Abstract
When cells of garlic (Allium sativum) are disrupted by wounding, they produce the defense substance allicin (diallylthiosulfinate). Allicin is an efficient thiol trap and readily passes through cell membranes into the cytosol, where it behaves as a redox toxin by oxidizing the cellular glutathione (GSH) pool and producing S-allylmercaptoglutathione (GSSA). An N-cyanosulfilimine analogue of allicin (CSA), which was predicted to have similar reactivity towards thiol groups but be more stable in storage, was synthesized and its properties investigated. Similarly to allicin, CSA was shown to inhibit the growth of various bacteria, a fungus (baker’s yeast), and Arabidopsis roots. A chemogenetic screen showed that yeast mutants with compromised GSH levels and metabolism were hypersensitive to CSA. GSH reacted with CSA to produce allyltrisulfanylglutathione (GS3A), which was a white solid virtually insoluble in water. Yeast Δgsh1 mutants are unable to synthesize GSH because they lack the γ-glutamylcysteine synthetase (GSH1) gene, and they are unable to grow without GSH supplementation in the medium. GS3A in the growth medium supported the auxotrophic requirement for GSH in Δgsh1 mutants. This result suggests that GS3A is being reduced to GSH in vivo, possibly by the enzyme glutathione reductase (GR), which has been shown to accept GSSA as a substrate. The results suggest that CSA has a mode of action similar to allicin and is effective at similar concentrations.
Keywords: allicin analogue, N-cyanosulfilimine, glutathione reductase
1. Introduction
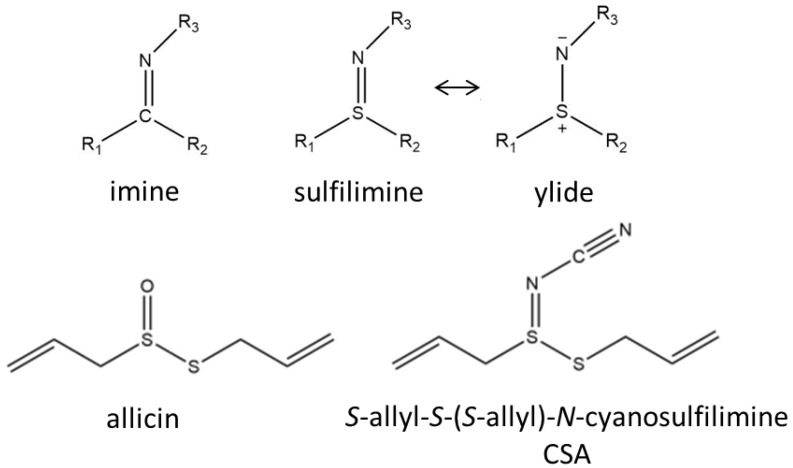
Sulfilimines are a group of organic sulfur compounds in which the sulfur atom is linked to a nitrogen atom via a formal double bond (Scheme 1). In the literature such structures have also been named sulfimides, sulfilimides, or iminosulfurans [1]. The sulfilimine molecule is mesomerically stabilized and in the ionic form can be regarded as a sulfur/nitrogen ylide (Scheme 1) [2]. There is much interest in sulfilimines because they have been discussed as potential anticancer drugs [3], and ylides are the starting point for many organic syntheses. The stability of sulfilimines is greatly influenced by the nature of the R3 substituent group; thus, electronegative carbonyl, sulfonyl, or cyano groups stabilize the S=N bond. In this context, N-cyanosulfilimines proved particularly interesting, and, consequently, their synthetic accessibility has recently been greatly improved [4,5,6]. Furthermore, significant bioactivities of such compounds have been revealed [7,8].
Scheme 1.
Shows the imine/sulfilimine structures and the mesomeric sulfilimine/ylide forms, as well as the structural formula of allicin and the N-cyanosulfilimine allicin analogue CSA (S-allyl-S-(S-allyl)-N-cyanosulfilimine).
Allicin (S-prop-2-en-1-yl prop-2-ene-1-sulfinothioate, Scheme 1) was shown by Cavallito to be the antimicrobial substance produced by garlic [9,10], and it was later demonstrated to deplete the cellular GSH pool, react with bacillothiol in Gram positive Bacillus, and S-thioallylate accessible cysteines in proteins [11,12,13]. Allicin is produced by damaged cells in which the enzyme alliinase comes together with its substrate alliin, both of which are usually separately compartmentalized in the cell. Allicin is a major secondary metabolite produced by garlic and a single clove can yield up to 5 mg [14]. Allicin gives fresh garlic its characteristic odor [15]. Allicin has been shown to kill many pathogenic bacteria in vitro, even as a vapor [16,17,18]. The electron-withdrawing nature of the O-atom in the thiosulfinate group in allicin leads to an electrophilic sulfur center, which reacts readily with thiols without the need for enzymic catalysis [11,19,20]. Furthermore, allicin can react with accessible cysteine thiols in proteins by S-thioallylation causing disulfide stress in cells [13,21,22].
Previously, we investigated the properties of thiosulfinate analogues of allicin by varying the alkyl groups attached to the thiosulfinate functional group. Here, we report the replacement of the thiosulfinate functional group in the synthesis of the S-allyl-S-(S-allyl)-N-cyanosulfilimine analogue of allicin (CSA), which, to our knowledge, has never been synthesized before. Because of the electron-withdrawing properties of the cyanosulfilimine group, we predicted that CSA would also have an electrophilic sulfur-center and be a potent thiol-trapping reagent similar to allicin but with a better thermal stability. Therefore, the antimicrobial activity of CSA was investigated and a yeast-based chemogenetic screen was performed to gain an insight into its potential mode of action [23].
Glutathione (GSH) is the major low molecular weight thiol in most eukaryotic and many prokaryotic cells, and it buffers and protects cells against oxidative stress [24,25] and is an electron donor for several reducing enzyme systems. GSH metabolism and the GSH pool have been shown previously to be particularly important in protecting cells against allicin action [26,27,28] and, thus, might be expected to similarly protect cells against the effects of CSA.
