used the Efficient Mixed Model Association eXpedited (EMMAX) algorithm for the mixed model analysis of common variants (minor allele frequency > 0.01) [37]. The population structure and sample relatedness were modeled using a kinship matrix estimated with the Balding–Nichols algorithm, which is the default method of the ‘make-kin’ function in the EPACTS pipeline that includes the EMMAX software (Hyun Min Kang, Ann Arbor, MI, USA) (https://genome.sph.umich.edu/wiki/EPACTS). Variants with minor allele frequencies greater than 0.01 and call rates greater than 95% were used to construct the kinship matrix. Modeling this genetic covariance as a random effect, the fixed SNP effect is tested for association with the binary cleft trait. The EPACTS pipeline includes a suite of association tests for genetic association analyses for population-level samples including EMMAX. An association p-value smaller than 5.0 × 10−8 was considered as genome-wide significant. To investigate the dependence among risk loci, we also performed conditional GWAS, adjusting for the effect of the sentinel association. This process was repeated for nine rounds in order to identify ten independent loci.
2.3. Transcriptome-Wide Association Study (TWAS) Using Predicted Expression Levels
We estimated gene expression levels from our genotype data based on the publicly available tissue-specific gene expression database (i.e., GTEx version 6 data) (Broad Institute, Boston, MA, USA), and we performed a transcriptome-wide association study of OFC using PrediXcan (Eric R Gamazon, Chicago, IL, USA) [38,39]. We chose four tissue types (i.e., whole blood, skeletal muscle, visceral adiposity and subcutaneous adiposity) that were relatively more relevant to OFC. Since the current gene expression prediction model cannot model sample relatedness and most of the GTEx samples were from Caucasians [38], we applied PrediXcan analyses on unrelated Caucasian CCDG samples only (n = 1220). To adjust for the cryptic population structure in this population, 10 principal components (PCs) were calculated and adjusted for in the following association analysis. Association tests were performed using the ‘glm’ function of the software R (Ross Ihaka and Robert Gentleman, Auckland, New Zealand). The number of genes included in four tissue-specific TWASs were 6736 in whole blood, 7220 in subcutaneous adipose tissue, 4527 in visceral adipose tissue and 6611 in skeletal muscle tissue. The significance thresholds for TWASs were set at Bonferroni levels with the number of genes in each tissue-specific transcriptome (p-value < 7.42 × 10−6 for a whole-blood TWAS, 6.93 × 10−6 for a subcutaneous adipose tissue TWAS, 1.10 × 10−5 for a visceral adipose tissue TWAS and 7.56 × 10−6 for a skeletal-muscle tissue TWAS). Significant genes were tested for enrichment based on the gene ontology data (MSigDB c5) using the GENE2FUNC function of FUMA GWAS [40,41]. For the candidate gene analysis, we used all genes reported from previous linkage and association studies (Table S1). We searched the 27 overlapping candidate genes from our PrediXcan analysis results. The thresholds used in the candidate gene analyses were adjusted with a Bonferroni correction based on the number of candidate genes in each tissue data, so significance thresholds were 0.005 for skeletal muscle and 0.003 for the remaining three tissues.
3. Results
We included 10,542 subjects in the mixed model association analysis, and the demographic characteristics are described in Table 1. The male-to-female ratio is similarly balanced in both the case and control groups (43.3% and 58.5%, respectively). The mean ages of the case and the control groups are 12.06 and 28.66 years (standard deviations of 13.03 and 16.95 years), and the control group is significantly older because it includes parents without cleft. This study population contains nine different ethnic groups: Caucasians and Asians largely represent the population (43.93% and 19.13%, respectively), while Africans, Native Americans and mixed populations compose the remaining 36.94%.
Table 1.
Demographic Information.
| Characteristics | Categories | Control (n = 7839) | Case (n = 2703) |
|---|---|---|---|
| Cleft Type | Cleft Lip only | - | 620 |
| Cleft Lip and Palate | - | 1771 | |
| Cleft Palate only | - | 312 | |
| Sex | Male | 3397 (43.3%) | 1581 (58.50%) |
| Female | 4442 (56.7%) | 1122 (41.50%) | |
| Age | 28.66 (±16.95) | 12.06 (±13.03) | |
| Race | African | 219 (2.79%) | 160 (5.92%) |
| Asian | 1433 (18.28%) | 584 (21.60%) | |
| Caucasian | 3563 (45.45%) | 1068 (39.51%) | |
| Caucasian and African | 33 (0.42%) | 11 (0.41%) | |
| Caucasian and Asian | 2 (0.026%) | 2 (0.074%) | |
| Caucasian and Native American | 768 (9.80%) | 171 (6.33%) | |
| Caucasian, African and Native American | 1617 (20.63%) | 665 (24.60%) | |
| Native American | 152 (1.94%) | 37 (1.37%) | |
| Unspecified | 52 (0.66%) | 5 (0.18%) | |
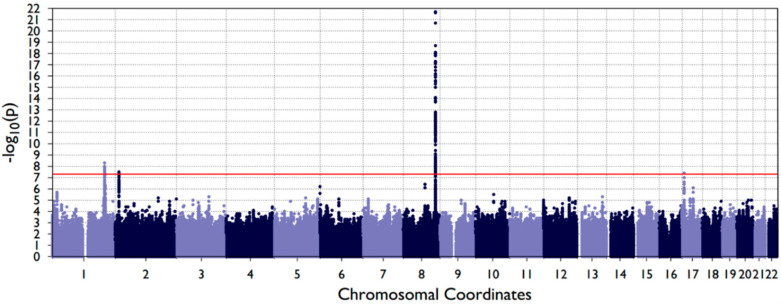
We identified 289 genome-wide significant associations for OFC (Figure 1) at previously reported loci [19]. The population structure was adequately controlled, and no systematic inflation was observed in the Q-Q plot (Figure S1). The majority of significant associations, 240 out of 289, were from the genomic region called the “gene desert” (8q24) [42]. Other genome-wide significant associations were mapped to IRF6, TRAF3IP3, FAM49A and NTN1 (Table 2). The sentinel SNPs in TRAF3IP3 (rs2235370, p-value = 5.15 × 10−9) were reported collectively as IRF6 in the previous GWAS [19]. The sentinel SNP for TRAF3IP3 (rs2235370) was not in LD with the sentinel SNP for IRF6 (rs2235373) in any population (r2 = 0.07 in European, 0.31 in Asian, 0.05 in African and 0.24 in Hispanic), indicating that the two association signals were independent. At a less stringent significance threshold (p-value < 1.00 × 10−5), 21 association signals, including loci in PAX7 and DCAF4L2, were identified.
