Abstract
The symptoms associated with Covid-19 caused by SARS-CoV-2 in severe conditions can cause multiple organ failure and fatality via a plethora of mechanisms, and it is essential to discover the efficacious and safe drug. For this, a successful strategy is to inhibit in different stages of the SARS-CoV-2 life cycle and host cell reactions. The current review briefly put forth the summary of the SARS-CoV-2 pandemic and highlight the critical areas of understanding in genomics, proteomics, medicinal chemistry, and natural products derived drug discovery. The review further extends to briefly put forth the updates in the drug testing system, biologics, biophysics, and their advances concerning SARS-CoV-2. The salient features include information on SARS-CoV-2 morphology, genomic characterization, and pathophysiology along with important protein targets and how they influence the drug design and development against SARS-CoV-2 and a concerted and integrated approach to target these stages. The review also gives the status of drug design and discovery to identify the drugs acting on critical targets in SARS-CoV-2 and host reactions to treat Covid-19.
Keywords: SARS-CoV-2, Covid-19, Repurposed drugs, Drug design, Vaccines, Biologics, Natural products, Food and nutrients
Graphical abstract
List of associated abbreviations
- A3AR
A3 adenosine receptor
- ACE-RAAS
Angiotensin Converting Enzyme-Renin-angiotensin-aldosterone
- ACE2
Angiotensin-converting enzyme 2
- ADAM17
Disintegrin and metalloproteinase domain
- Ads-nCoV
Adenovirus type-5 vectored Covid-19
- AI
Artificial Intelligence
- ALI
Acute lung injury
- ARBs
Angiotensin receptor blockers
- ARDS
Acute Respiratory Distress Syndrome
- CADD
Computer-assisted drug and designing
- CCR5
Chemokine receptor 5
- CEPI
Coalition for Epidemic Preparedness Innovations
- CGRP
Calcitonin gene-related peptide;
- 3CLpro
Cathepsin-like protease
- CSF
Granulocyte macrophage-colony stimulating factors
- CT scan
Computed Tomography imaging technology
- DNase 1
Deoxyribonuclease-1
- dATP
Deoxyadenosine triphosphate
- dCTP
deoxycytidine 5′-triphosphate
- EBV
Epstein-Barr virus
- ED
Ectodomain
- FDA
Food and Drug Administration
- H-CoV-229E
Human coronavirus 229E
- HCQ
Hydroxychloroquine
- HTS
High Throughput Screening
- IFN
Interferon
- IgG4
Human immunoglobulin G4
- IL-6
Interleukin 6
- NO
Nitric oxide
- JAK-STAT
Janus Kinase/Signal Transducer and Activator of Transcription
- LUNAR
Lipid enabled and unlocked nucleo-monomer agent modified RNA
- MAPK
Mitogen-activated protein kinase
- MERS-CoV
Middle East respiratory syndrome coronavirus
- ML
Machine learning
- mTOR
Mammalian target of rapamycin
- modRNA
Nucleoside modified mRNA
- NF-κB
Nuclear factor kappa-light-chain-enhancer of activated B cells
- NK
Natural Killer
- NK-1
Neurokinin-1
- NORS
Nitric Oxide Releasing Solution
- NRTI/NtRTI
Nucleoside/nucleotide reverse transcriptase inhibitors
- NTD
N-terminal Domain
- PDB
Protein Data Bank
- PLpro
Papain-like protease
- pp1a and pp1ab
Polyproteins
- RAAS
Renin-angiotensin-aldosterone system
- RBD
Receptor-binding domain
- RBD-CuMVTT
Recombinant receptor binding domain optimized virus-like particles based on cucumber mosaic virus
- RdRp:
RNA-dependent RNA polymerase
- rMV
Recombinant measles virus
- ROS
Reactive oxygen species
- RT-PCR
Reverse transcription-polymerase chain reaction
- SGLT2
Sodium-glucose cotransporter-2
- S1P
Sphingosine 1-phosphate receptor
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SD1, SD2
Sub-Domain 1 and 2
- SINE
Selective inhibitor of nuclear export
- SSRI
Serotonin reuptake inhibitor
- STARR
Self-transcribing and replicating RNA
- TMPRSS
Transmembrane protease serine 2
- TNF
Tumor necrosis factor
- VEGF
Vascular endothelial growth factor
- VIP
Vasoactive intestinal polypeptide;
- uRNA
Uridine containing mRNA
- XPO1
Exportin 1
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) is a causative virus for Covid-19 or coronavirus disease (Lai et al., 2020). The novel strain was first identified in December 2019 (Bittmann et al., 2020), and it is the seventh coronavirus strain that has been reported to infect Homo sapiens (Poduri et al., 2020). During the writing of this manuscript, Covid-19 has led to the mortality of 1, 453,355 as of November 30, 2020 (https://covid19.who.int/). SARS-CoV-2 infects the lower respiratory system like that of SARS-CoV and MERS-CoV and cause viral pneumonia. Additionally, SARS-CoV-2 may also affect the kidney, liver, central nervous system, and gastrointestinal system leading to multiple organ failure (Su et al., 2016; Zhu et al., 2020). Apart from lungs, Cardiovascular complications are recently seen as a new threat in Covid-19 along with blood clotting incidences (Escher et al., 2020; Varga et al., 2020). Further, the epidemiological data confirm that Covid-19 mortality is higher in men than in women attributed via significant role mediated by androgens (Goren et al., 2020; Sharifi and Ryan, 2020). Some studies have also highlighted the role of ABO system with the severity of Covid-19 suggesting A+ /AB system are more infected (Latz et al., 2020). The speedy spread of the disease and the mortality associated has caused havoc among humans all over the Globe, which has further worsened with the unavailability of some putative chemical bullets in forms of drugs and biologics (Pillaiyar et al., 2020; Xu and Dang, 2020). Covid-19 is now being considered as the second leading cause of death after cardiovascular diseases (Balakumar et al., 2016). The current scenario has led to the rapid repurposing of drugs for this pandemic. A data insight at Clinical Trials (https://clinicaltrials.gov/) informed that 2427 trials are being conducted around the Globe to find a putative lead. The data suggested that among all the studies, 25 studies are undergoing at preclinical phase, 141 in Phase 1, 539 in Phase 2, 327 in Phase 3, and 71 in Phase 4. Further data also suggested that among all the studies 182 have been completed, 1421 are either Ongoing/Recruiting/Active, not recruiting/Enrolling by invitation, whereas 41 have been either suspended/terminated or have withdrawn status, while remaining have unknown status. However, no apparent success has been seen so far. So, it is imperative to recognize the target to get high target specificity drugs that may be existing or designing new drug molecules to treat SARS-CoV-2 infection. Previously, during the pandemic of SARS-CoV and MERS various drugs including hydroxychloroquine (Zhou et al., 2020), nitazoxanide (Rossignol, 2016), terconazole (Omrani and Memish, 2015), ritonavir (Chan et al., 2015), homoharringtonine (Mustafa et al., 2018), lopinavir (Chan et al., 2015), ribavirin (Khalili et al., 2020), emetine (Sharif-Yakan and Kanj, 2014), and many more were repurposed using various methodologies. The drugs for current pandemic under repurposed category include drugs from broad groups encompassing antimalarials, interleukin inhibitor, serine protease inhibitor, antifibrinolytic, hemostatic, Antivirals, AT-1 receptor blocker, anticancer, antiretroviral, non-nucleoside reverse transcriptase inhibitor, anti-inflammatory, immunosuppressant, glucocorticoid, immunomodulator, anti-angiogenic, anti-inflammatory, anti-fibrotic gasotransmitter, an antigout, acetylcholinesterase inhibitor, Calcitonin gene-related peptide (CGRP) receptor antagonist, neurokinin1 receptor antagonist and many more. Various repurposing methodologies involved in identifying these drugs include, i. Computational approaches, that importantly covers various important tools including molecular docking, signature matching, pathway mapping, genetic association, novel data sources, and retrospective clinical analysis; ii. the experimental approaches includes methods such as phenotypic screening (Moffat et al., 2017) and binding biological assays which have eased the drug development process in SARS-CoV-2.
This review is therefore kept forth to compile the information on i. SARS-CoV-2 morphology, genomic characterization, and pathophysiology; ii. Important protein targets and how they influence the drug design and development against SARS-CoV-2; iii. Understand how the virus affects the major body organs after its induction inside the body, along with its interaction with the ACE-RAAS system; iv. Discussion on the dysregulated inflammatory response as a consequence, and iv. a concerted and integrated approach to target these stages of the SARS-CoV-2 life cycle with the use of upcoming repurposed drugs along their mechanisms involved. The current section(s) also presents the argument that multiple stages of the life cycle should be targeted in the quest for a better drug regimen in the future. The review also gives a strategic background to the scientist working on drug design and discovery to identify the essential targets in SARS-CoV-2 and host explore the active sites within them to facilitate them in rational drug development for Covid-19. The discussion on natural products derived molecules for Covid-19 will give a boost to natural products chemists to explore mother nature for finding underlying cure within nature only. Further, the review has also been extended to cover the role of physical science-based tools, artificial intelligence in assisting the drug discovery for SARS-CoV-2. A section has also been added to cover important in vitro and in vivo models along with upcoming facilities to test the alleged drug or drug-like candidates along with the brief discussion on the advancement in vaccines and monoclonal antibodies against SARS-CoV-2.
2. Genomic and morphological insights towards SARS-CoV-2
The study on Malayan pangolins highlighted that these animals host a virus similar to SARS-CoV-2 and can develop antibodies recognizing viral S protein. The findings, thus provide the evidence that SARS-CoV-2 share their origin from a bat virus which got recombined with a pangolin virus (Xiao et al., 2020). Further, given SARS-CoV-2 morphological characteristics and structural insights, it is pleomorphic in shape with a size of about ~125 nm, with a 30 kb (+) RNA genome. Considering the mutations, the virus may be classified into two lineages, S (Serine) (~30%) and L (Leucine) (~70%) types. In both lineage S type contributes to 3.7% of viral isolates compared to 96.3% of L type isolated from Wuhan, China. However, outside Wuhan, the percentage was 61.3% for L type and 38.4% S type. The L type was stated to be more rampant and termed as “aggressive type.” However, this classification system has been interrogated by the scientific community, since they have no documented biological differences and involves only sequence difference (Lu et al., 2020). SARS-CoV-2 genomes may be categorized into at least ten distinct groups based on phylogenetic relationships between various strain and are termed as clades.
Further, according to specific variations in different viral proteins, GISAID (global science initiative and primary source) identified clades of SARS-CoV-2 as, i. G clade (D614G in S protein) and mainly encompasses sequences from Europe, ii. V clade (G251V in ORF3) which mostly includes Asian and European sequences, iii. S clade (L84S in ORF8) and mostly comprises sequences from North America. A new clade of SARS-CoV-2 carrying V378I in ORF1ab (Open Reading Frame) has been linked to travellers returning from Iran to Australia. Based on Next strain analysis and nomenclature, original isolates from China are part of the B clade, whereas isolates from across the world belong to the A clade. A2a clade is the most recent and common sequenced clade (D614G on the Spike protein) (Forster et al., 2020; Li et al., 2020). In India, A2a (48.6%) clade was found to be dominant (Mondal et al., 2020). These clades further diverge in virulence which might affecting the efficacy of repurposed drugs and may dampens the effectiveness of vaccines/biologics when available in future.
3. Protein assembly and druggable targets to explore in SARS-CoV-2
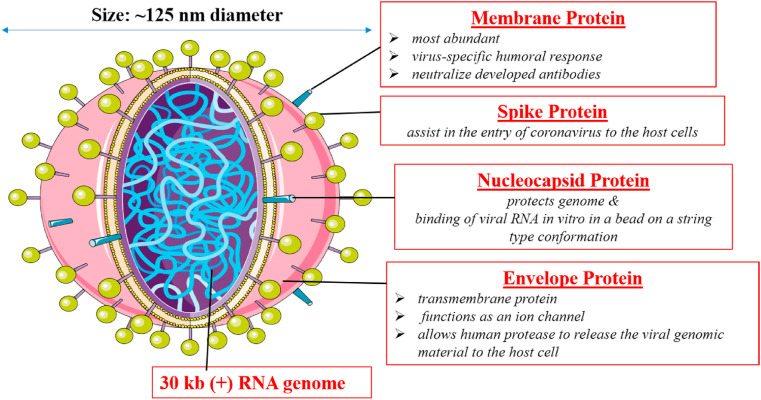
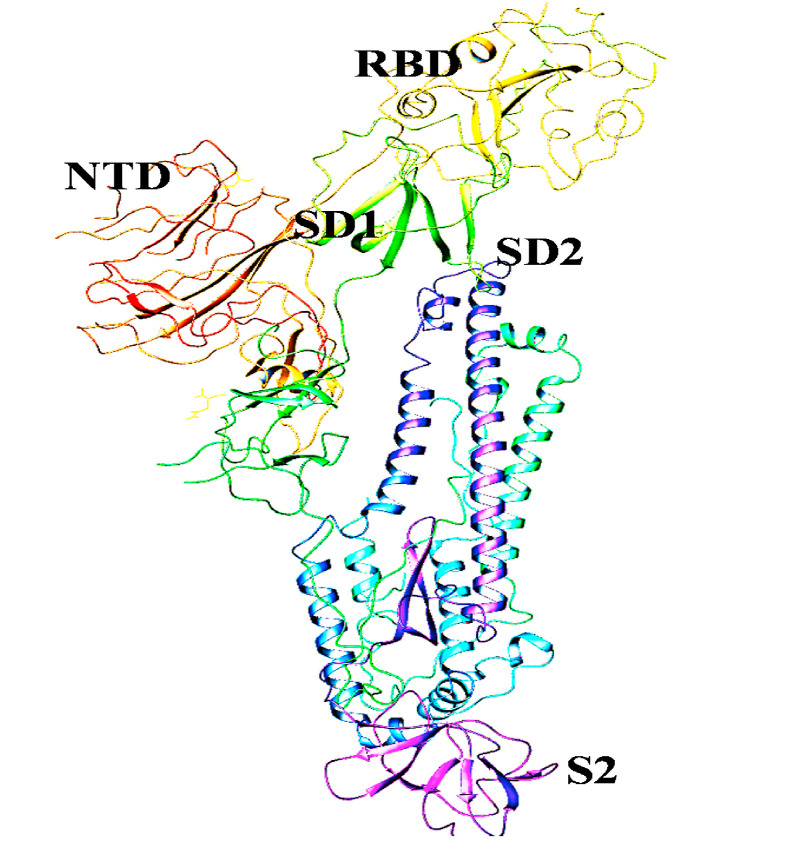
SARS-CoV-2 comprise of some essential proteins that include, Membrane Protein (M), which is most abundant and is responsible for virus-specific humoral response and also plays a vital role in neutralizing developed antibodies. Another protein is Spike (S) protein, which assists in the entry of coronavirus by binding to ACE2 (Angiotensin-converting enzyme) receptor of the host cell. Further few studies have also investigated the role of Neuropilin-1 in SARS-CoV-2 entry inside host cell along with ACE2 (Cantuti-Castelvetri et al., 2020) Then comes Nucleocapsid (N) protein, which protects the genome and assists in the binding of viral RNA in vitro in a bead on a string type conformation. Next is the Envelope (E) protein, a transmembrane protein which acts as an ion channel allowing the release of viral genomic material to the host cell via human protease. Further, the novel virus also possesses a lesser-known Hemagglutinin-esterase dimer protein (HE) that is known to binds to sialic acids and allows virus release from the infected cell after their hijack (Fig. 1 ). The S Protein of SARS-CoV-2 subunit consists of two major domains, S1 or N-terminal (upper domain) and S2 or C- terminal (lower domain). The upper domain is also called the receptor-binding domain (RBD) and is responsible for receptor binding. The S1 domain is further divided into two sub-domains (SD-1 and SD-2) which are thought to allow receptor-induced conformational changes in the C-terminal domain (Kirchdoerfer et al., 2016). The amino acids in this region are variable as they are under evolutionary pressure because of constant interaction with the immune system.