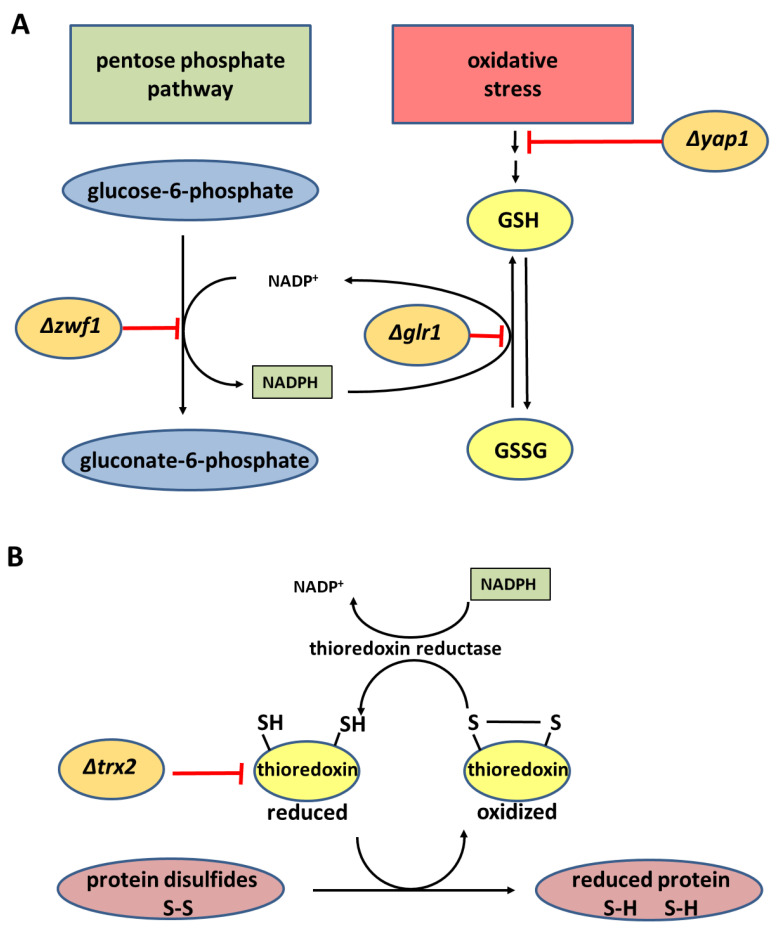
Under oxidative stress conditions GSH dimerizes to glutathione disulfide (GSSG). Furthermore, cysteine thiols in proteins may be reversibly glutathiolated to protect them from over-oxidation [21]. The GSH pool within the cell is restored by the reduction of GSSG to GSH by the NADPH-dependent enzyme glutathione reductase (Glr, E.C. 1.8.17) [24]. Glr1 not only reduces GSSG to GSH but also can accept the reaction product of allicin and glutathione (S-allylmercaptoglutathione, GSSA) as a substrate [11,12]. Scheme 2 shows the positions where the various mutants tested affect glutathione synthesis and metabolism. The Yap1 transcription factor coordinates the oxidative stress response (OSR) in yeast, including the activation of the Gsh1 gene for the rate-limiting Gsh1 enzyme in GSH synthesis. The Yap1p transcription factor activates several genes, enabling challenged cells to resist the effects of oxidative stress. Thus, as well as genes on the GSH biosynthetic pathway, glutathione reductase, glutaredoxins, thioredoxins, and thioredoxin reductase genes are activated. As a result of oxidative stress, protein disulfides form from cysteine thiols in proteins. The small (~12 kDa) protein thioredoxin reduces protein disulfides and, in doing so, forms an internal disulfide bond between two cysteines. Thioredoxin reductase regenerates thioredoxin by using NADPH as a reductant to reduce the disulfide bond. Thus, the production of NADPH is of major importance in providing reducing equivalents to protect against oxidative stress and the pentose phosphate pathway (PPP) is the major source of NADPH in eukaryotic and prokaryotic cells. Zwf1 (glucose-6-phosphare dehydrogenase) catalyzes the first step in the PPP (Scheme 2). In addition to serving as a reductant for protein disulfides, thioredoxin2 is important for resistance to H2O2-derived oxidative stress [29,30,31,32].
Scheme 2.
(A) The positions where the Δyap1, Δzwf1, and Δglr1 mutants affect glutathione synthesis and metabolism. (B) The effect due to lack of thioredoxin in the Δtrx2 mutant in compromising the reduction by thioredoxin-dependent mechanisms of protein disulfides produced under oxidative stress conditions. NADPH produced in the pentose phosphate pathway serves as a reductant for both glutathione reductase and thioredoxin reductase.
The goal of this investigation was to test whether CSA, which although based on the allicin molecule no longer retains the thiosulfinate functional group, has antimicrobial properties and whether its mode of action might be similar to that of allicin.
2. Materials and Methods
2.1. Synthesis and LC-MS of S-allyl-S-(S-allyl)-N-cyanosulfilimine
The synthesis followed a reaction protocol developed for sulfide imidations [4].
Into a 15 mL sealed-tube were added 0.39 mL of diallyl disulfide (with a technical grade of 80%; Sigma-Aldrich, Steinheim, Germany; 365.7 mg, 2.5 mmol, 1.0 equiv) and 2.5 mL of MeCN. Under stirring at room temperature, cyanamide (375 mg, 9.0 mmol, 3.6 equiv) and (diacetoxyiodo)benzene (2.67 g, 8.4 mmol, 3.3 equiv) were added to the reaction mixture in three portions every 40 min. The reaction was monitored by thin-layer chromatography (TLC). After consumption of the starting material, the solvent was removed by evaporation, and the product was purified by flash column chromatography over silica (pentane to pentane:dichloromethane 4:1), yielding 223 mg (1.20 mmol, 60% based on the listed purity of the starting material of 80%) of the product (CSA) as a colorless liquid. 1H-NMR (400 MHz, CDCl3) δ = 5.95–5.80 (m, 2H), 5.42–5.24 (m, 4H), 3.89 (d, J = 7.0 Hz, 2H), 3.71 (d, J = 7.0 Hz, 2H) ppm; 13C{1H}-NMR (101 MHz, CDCl3) δ = 132.1, 131.0, 121.9, 120.6, 116.1, 59.6, 42.8 ppm; IR (ATR) ν [cm–1] = 3367 (vw), 3084 (w), 2981 (w), 2923 (w), 2660 (vw), 2330 (vw), 2210 (vs), 2090 (vw), 1994 (vw), 1857 (vw), 1738 (vw), 1636 (m), 1571 (vw), 1422 (s), 1338 (m), 1288 (m), 1222 (m), 1170 (m), 1127 (m), 1078 (w), 987 (vs), 927 (vs), 721 (s); MS (EI, 70 eV): m/z (%) = 187.0 (3), 144.9 (12), 121.0 (100), 73.0 (12), 45.2 (11); high-resolution mass spectrometry HRMS (ESI): C7H10N2NaS2 Calcd. 209.0178 Found 209.0173.