Figure 1.
Manhattan plot of OFC GWAS using the EMMAX model. SNPs are positioned along the x-axis according to chromosomal position, with −log10 (p-value) along the y-axis. The genome-wide significance threshold (p-value = 5 × 10−8) is presented as a red horizontal line.
Table 2.
Genome-wide significant loci from the EMMAX analysis and their significance in the original multiethnic analysis.
| Locus | EMMAX Analysis | The Multiethnic Meta-Analysis [19] | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sentinel SNP | Chr:Pos | Ref/Alt | MAF | β (S.E.) | p-Value | Gene | Sentinel SNP | Meta OR † | p-Value | r2 ‡ | |
| 8q24 | rs72728755 | 8:129990382 | T/A | 0.16 | 0.08 (0.008) | 1.94 × 10−22 | Gene desert | rs17242358 | 2.00 [1.78–2.26] | 5.63 × 10−30 | 0.97 |
| 1q32.2 | rs2235370 | 1:209946027 | C/A | 0.13 | −0.05 (0.009) | 5.15 × 10−9 | TRAF3IP3 | rs11119345 | 1.81 [1.57–2.07] | 2.52 × 10−17 | 0.84 |
| rs2235373 | 1:209963803 | G/A | 0.26 | −0.04 (0.007) | 2.12 × 10−8 | IRF6 | 0.30 | ||||
| 2p24.2 | rs7552 | 2:16733928 | A/G | 0.50 | 0.04 (0.006) | 3.40 × 10−8 | FAM49A | rs7552 | 1.28 [1.17–1.40] | 4.22 × 10−8 | 1.00 |
| 17p13.1 | rs11273201 | 17:8930219 | AACCCAAAACCCAC/A | 0.27 | −0.04 (0.007) | 3.58 × 10−8 | NTN1 | rs11273201 | 1.44 [1.29–1.59] | 7.84 × 10−12 | 1.00 |
Chr = chromosome; Pos = position in base pairs (hg19); Ref = reference allele; Alt = alternate allele; MAF = minor allele frequency, S.E. = standard error of β. † Odds ratio from the meta-analysis of the case-parent trio analysis and case-control analysis. ‡ r2 LD statistics against all populations of 1000 Reference Genome.
To further understand the individual genetic contributions of these markers, we performed conditional analyses. In Table 3, the top ten associations from each sequential conditional GWAS are presented. Manhattan plots from individual conditional analyses are shown in Figure S2. The first genome-wide analysis conditioned on the most significant marker (rs72728755 at 8q24, p-value = 2.70 × 10−22) identified the next independently significant association, rs7552, which was in the 3′-UTR of FAM49A, a protein-coding gene at 2p24.2 (p-value = 2.67 × 10−8). This signal was also genome-wide significant but was the fifth significant signal in the first analysis. This may indicate that 8q24 biases the effect of 2p24.2 toward the null association when tested together. Conditioning on the first two variants, rs72728755 and rs7552 (8q24 and 2p24.2), the next most significantly associated variant was rs2235370 at 1q32.2 in the intron of TRAF3IP3 (p-value = 1.33 × 10−8). When we adjusted for the 8q24 effect, the associations in IRF6 became insignificant, although a genome-wide significance was still identified in TRAFIP3. This may indicate that the effect of 8q24 was highly correlated with IRF6 but not with TRAFIP3.
Table 3.
Top association from each step of the EMMAX conditional association analysis.
| No. | SNP ID | Chr:Pos | Ref/Alt | MAF | β (S.E.) | p-Value | Locus/Gene |
|---|---|---|---|---|---|---|---|
| 1 | rs72728755 | 8:129990382 | T/A | 0.16 | 0.08 (0.008) | 1.94 × 10−22 | 8q24 |
| 2 | rs7552 | 2:16733928 | A/G | 0.50 | 0.03 (0.006) | 2.67 × 10−8 | FAM49A |
| 3 | rs2235370 | 1:209946027 | C/A | 0.13 | −0.05 (0.009) | 1.33 × 10−8 | TRAF3IP3 |
| 4 | rs16957821 | 17:8948104 | C/G | 0.24 | 0.04 (0.007) | 1.40 × 10−7 | NTN1 |
| 5 | rs12600562 | 17:44977040 | G/T | 0.47 | −0.03 (0.006) | 7.06 × 10−7 | LRRC37A2 |
| 6 | rs12543318 | 8:88868340 | C.A | 0.45 | −0.03 (0.006) | 5.43 × 10−7 | 8q21 |
| 7 | rs3845903 | 3:66517670 | C/T | 0.03 | −0.10 (0.020) | 2.79 × 10−6 | LRIG1 |
| 8 | rs10778143 | 12:102094643 | A/C | 0.33 | 0.03 (0.007) | 1.76 × 10−6 | CHPT1 |
| 9 | rs6929507 | 6:1632072 | T/C | 0.01 | 0.13 (0.027) | 2.14 × 10−6 | GMDS |
| 10 | rs138322543 | 9:100641771 | CCACCA/C | 0.27 | 0.03 (0.007) | 4.82 × 10−6 | TRMO |
Abbreviations: No. = The sequential order of the conditional analysis; Chr = chromosome; Pos = position in base pairs; Ref = reference allele; Alt = alternate allele; MAF = minor allele frequency.