Fig. 1.
Pictorial representation of SARS-CoV-2 structure.
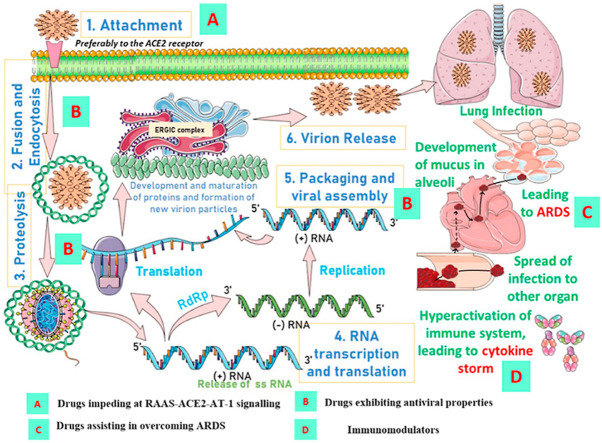
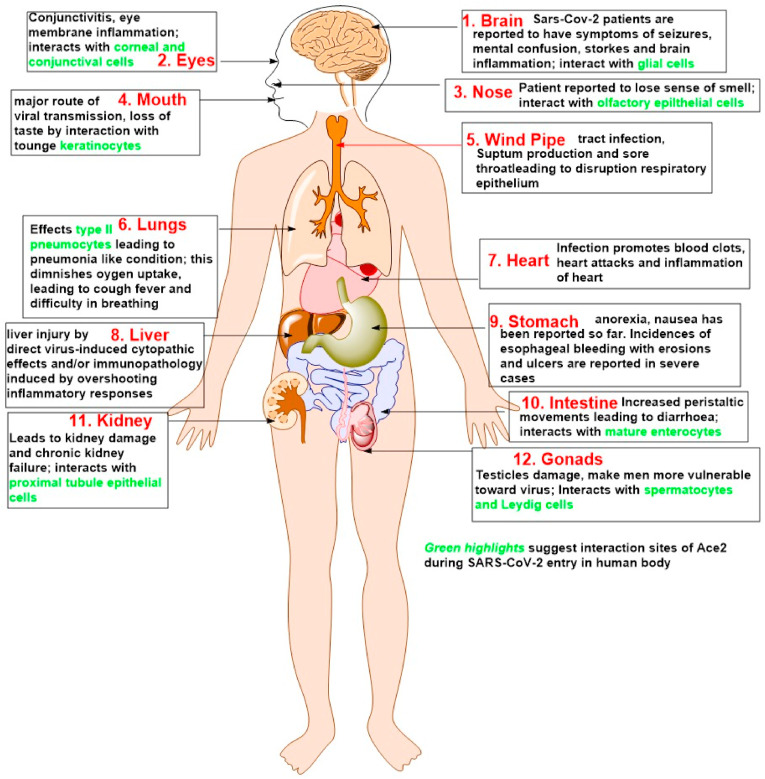
Further, the lower domain (S2) comprises of a fusion machinery, with a hydrophobic fusion peptide. Further, the interaction of SARS-CoV-2 with host cells occurs via various multistep pathways. The primary pathway is believed to occur via the following five routes, i. virus spike recognition by ACE2 protein (Attachment); ii. TMPRSS2 mediated cleavage of spike protein (Activation); iii. Virus endocytosis (Membrane Fusion); iv. The hijacking of host cell machinery by the virus (Infection); v. ADAM17-dependent ACE2 shedding leading to lung pathogenesis, ARDS followed by a cytokine storm. The pathway begins with the binding or fusion of the virus with the host cell. Mechanistically, it is very clearly known now that SARS-CoV-2 uses TMPRSS2, which is a known transmembrane serine protease (type II) expressed in epithelial cells of respiratory, gastrointestinal, and urogenital tracts (Hussain et al., 2020) for its interaction and, consequently, its entry to host cells. TMPRSS2 cleaves the S protein at the junction of hydrophobic fusion peptide junction into two domains, S1 and S2. Furin-mediated pre-cleavage at the S1/S2 site (cleavage site is 685/686 junction) in infected cells might promote subsequent TMPRSS2 action. The S1/S2 region is further known to possess numerous arginine or lysine residues that lead to cleavage of S protein catalysed by TMPRSS2 at these monobasic cleavage sites (Hofmann and Pohlmann, 2004). The S1 domain further interacts with the host ACE2 receptor and leading to SARS-CoV-2 entry inside the host cell(s) (Hoffmann et al., 2020; Lukassen et al., 2020). It is now known that SARS-CoV-2 has the potential to affect numerous body parts and organs along with lungs. The probable reason for this is attributed to the expression of ACE2 receptors throughout the body ( Fig. 2 ). ACE2 is widely distributed starting from the nasal epithelium and extending to the oral mucosa, brain, eyes, windpipe, lungs, heart, liver, stomach, intestine, kidney to gonads (Hikmet et al., 2020; Venkatakrishnan et al., 2020) The infection, therefore, starts with respiratory tract importantly lungs leading to pneumonia to severe acute respiratory distress syndrome (ARDS), followed by cytokines storm which further fuels significant organ dysfunctions eventually leading to death in severe cases. Studies also suggest 32 variants of ACE2 across populations defining the differential severity (Renieri et al., 2020). Before delving deeper into the role of ACE2 and their role in worsening the outcomes of SARS-CoV-2, let us understand in brief the underlying mechanisms of the virus inside the host cell(s).
Fig. 2.
Illustration depicting the major expression sites of ACE2 in the human body. The boxes briefly explain the consequence after the interaction of SARS-CoV-2. Green highlight corresponds to the interaction site(s) of ACE2 in those organs or organ systems. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
After interaction with the host cell, the next step is the translocation of the virus within the host cells. As per reports, SARS-CoV-2 is reported to translocate via two major pathways, endocytic and non-endocytic or non-endosomal. Endocytic pathway is more accepted and is believed to proceed with the assistance of lysosomes. The pathway has been traced based on positive outcomes of lysosomotropic agents, protease inhibitors, and clathrin-mediated endocytosis inhibitors, which favours the non-endosomal pathway (Wang et al., 2008). Once inside the endosomes, proteolysis mechanism activates. This involves the release of the viral genome as a single-stranded positive RNA. The RNA acts as messenger RNA and undergoes translation/replication by using host cell machinery. The critical steps involved are RNA synthesis, proofreading of template, capping followed by protein synthesis catalysed by Non-structural protein complexes (Nsp1-16). The important Nsp's identified and targeted to date are papain-like protease (PLpro (Nsp3)) and main protease (3CLpro (Nsp5)) which are which are also involved in the cleavage of polyproteins translated from viral mRNA. Thus, leading to the formation of functional or effector proteins. Further, 3CLpro also acts as a deubiquitinase and suppresses the innate immune system of the host (Yang et al., 2020b). Next is, primase complex (Nsp7-Nsp8) capable of both de novo initiation and primer extension; RNA dependent RNA polymerase; RdRp (Nsp12)), involved in viral transcription and replication; zinc-binding helicase (HEL (Nsp13)), involved in unwinding of duplex RNA/DNA with 5′ss tail in 5′–3′ direction; 2′O-methyltransferases (Nsp10/Nsp16). Further, the mRNA capping is done by Nsp14-16 complex, also, Nsp14 performs RNA proofreading. Besides, the host cell machinery (ER and Golgi apparatus; ERGIC complex) also plays a vital role in the generation and assembly of new viral particles (da Silva et al., 2020; Jang et al., 2020; Te Velthuis et al., 2012; Wang et al., 2020).
Since the template involves ss-positive sense RNA, it importantly acts as messenger RNA (mRNA) and possesses the ability to undergo translation. The translation leads to the generation of two polyproteins (pp1a and pp1ab) via ribosomal frameshift mechanism. These polyproteins encode for Nsp3 and Nsp5 along with forming Replication complex and Non-structural proteins (1–16). Nsp12 from this complex further catalysis replication process, followed by translation leading to the formation of sgRNA, structural, and accessory proteins. These developed proteins further assembles and matures in the Golgi complex of host cells to form matured or semi matured virion particals. The matured viruses than evades (virion release) the host cell via exocytosis or cell death, semi matured viruses then evades via the process of budding from the infected host cell and infects other healthy cells. The N-protein assist virion release by developing a favourable orientation for virion at perinuclear membranes allowing the release of virus particles. Once the infections spread, it starts with lung damage by pneumonia, followed by ARDS and cytokine rampage, thus destroying the host cell immune system and eventually leading to death in severe cases (Kim et al., 2020; Shang et al., 2020; Singh et al., 2020; Viswanathan et al., 2020; Wang et al., 2020). The brief mechanism is represented in Fig. 3 .
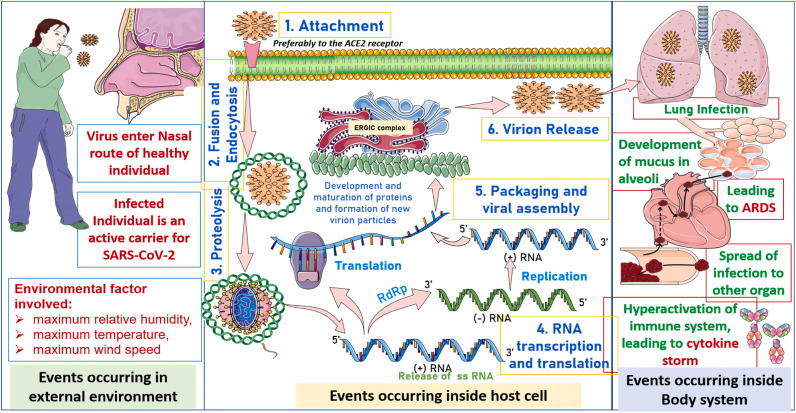
Fig. 3.
Illustrations depict three critical pathways concerning the SARS-CoV-2 infection. The pathways include transmission of the virus from an infected individual to healthy individual, the life cycle of virus development within the host cell and its progression within body system precipitating conditions like ARDS and cytokine storm.
Coming back to ACE2, it exists in two forms. The first form is insoluble with full length and is anchored to the extracellular domain. The second form consists of a soluble and cleaved form that circulates in varying concentrations in blood. Both these forms possess a protease domain and the catalytic domain. The protease domain is involved with S1 binding, which results in the downregulation of ACE2, thus diminishing its protective role in heart and lungs, which is further worsened by ACE2 internalization (Hamming et al., 2004; Yan et al., 2020). The second unit is the catalytic site and involved in the removal of an amino acid (phenylalanine) from octapeptide AngII (pressor hormone) to form Ang1-7. Ang1-7 also gets converted to Ang1-9 (cleaved product of AngI) in the presence of ACE2 and its endopeptidase, which is neutral. The physiological functions of AngII produce via AT-1 and 2 receptors. These promote vasoconstriction, proliferation, oxidative stress, fibrosis, thrombosis, and inflammation, while Ang1-7 signals via Mas receptor (GPCR mediated) and promotes cardioprotective, anti-inflammatory, vasodilatory, anti-thrombosis, and anti-proliferative properties (Donoghue et al., 2000).
Further, elevated Ang II levels during the infection are also associated with the disease progression leading to the stage of cytokine storm (Kuba et al., 2005). Though in contrast to this, there are no conclusive studies made to prove the hypothesis (Amraei and Rahimi, 2020; Misra et al., 2020). A few studies, however, have implicated the crosstalk of IL-6 and TNF-α with Ang II. The Cytokine Storm, in general, is an exuberant inflammatory response associated with advanced stages of Covid-19. It allows hyperactivation of the immune system leading to conditions like Acute lung injury (ALI) and collagen disposition in lungs (Kuba et al., 2005) Dozens of cytokines are released and are implicated in the pathogenesis of SARS-CoV-2 infection. Such cytokines are widely known to enhance neutrophil infiltration, which, in turn, induce lung inflammation and respiratory distress (ARDS) (Shi et al., 2020; Tay et al., 2020). Moreover, redox signalling events, including Activation of Transcriptional Factors (NFκB), Induction of Pro-inflammatory Cascades, NO Depletion, Vascular Endothelial Dysfunction, Apoptosis, Necrosis, activates a variety of intracellular protein kinases-MAPKinase family (ERK1/2, p38MAPK, PKC isoforms, JNK, STAT/JAK) Receptor and non-receptor tyrosine kinases and serine/threonine kinases, leading to vasoconstriction, platelet activation, aggregation and adhesion resulting in blood clot (inflammatory thrombocytosis); neutrophil recruitment, fibrosis, endothelial dysfunction, inflammation are associated lung damage (Huertas et al., 2020; Mehta et al., 2020; Pan et al., 2020). The studies conducted so far finds increased plasma concentration and decreased lymphocyte counts in Covid-19 patients along with elevation of inflammatory cytokines (IL-6 and TNF). The level of CD4+ T cells, CD8+ T cells, natural killer cells, perforin and granulysin-expressing cytotoxic T cells were also found to be downregulated (Diao et al., 2020; Zhong et al., 2020).
Therefore, in a nutshell, numerous protein/enzymes of the host, as well as virus system, leads to a complex signalling cascade directly from the interaction of the virus with the host cell, provoking the infection and hijacking the host machinery and leading to organ damage or even death at the very severe condition. These proteins/enzymes also allow targeting them by the development of some putative inhibitor based on the mechanism of viral transmission and infection. What is the recent information and what could be explored to develop some rational therapies in the future, is a question we will try to answer in the next section?
4. Key druggable targets available: exploring the binding pocket
The continuous efforts have been made in the search for druggable targets in SARS-CoV-2 virus. The important one includes spike protein, membrane protein, protease, envelope protein, hemagglutinin esterase, nucleocapsid protein, and helicase proteins (Prajapat et al., 2020). The majority of drugs explored so far includes drugs that either targets and inhibits viral replication or viral proteases (Joshi et al., 2020). The subsequent section of the article includes a detailed description of these target proteins, about the anti- SARS-CoV-2 drug development process.
4.1. Targets for SARS-CoV-2 entry for binding/translocation
The vital target in this category includes S-protein of the virus, ACE2 receptors of host cells, CD147 receptor and GRP78 receptors.
4.1.1. Spike protein
It is the most crucial SARS-CoV-2 druggable target, having a clove-like shape, and belongs to the type I transmembrane (TM) protein (Fig. 4 ). It reveals three segments, i.e., ectodomain (ED) region, TM region, and the intracellular domain (Belouzard et al., 2012; Prajapat et al., 2020). The ED region of this protein is further subdivided into two-part, i.e., the receptor-binding (RBD) S1 domain and the membrane fusion subunit S2 on the C-terminal domain. Spike protein and its role in facilitating the entry of the virus into the host cell is very critical, as its S1 domain starts an initial interaction with the host cells viz the ACE2 protein. Subsequently, the S2 part facilitates the fusion process and thus allowing the host cell internalization of the virus genome(Li, 2016; Liu et al., 2020c). Therefore, the role of Spike protein is very critical, and hence, it is a substantial target receptor for the development of anti -SARS-CoV-2 drug molecules (Prajapat et al., 2020). To understand it, we have several pieces of evidence, showing its importance in targeting SARS-CoV-2 propagation. The particular type of amino acid sequence is observed in the host receptor-binding part of the spike protein, and interestingly, a free peptide sequence, with sequence similarity to the host receptor-binding part, was found hampering the host cell penetration process of SARS-CoV-2 virus strain (Prajapat et al., 2020). The chloroquine and its impact in rendering the virus propagation are also well known. The action of chloroquine on SARS-CoV-2 is also related to the spike protein. It increases the pH of the endosomes and alters the ACE-2 terminal glycosylation. Thus, ultimately rendering the spike protein-mediated host cell entry of the viral genome. Furthermore, multiple small molecules are also reported, interfering with the Spike protein-ACE-2 interactions and found successful in retarding the propagation of the viral genome (Prajapat et al., 2020).