2.2. Stability of CSA Compared to Allicin
Aqueous 20 mM CSA and allicin solutions: (1.5 mL) each were transferred to a reaction vessel and incubated at 100 °C in a heating block. A sample was taken every 10 min, and the relative amount of substance was quantified by high-pressure liquid chromatography (HPLC). Separation was performed using H2O as mobile phase A and methanol as mobile phase B with the following gradient: 56% A (pre-run); 53% A (10 min); 7% A (15 min); 7% A (30 min); 56% A (31 min); 56% A (35 min) at a flow rate of 1 mL/min on a C18-reverse phase column. The detection was performed with a UV detector (Jasco GmbH, Groß Umstadt, Germany) at 254 nm.
2.3. Reaction of GSH with CSA and LC/MS of the GS3A Product
l-glutathione (400 mg, Carl Roth, GmbH, Karlsruhe, Germany) was dissolved in up to 4 mL of distilled H2O. CSA (280 mg) were dissolved in up to 12 mL in methanol. Under stirring, the methanolic solution was slowly added to the aqueous solution, and, subsequently, the stirring was continued for 2 h at room temperature. The insoluble product was collected by filtration and washed with 3 × 5 mL of dichloromethane and finally with 5 mL of distilled H2O. Subsequently, the product was dried to constant weight at 37 °C.
ESI-MS measurements were performed on a Thermo Fisher Scientific Orbitrap XL mass spectrometer (Bremen, Germany) in high resolution FT-mode. The sample was diluted in acetonitrile (AcCN)/Water (H2O) v/v 50:50 with HCl-solution added and introduced via direct flow using a syringe pump with a flow of 5 μL/min.
2.4. Experiments with Microorganisms
Bacillus subtilis, Escherichia coli, and Photorhabdus luminescens were cultivated on LB medium 5 g/L yeast extract (Duchefa, Haarlem, Netherlands), 10 g/L peptone (Duchefa, Haarlem, Netherlands), 10 g sodium chloride (Applichem, Darmstadt, Germany), and 1.5% agar (Carl Roth, Karlsruhe, Germany). The pseudomonads (Pseudomonas fluorescens AR-1, an allicin resistant strain and Pseudomonas syringae 1446A) were cultivated on King’s B medium (20 g/L peptone (Duchefa, Haarlem, Netherlands), 1.5 g/L anhydrous K2HPO4 (Applichem, Darmstadt, Germany), 12.6 g/L glycerol (Applichem, Darmstadt, Germany), and 1.5% agar (Carl Roth, Karlsruhe, Germany). With the exception of E. coli, which were cultivated at 37 °C, all bacteria were cultivated at 28 °C.
The plate inhibition assay was performed by diluting cultures overnight to an OD600 of 0.2. One hundred and twenty microliters of these cultures were plated onto a 20 mL LB agar plate (Pseudomonas: 20 mL KB agar plate) and uniformly distributed on the plate using sterile glass beads (ø = 4 mm). The test was evaluated after 48 h of cultivation under appropriate conditions as described above.
The haploid Saccharomyces cerevisiae yeast strain BY4742 (Matα; his3∆1; leu2∆0, lys2∆0, ura3∆0) [33] was used. The BY4742 mutant ∆gsh1 (Y17097) used in this study lacks the gene for γ-glutamylcysteine synthetase (YJL101C), which catalyzes the first step in glutathione biosynthesis. The mutant was obtained from the EUROSCARF Collection, University of Frankfurt (Main), Germany (http://www.euroscarf.de/).
Yeast was grown in complete synthetic mixture (CSM) medium (0.79 g L−1 CSM Drop-Out: Complete (ForMedium, Norwich, UK); 6.9 g L−1 Yeast Nitrogen Base (ForMedium, Norwich, UK); 40 g L−1 d-Glucose (Carl Roth, Karlsruhe, Germany); and 15 g L−1 agar for solid medium.
2.5. Experiments with Arabidopsis Roots
The Arabidopsis seedling root assay was performed after Reference [34]. Surface-sterilized Arabidopsis thaliana seeds (Col-0, pad2 and gr1) were sown on Murashige & Skoog (MS) solid medium and were grown under short day conditions (8 h light) at 21 °C. The Petri plates were tilted to an angle of approximately 70° to ensure root growth according to root gravitropism. After three days of cultivation, seedlings were transferred to MS medium that contained different amounts of CSA or allicin. After three days treatment, seedlings were photographed, and the root length was measured.
3. Results and Discussion
3.1. Stability of CSA Compared to Allicin
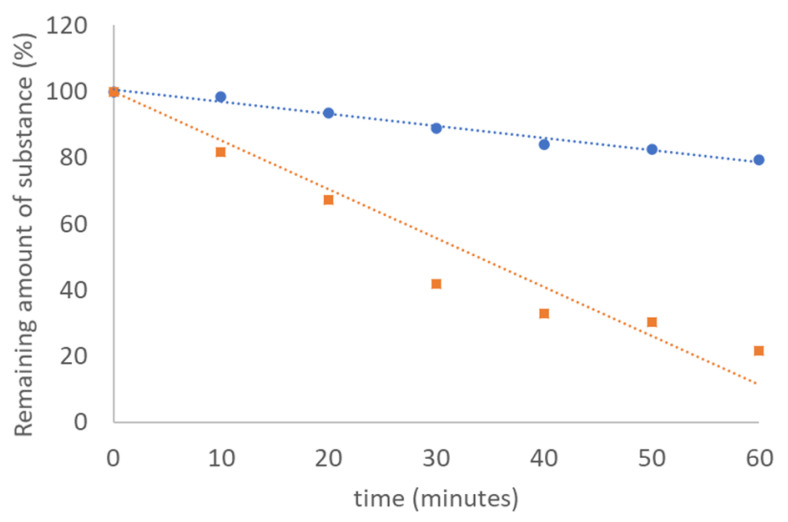
CSA is an analogue of allicin, which, however, does not have a thiosulfinate as a functional group, like allicin, but a sulfilimine group (Scheme 1). Allicin is a highly potent antibiotically active substance, but has relatively low thermal stability. To test whether CSA offers an advantage over allicin in this respect, both substances were incubated at 100 °C for one hour, and the remaining amount of substance was determined using HPLC. It is shown that allicin is degraded very quickly, as is already known, whereas CSA is more persistent in terms of thermal stability (Figure 1). This means that there is an advantage of CSA over allicin in this respect, and we, therefore, investigated its antibiotic effectiveness.