We additionally found three independent associations at a suggestive significance level (p-value < 1.00 × 10−6). Conditioning on three genome-wide significant loci, rs16957821 was identified within NTN1 at 17p13.1 (p-value = 1.40 × 10−7), which had been previously reported by CL/P and OFC (cleft lip and/or palate) GWAS and replication studies [15,16,19,22,36]. The tenth independently associated variant (rs138322543, p-value = 5.43 × 10−7) is not within a gene, but the nearest gene is DCAF4L2, which was previously identified by OFC GWAS [15,16,19].
From our TWAS analyses on four tissue types (whole blood, skeletal muscle, subcutaneous adipose and visceral adipose tissues), we found nine genes with transcriptome-wide significant associations (p-value < 2.00 × 10−6) (Table 4 and Figure S3). THEM79 and GLMP were predicted to be significantly associated with OFC in whole blood, DDT in skeletal muscle and subcutaneous adipose tissues, and DDTL, MIF, MIF-AS1, GSTT2 and USP2 in subcutaneous adipose tissue. Previously, MIF was reported in association with susceptibility to juvenile arthritis [43], GSTT2 with S-phenylmercapturic acid levels in smokers [44], and USP2 suggestively with urate levels [45]. In the visceral adipose tissue, we identified four genes that overlapped or related to the subcutaneous adipose tissue result: MIF, MIF-AS1, DDTL, GSTT2, with the addition of GSTT2B. More genes with suggestive evidence of associations are described in Table S2. From the gene ontology analyses, we found that MIF and DDT were enriched in terms of intramolecular oxidoreductase activity (adjusted p-value = 1.31 × 10−2) and the activity transposing carbon-carbon double bonds (adjusted p-value = 3.11 × 10−3); GSTT2B and GSTT2 were enriched in terms of glutathione transferase activity (adjusted p-value = 7.00 × 10−3), as well as in terms of the alkyl or aryl group transferase activity (adjusted p-value = 1.59 × 10−2)
Table 4.
Genes identified from the association analysis of the predicted expression levels.
| Gene | Ensembl ID | GRCh37 Coordinate | β (S.E.) | z | p-Value |
|---|---|---|---|---|---|
| Whole blood | |||||
| THEM79 | ENSG00000163472 | 1: 156,252,726–156,262,976 | 2.71 (0.56) | 4.87 | 1.13 × 10−6 |
| GLMP | ENSG00000198715 | 1: 156,259,880–156,265,480 | 1.57 (0.33) | 4.83 | 1.37 × 10−6 |
| Skeletal muscle | |||||
| DDT | ENSG00000099977 | 22: 24,313,554–24,322,660 | 1.12 (0.21) | 5.25 | 1.52 × 10−7 |
| Adipose (Subcutaneous) | |||||
| MIF | ENSG00000240972 | 22: 24,236,191–24,237,414 | 1.79 (0.28) | 6.39 | 1.62 × 10−10 |
| MIF-AS1 | ENSG00000218537 | 22: 24,236,613–24,241,117 | 1.39 (0.24) | 5.87 | 4.25 × 10−9 |
| DDT | ENSG00000099977 | 22: 24,313,554–24,322,660 | 2.94 (0.54) | 5.43 | 5.66 × 10−8 |
| GSTT2 | ENSG00000099984 | 22: 24,322,249–24,326,106 | −1.42 (0.28) | −5.16 | 2.46 × 10−7 |
| USP2 | ENSG00000036672 | 11: 119,225,925–119,252,436 | −1.75 (0.38) | −4.57 | 4.93 × 10−6 |
| DDTL | ENSG00000099974 | 22: 24,309,089–24,314,721 | −1.46 (0.32) | −4.54 | 5.68 × 10−6 |
| Adipose (Visceral) | |||||
| MIF | ENSG00000240972 | 22: 24,236,191–24,237,414 | 1.66 (0.26) | 6.52 | 7.09 × 10−11 |
| DDTL | ENSG00000099974 | 22: 24,309,089–24,314,721 | −2.60 (0.45) | −5.81 | 6.37 × 10−9 |
| MIF-AS1 | ENSG00000218537 | 22: 24,236,613–24,241,117 | 1.10 (0.19) | 5.78 | 7.31 × 10−9 |
| GSTT2 | ENSG00000099984 | 22: 24,322,249–24,326,106 | −1.05 (0.19) | −5.53 | 3.27 × 10−8 |
| GSTT2B | ENSG00000133433 | 22: 24,299,601–24,303,373 | −1.36 (0.28) | −4.94 | 7.92 × 10−7 |
We also looked up previously reported candidate genes in our four tissue-specific TWAS (Table S3). We listed 34 OFC loci reported from previous GWASs, including suggestive evidence in the largest and most recent meta-analysis [22,46]. Of these 34 candidate genes, the GTEx database contained 27 genes at 25 loci (Table S1); the number of available candidate genes varied by tissues (12 in whole blood, 11 in skeletal muscle tissue, seven in subcutaneous adipose tissue and eight in visceral adipose tissue). Bonferroni significance levels were also differently set depending on the tissues, with 0.004 for whole blood and skeletal muscle tissue, 0.007 for subcutaneous adipose tissue and 0.006 for visceral adipose tissue. RHPN2 at 19q13.11 was identified as being Bonferroni-significant from skeletal muscle and subcutaneous adipose tissue transcriptome association tests (p-value = 1.90 × 10−3 and 1.45 × 10−3). In both tissues, the upregulation of RHPN2 had a beneficial effect on the risk of OFC.