Fig. 4.
Structural feature representation showing receptor-binding domain (RBD), N terminal Domain (NTD) and Sub-Domain 1 and 2 (SD1, SD2) as a part of S1 subunit and S2 subunit in case of SARS-CoV-2 spike protein.
4.1.2. ACE2 receptors
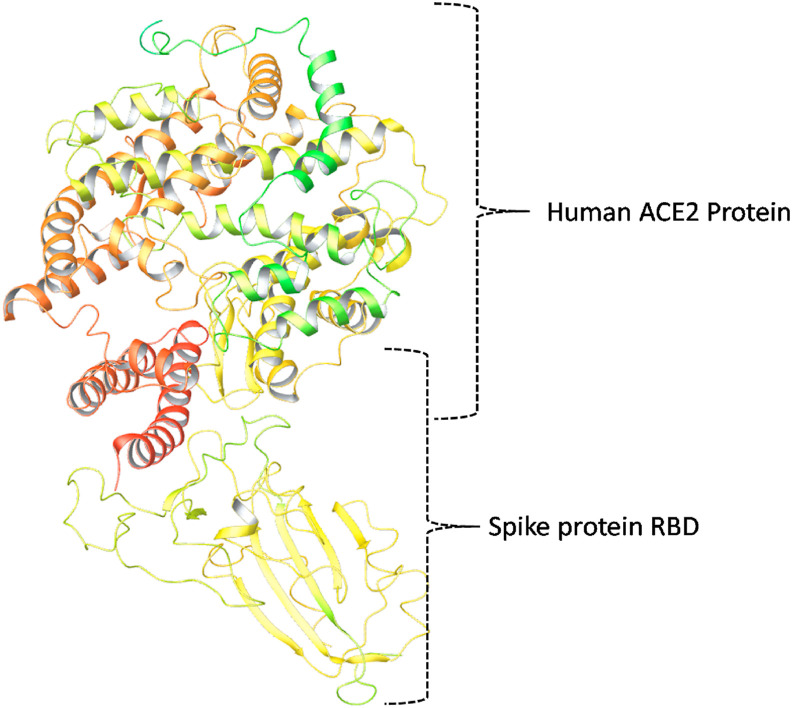
Angiotensin-converting enzyme 2 (ACE2) is the protein, affecting the initiation and propagation of the Covid-19 disease in multiple ways. At the initial stage, it facilitates the entry of the SARS-CoV-2 virus into the host cell, and that is mediated by Spike protein-ACE2 interaction phenomenon (Huang et al., 2020). Other than that, ACE2 is also involved in critical Covid-19 pathophysiological processes, such as virus-induced acute lung injury, and other organ damage (Huang et al., 2020). The interaction between ACE2 and spike protein is the most crucial step in the SARS-CoV-2 pathology, and thus the development of small molecules, monoclonal antibodies towards RBD, and recombinant human ACE2 protein seem very promising to retard the start and propagation of Covid-19 disease (Huang et al., 2020). The interaction pattern of ACE-2 and Spike protein RBD is as represented in Fig. 5 .
Fig. 5.
Structural representation of the interaction pattern of Human ACE2 and the SARS-CoV-2 Spike protein RBD domain.
4.1.3. CD147 and GRP78 receptor
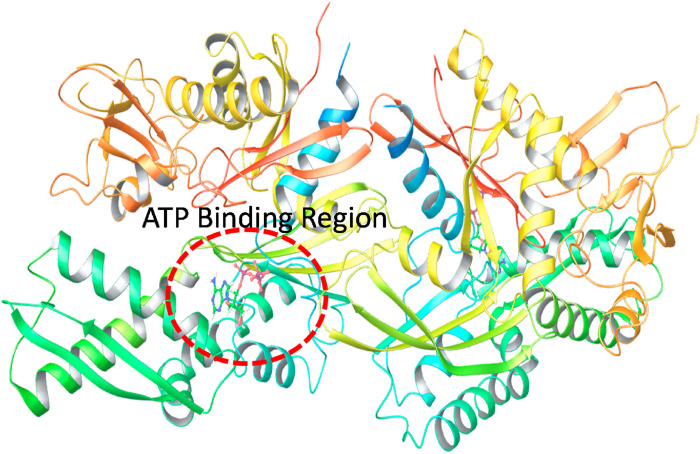
The CD147 receptor that is present on the host cell, also play a significant role in SARS-CoV-2 invasion (Ulrich and Pillat, 2020). The CD147-Spike protein interaction is yet another critical step in the propagation of the disease. Therefore, small drug-like molecules, and/or other therapeutic entity that either interfere with the CD147-Spike protein interaction or downregulate the CD147 expression, seems useful in reducing the propagation rate of the virus entity. Several clinical studies related to azithromycin in hospitalized patients revealed substantial beneficial effect, and it was suggested to be due to the interfering effect of azithromycin on CD147 related invasion phenomenon. Therefore, very similar to ACE2, CD147 can also be considered as an essential target to target SARS-CoV-2 propagation phenomenon (Ulrich and Pillat, 2020). Similarly, GRP78 is also known to bind with the spike protein and helps in the propagation of the disease. A structural feature of GRP78 with ATP as a co-crystallized ligand is represented in Fig. 6 . Herein, the position of the receptor where the ATP binds seems an important binding cavity that may be targeted for producing the antiviral activity against the disease.
Fig. 6.
Structural features of the GRP78 showing the position of the ATP binding site within its structure.
4.2. Protease as a drug target
Like many other viruses, the role of proteases is critical for the proper replication of the SARS-CoV-2 viral genome. The major component of the SARS-CoV-2 viral genome is the replicase gene (Lindner et al., 2005). It holds information to produce 16 most important non-structural proteins. As a precursor of these 16 non-structural proteins, it initially makes two large polyproteins named as polyproteins 1a and polyproteins 1 ab (Lindner et al., 2005; Prajapat et al., 2020). However, subsequently, after the translation process, a couple of cysteine proteases act on these two polyproteins, cleave them carefully, to release the non-structural proteins. Designing and development of SARS-CoV-2 specific protease inhibitors are thus considered very promising strategy against this infectious disease (Prajapat et al., 2020). The important proteases associated with the virus are discussed below.
4.2.1. Host protease
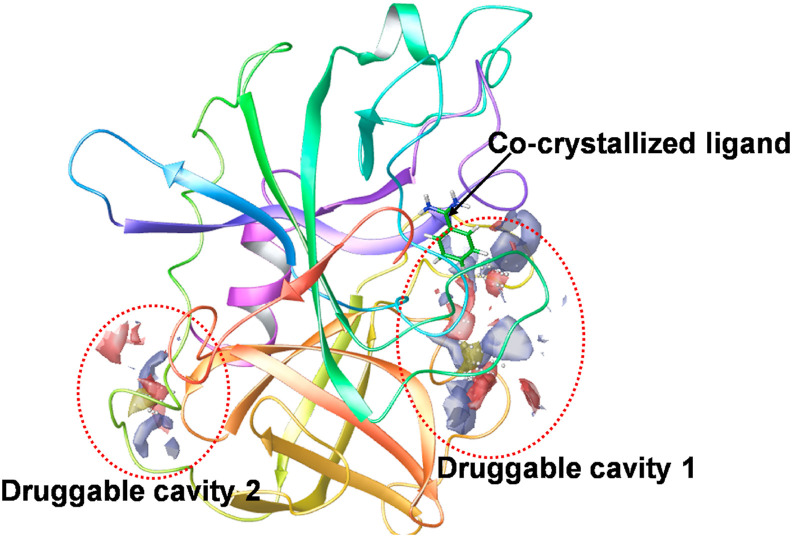
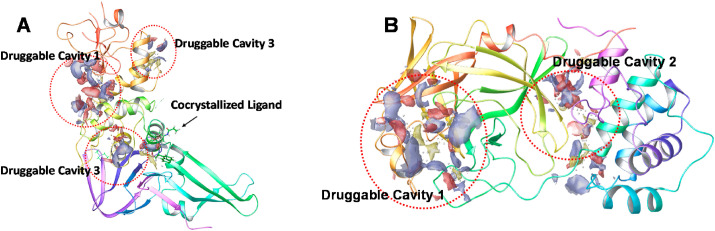
These are a group of host proteases such as trypsin-like peptides, furin, cathepsin L, transmembrane proteases (TMPRSS) and others, that plays a critical role in cell entry of the SARS-CoV-2 virus (Bittmann et al., 2020). Among these, two important proteases viz TMPRSS2 and TMPRSS11a are abundantly observed in the respiratory tract, and facilitate the virus-host cell entry and may seem responsible for the respiratory symptoms of the Covid-19. Moreover, TMPRSS 11D, i.e. human airway trypsin-like protease, works as a proteolytic activator of the spike protein as observed in studies. Other than that, TMPRSS2 tends to form a complex with the ACE2 receptor, that ultimately results in facilitating the efficient penetration of the virus within the host cell (Bittmann et al., 2020). In the end, both the proteases viz TMPRSS2 and TMPRSS 11D tends to cleave the spike protein, to S1 and S2 subunits, and this process of activation facilitates the endosome-independent intracellular entry of the virus, through the cellular membrane. The crystal structure of TRPRSS2 with its most promising binding cavity is as represented in Fig. 7 . This will provide with an insight to design and develop Anti-SARS-CoV-2 drug molecules.
Fig. 7.
Crystal structure of TMPRSS2 (PDB ID 2OQ5), representing the co-crystallized ligand as well as two most promising druggable cavities as predicted by SiteMap application of Schrodinger Maestro.
4.2.2. Viral protease
The papain-like protease (PLpro) and 3C-like protease (3CLpro), are the two crucial viral proteases that play a critical role in SARS-CoV-2 replication process (Báez-Santos et al., 2015). The primary role of both proteins is to process viral polyproteins. In addition to that, PLpro also performs a critical function to facilitate the coronaviruses to evade the host innate immunity response. Therefore, targeting these viral proteases via small drug-like molecules/therapeutic agents seems very promising in curing the viral disease (Báez-Santos et al., 2015). The crystal structure of PLpro (A) and 3CLpro of SARS-CoV-2 (B) with a crucial druggable cavity is represented in Fig. 8 .
Fig. 8.
(A) Crystal structure of PLpro (PDB ID, 7JIW) and (B) (PDB ID 2DUC) of SARS-CoV-2, showing important druggable cavities as predicted from SiteMap application of Schrodinger Maestro.
4.3. Viral replication machinery as drug targets
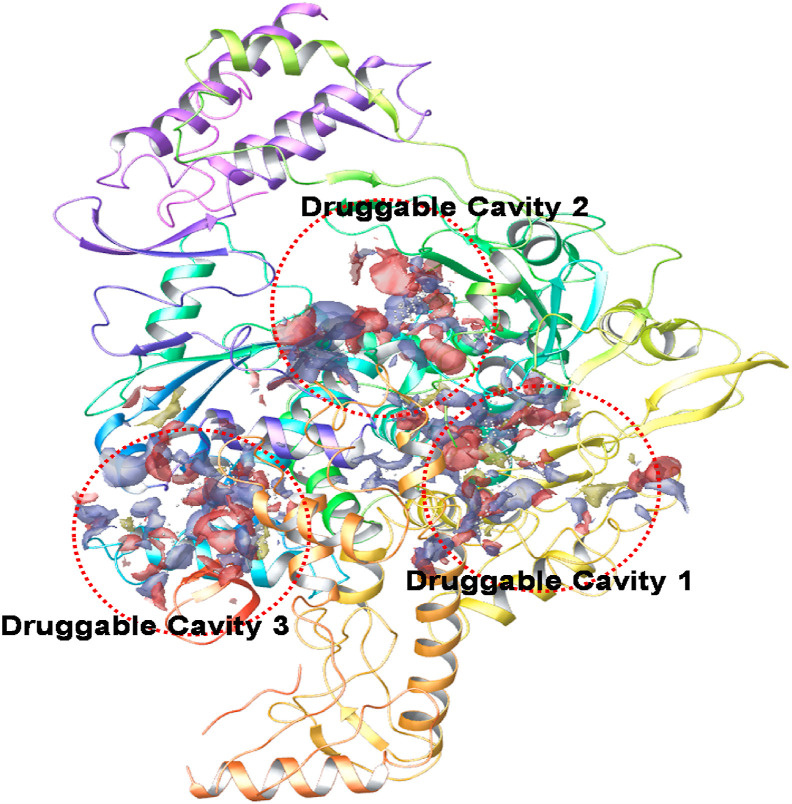
The RNA dependent RNA polymerase (RdRp) is a vital enzyme that facilitates the viral replication process within the host cell (Liu et al., 2020c). RdRp, therefore, seems very important for proper replication process of the SARS-CoV-2 viral genome. Fascinatingly, it is a virus-specific protein, and host cells do not express it, and therefore it can be considered as a selective target for the virus entity. RdRp inhibitors thus seem to have very high potency to selectively target virus without affecting the host cell replication machinery (Liu et al., 2020c). The crystal structure of the RdRp from SARS-CoV-2 with important druggable cavities is as represented in Fig. 9 .
Fig. 9.
Crystal structure od SARS-CoV-2 RdRp protein (PDB ID 6M71) with important druggable cavities as predicted from SiteMap application of Schrodinger Maestro.
4.4. Other targets currently being explored
4.4.1. Membrane protein
The membrane protein is an essential protein that helps in maintaining the proper shape of the envelope of the virus (Schoeman and Fielding, 2019). It also helps in establishing interactions with other proteins, satisfactory assembly of the Golgi complex into the new virion, and in maintaining the stability of the nucleocapsid protein (Schoeman and Fielding, 2019). On a structural basis, membrane protein holds three transmembrane domains inside the long C-terminal domain and outside the short N-terminal domain. Because of its critical role in multiple protein-protein interactions, it is a protein that is crucially involved in maintaining the intracellular homeostasis phenomena (Hogue and Machamer, 2007). Owing to its vital role, the membrane protein can also be regarded as an important target to develop antiviral drug molecules.
4.4.2. Envelope protein
The envelope protein is the comparatively smaller transmembrane structure proteins. It has a size range of about 8.4–12 kDa and contains two distinct domains (Kuo et al., 2007).The two of its domains are the hydrophobic domain and the charged cytoplasmic tail. High variations in its structure feature were observed for the various members of the coronavirus family (Venkatagopalan et al., 2015). The functional role of the envelope protein is very critical; it performs vital functions during morphogenesis of the virus, especially during its assembly and egress phenomena (Prajapat et al., 2020). Based on several studies, it has been further concluded that the envelope protein also has a central role in coordinating with other intracellular protein, in modulating the physiological role of these proteins. Therefore, because of the central coordinating roles of the envelope protein, it is also considered as an essential target to retard the virulent growth of the SARS-CoV-2 entity (Prajapat et al., 2020).