Figure 1.
Comparative stability of allicin and CSA at 100 °C over a period of one hour. The relative amount of substance was determined by HPLC.
3.2. CSA Inhibits the Growth of Bacteria and Yeast in Agar-Diffusion Tests
In agar diffusion assays, solutions of a test substance are applied to wells and the substance diffuses into the agar medium giving a gradient of decreasing concentration away from the well. The test organisms are applied evenly over the Petri plate and can grow up to the threshold inhibitory concentration around each well. Thus, the relative sensitivities of various microorganisms to a given test substance can be demonstrated. However, the relative effectivities of different test substances against a specific microorganism cannot be compared in this assay because of the different diffusion behavior of the test molecules. To compare the relative effectivities of different test substances, fixed concentrations are incorporated into the growth medium and a dilution series of a cell-suspension culture pipetted onto the medium in a so-called ‘drop test’ [23], as in Section 3.3.
Bacillus subtilis (Gram positive), Escherichia coli, Photorhabdus luminescens, and Pseudomonas syringae pv. phaseolicola 1446A (all Gram negative) showed a dose-dependent inhibition of growth in the presence of CSA. In contrast, the highly allicin-resistant Pseudomonas fluorescens isolate PfAR-1 showed only an extremely small inhibition zone at the highest concentration of CSA tested (Figure 2). PfAR-1 was isolated from garlic and has 3 genomic islands carrying repeats containing genes which confer allicin resistance [28]. The insensitivity of PfAR-1 to CSA, as well as allicin, suggests that the mode of action of both compounds might be similar, i.e., targeting the GSH pool and causing oxidative disulfide stress [13,27,28]. Instead of GSH, Bacillus spp. contain bacillothiol as a redox-buffer and protectant against oxidative stress. We have previously shown that allicin targets bacillothiol, and, presumably, this is similarly the case for CSA [13].
Figure 2.
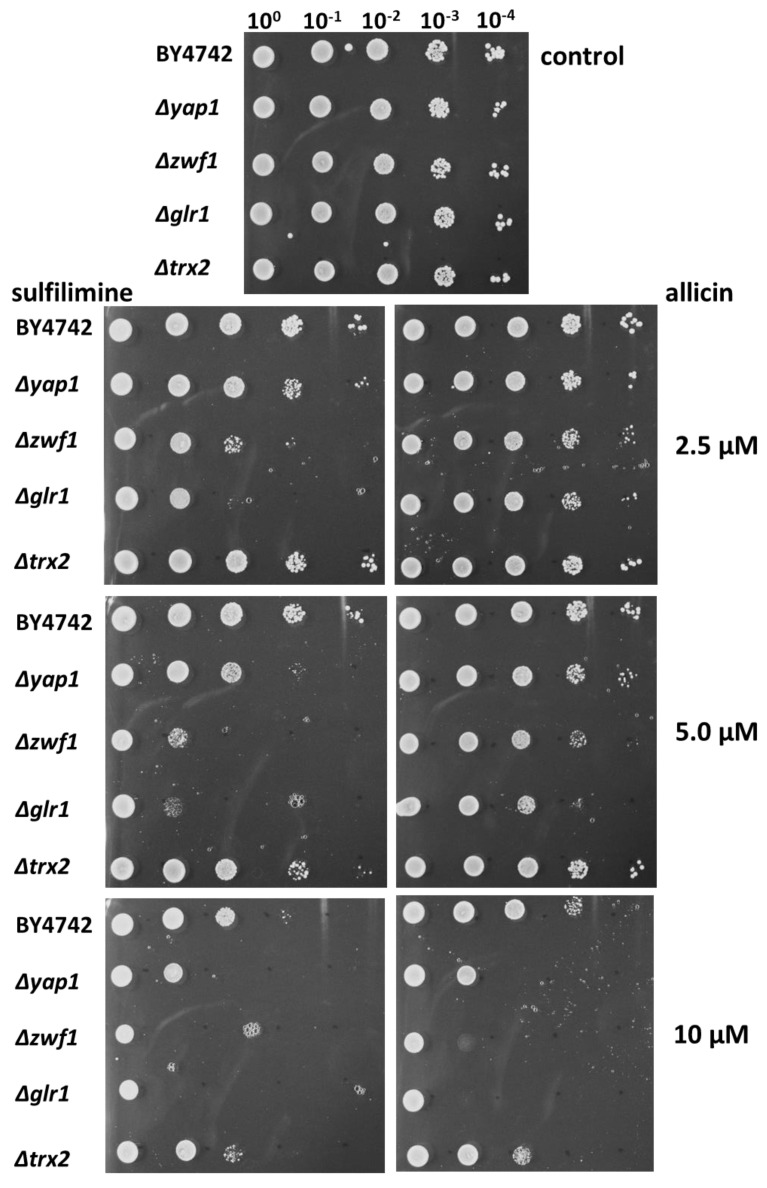
CSA (S-allyl-S-(S-allyl)-N-cyanosulfilimine) inhibits the growth of bacteria and yeast. Bacteria were spread on the surface of the agar medium and 20 µL of 10, 20, 40, and 60 mM CSA in dimethylsulfoxide (DMSO) pipetted into wells cut out of the agar, giving total amounts of 37, 75, 149, and 223 µg CSA, respectively. For the wildtype yeast strain BY4742 and the Δyap1, Δzwf1, Δglr1, and Δtrx2 mutants, 20 µL of 1, 2, 3, and 4 mM CSA in DMSO was used, giving total amounts of 4, 8, 12, and 16 µg, respectively. The control was 20 µL of DMSO.
As previously demonstrated for allicin, the fungus Saccharomyces cerevisiae (baker’s yeast) was much more sensitive to CSA than were bacteria, and comparable inhibition zones were observed with much lower amounts of active substance (Figure 2).