4. Discussion
Since we performed association analyses on OFC (cleft lip and/or palate), including “cleft palate only” cases, a direct comparison to GWASs of “cleft lip with or without cleft palate case” may not be eligible. However, we hypothesize that there is a genetic variant that has influence on the shared genetics between two related phenotypes. A recent meta-analysis performed GWAS on three cleft phenotypes, including two well-studied phenotypes, CL/P and CP, and additionally a constitute phenotype, OFC (the same as “OFC” in the study) [22], where they identified a novel locus, FOXE1, in association with OFC but not with CL/P or CP. FOXE1 had been reported in linkage studies and a candidate gene setting [8,9,10] but had never been identified by prior GWAS. This study showed that seven out of ten genome-wide association signals from CL/P and CP studies could be captured with the OFC study. Three out of the ten genome-wide significant signals were not recaptured by the OFC study, and two of these (3q28 and 15q24) were from the whole population analysis, while the other one (17q23.2) was from the European analysis; however, only the last one was replicated by a multiethnic GWAS [19].
A mixed-model analysis can not only adjust for population structures, but it can also adjust for sample relatedness, making the approach especially useful for multiethnic family studies. To account for the population structure, the previous CCDG GWAS study [19] estimated the principal components for the pooled ancestry based on the subset of subjects and SNPs. On the other hand, the EMMAX analysis accounts for the population structures, including those of excluded ancestries, by building up genetic relationships between all pairs of subjects based on all available SNPs. This additional information on the genetic covariance between subjects and the population structure enabled our study to identify a comparable set of loci associated with OFC. At the genome-wide significant level (p = 5.00 × 10−8), the CCDG GWAS [19] identified seven loci in the pooled population, i.e., 19q13 near RHPN2, PAX7 at 1p36, ARHGAP29 at 1p22, IRF6 at 1q32, 8q24 and NTN1 at 17p13, and a novel locus 2p24 near FAM49A. In this study, we identified five genes at four loci (8q24, IRF6 and TRAF3IP3 at 1q32, FAM49A at 2p24 and NTN1 at 17p13). All four loci identified by the CCDG GWAS [19] were recapitulated, and the other three genes, 19q13 near RHPN2, PAX7 at 1p36 and ARHGAP29 at 1p22, were not. However, we suggest that TRAF3IP3 should be considered as a novel finding, one independent from IRF6. Compared to the traditional linkage or trio-based methods, the linear mixed-model analysis can be beneficial to a larger sample size for variants with a lower penetrance. At the suggestive significance level (p-value < 1.00 × 10−6), we identified several novel genes, DIEXF, another gene at 1q32.2 (p-value = 5.51 × 10−8), DCAF4L2 at 8q21.3 (p-value = 3.64 × 10−7), and a novel locus GMDS at 6p25.3 (p-value = 6.70 × 10−7).
We observed that many association signals showed different allele frequency distributions between subpopulations (Table S4), which supports the idea that we need to pay extensive attention to controlling the population structure in genetic analyses. Recently, meta-analyses of multiple races showed that a set of genome-wide significant loci considerably differed by racial groups [22]. Furthermore, a GWAS in a Chinese population suggested that the roles of SNPs at the well-known 1q32.2 locus were different in the Chinese haplotype structure [17]. In this study, the most significant SNP was at the well-known 8q24 locus, and allele frequencies for this SNP (rs72728755) varied in subpopulations; the alternate allele (A) is rare in Asians and Native Americans but is quite prevalent in other populations. Allele frequencies of the top SNP at the 2p24.2 locus show that the ‘G’ allele is minor only in Caucasians but major in all other populations. In a previous CCDG GWAS [19], stratified analyses were performed on Asians and Caucasians, but this locus was not reported, which means that the 2p24.2 signal was detectable by the addition of other population samples in our mixed-model analysis. At the locus of 1q32.2, the alternate allele does not exist in Africans and is uncommon in Caucasians, while it is common in Asian, Native American and other mixed populations. The TRAF3IP3 gene was also not previously identified from the previous CCDG analysis, but it is likely that adding other populations improved the statistical power to detect this locus.
While our initial GWAS results were consistent with previous findings [46], some signals were not independent of each other. Conditioning on 8q24, the effect of 1q32.2 diminished and strengthened another signal at 2p24.2 (FAM49A) which had been less significant than that from 1q32.2. This could indicate that 8q24 is strongly associated with 1q32.2 and that the effect from these loci masks the effect from 2p24.2. Interestingly, the effect of one of the most well-established loci in the OFC association, IRF6, was removed after adjusting for the 8q24 effect, but the effect of a marker in TRAF3IP3 at the same locus (1q32.2) survived. TRAF3IP3 is a gene encoding the TRAF3-interacting protein 3, and TRAF3 encodes a TNF receptor-associated factor 3. Although TRAF3IP3 has never been identified by previous linkage and GWA studies, the gene is proximate to IRF6 at the 1q32.2 locus, suggesting that TRAF3IP3 may play a regulatory role for IRF6. From our analysis, this TRAF3IP3 gene was associated with a lower risk of cleft (β = −0.0518). Additionally, after adjusting the effects of 8q24, 2p24.2 and 1q32.2, one of the genome-wide significant loci from the initial analysis, 17p13.1 (NTN1), became insignificant. This can also suggest that the 17p13.1 locus is closely associated with the three loci adjusted in the step, especially with 8q24 and 1q32.2, which had been reported together with 17p13.11 by Leslie et al. [19].