4.4.3. Hemagglutinin esterase
The hemagglutinin esterase is yet another enzyme, that is present in the viruses of this family. It is most commonly observed among the beta-coronavirus family (Prajapat et al., 2020; Zeng et al., 2008). The hemagglutinin activity of this protein tends to facilitate virus propagation through the mucosa. Herein, this protein was found binding to binding to the sialic acid on glycoprotein's surface and produced acetyl-esterase activity (Prajapat et al., 2020; Zeng et al., 2008). The above activity ultimately results in enhanced spike protein-mediated entry and spread of viral infection through the mucosa. Therefore, downregulating the activity of hemagglutinin esterase is also a promising strategy for reducing the propagation tendency of the Covid-19 disease (Prajapat et al., 2020; Zeng et al., 2008).
4.4.4. Nucleocapsid protein
The structural feature of the nucleocapsid protein was found conserved among the different members of the CoV family (Chang et al., 2016). The structural feature of the nucleocapsid protein includes three characteristic intrinsically disordered regions viz, N-arm, central linker, and the C-tail. However, N-terminal and C-terminal domains are the major functionally essential domains available in this protein (Prajapat et al., 2020). The N-terminal domain performs the function of the RNA binding, and C-terminal domain executes the function of dimerization. The C-terminal intrinsically disordered regions further take part in the nucleocapsid protein oligomerization, and also in nucleocapsid protein-membrane protein interactions (Prajapat et al., 2020). This protein has several critical roles and considered as a crucial perspective target for the development of antiviral drug moieties.
4.4.5. NTPase/helicase
Like other viruses, NTPase/helicase also plays an essential role in SARS-CoV-2 growth and propagation (Mayank et al., 2020). The helicase enzyme of SARS-CoV-2 is a member of SF1 (Superfamily 1) that generally works with ATP, dATP, and dCTP based substrates (Karpe and Lole, 2010; Mayank et al., 2020). However, it also can hydrolyse all other forms of the NTPs to release the energy required for the process (Karpe and Lole, 2010; Prajapat et al., 2020). It is one of the most focused targets for producing antiviral drug molecules. Producing SARS-CoV-2 specific NTPase/helicase inhibitor molecules is a significant challenge. However, it has now been recognized as a critical druggable target against the Covid disease (Zeng et al., 2008).
Further, the effect of the virus on the body can be examined, and information is regarding drug targets is obtained via using biophysics or physics-based tool assisting drug discovery for Covid-19. The details are discussed in subsequent sections.
5. Biophysics or physics-based tool assisting drug discovery for Covid-19
To get insights into the structural features of biological molecules, e.g. virus, Physics-based techniques (X-ray crystallography, Cryo-Electron Microscopy, etc.) provides quick results. The effect of the virus on the body can be examined, and information is regarding drug targets is obtained with these techniques. The essential tools for assisting includes:
5.1. X-ray crystallography
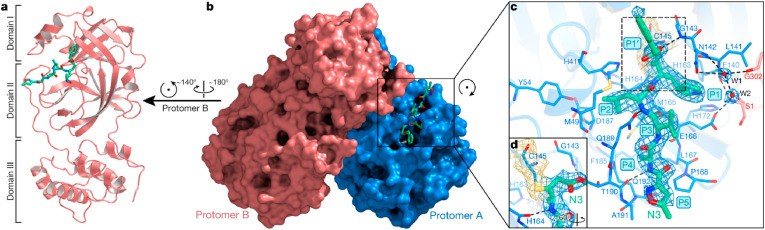
The 3D structure of the virus can be obtained via X-ray crystallography combined with high-quality X-rays, automated tools, and fast computing. The first high-resolution structure (with a resolution of 2.1 Å) of the virus's main protease SARS-CoV-2 Mpro (that ultimately enables the replication of the virus) has been obtained with X-ray crystallography. It allows easier indentification of drug targets for developing drugs to suppress the possibility of virus replication.
Fig. 10(a-d) illustrates the representation of one protomer of the dimeric Mpro–inhibitor complex. Each protomer possesses three domains Domain I (residues 8–101), followed by domain II (residues 102–184; antiparallel β-barrel structure), and Domain III (residues 201–303; five α-helices forming antiparallel globular cluster). The Domain III is linked to Domain II via an extended loop region having residues 185–200. The polypeptides are designated as protomer A (blue), B (salmon) and form a dimer by a crystallographic two-fold axis of symmetry. In SARS-CoV-2 the substrate-binding site is located in a cleft between domain I and domain II. Fig. 10 shows the electron density maps and all residues (1–306) can be identified. Here, N3 (designated by green sticks) binds in the substrate-binding pocket in an extended conformation (Yang et al., 2020a).
Fig. 10.
a. Illustration of protomer of dimeric Mpro–inhibitor complex. b. Surface with protomer A and B of Mpro. c. substrate-binding pocket suggesting essential residual proteins. d. The C–S covalent bond [Reprinted by permission from Ref #(Yang et al., 2020a), © 2020, Springer Nature].
5.2. Cryo-Electron Microscopy
To support and boost the drug design for SARS-CoV-2 Cryo-Electron Microscopy has been adopted to obtain structural information of spike protein. The spike protein of SARS-CoV-2 is a significant target for creating drugs and vaccines. Song et al. used Cryo-EM to resolve the structure of an outer “spike” protein (glycoprotein) of SARS-CoV-2. This spike protein is responsible for host cell attachment and mediating host cell membrane and viral membrane fusion during infection (Song et al., 2018).
Recently, Cai et al. (2020), reported the two new structures of the SARS-CoV-2 spike protein (wild-type S protein) before and after fusion to human cells with Cryo-EM. After Covid-19 infection, the S proteins on the surface of SARS-CoV-2 bind to human cells via the ACE2 receptor and change their shape. It was also noted from the study that two possible conformational changes routes exist. In first route spike protein shows spontaneously transition from the pre-fusion to post-fusion state prematurely, i.e. without binding to the ACE2 receptor. In second route spike protein binds with the ACE2 receptor and facilitates the virus to enter in the host cell. Authors found that the post-fusion structure was rigid and is decorated by N-linked glycan sugar molecules, forming spikes. The rigid nature and presence of spikes prevent the virus against host immune responses and harsh external conditions. So, to accelerate vaccine development and make effective use of antibodies to constraint virus spread, spike protein stabilization needs to be examined.
5.3. CT scans
Another crucial technology is Computed Tomography imaging technology (CT scan) and is generally used in cancer screenings. It uses a narrow beam of x-rays to produce cross-sectional images and stacked together to form a 3D image. It provides insights about the bones, soft tissue, and blood vessels. As, SARS-CoV-2 first attacks on the lungs, and becomes inactive due to inflammation (lack of oxygen in the blood, and hinders CO2 filtration). So, CT scans information about the opaque spots within the lungs could be vital (Zhao et al., 2020).
5.4. Mass spectrometry
Mass spectrometry (MS) is an important technique that has the potential to provide genomic information related that will help in fighting against the Covid-19 disease. It gives exact details about the dynamics of the protein. MS is complementary to the reverse transcription-polymerase chain reaction (RT-PCR) test. Former one measures proteins of virus directly, while genetic material is identified by later. The MS is particular and targets a protein that is only present in SARS-CoV-2. MS is superior to PCR due to a shortage of RNA extraction kits, RT-PCR reagents for the PCR test. Ihling et al. illustrated the MS-based method to detect the SARS-CoV-2 virus protein from gargle solution samples of Covid-19 patients (Ihling et al., 2020). The analysis enabled the identification of unique peptides (comprising the sequence RPQGLPNNTASWFTALTQHGK) originating from SARS-CoV-2 nucleoprotein in two samples out of three. The outcomes were in absolute agreement with the PCR analyses, exhibiting a higher viral load in the samples. The only drawback with the MS approach is that it takes approximately 3 h for one sample, and resolution using triple-quadrupole instruments. Another report by Gouveia et al. presented the use of tandem mass spectrometry to detect SARS-CoV-2 marker peptides in nasopharyngeal swabs without using any specific reagents (Gouveia et al., 2020). This test takes only 3 min for one sample. The mass spectra were obtained for m/z = 35–1500 ions with positive charges 2+ and 3+ were considered. The authors concluded that the peptides ADETQALPQR and GFYAQGSR from the nucleocapsid protein are essential due to their high signal. Also, the peptides elution can be obtained within a 3 min window in the tested conditions.
5.5. Particle physics contribution
Particle physics also contributed to determining the protein structures of the virus. A particle accelerator, i.e., synchrotron (gives information about atoms), was used to examine the virus. In this, particles with the speed of light are accelerated, and an intense light beam is created that allows the immediate investigation of the virus. Researchers obtained the structure of protease (Mpro) with very high resolution, better than 1.3 Å. It opened doors for developing drugs by analyzing the interactions between drug molecules and protease. In crystallography, one drawback is that positions of H-atoms cannot be identified. It is well known that the hydrogen atoms play a crucial role in the bindings of drugs to target enzymes (via H-bonding), and crystallography fails to give exact details. This problem can be resolved by using the Neutron crystallography where neutrons interact with atomic nuclei to provide H-bonding details comparable to those of the protein's nitrogen, oxygen, and carbon atoms. Also, neutrons penetrate deep without damaging the protein samples so that it can be performed at room temperature also. As the Covid-19 virus have a viral envelope (of lipids, proteins, and sugars) outside and its molecular structure and composition need to be examined for developing appropriate drugs. It can be obtained by neutron reflectometry that enables to obtain information about the mechanism used by the virus for penetration inside the cell/plasma membrane. It will also give detail about the interaction of peptides that allows binding with the receptor of the host cell.
5.6. Laser physics
Laser physics emerged as a practical approach to reduce the coronavirus spread. A group led by Chris Barty (University of California), used diodes from Blu-ray digital video disc devices to rapidly disinfect surfaces and the indoor air by modulating them to work as deep-ultraviolet laser photon sources. The UV-radiation in wavelength between 200 and 260 nm is effective in killing the viruses with negligible risk to humans (García de Abajo et al., 2020).
5.7. Artificial intelligence, machine learning, deep learning
We think we will not justify the manuscript if we exclude artificial intelligence from the Covid-19 drug search. The artificial intelligence has been used to process the visual or audio data by various projects. CAD4COVID builds off existing technology that has been highly successful in diagnosing tuberculosis through the analysis of chest X-rays. Another project‘Covid Voice Detector’ collects the audio recordings, and AI will help in recognizing signs of Covid-19 infection in a person's voice. Machine learning (ML) can be used to examine the target community for Covid-19 by recognizing markers that appear in blood tests. ML is also being used to identify the drugs via the drug repurposing approach (reuse of existing drug for another purpose) to treat Covid-19. Deep learning has the potential to extract information from image analysis. ML can be collaborated with deep learning to predict the interactions of drugs in the commercial market with the coronavirus. For effective treatment of the Covid-19, the physical structure of the virus needs to understand for drug development. The computer simulations by physicists with the main focus on protein-folding simulations are being used to identify the targets for the drug.
Along with this, physicists can process the data related to the SARS-CoV-2 genome. This will be beneficial for identifying the molecules that are contributing to virus growth as well as spread. Along with this, the virus mutations can be understood. Recently deep learning-based high resolution Computed tomography (CT) has been proposed for detecting and diagnosing the patient's Covid-19 and pneumonia (Chen et al., 2020a).
The subsequent section of the article includes a detailed description of how these target proteins are explored in the anti- SARS-CoV-2 drug development process.
6. Small molecule inhibitors targeting various stages of SARS-CoV-2 life cycle
As discussed in previous sections, the journey of SARS-CoV-2 initiates once it enters the respiratory tract and binds to ACE2 called fusion. After successful fusion, it undergoes a subsequent journey, as described in section 3. The crucial steps are translocation, which involves virus entry inside cells, followed by proteolysis leading to release of viral genomic material, RNA replication, and translation to create new virus genomic content followed by packaging and endocytosis. Various proteins and enzymes (discussed above) are the sites of targets by the drug repurposed recently for Covid-19. The drugs thus identified for their potential to treat Covid-19 is divided into four categories. The vital category includes drugs, exhibiting antiviral properties, immunomodulators, modulating the cytokine storm syndrome, assisting in overcoming ARDS and those impeding at RAAS-ACE2-AT-1 signalling.