The behavior of the Δyap1, Δzwf1, Δglr1, and Δtrx2 yeast mutants to CSA was also similar to that for allicin [27], suggesting that CSA behaves similarly in cells by targeting thiols. Thus, as for allicin, the Δyap1, Δzwf1, and Δglr1 mutants were all hypersensitive to CSA compared to wildtype cells, while the effect of the Δtrx2 mutation was marginal. The lack of hypersensitivity in the trx2 mutant suggests that, as for allicin, H2O2-generation is not an important factor in the oxidative stress caused by CSA. The Yap1 transcription factor coordinates the oxidative stress response in yeast, and Δyap1 yeast cells were more sensitive to CSA than the wildtype, indicating that as for allicin, CSA caused oxidative stress in the cells. It was previously shown that allicin directly targets specific cysteines in Yap1 [26], and it is likely that CSA has a similar mechanism of action. As previously reported for allicin, the Δzwf1 and Δglr1 mutants showed extreme sensitivity to CSA. Indeed, the growth of Δglr1 was inhibited over almost the entire Petri plate. Thus, GSH metabolism appears to be important for protecting the cells against CSA, and this suggests that, similarly to allicin, CSA works by causing disulfide stress via the oxidation of thiols [22,27,28].
3.3. Chemogenetic Screen of CSA Compared to Allicin in the Yeast Mutants Δglr1, Δtrx2, Δyap1, and Δzwf1
The inhibitory activity of CSA was compared directly to that of allicin to assess the relative effectiveness of the two substances using a sensitive drop test where fixed concentrations of the test substance are incorporated into the medium and a dilution series of cells plated out [19]. Furthermore, the role of GSH metabolism and the oxidative stress response (OSR) in protecting yeast against CSA described in Section 3.2 was here examined in more detail in a chemogenetic screen with selected yeast mutants [27,35].
In the absence of stress, the yeast BY4742 wildtype and the mutants all grew similarly down to the 10−4 dilution of the original culture (Figure 3). The BY4742 wildtype yeast strain showed similar sensitivity to both allicin and CSA over the 2.5–10 µM range; suggesting that the two compounds are similarly active on a mol-for-mol basis in yeast, with perhaps a slightly greater degree of inhibition visible at 10 µM CSA at the 10−3 dilution (Figure 3). However, although BY4742 was not inhibited by either CSA or allicin at 2.5 µM, the Δglr1 and Δzwf1 mutants were inhibited by CSA but not allicin at this concentration (Figure 3). The higher sensitivity of Δglr1, Δzwf1 and the Δyap1 mutant for CSA over allicin is very clear at 5 µM, but, at 10 µM, sensitivities are again similar (Figure 3). Taken together, these results suggest that CSA is slightly more active on an equimolar basis than allicin against yeast, but that the GSH and NADPH-based defence responses can cope equally well against oxidative stress caused by both compounds in the wildtype strain.
Figure 3.
Chemogenetic screen of CSA (S-allyl-S-(S-allyl)-N-cyanosulfilimine) and allicin against a serial dilution of selected yeast mutants in a drop test.
The results emphasize the importance of GSH metabolism in overcoming CSA stress. The glutathione reductase (Glr) enzyme, that is absent in the Δglr1 mutant, uses NADPH as a cosubstrate and reductant. The major source of NADPH in the cell is the Zwf1 gene-encoded glucose-6-phosphate dehydrogenase, which catalyzes the first step in the pentose phosphate pathway (Scheme 2). Thus, the Δzwf1 mutant is compromised in the production of NADPH for Glr, and both the Δglr1 and Δzwf1 mutants showed a high degree of inhibition by CSA (Figure 3).
The OSR in yeast is coordinated by the Yap1p transcription factor, and it was shown previously that allicin activates Yap1 and that a Δyap1 mutant was hypersensitive to allicin [26]. The hypersensitivity of the Δyap1 mutant to CSA (Figure 2) shows that the protective OSR in yeast is needed for cells to work against the effects of CSA and implies that CSA, like allicin, causes oxidative stress in yeast.
It was previously shown that Trx2 plays only a minor role in protecting yeast cells against the effects of allicin [26,27], and this is also the case for CSA (Figure 3).
3.4. Effect of CSA on Arabidopsis Root Growth
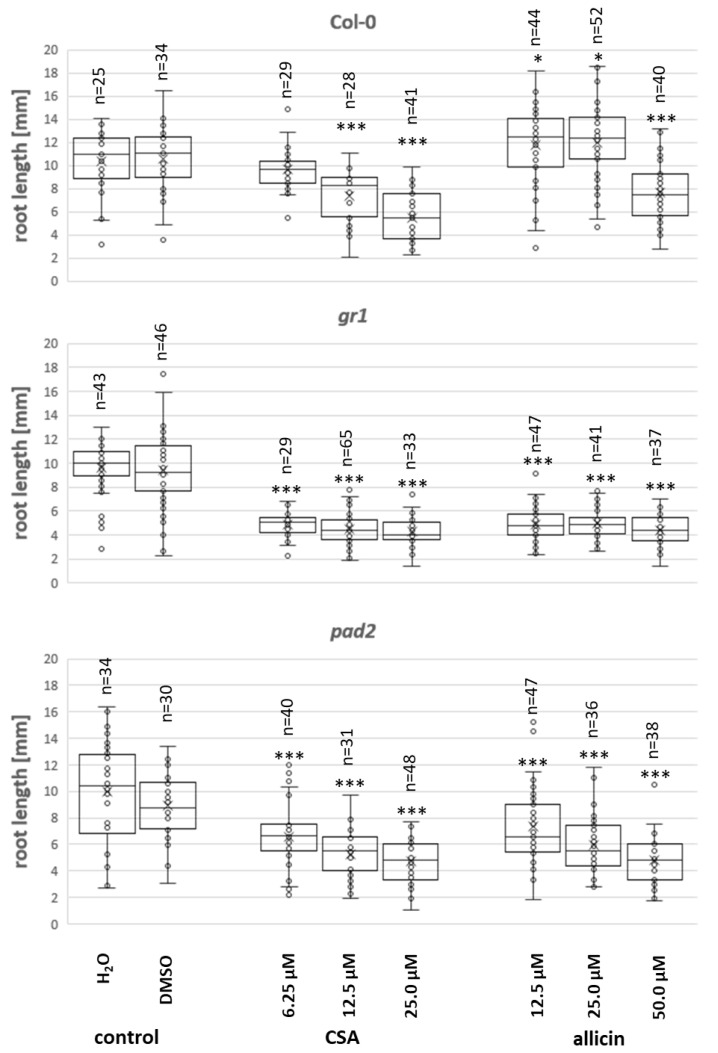
The importance of GSH levels and GSH metabolism in resistance against CSA was further supported by experiments using the model plant Arabidopsis thaliana. Arabidopsis Col-0 wildtype and pad2 and gr1 mutants in the Col-0 background were tested. The pad2 mutant has only approximately 20% of the GSH level found in the wildtype [36]. The gr1 mutant line is a knockout mutant of glutathione reductase and has a higher proportion of GSSG in the glutathione pool because it cannot reduce GSSG back to GSH [37]. Seeds were germinated for three days before transplanting onto medium containing CSA. Root length was measured after a further three days of growth. As can be seen in Figure 4, CSA impaired root growth in the Col-0 wildtype in a concentration-dependent manner. Wildtype Col-0 seedlings were clearly more sensitive to CSA than to allicin. Thus, significant root growth inhibition (p < 0.001) was observed at 12.5 µM CSA whereas significant inhibition occurred between 25–50 µM allicin and <25 µM allicin was slightly stimulatory (p < 0.05) to root growth in the wildtype. The gr1 and pad2 mutants were both very sensitive to CSA and allicin at concentrations of 6.25–12.5 µM (p < 0.001, Figure 4). These results further confirm the important role observed in yeast (Figure 2 and Figure 3), of GSH metabolism in countering CSA-induced oxidative stress.