We identified significant associations in the predicted expression TWAS, and our result may add novel evidence to OFC etiology. Two human h-class isoenzymes, GSTT1 and GSTT2, are both located at 22q11.2 and are separated by approximately 50 kb. Previously, copy-number variations (CNVs) at 22q11.2 were reported in association with a low birth weight and preterm delivery outcomes [47]. An increased risk of cleft lip with or without a cleft palate has been associated with maternal smoking in children with null polymorphisms in GSTT1, implicating that it may modify the effect of maternal smoking by involving the removal of toxic compounds generated from smoking [48,49,50]. The expression level for GSTT1 was not included in our TWAS, but the isoenzyme coding gene GSTT2 was identified with a significance. The messenger RNA expression of GSTT2 was reported as being associated with Charcot–Marie–Tooth disease, which is one of the most common inherited neurological disorders [51]. GSTT1 and GSTT2 genes are known to be involved in glutathione synthesis and drug detoxification [52,53]. Additionally, a GWAS of liver enzymes reported that a SNP (rs2739330) associated with a serum γ–glutamyl transferase enzyme was an eQTL for DDT, DDTL, GSTT1 and MIF [54], suggesting a shared genetic control over our TWAS genes. By including another closely located TWAS significant gene, GSTT2B, our TWAS result may imply glutathione synthesis and a drug detoxification-related OFC etiology.
Our study exhibits several limitations. OFC prevalence varies among populations, and some genes identified from previous GWASs showed distinct patterns by population in our analyses (e.g., the desert region in 8q24) [13]. This might indicate a differential transcriptomic variation of OFC-related genes in populations. Since the GTEx database has mainly developed expression datasets for Caucasians, this study also included subjects with Caucasian ancestry only. Thus, findings from our TWAS analysis would not apply to races other than Caucasians. Furthermore, due to a lack of tissues directly related to orofacial clefts, this study carefully selected four tissues that were relatively more relevant to our biological understandings. In addition to whole blood and skeletal muscle tissue, we have taken subcutaneous and visceral tissues into account. In the fusion process developmental theory, insufficient penetration of the middle layer of the embryo (“mesoderm”) causes cleft [55,56]. A study that tracked the fate of major mesodermal subcompartments in mice demonstrated that visceral and subcutaneous adipocytes were segregated from posterior lateral plate mesoderm by late embryogenesis [5]. Visceral and subcutaneous adipose tissues were used as surrogates for the mesodermal lineage; however, studies with eQTL from orofacial tissues will provide more fundamental evidence. Last, not all of our candidate genes were included in the GTEx eQTL data. More findings on eQTLs associated with those candidate genes will help us to better understand OFC etiology. It is likely that candidate genes of interest did not show a significant association with expression levels in the GTEx database, so no eQTLs were identified. However, the GTEx project is ongoing, and more samples from diverse populations may add eQTLs associated with the candidate genes. For example, a COGENE (Craniofacial and Oral Gene Expression Network) project reported an elevated expression profile for GSTT1 in craniofacial tissues during embryonic development, but GSTT1 was not included in the trained expression dataset. Instead, genome-wide significant signals were identified from GSTT2 and GSTT2B in this study.
5. Conclusions
Using a mixed-model association analysis, we confirmed previous findings for OFC as well as independence among risk loci. We have identified that unlike IRF6, the genome-wide significant association of TRAF3IP3 is independent from the association signal at 8q24. Additionally, TWAS using predicted expression levels identified novel genes associated with OFC that implicate the etiology related to glutathione synthesis and drug detoxification. In conclusion, in spite of several limitations, we added meaningful evidence to the OFC etiology using novel statistical strategies.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/11/1280/s1, Figure S1: Quantile-quantile (QQ) plot of OFC GWAS using EMMAX model, Figure S2: QQ Plots and Manhattan Plots from Nine Sequential Conditional GWASs, Figure S3: QQ Plots of OFC TWASs Using Predicted Expression Levels in Four Tissues. Table S1: Candidate Genes in the GTEx database, Table S2: Results with suggestive evidence of association from the gene-based association study, Table S3: Candidate Gene Study Using Predicted Expression Levels, Table S4: Alternate Allele Frequencies of the Most Significant Markers from a Series of Conditional GWAS in Sub-populations.
Author Contributions
Conceptualization, G.J.; Methodology, G.J.; Resources, G.J.; Formal Analysis, Y.Y.; Writing—Original Draft Preparation, Y.Y.; Writing—Review & Editing, G.J., A.S., J.I.; Supervision, G.J.; Funding Acquisition, G.J., J.I. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by NIDCR grants R03DE026208, R01DE026767, R03DE026509, R03DE028340 and by NICHD grant R03HD098552.
Conflicts of Interest
The authors declare no conflict of interest.