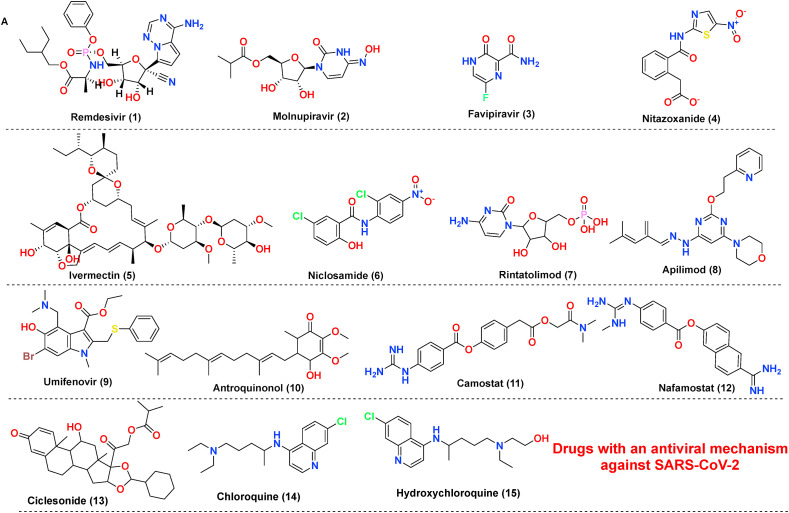
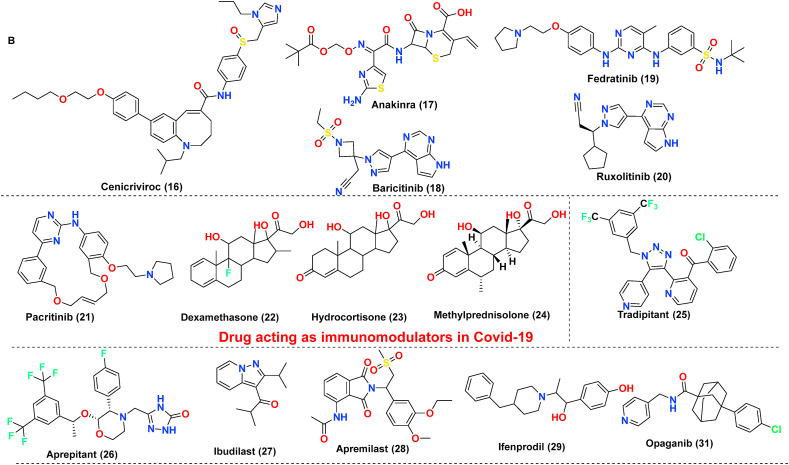
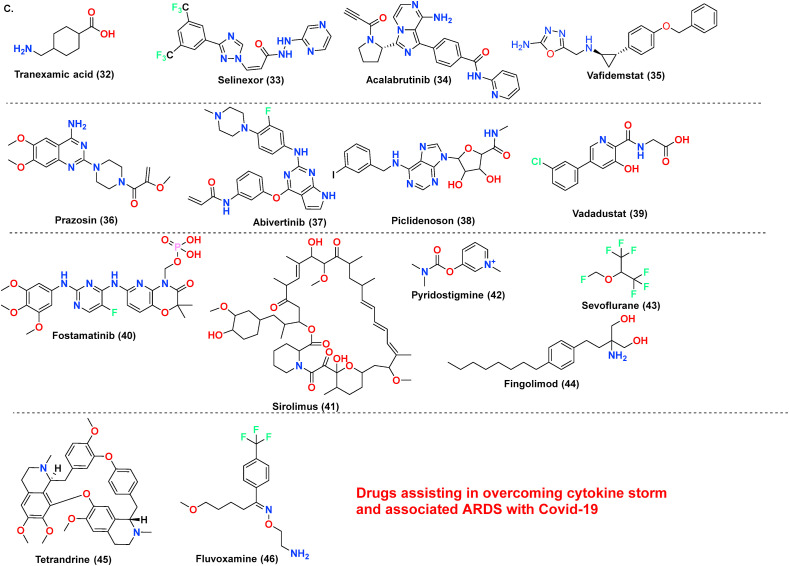
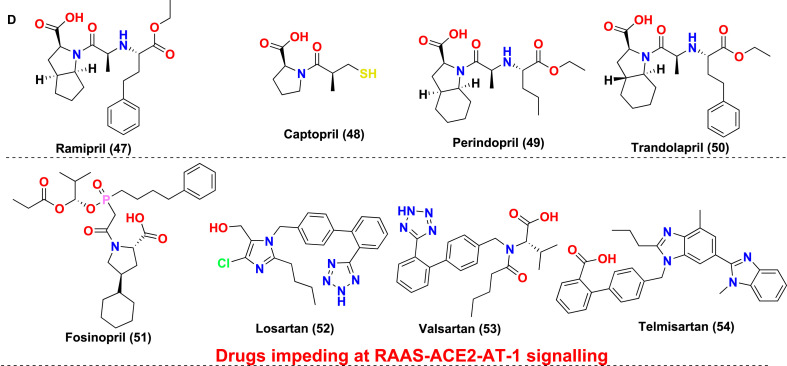
The antiviral drug consists of ‘magic bullets’ that can obstruct or impede i. virus binding or interfere with the fusion of viral domain with the host cell; ii. virus translocation within host cells; iii. virus proteolysis mechanism within host cells and iv. molecular development of virus genomic material. The choice of drugs includes classical antivirals (Remdesivir, Favipiravir) or other categories of drugs (eg., Niclosamide, Bemcentinib) exhibiting antiviral action against SARS-CoV-2 (Poduri et al., 2020). Till date Remdesivir is the only drug approved by USFDA on October 22, 2020, to treat Covid-19 disease. Covid-19 is associated with hyper inflammation importantly caused by cytokine release mediated chiefly via Interleukins (ILs), Janus kinase pathways, monocyte chemoattractant protein-1 (MCP1) numb-associated kinase (NAK), macrophage inflammatory protein-1 (MIP-1), GCSF, INF-gamma, and TNF, and inhibition of these pathways is one of acclaimed therapeutic option to treat mild or severe cases. The essential drugs in this category include biologics (eg., Infliximab, Abatacept); IL inhibitors (eg., tocilizumab, anakinra); JAK and NAK inhibitors (eg., baricitinib, fedratinib, ruxolitinib); Corticosteroids (eg., dexamethasone and hydrocortisone) along with various miscellaneous therapies including Nitric oxide and Statins (Remy et al., 2020). In severe cases of Covid-19, inflammation may be hyperactivated by cytokine release and precipitating a condition called a cytokine storm, eventually leading to pneumonia or ARDS. The most common receptors affecting this includes Neurokinin-1 (NK-1) receptors, Colony-stimulator factors (CSFs), and Phosphodiesterase (PDE) receptors. Therefore, treatment of these conditions with NK-1 inhibitors (eg., Tradipitant), mabs affecting CSFs (Sargramostim) or PDE inhibitors (eg., Ibudilast, Apremilast) are highly recommended. The inhibitors to treat cytokine storm or ARDS further also involve other categories of drugs, importantly Ifenprodil (NMDA antagonist), Opaganib (Sphingosine kinase-2 (SK2) inhibitor), CM4620-IE (Calcium release-activated calcium (CRAC) channel inhibitor) and many other (Ye et al., 2020). Another essential category includes the drugs interfere at RAAS-ACE2-AT-1 signalling. As discussed previously, ACE2 possess two significant sites, catalytic site and N-terminal protease domain. N-terminal protease domain assists in the binding of spike glycoprotein S1 with host cell whereas the catalytic site is involved in catalyzes of Ang I and Ang II. However, there exist a controversial data whether to use or discontinue the drug affecting RAAS-ACE2-AT-1. The primary drug class includes angiotensin-converting enzyme inhibitors (ACEIs) and angiotensin receptor blockers (ARBs). The central hypothesis is, expression of ACE2 receptors could enhance viral entry. In contrast, a contrasting hypothesis was put forth which states ACE inhibitors or ARBs will reduce the production of Ang II allowing the enhanced generation of Ang 1–7 which could attenuate lung injury by decreasing inflammation and fibrosis associated with Covid-19 (Poduri et al., 2020). Lately, the sepsis-induced by Covid-19 and its correlation with the coagulation system has led to a few retrospective cohort studies claiming prophylactic or therapeutic use of anticoagulants in critically ill patients. Currently, a recombinant nematode anticoagulant protein c2 (rNAPc2), AB201 is under investigation, which is thought to inhibit tissue factor (VIIa complex). However, the exacts mechanism of how anticoagulants play a role in Covid-19 is unclear (Maldonado et al., 2020). The brief discussion on drugs based on these categories is made in Table 1 . The table importantly covers drugs, their original target, target employed against SARS-CoV-2 along with their mechanism of action. The chemical structures of the discussed drugs are represented in Fig. 11 (A-C).
Table 1.
Compilation of small molecule inhibitors targeting various stages of SARS-CoV-2 life cycle.
| Drug (Structure entry no.) |
Original Target |
Target in SARS-CoV-2 |
Mechanism of action |
NCT |
|---|---|---|---|---|
| Drugs with an antiviral mechanism against SARS-CoV-2 | ||||
| Remdesivir (1) | Nucleotide analog prodrug | Viral RNA-dependent RNA polymerase (RdRp) | The drug interferes with RdRp action and thereby interfere with the proofreading by viral exoribonuclease, leading to decreased viral replication and consequently viral load. | NCT04292730 |
| Molnupiravir (2) | Nucleoside analog | RdRp | Interfere with the viral genome proofreading leading to copying errors during RNA replication | NCT04575584 |
| Favipiravir (3) | RNA polymerase inhibitor | RNA polymerase | Block the negative-sense RNA strand involved in the termination of RNA transcription | NCT04402203 |
| Nitazoxanide (4) | Antiprotozoal | Viral hemagglutinin | Potentiate interferon production allowing the activation of eukaryotic translation initiation factor 2α, which inhibits viral hemagglutinin maturation | NCT04552483 |
| Ivermectin (5) | Antiparasitic | importin α/β1 | Inhibits the importin (IMP) α/β1 that is responsible for integrase protein nuclear import | NCT04445311 |
| Niclosamide (6) | Anthelmintic | Not known | Drug decrease prolonged infection and transmission by targeting the viral reservoir in the gut region | NCT04399356 |
| Rintatolimod (7) | Toll-like receptor 3 (TLR-3) agonist | Positive RNA strand | Interferes with the formation of pp1a and 1b | NCT04379518 |
| Apilimod (8) | Interleukin inhibitor | PIKfyve inhibitor | Inhibits lipid kinase enzyme, PIKfyve which plays a pivotal role in cellular regulation thus denying the entry of SARS-CoV-2 inside the host cell | NCT00642629 |
| Umifenovir (9) | Antiviral | hemagglutinin protein | Targets the viral glycoproteins, involved in recognition of S protein allowing its membrane fusion with ACE2 | NCT04350684 |
| Antroquinonol (10) | Antiviral/anti-inflammatory agent | Unknown | Inhibits viral replication and viral protein synthesis | NCT04523181 |
| Camostat (11) Nafamostat (12) |
Serine protease inhibitor | TMPRSS2 inhibitor | Blocks the binding of SARS-CoV-2 by interfering with RBD |
NCT04321096 NCT04352400 |
| Ciclesonide (13) | Glucocorticoids | Nsp15 | Inhibits the viral replication of coronavirus directly or indirectly by interfering with Nsp15 | NCT04435795 |
| CQ/HCQ (14–15) (discontinued now by USFDA) |
Antimalarial agents | Endosomes | Interrupt the intracellular trafficking of the virus by increasing the endosomal pH. Also, interfering with glycosylation of ACE2 receptors. The immunomodulatory mechanism is also reported. | (Andreania et al., 2020; Gautret et al., 2020) |
| Drug acting as immunomodulators in SARS-CoV-2 | ||||
| Cenicriviroc (16) | CCR2 and CCR5 antagonist | Not known | Involved with respiratory injury developed as a consequence of Covid-19. The exact mechanism as antiviral not known. | NCT04500418 |
| Anakinra (17) | IL antagonist | IL-1 inhibitor | Blocks the production of pro-inflammatory cytokines interleukin (IL)-1α and IL-1β | NCT04341584 |
| Baricitinib (18) | JAK inhibitor | AP2-associated protein kinase-1 (AAK1) | AAK1 is a known regulator of endocytosis, and its inhibition is reported to intrude intracellular virus entry | NCT04358614 |
| Fedratinib (19) Ruxolitinib (20) |
Anticancer | JAK-STAT inhibitors | Target JAK-1 and 2 and halt the release of a proinflammatory mediator such as cytokines and growth factor |
NCT04345289 NCT04359290 |
| Pacritinib (21) | Myelofibrosis | AK2, IL-1, IL-6 CSF-1 inhibitor | Decreases immune system hyperactivation via inhibition of target receptors | NCT04404361 |
| Corticosteroids (dexamethasone (22) hydrocortisone (23) methylprednisolone (24) |
Glucocorticoid and mineralocorticoid receptor | Acute fibrinous and organizing pneumonia (AFOP) | Play a role in the trafficking of T cells from the blood tissues, decrease the inflammatory cytokines, extravasation of immune cells, and causes destruction through apoptosis and indirectly decrease the cytotoxic effects of nitric oxide, TNFα expression and NF-kβ activation | NCT04273321 |
| Nitric Oxide | Vasodilator | VEGFR agonist | Drug increases the blood flow in the lungs, improves the condition of hypoxia, and regulates the blood pressure, which reverses pulmonary hypertension | NCT04476992 |
| Drugs assisting in overcoming cytokine storm and associated ARDS with Covid-19 | ||||
| Tradipitant (25) Aprepitant (26) |
NK-1 receptor | NK-1 receptor antagonist | NK-1 receptor is involved with the release of substance P, associated with neuroinflammatory processes causing serious lung injury | NCT04326426 |
| Ibudilast (27) Apremilast (28) |
Phosphodiesterase inhibitors | PDE- 4,10 and MIF inhibitor | PDE inhibition allows elevation in intracellular cAMP levels allowing suppression of proinflammatory cytokines and endorses neurotrophic factors development | NCT04470622 |
| Ifenprodil (29) Aviptadil (30) |
NDMA and glutamate receptor antagonist | NDMA and glutamate receptor antagonist | The drug binds to the vasoactive intestinal polypeptide (VIP) found in the lungs and selectively and blocks NMDA induced caspase-3 activation in the lungs, along with blocks the production of TNFα and IL6 | NCT04382924 |
| Opaganib (31) | Anticancer | Sphingosine kinase-2 (SK-2) inhibitor | SK-2 receptor allows activation and regulation of mast cells which mediates sphingosine-1-phosphate production thus allowing calcium influx, cytokine production, NF-kappa-B activation | NCT04414618 |
| Tranexamic acid (32) | Antifibrinolytic hemostatic agent | Kringle domain of plasminogen | Inhibits activation of plasminogen and, consequently, the conversion of plasminogen to plasmin reduces which catalyzes and assist in cleavage of S-protein at the site of furin | NCT04338126 |
| Selinexor (33) | Anticancer | A selective inhibitor of nuclear export protein (XPO1) | Inhibits the cellular protein XPO1, which assist in the transportation of viral protein from the host cell nucleus to the cytoplasm. Beside this drug decreases inflammatory cytokines | NCT04349098 |
| Acalabrutinib (34) | Anticancer | Bruton Tyrosine Kinase (BTK) inhibitor | Binds covalently to Cys481 in the ATP domain of BTK and involved in the downregulation of inflammatory cytokines in patients with respiratory complications | NCT04346199 |
| Vafidemstat (35) | CNS optimized inhibitor of lysine-specific demethylase (LSD1) | LSD1 inhibitor | Reduces the patient inflammatory response thereby preventing progression of pneumonia to ARDS in severely ill Covid-19 patients | NCT03867253 |
| Prazosin (36) | Alpha-blockers | Alpha1 receptor antagonist, | Increase in catecholamine release is associated with cytokine storm via IL-6 overproduction mediated by a signalling loop that entails alpha1 adrenergic receptor | NCT04365257 |
| Abivertinib (37) | Anticancer | EGFR and BTK | involved in the downregulation of inflammatory cytokines in patients with respiratory complications | NCT04440007 |
| Piclidenoson (38) | Autoimmune inflammatory diseases | A3 adenosine receptor (A3AR) agonist | Affect modulation of signalling proteins such as PKA, P13K, PKB/AKT, NF-kβ and IKK leading in deregulation of the Wnt/β-catenin pathway allowing anti-inflammatory response | NCT04333472 |
| Vadadustat (39) | Anaemia associated with chronic kidney disease | Hypoxia-inducible factor prolyl hydroxylase (HIF-PH) inhibitor | HIF-PH inhibition increases endogenous production of erythropoietin which indirectly allows the production of haemoglobin and red blood cells, vital in associated ARDS | NCT04478071 |
| Fostamatinib (40) | Tyrosine kinase inhibitor | Spleen tyrosine kinase (SYK) inhibitor | SYK receptor is associated with the signalling cascade of Fc gamma (Fcγ) and c-type lectin (CL) receptor allowing the cytokine release. The drug also decreases mucin-1 protein, a biomarker used for ARDS development prediction | NCT04579393 |
| Sirolimus (41) | Antifungal | PI3K/mTOR inhibitor | Drug target rapamycin protein that plays a role in cell division, viral replication, and viral release. It inhibits the antigen-induced T and B cells activation control and possesses immunosuppressive activity | NCT04341675 |
| Pyridostigmine (42) | Myasthenia gravis | Acetylcholine-esterase | Inhibits acetylcholine-esterase, which degrades acetylcholine (Ach), sequentially upsurges Ach bioavailability. Ach ligates to a nicotinic-alpha7 receptor present in macrophages and T cells, thereby resulting in a reduction of overactivated immune cells | NCT04343963 |
| Sevoflurane (43) | Anaesthetic agent | HIF-1 alpha | Improves oxygenation and vascular dilation. The drug is also reported to reduce cytokines in ARDS. | NCT04355962 |
| Fingolimod (44) | Multiple sclerosis | Sphingosine 1-phosphate receptor | The drug acts as a modulator on sphingosine 1-phosphate receptor that sequesters T and B lymphocyte in lymph nodes, thus inhibiting the pulmonary edema and hyaline membrane formation thus overcoming ARDS in the severe patients | NCT04280588 |
| Tetrandrine (45) | anti-inflammatory, anti-tumor | AAK1 | Inhibits fibroblasts resulting in the reduction of pulmonary fibrosis | NCT04308317 |
| Fluvoxamine (46) | selective serotonin reuptake inhibitor (SSRI) | SSRI | Cytokines storms drive mechanism not known, used to prevent sepsis, and other inflammatory mediator associated conditions | NCT04342663 |
| Drugs impeding at RAAS-ACE2-AT-1 signalling | ||||
| Ramipril (47) Captopril (48) Perindopril (49) Trandolapril (50) Fosinopril (51) |
Hypertension | ACE inhibitors | These drugs reduce the production of Ang II, allowing the enhanced generation of Ang 1–7, which could attenuate lung injury by decreasing inflammation and fibrosis associated with Covid-19. They are also reported to mask the ADAM17 inhibitory activity on ACE2 | NCT04330300 |
| Losartan (52) Valsartan (53) Telmisartan (54) |
Hypertension | AT-1 receptors inhibitors | Prevent Ang II-AT-1 receptor-mediated cytokine release by competitively inhibiting Ang II binding to AT-1 receptor. Increases ACE2 concentration by inhibiting ADAM17 | NCT04318418 |
Fig. 11.