Figure 4.
Effects of CSA and allicin on seedling root growth of Arabidopsis thaliana Col-0 (wildtype), a mutant with reduced glutathione (pad2), and a mutant lacking glutathione reductase (gr1). The box plots show the range of the data, the mean (x), the median (horizontal line), and standard deviation (box). A t-test of each sample-group against the untreated control was performed. For non-normally distributed data, the test was carried out on ranks (Wilcoxon–Mann–Whitney test): *: p < 0.05; ***: p < 0.001.
3.5. Reaction of CSA with GSH and Characterization of GS3A as the Reaction Product
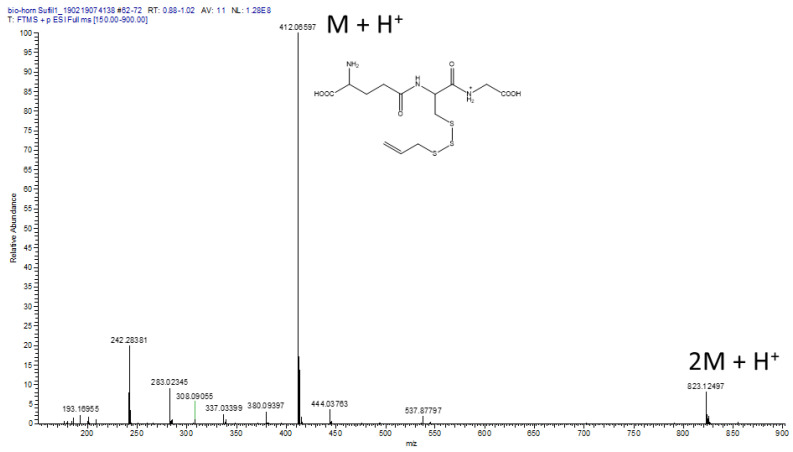
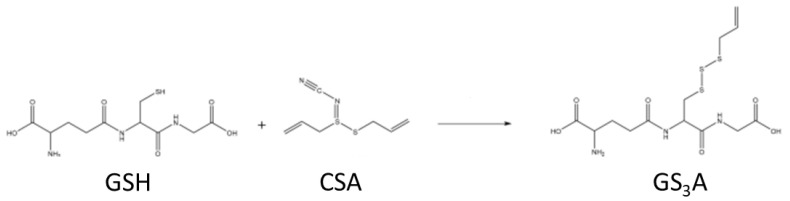
CSA reacted with GSH at room temperature to produce a highly water-insoluble product. ESI-MS of this product (diluted in AcCN/H2O, HCl, measured on a Thermo Fisher Scientific Orbitrap XL) revealed a major signal at M + H+ = 412 Mass Units and a second signal for 2M + H+ = 823 Mass Units (Figure 5). HR-ESI-MS (Bremen, Germany) measurements proved the identity of the compound by exact mass measurements (HRMS (ESI): C13H22N3O6S3 Calcd. 412.06652 Found 412.06597). We have previously shown that allicin reacts with GSH to produce S-allylmercaptoglutathione (GSSA, M + H+ = 380). The reaction product of CSA with GSH has a mass compatible with an additional sulfur atom in the structure, i.e., allyltrisulfanylglutathione (GS3A, Scheme 3).
Figure 5.
LC/MS of GS3A, the putative reaction product of CSA with GSH.
Scheme 3.
Shows the reaction of glutathione (GSH) with CSA to make allyltrisulfanylglutathione (GS3A).
3.6. A Δgsh1 Mutant can Grow on Medium without GSH when Supplemented with GS3A
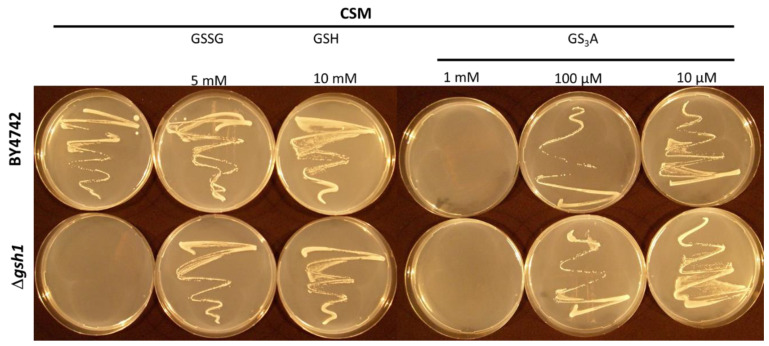
Δgsh1 yeast mutants lacking the Gsh1 enzyme (γ-glutamylcysteine synthetase) are auxotrophic for GSH, i.e., they cannot grow without GSH-supplementation. GSSG can serve as a source of GSH because it can be reduced by the NADPH-dependent enzyme glutathione reductase (Glr). In Figure 6 it can be seen that S-allyltrisulfanylglutathione (GS3A) can also serve as a source of GSH for the Δgsh1 mutant, indicating that it is possibly a substrate for GR. Obviously, GS3A can inhibit growth of the diploid wildtype BY4742 yeast cells at 1 mM and appears to be partially inhibitory at 100 µM, while still being able to substitute for GSH at that concentration. At 10 µM GS3A, it does not appear to inhibit BY4742 cells, and it fully complements the Δgsh1 mutant. Therefore, it seems likely that GS3A might also be a substrate for Glr. Trisulfides as substrates for Glr have been reported [38]. However, due to the extreme insolubility of GS3A we could not test this directly with GR in vitro. Nevertheless, this observation is of importance for in vivo considerations of the consequences of CSA reacting with GSH. Thus, at sublethal CSA doses, it seems that GS3A might be further metabolized by Glr, releasing GSH in the process.