Data Availability
Center for Craniofacial and Dental Genetics (CCDG) data on dbGaP (accession: phs000774.v2.p1).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Stanier P., Moore G.E. Genetics of cleft lip and palate: Syndromic genes contribute to the incidence of non-syndromic clefts. Hum. Mol. Genet. 2004;13:R73–R81. doi: 10.1093/hmg/ddh052. [DOI] [PubMed] [Google Scholar]
- 2.Rahimov F., Jugessur A., Murray J.C. Genetics of nonsyndromic orofacial clefts. Cleft Palate-Craniofacial J. 2011;49:73–91. doi: 10.1597/10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berk N.W., Marazita L.M. Cleft Lip and Palate: From Origin to Treatment. Oxford University Press; New York, NY, USA: 2002. Chapter 36. Costs of cleft lip and polate: Personal and societal implications; pp. 458–467. [Google Scholar]
- 4.Grosen D., Chevrier C., Skytthe A., Bille C., Mølsted K., Sivertsen Å., Murray J.C., Christensen K. A cohort study of recurrence patterns among more than 54,000 relatives of oral cleft cases in Denmark: Support for the multifactorial threshold model of inheritance. J. Med. Genet. 2009;47:162–168. doi: 10.1136/jmg.2009.069385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sivertsen Å., Wilcox A.J., Skjaerven R., Vindenes H.A., Åbyholm F., Harville E., Lie R.T. Familial risk of oral clefts by morphological type and severity: Population based cohort study of first degree relatives. BMJ. 2008;336:432–434. doi: 10.1136/bmj.39458.563611.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosen D., Bille C., Petersen I., Skytthe A., Hjelmborg J.V.B., Pedersen J.K., Murray J.C., Christensen K. Risk of Oral Clefts in Twins. Epidemiology. 2011;22:313–319. doi: 10.1097/EDE.0b013e3182125f9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marazita M. Cleft Lip and Palate: From Origin to Treatment. Oxford University Press; New York, NY, USA: 2002. Chapter 18. Segregation Analyses; pp. 222–233. [Google Scholar]
- 8.Marazita M.L., Murray J.C., Lidral A.C., Arcos-Burgos M., Cooper M.E., Goldstein T., Maher B.S., Daack-Hirsch S., Schultz R., Mansilla M.A., et al. Meta-Analysis of 13 Genome Scans Reveals Multiple Cleft Lip/Palate Genes with Novel Loci on 9q21 and 2q32-35. Am. J. Hum. Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marazita M.L., Lidral A.C., Murray J.C., Field L., Maher B.S., McHenry T.G., Cooper M.E., Govil M., Daack-Hirsch S., Riley B., et al. Genome Scan, Fine-Mapping, and Candidate Gene Analysis of Non-Syndromic Cleft Lip with or without Cleft Palate Reveals Phenotype-Specific Differences in Linkage and Association Results. Hum. Hered. 2009;68:151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig K.U., Böhmer A.C., Rubini M., Mossey P.A., Herms S., Nowak S., Reutter H., Alblas M.A., Lippke B., Barth S., et al. Strong Association of Variants around FOXE1 and Orofacial Clefting. J. Dent. Res. 2014;93:376–381. doi: 10.1177/0022034514523987. [DOI] [PubMed] [Google Scholar]
- 11.Birnbaum S., Ludwig K.U., Reutter H.M., Herns S., Steffens M., Rubini M., Baluardo C., Ferrian M., De Assis N.A., Alblas M.A., et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat. Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 12.Grant S.F., Wang K., Zhang H., Glaberson W., Annaiah K., Kim C.E., Bradfield J.P., Glessner J.T., Thomas K.A., Garris M., et al. A Genome-Wide Association Study Identifies a Locus for Nonsyndromic Cleft Lip with or without Cleft Palate on 8q24. J. Pediatr. 2009;155:909–913. doi: 10.1016/j.jpeds.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Beaty T.H., Murray J.C., Marazita M.L., Munger R.G., Ruczinski I., Hetmanski J.B., Liang K.Y., Wu T., Murray T., Fallin M.D., et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat. Genet. 2010;42:525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H.M., De Assis N.A., Al Chawa T., Mattheisen M., et al. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2009;42:24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig K.U., Mangold E., Herms S., Nowak S., Reutter H., Paul A., Becker J., Herberz R., AlChawa T., Nasser E., et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat. Genet. 2012;44:968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaty T.H., Taub M.A., Scott A.F., Murray J.C., Marazita M.L., Schwender H., Parker M., Hetmanski J.B., Balakrishnan P., Mansilla M.A., et al. Confirming genes influencing risk to cleft lip with/without cleft palate in a case-parent trio study. Hum. Genet. 2013;132:771–781. doi: 10.1007/s00439-013-1283-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y., Huang Y., Yin A., Pan Y., Wang Y., Wang C., Du Y., Wang M., Lan F., Hu Z., et al. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat. Commun. 2015;6:6414. doi: 10.1038/ncomms7414. [DOI] [PubMed] [Google Scholar]
