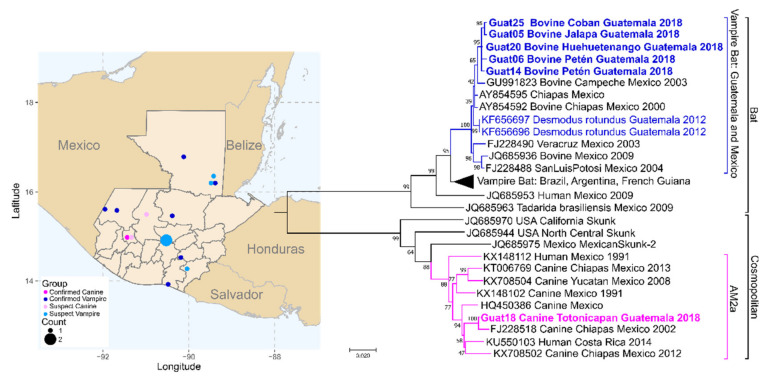
Figure 4.
Phylogenetic analysis of MinION sequences generated in Guatemala City, Guatemala. Complete nucleoprotein gene sequences from Guatemala rabies virus isolates were compared to publicly available sequences. Reference sequences were chosen based on sequence similarity to the new Guatemala sequences, isolation from vampire bat or bovine in Mexico or Guatemala, or inclusion Cosmopolitan Americas-2a (AM2a) rabies phylogenetic clade based on Troupin et al. [31]. Accession number, host animal, location, and collection year are included for reference, when available. Sequences generated in this study are highlighted in bold; additional information can be found in Table S1. Colored points on the map correspond to the location of LN34 positive samples; confirmed canine or vampire bat lineage isolates are colored dark blue or dark pink on the map and phylogenetic tree. Suspected lineage was based on host (canine or bovine). Phylogenetic analysis was performed by maximum likelihood based on the GTR+G+I model in Mega7. Number of changes per site is shown along the horizontal axis. Bootstrap values near the branch points represent the percentage of trees that had the same clustering out of 1000 replicates.