Figure 6.
Growth of the wildtype diploid BY4742 yeast strain compared with the Δgsh1 mutant on complete synthetic mixture (CSM) and CSM supplemented with 10 mM glutathione (GSH), 5 mM glutathione disulfide (GSSG), or 10 mM S-allylmercaptoglutathione (GSSA). A 100 µL aliquot of a logarithmically growing yeast culture adjusted to OD600 = 0.2 (OD = optical density) was spread onto the surface of each plate with an inoculation loop spatula.
4. Conclusions/Highlights
CSA is antimicrobial in a dose-dependent manner.
CSA probably has a similar mechanism of action to allicin.
CSA reacts with GSH to make GS3A.
GS3A can complement a Δgsh1 yeast mutant and, therefore, may be a substrate for Glr.
Acknowledgments
Financial support from RWTH Aachen University is acknowledged. M.-R.H. thanks MOST-DAAD for a sandwich scholarship.
Author Contributions
Conceptualization, A.J.S., C.B., and M.C.H.G.; Investigation, T.H., U.N., F.K., M.-R.H., W.B.; Methodology A.J.S., M.C.H.G., C.B.; Mass Spectrometry, W.B.; Resources, A.J.S., W.B., C.B.; Supervision, A.J.S., C.B., and M.C.H.G.; Writing—original draft A.J.S., C.B., and M.C.H.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by RWTH Aachen University.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kohrt E., Hartke K. Neuartige sulfimide. Arch. Pharm. 1993;326:267–270. doi: 10.1002/ardp.19933260504. [DOI] [Google Scholar]
- 2.Gilchrist T.L., Moody C.J. The chemistry of sulfilimines. Chem. Rev. 1977;77:409–435. doi: 10.1021/cr60307a005. [DOI] [Google Scholar]
- 3.Block E. Sulfilimines and Related Derivatives. In: Oae S., Furukawa N., editors. ACS Monograph Series. Vol. 179. ACS; Washington, DC, USA: 1983. p. 340. [Google Scholar]
- 4.Garciá Mancheño O., Bolm C. Synthesis of N-(1H)-tetrazole sulfoximines. Org. Lett. 2007;9:2951–2954. doi: 10.1021/ol071302+. [DOI] [PubMed] [Google Scholar]
- 5.Garciá Mancheño O., Bistri O., Bolm C. Iodinane- and metal-free synthesis of N-cyano sulfilimines: Novel and easy access of NH-sulfoximines. Org. Lett. 2007;9:3809–3811. doi: 10.1021/ol7016577. [DOI] [PubMed] [Google Scholar]
- 6.Pandey A., Bolm C. Metal-free synthesis of N-cyano-substituted sulfilimines and sulfoximines. Synthesis. 2010:2922–2925. doi: 10.1055/s-0030-1258192. [DOI] [Google Scholar]
- 7.Steinkamp A.-D., Seling N., Lee S., Boedtkjer E., Bolm C. Synthesis of N-Cyano-substituted sulfilimine and sulfoximine derivatives of S0859 and their biological evaluation as sodium bicarbonate Co-transport Inhibitors. Med. Chem. Commun. 2015;6:2163–2169. doi: 10.1039/C5MD00367A. [DOI] [Google Scholar]
- 8.Karpel-Massler G., Kast R.E., Siegelin M.D., Dwucet A., Schneider E., Westhoff M.-A., Wirtz C.R., Chen X.Y., Halatsch M.-E., Bolm C. Anti-glioma activity of dapsone and its enhancement by synthetic chemical modification. Neurochem. Res. 2017;42:3382–3389. doi: 10.1007/s11064-017-2378-6. [DOI] [PubMed] [Google Scholar]
- 9.Cavallito C.J., Bailey J.H. Allicin, the antibacterial principle of Allium sativum I.; isolation, physical properties and antibacterial action. J. Am. Chem. Soc. 1944;66:1950–1951. doi: 10.1021/ja01239a048. [DOI] [Google Scholar]
- 10.Cavallito C.J., Buck J.S., Suter C.M. Allicin, the antibacterial principle of Allium sativum. II. Determination of the chemical structure. J. Am. Chem. Soc. 1944;66:1952–1954. doi: 10.1021/ja01239a049. [DOI] [Google Scholar]
- 11.Rabinkov A., Miron T., Mirelman D., Wilchek M., Glozman S., Yavin E., Weiner L. S-allylmercaptoglutathione: The reaction product of allicin with glutathione possesses SH-modifying and antioxidant properties. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2000;1499:144–153. doi: 10.1016/S0167-4889(00)00119-1. [DOI] [PubMed] [Google Scholar]
- 12.Horn T., Bettray W., Slusarenko A.J., Gruhlke M.C.H. S-allylmercaptoglutathione Is a substrate for glutathione reductase (EC. 1.8.1.7) from Yeast (Saccharomyces cerevisiae) Antioxidants. 2018;7:86. doi: 10.3390/antiox7070086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi B.K., Huyen N.T.T., Van Loi V., Gruhlke M.C.H., Mäder U., Maaß S., Becher D., Bernhardt J., Arbach M., Hamilton C.J. The disulfide stress response and protein S-thioallylation caused by allicin and diallyl polysulfanes in B. subtilis as revealed by transcriptomics and proteomics. Antioxidants. 2019;8:605. doi: 10.3390/antiox8120605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson L.D., Wood S.G., Hughes B.G. HPLC analysis of allicin and other thiosulfinates in garlic clove homogenates. Planta Med. 1991;57:263–270. doi: 10.1055/s-2006-960087. [DOI] [PubMed] [Google Scholar]
- 15.Block E. Garlic and Other Alliums—The Lore and the Science. RSC Publishing; Cambridge, UK: 2010. [Google Scholar]
- 16.Reiter J., Levina N., Van der Linden M., Gruhlke M., Martin C., Slusarenko A.J. Diallylthiosulfinate (Allicin), a volatile Antimicrobial from GARLIC (Allium sativum), Kills human lung pathogenic Bacteria, Including MDR strains, as a Vapor. Molecules. 2017;22:1711. doi: 10.3390/molecules22101711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiter J., Hübbers A.M., Albrecht F., Leichert L.I.O., Slusarenko A.J. Allicin, a natural antimicrobial defence substance from garlic, inhibits DNA gyrase activity in bacteria. Int. J. Med. Microbiol. 2020;310:151359. doi: 10.1016/j.ijmm.2019.151359. [DOI] [PubMed] [Google Scholar]