- 18.Leslie E.J., Liu H., Carlson J.C., Shaffer J.R., Feingold E., Wehby G., Laurie C.A., Jain D., Laurie C.C., Doheny K.F., et al. A Genome-wide Association Study of Nonsyndromic Cleft Palate Identifies an Etiologic Missense Variant in GRHL3. Am. J. Hum. Genet. 2016;98:744–754. doi: 10.1016/j.ajhg.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leslie E.J., Carlson J.C., Shaffer J.R., Feingold E., Wehby G., Laurie C.A., Jain D., Laurie C.C., Doheny K.F., McHenry T., et al. A multi-ethnic genome-wide association study identifies novel loci for non-syndromic cleft lip with or without cleft palate on 2p24.2, 17q23 and 19q13. Hum. Mol. Genet. 2016;25:2862–2872. doi: 10.1093/hmg/ddw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangold E., Böhmer A.C., Ishorst N., Hoebel A.-K., Gültepe P., Schuenke H., Klamt J., Hofmann A., Gölz L., Raff R., et al. Sequencing the GRHL3 Coding Region Reveals Rare Truncating Mutations and a Common Susceptibility Variant for Nonsyndromic Cleft Palate. Am. J. Hum. Genet. 2016;98:755–762. doi: 10.1016/j.ajhg.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leslie E.J., Carlson J.C., Shaffer J.R., Buxó C.J., Castilla E.E., Christensen K., Deleyiannis F.W.B., Field L.L., Hecht J.T., Moreno L., et al. Association studies of low-frequency coding variants in nonsyndromic cleft lip with or without cleft palate. Am. J. Med Genet. Part A. 2017;173:1531–1538. doi: 10.1002/ajmg.a.38210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie E.J., Carlson J.C., Shaffer J.R., Butali A., Buxó C.J., Castilla E.E., Christensen K., Deleyiannis F.W.B., Field L.L., Hecht J.T., et al. Genome-wide meta-analyses of nonsyndromic orofacial clefts identify novel associations between FOXE1 and all orofacial clefts, and TP63 and cleft lip with or without cleft palate. Hum. Genet. 2017;136:275–286. doi: 10.1007/s00439-016-1754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Y., Zuo X., He M., Gao J., Fu Y., Qin C., Meng L., Wang W., Song Y., Cheng Y., et al. Genome-wide analyses of non-syndromic cleft lip with palate identify 14 novel loci and genetic heterogeneity. Nat. Commun. 2017;8:14364. doi: 10.1038/ncomms14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eshete M., Liu H., Li M., Adeyemo W., Gowans L., Mossey P., Busch T., Deressa W., Donkor P., Olaitan P., et al. Loss-of-Function GRHL3 Variants Detected in African Patients with Isolated Cleft Palate. J. Dent. Res. 2017;97:41–48. doi: 10.1177/0022034517729819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaty T.H., Ruczinski I., Murray J.C., Marazita M.L., Munger R.G., Hetmanski J.B., Murray T., Redett R.J., Fallin M.D., Liang K.Y., et al. Evidence for gene-environment interaction in a genome wide study of nonsyndromic cleft palate. Genet. Epidemiol. 2011;35:469–478. doi: 10.1002/gepi.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haaland Ø.A., Lie R.T., Romanowska J., Gjerdevik M., Gjessing H.K., Jugessur A. A Genome-Wide Search for Gene-Environment Effects in Isolated Cleft Lip with or without Cleft Palate Triads Points to an Interaction between Maternal Periconceptional Vitamin Use and Variants in ESRRG. Front. Genet. 2018;9:60. doi: 10.3389/fgene.2018.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curà F., Palmieri A., Girardi A., Carinci F., Morselli P.G., Nouri N., Pezzetti F., Scapoli L., Martinelli M. Possible effect of SNAIL family transcriptional repressor 1 polymorphisms in non-syndromic cleft lip with or without cleft palate. Clin. Oral Investig. 2018;22:2535–2541. doi: 10.1007/s00784-018-2350-0. [DOI] [PubMed] [Google Scholar]
- 28.Estandia-Ortega B., Velázquez-Aragón J.A., Alcántara-Ortigoza M.A., Reyna-Fabián M.E., Villagómez-Martínez S., Angel A.G.-D. 5,10-Methylenetetrahydrofolate reductase single nucleotide polymorphisms and gene-environment interaction analysis in non-syndromic cleft lip/palate. Eur. J. Oral Sci. 2014;122:109–113. doi: 10.1111/eos.12114. [DOI] [PubMed] [Google Scholar]
- 29.Wu T., Fallin M.D., Shi M., Ruczinski I., Liang K.Y., Hetmanski J.B., Wang H., Ingersoll R.G., Huang S., Ye X., et al. Evidence of gene-environment interaction for the RUNX2 gene and environmental tobacco smoke in controlling the risk of cleft lip with/without cleft palate. Birth Defects Res. Part A Clin. Mol. Teratol. 2012;94:76–83. doi: 10.1002/bdra.22885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niktabar S.M., Aarafi H., Dastgheib S.A., NooriShadkam M., Mirjalili S.R., Lookzadeh M.H., Akbarian-Bafghi M.J., Morovati-Sharifabad M., Neamatzadeh H. Association of MTHFR 1298A > C Polymorphism with Susceptibility to Non-Syndromic Cleft Lip with or without Palate: A Case-Control Study and Meta-Analysis. Fetal Pediatr. Pathol. 2019:1–17. doi: 10.1080/15513815.2019.1683918. [DOI] [PubMed] [Google Scholar]
- 31.Li W., Wang M., Zhou R., Wang S., Zheng H., Liu D., Zhou Z., Zhu H., Wu T., Beaty T.H. Exploring the interaction between FGF Genes and T-box genes among chinese nonsyndromic cleft lip with or without cleft palate case-parent trios. Environ. Mol. Mutagen. 2019;60:602–606. doi: 10.1002/em.22286. [DOI] [PubMed] [Google Scholar]