(A–D) The chemical structures of repurposed drugs candidates. These drugs have been exploited to act on the various life cycle of SARS-CoV-2 along with affecting its post consequence, which includes cytokine storm and ARDS.
Further, various regimens are proposed for the therapeutic management of patients with Covid-19. In the current work, we have focused on treatment strategy prescribed by the National Institutes of Health's (NIH), which classifies the medication based on severity of Covid-19 patient (COVID-19 Treatment Guidelines) and the MATH+ protocol (Marik et al., 2020), which classifies management of each phase of Covid-19. The vital phase in MATH+ protocol includes incubation, symptomatic, early pulmonary and late pulmonary stages. The brief recommended regimens are compiled in Table 2 .
Table 2.
Drug regimens are proposed for the therapeutic management of patients with Covid-19.
| Patient Severity |
Recommended drug regimen |
|---|---|
| RECOMMENDATION BASED ON NIH GUIDELINES | |
| The hospitalized patient requiring invasive mechanical ventilation | The regimen includes remdesivir (200 mg via IV route for day 1, followed by 100 mg IV for 4 days or until hospital discharge, whichever comes first; OR Combination of remdesivir (doses as mentioned above) along with dexamethasone 6 mg via IV or orally route for up to 10 days or until hospital discharge |
| The hospitalized patient requiring oxygen delivery | A combination of dexamethasone plus remdesivir or Dexamethasone alone (doses as mentioned above) |
| Hospitalized patient and requires supplement oxygen | Remdesivir or combination of remdesivir plus dexamethasone (doses as mentioned above) |
| Not hospitalized or hospitalized but do not require oxygen | Any specific antiviral or immunomodulatory therapy is not recommended |
| Recommendation on the use of antivirals therapy | |
| CQ or HCQ with or without azithromycin | Not recommended in any stage |
| Lopinavir/Ritonavir and other HIV Protease | Not recommended for treatment except for clinical trial |
| Ivermectin | Not recommended for treatment except for clinical trial |
| Recommendation on the use of immunomodulatory therapy | |
| Anakinra and Interferon beta | Treatment of mild and moderate Covid-19 |
| Sarilumab, tocilizumab | Not recommended for treatment except for clinical trial |
| Acalabrutinib, ibrutinib, zanubrutinib | Not recommended for treatment except for clinical trial |
| Baricitinib, ruxolitinib, tofacitinib | Not recommended for treatment except for clinical trial |
| Recommendation on the use of Adjunctive Therapy | |
| Anticoagulant therapy | Recommended at therapeutic doses to patients with Covid-19 having an incident of the thromboembolic event or those highly suspected to have the thromboembolic disease |
| Vitamin C | Insufficient data to recommend either for or against the use of vitamin C for the treatment of Covid-19 in non-critically or critically ill patients |
| Vitamin D | Insufficient data to recommend either for or against the use of vitamin D for the prevention or treatment of Covid-19 |
| Zinc | Recommends against using zinc supplementation above the recommended dietary allowance for the prevention of Covid-19, except in a clinical trial. Further, there is insufficient clinical data to recommend for or against the use of zinc supplement for Covid-19 treatment |
| Recommendation on the use of Concomitant Medications | |
| ACE Inhibitors and ARBs | Recommends against the use of ACE inhibitors or ARBs for the treatment of Covid-19, except in a clinical trial; and persons with Covid-19 and cardiovascular disease prescribed with these drugs should continue to use |
| Statins | Recommends against the use of statins for the treatment of Covid-19, except in a clinical trial; and persons with Covid-19 and cardiovascular disease prescribed with these drugs should continue to use |
| NSAIDs | May be given at therapeutic dose to Covid-19 patients with or without comorbid state |
| RECOMMENDATIONS BASED ON MATH + PROTOCOL | |
| Methylprednisolone (IV) | A loading dose of 80 mg followed by 40 mg after every 12 h for 7 days in ICU or until discharged. In worsening stages 80 mg after every 12 h followed by 120 mg and titrated appropriately to maintain doses under control |
| Ascorbic Acid (Infusion) | 3 gm/100 mL after every 6 h for 7 days or until discharged |
| Thiamine (IV) | 200 mg after every 12 h for at least 7 days or until discharged |
| Heparin | For critically ill patients: 1 mg/kg every 12 h Stable patient: 0.5 mg/kg every 12 h Periodically check for Serum creatinine level |
| Adjuvant | Zinc, magnesium, atorvastatin, famotidine, melatonin and Vitamin D |
Further, the crucial drugs under considerations are hampered by their known adverse drug reactions. Among them HCQ, originally an antimalarial drug was issued the USFDA Emergency use authorization on March 28, 2020 (Food and Administration, 2020). However, with the span of fewer than 3 months, the USFDA has revoked the authorized use of HCQ/CQ recently on June 15, 2020. However, still, there is much debate on whether to use this drug for Covid-19 treatment or prophylactic use. Numerous reports point toward the failure of using HCQ therapy for treating Covid-19. In this context, Ferner and Aronson report the use of CQ/HCQ in Covid-19 is “premature and potentially harmful.” However, they agreed that disparity between laboratory and clinical experiments is mainly due to complex pharmacokinetics and hence makes it difficult to extrapolate drug concentrations in culture media to human doses (Ferner and Aronson, 2020). The major drawback associated with HCQ use includes effects on the heart leading to QRS and QT interval prolongation via sodium and potassium channel blockade, causing dysrhythmias and ventricular fibrillation. Besides some ophthalmologic complications (retinopathy), hypoglycemia and death at overdoses are reported (Downes et al., 2020). The major complication, however, lies with improper monitoring of doses as it primarily depends on HCQ pharmacokinetics (Liu et al., 2020b). Another drug and only approved drug by USFDA for Covid-19, remdesivir has also not remained untouched with the controversy associated with its adverse effects. The drug is reported to be associated with increased risk of liver impairment (Montastruc et al., 2020), constipation, hypoalbuminemia, hypokalemia, hypotension respiratory failure as per evidence of the clinical trials for its efficacy against Covid-19 (Chaplin, 2020). However, as the drug is prescribed for severely ill patients in the trials who also possess various comorbid states, it's difficult to certain the exact mechanism for induced ADRs (Fan et al., 2020).
Beside small molecules, drugs derived from nature or herbal formulations are also exploited for their alleged role against SARS-CoV-2. Numerous research has evidenced the naturally derived product and their remedies as a promising tool against different viral infections, and thus naturally derived products may be an essential tactic for Covid-19 treatment (Ganjhu et al., 2015). Some of the tested natural product(s) regimen is discussed in the subsequent section.
7. Natural products and natural products derived drugs reported so far to treat or for prophylactic use in Covid-19
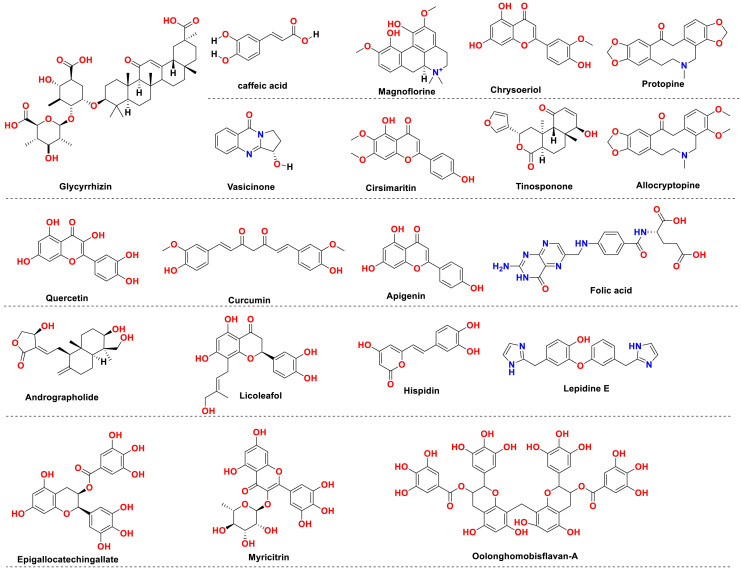
Natural products have shown well established antiviral pharmacology and are found to be effective against SARS-CoV-2 growth and propagation. Many of the naturally originated molecules (Fig. 12 ) have found extensively useful against SARS-CoV-2 and therefore considered an essential tactic for Covid-19 treatment (Ganjhu et al., 2015). Glycyrrhizin is a natural product with pleasant pharmacological properties. The biological activities associated with it include its efficient binding against ACE2, proinflammatory cytokines downregulation, inhibition of intracellular reactive oxygen species accumulation, inhibitory effect on thrombin, retarding the hyperproduction of airway exudates, and facilitation of endogenous interferon-based activity. All these effects are considered very promising to retard the growth and propagation of the Covid-19 disease (LuoLiu and Li, 2020). The caffeic acid, i.e., yet another commonly available natural product, also found very promising against the disease. The in vitro study of the caffeic acid revealed its inhibitory potential against human coronavirus (HCOV) NL63. Its impact on retarding the virus's interaction with the ACE2 receptor is particularly documented concerning its antivirus potential. Because of the ACE2 related effects, caffeic acid is also considered a promising lead against this disease.
Fig. 12.
Chemical structures of some natural product derived drugs reported exhibiting potential against Covid-19.
Cinnamon, a natural product, also revealed very promising antiviral potential. The butanol extract of cinnamon, when investigated, showed promising activity against and SARS CoV related infections. These studies revealed that the naturally occurring chemical component within the cinnamon is very promising against Covid-19 disease. Moreover, various natural products including vasicinone, magnoflorine, cirsimaritin, chrysoeriol, tinosponone, quercetin, curcumin, apigenin, and epigallocatechin gallate were repoted to show Spike protein targeting potential and are essential leads against Covid-19 disease. Similarly, Protopine, allocryptopine, Oolonghomobisflavan-A, Andrographolide, folic acid Hispidin, lepidine E, were reported to show targeting potent against the viral Main Protease (Mpro) protein. However, flavone, coumarin, licoleafol, colistin, methyl rosmarinat, calceolarioside B and myricitrin, on the other hand, were reported effective against 3CLpro protein. Therefore, natural products are the rich source of highly promising SARS-CoV-2 leads, with immense clinical potential(Antonio et al., 2020; Boozari and Hosseinzadeh, 2020).
Apart from synthetic drugs and natural product derived regimen, a particular emphasis has been given to food or food supplements and nutrients. The sole purpose is to improve the existing immune system for prophylaxis or assist in speedy recovery by boosting the immune system. Some of the vital food or food supplements and nutrients identified so far for this purpose are compiled in Table 3 .
Table 3.
Identified nutrient and food supplements along with their sources and mechanisms improving the immunity against Covid-19.
| Nutrients | Proposed Mechanism | Sources | Ref |
|---|---|---|---|
| Micronutrients | The elements such as Zn, Se, Cu, Fe, and Mg, enhance both cell-mediated and humoral immunity. Also, they are associated with the production of Th1, Th17, Th9 having antioxidant properties along with the production of IgG |
Zn: red meat, shellfish Mg: dark chocolate, black beans, avocados and whole grains Se: red meat, shellfish, chicken, garlic, grains |
Gasmi et al. (2020) |
| Antioxidants | They possess redox mechanisms and allow in scavenging free radicals thus assisting in repairment of tissue and regulation the innate immune system | Green tea, black tea, chilli pepper, oregano, red onion, fennel leaf, grapes, apples, green vegetables, peppers, N-acetylcysteine | Nasi et al. (2020) |
| Probiotics | Consist of live microorganism (Lactobacillus, Bifidobacterium and Saccharomyces) that are intended to promote gut immunity and reduce the acute upper tract respiratory infections | Yoghurt, fermented oats, kefir, kombucha, sauerkraut, tempeh, sourdough bread, and some cheese | Baud et al. (2020) |
| Vitamin D | Enhance the innate immunity, through the production of antimicrobial peptides, supports in the production of monocytes, macrophages and dendritic cell, decrease the pro-inflammatory cytokines and enhance anti-inflammatory cytokines | Salmon, tuna, spinach, hazelnuts | Ilie et al. (2020) |
| Vitamin C and E | Both these vitamins possess antioxidant as well as anti-inflammatory properties that reduce oxidative stress. They have potential as an antiviral agent and supports host defence against the infection by affecting the growth and development of T lymphocytes and natural killer (NK) cells, thus preventing cellular damage |
Vit. C: lemon, grapefruits, oranges, tangerines, sweet red pepper, broccoli, papaya, eggplant Vit. E: nuts, sunflower seeds, avocados, spinach and hazelnuts |
Carr, 2020 |
| Vitamin A | Enhance both innate (through natural killer cells, macrophages, neutrophils) and adaptive immunity (by differentiation, proliferation, functioning and movement) as well as assist in the maintenance of mucosal epithelium integrity and possess anti-inflammatory properties. | Carrots, spinach, kale, apricots, sweet potato, squash and cantaloupe | Jovic et al. (2020) |
| Water content-rich | Water intake keeps the mucus membrane moist and produces lymph which circulates the white blood cells and immune cell throughout the body lower the chances of diseases such as cold and flu. | Cucumber, watermelon, celery, coconut water, | Kau et al. (2011) |
| Amino acids and proteins | They are used to boost the immune system and also possess antioxidant properties. | Soybean, seafood, meat, fish, eggs, dairy products, tofu, cereal and pulses | Ferrara et al. (2020) |
8. In vitro and in vivo models to test repurposed drugs against SARS-CoV-2
Tremendous development has been witnessed in a brief period in the field of Covid-19 research. During this period, tremendous progress has been recorded in the development of various research models viz. cell line, organoid, animal, and 3-D culture models. Human airway epithelial cells, caco-2 cells, calu-3 cells, Vero E6 cells (both wild type and TMPRSS2 overexpressing), HEK293T cells, and Huh7 cells used as in vitro models (Takayama, 2020a). It has been established in one of the studies that some cell lines are quite accommodating to SARS coronavirus (SARS-CoV) contagion, and that comprises human lung epithelial cell lines NCI-H1650 and NCI-H1563, human intestinal cell line LOVO, and normal nasopharyngeal epithelial cell lines NP69 and NP460 (To and Chan, 2009). The organoids' research models are made up of different types of cells, enabling it to mimic the physiological environments of human organs. (Lancaster and Knoblich, 2014). SARS-CoV-2 impartially affects the lung damage triggered by pneumonia, and it also distresses several organs such as the liver, kidney, and cardiovascular system. Piloting of the SARS-CoV-2 study led to the development of two organoids, i.e., human bronchial organoids or human lung organoids by Suzuki et al. and Han et al. respectively, (Clevers, 2020). For the administration of drug testing, numerous animal models such as mice, ferrets, cynomolgus macaques, rhesus macaques, cats, Syrian hamsters are being used for conducting in vivo studies to identify a potential drug candidate(Ou et al., 2020). Many 3D models have come into existence to overcome the ethical disputes involved in drug monitoring in in vivo and to name a few important ones developed by organoid technology are human blood vessel organoids, human bronchial organoids, human lung organoids, human intestinal organoids, human liver ductal organoids, and human kidney organoids (Jeon et al., 2020; Takayama, 2020b; Touret et al., 2020), Other than tests mentioned above for the testing of drug candidates against Covid-19 disease, new technologies are coming into the picture which comprises single-cell analysis, bioprinting, and bio-fabrication.