- 18.Reiter J., Borlinghaus J., Dörner P., Schröder W., Gruhlke M.C.H., Klaas M., Slusarenko A.J. Investigation of the deposition behaviour and antibacterial effectivity of allicin aerosols and vapour using a lung model. Exp. Ther. Med. 2020;19:1541–1549. doi: 10.3892/etm.2019.8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miron T., Rabinkov A., Mirelman D., Wilchek M., Weiner L. The mode of action of allicin: Its ready permeability through phospholipid membranes may contribute to its biological activity. Biochim. Biophys. Acta (BBA) Biomembr. 2000;1463:20–30. doi: 10.1016/S0005-2736(99)00174-1. [DOI] [PubMed] [Google Scholar]
- 20.Gruhlke M.C.H., Portz D., Stitz M., Anwar A., Schneider T., Jacob C., Schlaich N.L., Slusarenko A.J. Allicin disrupts the cell’s electrochemical potential and induces apoptosis in yeast. Free Radic. Biol. Med. 2012;49:1916–1924. doi: 10.1016/j.freeradbiomed.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 21.Gruhlke M.C.H., Slusarenko A.J. The biology of reactive sulfur species (RSS) Plant Physiol. Biochem. 2012;59:98–107. doi: 10.1016/j.plaphy.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 22.Müller A., Eller J., Albrecht F., Prochnow P., Kuhlmann K., Bandow J.E., Slusarenko A.J., Leichert L.I. Allicin induces thiol Stress in bacteria through S-allylmercapto modification of protein cysteines. J. Biol. Chem. 2016;291:11477–11490. doi: 10.1074/jbc.M115.702308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leontiev R., Slusarenko A.J. Finding the starting point for mode-of-action studies of novel selenium compounds: Yeast as a genetic toolkit. Curr. Org. Synth. 2017;14:1–7. doi: 10.2174/1570179414666170525112446. [DOI] [Google Scholar]
- 24.Noctor G., Foyer C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:249–279. doi: 10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 25.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/S0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 26.Gruhlke M.C.H., Schlembach I., Leontiev R., Uebachs A., Gollwitzer P., Weiss A., Delaunay A., Toledano M.B., Slusarenko A.J. Yap1p, the central regulator of the S. cerevisiae oxidative stress response, is activated by allicin, a natural oxidant and defence substance of garlic. Free Radic. Biol. Med. 2017;108:793–802. doi: 10.1016/j.freeradbiomed.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Leontiev R., Hohaus N., Jacob C., Gruhlke M.C.H., Slusarenko A.J. A comparison of the antibacterial and antifungal activities of thiosulfinate analogues of allicin. Sci. Rep. 2018;8:6763. doi: 10.1038/s41598-018-25154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borlinghaus J., Bolger A., Schier C., Vogel A., Usadel B., Gruhlke M.C.H., Slusarenko A.J. Genetic and molecular characterization of multi-component resistance of Pseudomonas against allicin. Life Sci. Alliance. 2020;3:e202000670. doi: 10.26508/lsa.202000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuge S., Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jamieson D.J. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 31.Toledano M.B., Delaunay A., Biteau B., Spector D., Azevedo D. Oxidative stress responses in yeast. In: Hohmann S., Mager W.H., editors. Yeast Stress Responses. Springer; Berlin/Heidelberg, Germany: 2003. pp. 241–303. [Google Scholar]
- 32.Morano K.A., Grant C.M., Moye-Rowley W.S. The response to heat Shock and oxidative stress in saccharomyces cerevisiae. Genetics. 2011;190:1157–1195. doi: 10.1534/genetics.111.128033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brachmann C., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 34.Borlinghaus J., Albrecht F., Gruhlke M.C.H., Nwachukwu I.D., Slusarenko A.J. Allicin: Chemistry and biological properties. Molecules. 2014;19:12591–12618. doi: 10.3390/molecules190812591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarfraz M., Nasim M.J., Jacob C., Gruhlke M.C. Yeast chemogenetic screening as a tool to unravel the antifungal mode of action of Two selected selenocyanates. Appl. Sci. 2019;9:3728. doi: 10.3390/app9183728. [DOI] [Google Scholar]
- 36.Parisy V., Poinssot B., Owsianowski L., Buchala A., Glazebrook J., Mauch F. Identification of PAD2 as a gamma-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 2007;49:159–172. doi: 10.1111/j.1365-313X.2006.02938.x. [DOI] [PubMed] [Google Scholar]
- 37.Mhamdi A., Hager J., Chaouch S., Queval G., Han Y., Taconnat L., Saindrenan P., Gouia H., Issakidis-Bourguet E., Renou J.-P. Arabidopsis glutathione reductase1 plays a crucial role in leaf responses to intracellular Hydrogen Peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010;153:1144–1160. doi: 10.1104/pp.110.153767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moutiez M., Aumercier M., Parmentier B., Tartar A., Sergheraert C. Compared recognition of di- and trisulfides substrates by glutathione and trypanothione reductases. Biochim. Biophys. Acta. 1995;1245:161–166. doi: 10.1016/0304-4165(95)00085-P. [DOI] [PubMed] [Google Scholar]