- 32.Jugessur A., Rahimov F., Lie R.T., Wilcox A.J., Gjessing H.K., Nilsen R.M., Nguyen T.T., Murray J.C. Genetic variants inIRF6and the risk of facial clefts: Single-marker and haplotype-based analyses in a population-based case-control study of facial clefts in Norway. Genet. Epidemiol. 2008;32:413–424. doi: 10.1002/gepi.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahimov F., Program N.C.S., Marazita M.L., Visel A., Cooper M.E., Hitchler M.J., Rubini M., Domann F.E., Govil M., Christensen K., et al. Disruption of an AP-2α binding site in an IRF6 enhancer is associated with cleft lip. Nat. Genet. 2008;40:1341–1347. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ingraham C.R., Kinoshita A., Kondo S., Yang B., Sajan S., Trout K.J., Malik M.I., Dunnwald M., Goudy S.L., Lovett M., et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6) Nat. Genet. 2006;38:1335–1340. doi: 10.1038/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J., Mendoza F., Tan E., Bertol J.W., Gaggar A.S., Jun G., Biguetti C., Fakhouri W.D. A cleft lip and palate gene, Irf6, is involved in osteoblast differentiation of craniofacial bone. Dev. Dyn. 2019;248:221–232. doi: 10.1002/dvdy.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leslie E.J., Taub M.A., Liu H., Steinberg K.M., Koboldt D.C., Zhang Q., Carlson J.C., Hetmanski J.B., Wang H., Larson D.E., et al. Identification of Functional Variants for Cleft Lip with or without Cleft Palate in or near PAX7, FGFR2, and NOG by Targeted Sequencing of GWAS Loci. Am. J. Hum. Genet. 2015;96:397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson G. GTEx detects genetic effects. Science. 2015;348:640–641. doi: 10.1126/science.aab3002. [DOI] [PubMed] [Google Scholar]
- 39.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J., et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe K., Taskesen E., Van Bochoven A., Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1–11. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ehuppi K., Pitt J.J., Wahlberg B.M., Caplen N.J. The 8q24 Gene Desert: An Oasis of Non-Coding Transcriptional Activity. Front. Genet. 2012;3:69. doi: 10.3389/fgene.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glass D.N., Giannini E.H. Juvenile rheumatoid arthritis as a complex genetic trait. Arthritis Rheum. 1999;42:2261–2268. doi: 10.1002/1529-0131(199911)42:11<2261::AID-ANR1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 44.Haiman C.A., Patel Y.M., Stram D.O., Carmella S.G., Chen M., Wilkens L.R., Le Marchand L., Hecht S.S. Benzene Uptake and Glutathione S-transferase T1 Status as Determinants of S-Phenylmercapturic Acid in Cigarette Smokers in the Multiethnic Cohort. PLoS ONE. 2016;11:e0150641. doi: 10.1371/journal.pone.0150641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koettgen A., Albrecht E., Teumer A., Vitart V., Krumsiek J., Hundertmark C., Pistis G., Ruggiero D., O’Seaghdha C.M., Haller T., et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat. Genet. 2012;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beaty T.H., Marazita M.L., Leslie E.J. Genetic factors influencing risk to orofacial clefts: Today’s challenges and tomorrow’s opportunities. F1000Research. 2016;5:2800. doi: 10.12688/f1000research.9503.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng X., Feingold E., Ryckman K.K., Shaffer J.R., Boyd H.A., Feenstra B., Melbye M., Marazita M.L., Murray J.C., Cuenco K.T. Association of maternal CNVs in GSTT1/GSTT2 with smoking, preterm delivery, and low birth weight. Front. Genet. 2013;4:196. doi: 10.3389/fgene.2013.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lammer E.J., Shaw G.M., Iovannisci D.M., Finnell R.H. Maternal Smoking, Genetic Variation of Glutathione S-Transferases, and Risk for Orofacial Clefts. Epidemiology. 2005;16:698–701. doi: 10.1097/01.ede.0000172136.26733.4b. [DOI] [PubMed] [Google Scholar]
- 49.Hozyasz K.K., Mostowska A., Surowiec Z., Jagodziński P.P. Genetic polymorphisms of GSTM1 and GSTT1 in mothers of children with isolated cleft lip with or without cleft palate. Przeglad Lek. 2005;62:1019–1022. [PubMed] [Google Scholar]
- 50.Shi M., Christensen K., Weinberg C.R., Romitti P., Bathum L., Lozada A., Morris R.W., Lovett M., Murray J.C. Orofacial Cleft Risk Is Increased with Maternal Smoking and Specific Detoxification-Gene Variants. Am. J. Hum. Genet. 2007;80:76–90. doi: 10.1086/510518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitani-Morii F., Imamura K., Kondo T., Ohara R., Enami T., Shibukawa R., Yamamoto T., Sekiguchi K., Toguchida J., Mizuno T., et al. Analysis of neural crest cells from Charcot–Marie–Tooth disease patients demonstrates disease-relevant molecular signature. NeuroReport. 2017;28:814–821. doi: 10.1097/WNR.0000000000000831. [DOI] [PubMed] [Google Scholar]
- 52.Bolt H.M., Thier R. Relevance of the Deletion Polymorphisms of the Glutathione S-Transferases GSTT1 and GSTM1 in Pharmacology and Toxicology. Curr. Drug Metab. 2006;7:613–628. doi: 10.2174/138920006778017786. [DOI] [PubMed] [Google Scholar]
- 53.Zhang H., Forman H.J., Choi J. γ-glutamyl transpeptidase in glutathione biosynthesis. Methods Enzymol. 2005;401:468–483. doi: 10.1016/s0076-6879(05)01028-1. [DOI] [PubMed] [Google Scholar]
- 54.Chambers J.C., Zhang W., Sehmi J., Li X., Wass M.N., Van Der Harst P., Holm H., Sanna S., Kavousi M., Baumeister S.E., et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat. Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra S., Sabhlok S., Panda P.K., Khatri I. Management of Midline Facial Clefts. J. Maxillofac. Oral Surg. 2015;14:883–890. doi: 10.1007/s12663-015-0763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sebo Z.L., Jeffery E., Holtrup B., Rodeheffer M.S. A mesodermal fate map for adipose tissue. Development. 2018;145:dev166801. doi: 10.1242/dev.166801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Center for Craniofacial and Dental Genetics (CCDG) data on dbGaP (accession: phs000774.v2.p1).