9. Biopharmaceuticals in the treatment of Covid-19
Biopharmaceuticals extend to one the critical class of engineered macromolecular product(s) as drug candidates. This class consists of two significant subclasses that include vaccines and biologics. The important biologics explored in SARS-CoV-2 includes the following:
9.1. Monoclonal/polyclonal antibodies
These biologics or macromolecules interferes with the most vulnerable sites in the virus and thus confer their mechanism of action (Rothlin et al., 2020). The vital drug include Clazakizumab (Lambert et al., 2020a; Russell et al., 2020), Tocilizumab (Ikonomidis et al., 2020; Luo et al., 2020; Radbel et al., 2020) and Sarimulab are monoclonal antibodies binds to the IL-6 receptor, i.e., the competitive inhibitor of IL-6 involve in the proliferation and differentiation of T lymphocytes, expression of vascular endothelial growth factor and vascular permeability. In SARS-CoV-2 patients, elevated concentration of pro-inflammatory mediators was seen; thus, IL-6 antagonists considered an effective treatment. Next, Nivolumab is a human immunoglobulin G4 (IgG4) monoclonal antibody inhibits the PD-1 receptor, i.e., checkpoint protein on immune cell (Remy et al., 2020). Leronlimb is a human immunoglobulin G4 (IgG4) monoclonal antibody which binds to human chemokines receptor CCR5 and blocks CCR5 mediated chemokines signalling (Lambert et al., 2020b; Philippidis, 2020). Canakinumab is a humanized monoclonal antibody inhibits IL-1B suppress the edema in SARS-CoV-2 patients (Liu et al., 2020a; Moutsopoulos, 2020). Eculizumab, monoclonal antibody blocks the C5 protein leads to the inactivation of complement-mediated immunity (Diurno et al., 2020). Bevacizumab inhibits the Vascular endothelial growth factor, thus reduce the edema in corona patients (Rosa and Santos, 2020). Further polyclonal antibodies include human immunoglobulin G, which blocks the Fc receptor of phagocytes, superantigen binding to T cells, prevents idiotype-anti-idiotype dimer formation, inhibits stimulation of T-cells and dendritic cells and suppress the platelets depletion may be effective against SARS-CoV-2 (Casadevall and Pirofski, 2020). Among these important Mabs, tocilizulmab was recently found to be not effective for preventing intubation or death in moderately ill hospitalized patients with Covid-19. The result came from few clinical trials that found negative regulation of IL-6 in Covid-19 patients which were earlier thought to be linked with cytokine storm. The trials suggested poor outcome of patients treated with tocilizumab and possessing high levels of IL-6. This might be plausibly attributed to the plethora of immune system overreactions which cascades with cooperation of numerous immune activators along with IL-6 contributing to lung infection and cytokine storm (Gupta et al., 2020; Stone et al., 2020). However, concerning the side effects or ADRs of tocilizumab in Covid-19 infected patient the drug is found to be relatively safe in initial studies of short durations (Samaee et al., 2020).
9.2. Interferons
These are involved in developing antiviral immunity and synergistically improving immunomodulatory effects. The category includes Peginterferon lambda-1a investigated for the SARS-CoV-2 virus show immunomodulatory effect in patients associated with Covid-19 disease (Andreakos and Tsiodras, 2020). Interferon-β-1a is used to stimulate macrophages that engulf antigens and natural killer (NK) cells (Sallard et al., 2020). Interferon beta-1b shows an immunomodulatory effect in patients associated with Covid-19 disease (Vanden Eynde, 2020).
9.3. Oligonucleotides
These biologics significantly interfere with the S-protein and/or replication machinery in SARS-CoV-2. This class covers defibrotide, which decreases the level of an inflammatory mediator such as PGI2, PGE2, and prostacyclins, leukocyte tissue infiltration as well as expression of adhesion molecules and epithelial destruction thus promote immune function may be effective against SARS-CoV-2 (Rossi and Rossi, 2020).
9.4. Interleukins
This class is involved in the inhibition of critical immune players involved in inflammation and precipitating cytokinin storm. This includes Anakinra is an interleukin antagonist that blocks the production of pro-inflammatory cytokines interleukin (IL)-1α and IL-1β consider an effective treatment for SARS-CoV-2 (Liu et al., 2020a).
9.5. Hematopoietic growth factors
This category biologics are involved in enhancing the production of hematopoietic cells (Terpos et al., 2020). Important one includes, Sargramostim (Leukins) binds to the granulocyte, macrophage-colony stimulating factors (GM-CSF), which trigger a JAK2/STAT pathway and enhance the production of white blood cells, red blood cells, and platelets (Damiani et al., 2020; Ito et al., 2020).
9.6. Recombinant enzyme and protein
The category includes Dornase is a deoxyribonuclease 1 (DNase 1) enzyme used as mucolytic in cystic fibrosis (CF) to revamp the pulmonary function. The DNase enzyme selectively breaks down DNA and enhance the clearance of neutrophils extracellular traps (NETs), addition also has anti-inflammatory activity may be effective treatment from SARS-CoV-2 (Barnes et al., 2020; Collier, 1995). Ad5-nCoV is a first recombinant novel coronavirus vaccine, i.e., adenovirus type-5 vectored Covid-19 (Ads-nCoV) stimulate the production of antibodies and T-cells (Le et al., 2020). Further, DAS-181 is a sialidase protein that cleaves sialic acid located on the surface of epithelial cell lining in the lungs and prevents the entry of SARS-CoV-2 into the respiratory epithelial cell and impedes the viral infection and spread could be effective against SARS-CoV-2 (Ito et al., 2020).
9.7. Vaccines
A second important highlight of this pandemic is the rapid speed in which research on the development of vaccines is going on. There are currently 133 vaccines in development with 10 in human trials as per data published by WHO (Mullard, 2020). These vaccines are being developed considering the host immune system to prepare and engage itself more efficiently to recognize the pathogenic SARS-CoV-2 strain and destroy them to allow herd immunity (Pulendran and Ahmed, 2011). A particular emphasis is given to the safety evaluations, which otherwise confer fatal consequences like immunopotentiation (Chen et al., 2020b). The vaccines are based on live/attenuated status of the virus also called as virus vaccines, vector-based/modified vaccines, vaccines involving the genomic or nuclear material involving DNA, plasmid, mRNA, siRNA, vaccines and protein preparations, involving Capsid-like particles, a recombinant protein or peptide antigen (Amanat and Krammer, 2020). The Coalition for Epidemic Preparedness Innovation (CEPI) is involved in harmonizing with health authorities across the globe for vaccine development. As per the updated data CoronaVac, AZD1222, mRNA-1273, BNT162, Sputnik V and COVAXIN have proceeded to phase III trial.
10. Conclusion
The world is currently experiencing a new Covid-19 pandemic outbreak by SARS-CoV-2, and the waiting for new or repurposed drugs for the treatment. Though, no drug (except remdesivir) or vaccine has been approved for the Covid-19 disease. Numerous clinical trials are undergoing in the quest to search for the best lead among the repurposed drugs. These drugs belong to diverse classes, which broadly include antiviral, anti-inflammatory, anticancer, antiparasitic, antiprotozoal, antifungal, antibacterial, RAAS-ACE-ARBs inhibitors, and natural products along with other vital classes. These drugs importantly target the virus life cycle at different stages, and some are used as immunomodulators for developed cytokine storm with the progression of Covid-19 disease. As of now, most of the drugs in trials have exhibited good potential in a limited observational or non-randomized clinical study. The potential benefits are still awaited to come in the form of a first approved drug marking its presence for Covid-19 disease. The drug(s) have further remained to be in enough controversies. USFDA has revoked authorized use of HCQ/CQ recently on June 15, 2020, which again has left many unasked questions. The RECOVERY TRIAL, (June 29, 2020), suggested, the combination of Lopinavir-ritonavir, an HIV drug, and other combination of hydroxychloroquine and lopinavir-ritonavir does not provide clinical benefit in hospitalized Covid-19 patients. The trial also has shown that dexamethasone reduces mortality in critically ill patients.
Further, Favipiravir has also received regulatory clearance in India and Russia for the treatment of Covid-19 and has been actively tested across the world, including these two countries (NCT04359615). Moreover, BTK inhibitor (acalabrutinib) and Remdesivir have shown promise in Covid-19 treatment. Further, the authors believe that a greater understanding of the genomic and protein machinery of SARS-CoV-2 is required to design and develop some putative drugs belonging to the natural and synthetic origin. Considering the current scenario, it warrants the need of a multi-targeted approach to inhibit multiple targets involved in the virus life cycle using a combination of repurposed/new drugs that would lead to significant results than monotherapy alone.
In a nutshell, we conclude that SARS-CoV-2 is an “evil genius.” It enters the host cell via spike protein like a lamb by ducking its head (RBD) for immune evasion, succeeds in binding ACE2, enters the cell, hijacks host translation cellular machinery for replicating itself (into millions of copies), tearing the organ system, resulting in its death and stalling the world economy by causing widespread unemployment, poverty, and hunger.
Author contributions
The manuscript has been prepared and drafted with mutual understanding and contributions. ST and Mayank have made an equal contribution toward the current work.
Funding source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
ClinicalTrials. gov, 2020; COVID-19 treatment guidelines panel. Coronavirus disease 2019 (COVID-19) treatment guidelines, 2020; DAILY UNM, 2020a; DAILY UNM, 2020b; Fact sheet for health care providers, 2020; How neutron science supports research on viruses such as Covid-19, 2020; Kaur et al., 2020; LENS and the fight against Covid-19, ; Scientists identify COVID-19 protein using Australian Synchrotron, .
CRediT authorship contribution statement
Shikha Thakur: Writing - original draft. Mayank: Writing - original draft. Bibekananda Sarkar: Writing - original draft. Arshad J. Ansari: Writing - original draft. Akanksha Khandelwal: Review & editing. Anil Arya: Methodology, Writing - review & editing. Ramarao Poduri: Ideation - review & editing. Gaurav Joshi: Writing - original draft, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
GJ thanks the Central University of Punjab, Bathinda, India for necessary resources and infrastructure.
References
- García de Abajo F.J., Hernández R.J., Kaminer I., Meyerhans A., Rosell-Llompart J., Sanchez-Elsner T. Back to normal: an old physics route to reduce SARS-CoV-2 transmission in indoor spaces. ACS Nano. 2020;14:7704–7713. doi: 10.1021/acsnano.0c04596. [DOI] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amraei R., Rahimi N. COVID-19, renin-angiotensin system and endothelial dysfunction. Cells. 2020;9:1652. doi: 10.3390/cells9071652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreakos E., Tsiodras S. COVID-19: lambda interferon against viral load and hyperinflammation. EMBO Mol. Med. 2020;12:e12465. doi: 10.15252/emmm.202012465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreania J., Le Bideaua M., Duflota I., Jardota P., Rollanda C., Boxbergera M., Khalila J.Y.B., Baudouinb J.-P., Wurtza N., Rolaina J.-M. In vitro testing of Hydroxychloroquine and Azithromycin on SARS-CoV-2 shows 1 synergistic effect 2. Microb. Pathog. 2020;145:104228. doi: 10.1016/j.micpath.2020.104228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio A.S.W., Larissa S.M.V.J, Valdir F. Natural products’ role against COVID-19. RSC Adv. 2020;10(39):23379–23393. doi: 10.1039/D0RA03774E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Báez-Santos Y.M., John S.E.S., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakumar P., Maung U.K., Jagadeesh G. Prevalence and prevention of cardiovascular disease and diabetes mellitus. Pharmacol. Res. 2016;113:600–609. doi: 10.1016/j.phrs.2016.09.040. [DOI] [PubMed] [Google Scholar]
- Barnes B.J., Adrover J.M., Baxter-Stoltzfus A., Borczuk A., Cools-Lartigue J., Crawford J.M., Dassler-Plenker J., Guerci P., Huynh C., Knight J.S., Loda M., Looney M.R., McAllister F., Rayes R., Renaud S., Rousseau S., Salvatore S., Schwartz R.E., Spicer J.D., Yost C.C., Weber A., Zuo Y., Egeblad M. Targeting potential drivers of COVID-19. Neutrophil. Extracellular Traps. J. Exp. Med. 2020:217. doi: 10.1084/jem.20200652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud D., Agri V.D., Gibson G.R., Reid G., Giannoni E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front. Publ. Health. 2020;8:186. doi: 10.3389/fpubh.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittmann S., Weissenstein A., Moschüring-Alieva E., Bittmann L., Luchter E., Villalon G. The role of TMPRSS2 and TMPRSS2-inhibitors in cell entry mechanism of COVID-19. J. Regen. Biol. Med. 2020;2:1–3. [Google Scholar]
- Boozari M., Hosseinzadeh H. Natural products for COVID‐19 prevention and treatment regarding to previous coronavirus infections and novel studies. Phytother. Res. 2020:1–13. doi: 10.1002/ptr.6873. [DOI] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Osterlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr A.C. A new clinical trial to test high-dose vitamin C in patients with COVID-19 24(1) (2020) 133. Crit. Care. 2020;24(1):133. doi: 10.1186/s13054-020-02851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Yao Y., Yeung M.-L., Deng W., Bao L., Jia L., Li F., Xiao C., Gao H., Yu P. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-k., Lo S.-C., Wang Y.-S., Hou M.-H. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov. Today. 2016;21:562–572. doi: 10.1016/j.drudis.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplin S.J.P. Remdesivir: an antiviral for the treatment of COVID-19. 2020;31:31–33. [Google Scholar]
- Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Hu S., Wang Y., Hu X., Zheng B. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. Sci. Rep. 2020;10:19196. doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.H., Strych U., Hotez P.J., Bottazzi M.E. The SARS-CoV-2 vaccine pipeline: an overview. Curr. Trop. Med. Rep. 2020;3:1–4. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. COVID-19: organoids go viral. Nat. Rev. Mol. Cell Biol. 2020;21:355–356. doi: 10.1038/s41580-020-0258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrials gov ClinicalTrials gov. (Accessed November 10 2020).
- Collier J. Dornase-alfa and orphan drugs. Lancet (London, England) 1995;346:633. doi: 10.1016/s0140-6736(95)91460-9. [DOI] [PubMed] [Google Scholar]
- COVID-19 Treatment Guidelines 2020. https://www.covid19treatmentguidelines.nih.gov/
- COVID-19 treatment guidelines panel Coronavirus disease 2019 (COVID-19) treatment guidelines 2020. https://www.covid19treatmentguidelines.nih.gov/ Accessed November 10 2020.
- da Silva S.J.R., Alves da Silva C.T., Mendes R.P.G., Pena L. Role of nonstructural proteins in the pathogenesis of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily Unm Global health COVID-19 briefing. 2020. https://vivo.health.unm.edu/display/n54896 Accessed April 16 2020.
- Daily Unm Global health COVID-19 briefing. 2020. https://digitalrepository.unm.edu/hsc_covid19_briefings/4/ Accessed April 13 2020.
- Damiani G., McCormick T.S., Leal L.O., Ghannoum M.A. Recombinant human granulocyte macrophage-colony stimulating factor expressed in yeast (sargramostim): a potential ally to combat serious infections. Clin. Immunol. 2020;210 doi: 10.1016/j.clim.2019.108292. [DOI] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diurno F., Numis F., Porta G., Cirillo F., Maddaluno S., Ragozzino A., De Negri P., Di Gennaro C., Pagano A., Allegorico E. Eculizumab treatment in patients with COVID-19: preliminary results from real life ASL Napoli 2 Nord experience. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4040–4047. doi: 10.26355/eurrev_202004_20875. [DOI] [PubMed] [Google Scholar]
- Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R. A novel angiotensin-converting enzyme–related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ. Res. 2000;87:e1–e9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- Downes S.M., Leroy B.P., Sharma S.M., Sivaprasad S., Dollfus H. Hydroxychloroquine hitting the headlines—retinal considerations. Eye. 2020;34:1158–1160. doi: 10.1038/s41433-020-0934-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escher R., Breakey N., Lämmle B. Severe COVID-19 infection associated with endothelial activation. Thromb. Res. 2020;190:62. doi: 10.1016/j.thromres.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fact sheet for health care providers Emergency use authorization (EUA) of hydroxychloroquine sulfate supplied from the strategic national stockpile for treatment of COVID-19 in certain hospitalized patients. 2020. https://www.fda.gov/media/136537/download Accessed June 15 2020.
- Fan Q., Zhang B., Ma J., Zhang S. Safety profile of the antiviral drug remdesivir: an update. Biomed. Pharmacother. 2020;130 doi: 10.1016/j.biopha.2020.110532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner R.E., Aronson J.K. Chloroquine and hydroxychloroquine in covid-19. Br. Med. J. 2020;369 doi: 10.1136/bmj.m1432. [DOI] [PubMed] [Google Scholar]
- Ferrara F., De Rosa F., Vitiello A. The Central Role of Clinical Nutrition in COVID-19 Patients During and After Hospitalization in Intensive Care Unit. SN Compr. Clin. Med. 2020;2:1064–1068. doi: 10.1007/s42399-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc. Natl. Acad. Sci. U.S.A. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganjhu R.K., Mudgal P.P., Maity H., Dowarha D., Devadiga S., Nag S., Arunkumar G. Herbal plants and plant preparations as remedial approach for viral diseases. Virus Dis. 2015;26:225–236. doi: 10.1007/s13337-015-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasmi A., Tippairote T., Mujawdiya P.K., Peana M., Menzel A., Dadar M., Benahmed A.G., Bjørklund G.J.C.I. Micronutrients as immunomodulatory tools for COVID-19 management. Clin. Immunol. 2020;220 doi: 10.1016/j.clim.2020.108545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J.-C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goren A., Vaño-Galván S., Wambier C.G., McCoy J., Gomez-Zubiaur A., Moreno-Arrones O.M., Shapiro J., Sinclair R.D., Gold M.H., Kovacevic M. A preliminary observation: male pattern hair loss among hospitalized COVID-19 patients in Spain–A potential clue to the role of androgens in COVID-19 severity. J. Cosmet. Dermatol. 2020;19:1545–1547. doi: 10.1111/jocd.13443. [DOI] [PubMed] [Google Scholar]
- Gouveia D., Miotello G., Gallais F., Gaillard J.C., Debroas S., Bellanger L., Lavigne J.P., Sotto A., Grenga L., Pible O., Armengaud J. Proteotyping SARS-CoV-2 virus from nasopharyngeal swabs: a proof-of-concept focused on a 3 min mass spectrometry window. J. Proteome Res. 2020;19:4407–4416. doi: 10.1021/acs.jproteome.0c00535. [DOI] [PubMed] [Google Scholar]
- Gupta S., Wang W., Hayek S.S., Chan L., Mathews K.S., Melamed M.L., Brenner S.K., Leonberg-Yoo A., Schenck E.J., Radbel J.J.J.i.m. Association between early treatment with tocilizumab and mortality among critically ill patients with COVID-19. JAMA Intern. Med. 2020 doi: 10.1001/jamainternmed.2020.6252. 2020.6252 10.1001/jamainternmed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikmet F., Mear L., Uhlen M., Lindskog C. The protein expression profile of ACE2 in human tissues. bioRxiv. 2020 doi: 10.1101/2020.03.31.016048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pohlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogue B.G., Machamer C.E. ASM Press; Washington, DC: 2007. Coronavirus Structural Proteins and Virus Assembly. [Google Scholar]
- How neutron science supports research on viruses such as Covid-19 2020. https://www.ill.eu/neutrons-for-society/science-at-the-ill/biology-and-health/how-neutron-science-supports-research-on-viruses-such-as-covid-19/ Accessed May 15.
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas A., Montani D., Savale L., Pichon J., Tu L., Parent F., Guignabert C., Humbert M. Endothelial cell dysfunction: a major player in SARS-CoV-2 infection (COVID-19)? Eur. Respir. J. 2020:56. doi: 10.1183/13993003.01634-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Jabeen N., Amanullah A., Baig A.A., Aziz B., Shabbir S., Raza F. Structural basis of SARS-CoV-2 spike protein priming by TMPRSS2. bioRxiv. 2020 doi: 10.1101/2020.04.21.052639. [DOI] [Google Scholar]
- Ihling C., Tanzler D., Hagemann S., Kehlen A., Huttelmaier S., Arlt C., Sinz A. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J. Proteome Res. 2020;19:4389–4392. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- Ikonomidis I., Pavlidis G., Katsimbri P., Lambadiari V., Parissis J., Andreadou I., Tsoumani M., Boumpas D., Kouretas D., Iliodromitis E.J.F., Toxicology C. Tocilizumab improves oxidative stress and endothelial glycocalyx. A mechanism that may explain the effects of biological treatment on COVID-19. 2020;145:111694. doi: 10.1016/j.fct.2020.111694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilie P.C., Stefanescu S., Smith L. The role of vitamin D in the prevention of coronavirus disease 2019 infection and mortality. Aging Clin. Exp. Res. 2020;32:1195–1198. doi: 10.1007/s40520-020-01570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Ohmagari N., Mikami A., Sugiura W. Major ongoing clinical trials for COVID-19 treatment and studies currently being conducted or scheduled in Japan. Glob. Publ. Health. 2020;2:96–101. doi: 10.35772/ghm.2020.01034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.J., Jeong S., Kang D.Y., Sp N., Yang Y.M., Kim D.E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA. Sci. Rep. 2020;10:4481. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob. Agents Chemother. 2020:64. doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S., Joshi M., Degani M.S. Tackling SARS-CoV-2: proposed targets and repurposed drugs. Future Med. Chem. 2020;12:1579–1601. doi: 10.4155/fmc-2020-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovic T.H., Ali S.R., Ibrahim N., Jessop Z.M., Tarassoli S.P., Dobbs T.D., Holford P., Thornton C.A., Whitaker I.S.J.N. Could Vitamins Help in the Fight Against COVID-19? 2020;12:2550. doi: 10.3390/nu12092550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpe Y.A., Lole K.S. NTPase and 5' to 3' RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J. Virol. 2010;84:3595–3602. doi: 10.1128/JVI.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau A.L., Ahern P.P., Griffin N.W., Goodman A.L., Gordon J.I. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayank, Kumar D., Kaur N., Giri R., Singh N. A biscoumarin scaffold as an efficient anti-Zika virus lead with NS3-helicase inhibitory potential: in vitro and in silico investigations. New J. Chem. 2020;44:1872–1880. doi: 10.1039/c9nj05225a. [DOI] [Google Scholar]
- Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu Y. Novel coronavirus treatment with ribavirin: groundwork for an evaluation concerning COVID-19. J. Med. Virol. 2020;92:740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J.Y., Yang J.S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921. doi: 10.1016/j.cell.2020.04.011. e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer, R.N., Cottrell, C.A., Wang, N., Pallesen, J., Yassine, H.M., Turner, H.L., Corbett, K.S., Graham, B.S., McLellan, J.S., Ward, A.B.J.N., 2016. Pre-fusion structure of a human coronavirus spike protein. 531, 118–121.Pubmed Partial stl. [DOI] [PMC free article] [PubMed]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo L., Hurst K.R., Masters P.S. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 2007;81:2249–2262. doi: 10.1128/JVI.01577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345 doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- Latz C.A., DeCarlo C., Boitano L., Png C.Y.M., Patell R., Conrad M.F., Eagleton M., Dua A. Blood type and outcomes in patients with COVID-19. Ann. Hematol. 2020;99:2113–2118. doi: 10.1007/s00277-020-04169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- LENS and the fight against Covid-19 How neutrons will contribute. https://www.lens-initiative.org/2020/03/27/lens-fighting-covid-19-how-neutrons-will-contribute/
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zai J., Zhao Q., Nie Q., Li Y., Foley B.T., Chaillon A. Evolutionary history, potential intermediate animal host, and cross-species analyses of SARS-CoV-2. J. Med. Virol. 2020;92:602–611. doi: 10.1002/jmv.25731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner H.A., Fotouhi-Ardakani N., Lytvyn V., Lachance P., Sulea T., Ménard R. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 2005;79:15199–15208. doi: 10.1128/JVI.79.24.15199-15208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Li M., Zhou Z., Guan X., Xiang Y. Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun. 2020;111 doi: 10.1016/j.jaut.2020.102452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Cui J., Qian Z., Wang Y., Zhang H., Duan Y., Wu X., Yao X., Song Y., Li X., Wu C., Tang X. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., Hennig B.P., Kreuter M., Conrad C., Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo P., Liu Y., Qiu L., Liu X., Liu D., Li J. Tocilizumab treatment in COVID-19: a single center experience. J. Med. Virol. 2020;92:814–818. doi: 10.1002/jmv.25801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LuoLiu P., Li J. Pharmacologic perspective: glycyrrhizin may be an efficacious therapeutic agent for COVID-19. Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado E, Tao D, Mackey K. Antithrombotic Therapies in COVID-19 Disease: a Systematic Review. J. Gen. Intern. Med. 2020;35(9):2698–2706. doi: 10.1007/s11606-020-05906-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marik P.E., Kory P., Varon J., Iglesias J., Meduri G.U. MATH+ protocol for the treatment of SARS-CoV-2 infection: the scientific rationale. Expert Rev. Anti Infect. Ther. 2020:1–7. doi: 10.1080/14787210.2020.1808462. [DOI] [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh Across Speciality Collaboration U.K. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra D.P., Agarwal V., Gasparyan A.Y., Zimba O. Rheumatologists' perspective on coronavirus disease 19 (COVID-19) and potential therapeutic targets. Clin. Rheumatol. 2020;39:2055–2062. doi: 10.1007/s10067-020-05073-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J.G., Vincent F., Lee J.A., Eder J., Prunotto M. Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat. Rev. Drug Discov. 2017;16:531–543. doi: 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- Mondal M., Lawarde A., Somasundaram K. Genomics of Indian SARS-CoV-2: implications in genetic diversity, possible origin and spread of virus. Curr. Sci. 2020:118. [Google Scholar]
- Montastruc F., Thuriot S., Durrieu G.J.C.G., Hepatology Hepatic disorders with the use of remdesivir for coronavirus 2019. Clin. Gastroenterol. Hepatol. 2020;18:2835–2836. doi: 10.1016/j.cgh.2020.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutsopoulos H.M. Anti-inflammatory therapy may ameliorate the clinical picture of COVID-19. Ann. Rheum. 2020;79:1253–1254. doi: 10.1136/annrheumdis-2020-217562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullard A. COVID-19 vaccine development pipeline gears up. Lancet. 2020;395:1751–1752. doi: 10.1016/S0140-6736(20)31252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafa S., Balkhy H., Gabere M.N. Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J. Infect. Publ. Health. 2018;11:9–17. doi: 10.1016/j.jiph.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasi A., McArdle S., Gaudernack G., Westman G., Melief C., Rockberg J., Arens R., Kouretas D., Sjölin J., Mangsbo S.J.T.r. Reactive oxygen species as an initiator of toxic innate immune responses in retort to SARS-CoV-2 in an ageing population, consider N-acetylcysteine as early therapeutic intervention. 2020;7:768–771. doi: 10.1016/j.toxrep.2020.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrani A.S., Memish Z.A. Therapeutic options for Middle East respiratory syndrome coronavirus (MERS-CoV) infection: how close are we? Curr. Treat Options Infect. Dis. 2015;7:202–216. doi: 10.1007/s40506-015-0048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Ye T., Sun P., Gui S., Liang B., Li L., Zheng D., Wang J., Hesketh R.L., Yang L. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology. 2020;295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippidis A. COVID-19: top 60 Drug Treatments in Development: the biopharma industry is ramping up the development of dozens of potential drug therapies and clinical testing in an all-hands effort to combat the pandemic. Genet. Eng. Biotechnol. News. 2020;40:10–13. [Google Scholar]
- Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov. Today Off. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poduri R., Joshi G., Jagadeesh G. Drugs targeting various stages of the SARS-CoV-2 life cycle: exploring promising drugs for the treatment of Covid-19. Cell. Signal. 2020;74:109721. doi: 10.1016/j.cellsig.2020.109721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapat M., Sarma P., Shekhar N., Avti P., Sinha S., Kaur H., Kumar S., Bhattacharyya A., Kumar H., Bansal S., Medhi B. Drug targets for corona virus: a systematic review. Indian J. Pharmacol. 2020;52:56–65. doi: 10.4103/ijp.IJP_115_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B., Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbel J., Narayanan N., Bhatt P.J. Use of tocilizumab for COVID-19 infection-induced cytokine release syndrome: a cautionary case report. Chest. 2020;158:e15–e19. doi: 10.1016/j.chest.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy K.E., Brakenridge S.C., Francois B., Daix T., Deutschman C.S., Monneret G., Jeannet R., Laterre P.F., Hotchkiss R.S., Moldawer L.L. Immunotherapies for COVID-19: lessons learned from sepsis. Lancet Respir. Med. 2020;8:946–949. doi: 10.1016/S2213-2600(20)30217-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renieri A., Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C. ACE2 variants underlie interindividual variability and susceptibility to COVID-19 in Italian population. Eur. J. Hum. Genet. 2020;28:1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S.G.V., Santos W.C. Clinical trials on drug repositioning for COVID-19 treatment. Rev. Panam. Salud Públic. 2020;44:e40. doi: 10.26633/RPSP.2020.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J.J., Rossi D. Oligonucleotides and the COVID-19 pandemic: a perspective. Nucleic Acid Therapeut. 2020;30:129–132. doi: 10.1089/nat.2020.0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect Publ. Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothlin R.P., Vetulli H.M., Duarte M., Pelorosso F.G. Telmisartan as tentative angiotensin receptor blocker therapeutic for COVID-19. Drug Dev. Res. 2020;81:768–770. doi: 10.1002/ddr.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B., Moss C., George G., Santaolalla A., Cope A., Papa S., Van Hemelrijck M. Associations between immune-suppressive and stimulating drugs and novel COVID-19—a systematic review of current evidence. Ecancermedicalscience. 2020:14. doi: 10.3332/ecancer.2020.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallard E., Lescure F.-X., Yazdanpanah Y., Mentre F., Peiffer-Smadja N., Florence A., Yazdanpanah Y., Mentre F., Lescure F.-X., Peiffer-Smadja N. Type 1 interferons as a potential treatment against COVID-19. Antivir. Res. 2020;178:104791. doi: 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaee H., Mohsenzadegan M., Ala S., Maroufi S.S., Moradimajd P. Tocilizumab for treatment patients with COVID-19: recommended medication for novel disease. Int. Immunopharm. 2020;89 doi: 10.1016/j.intimp.2020.107018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scientists identify COVID-19 protein using Australian Synchrotron https://www.neimagazine.com/news/newsscientists-identify-covid-19-protein-using-australian-synchrotron-7866759
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif-Yakan A., Kanj S.S. Emergence of MERS-CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog. 2014;10:e1004457. doi: 10.1371/journal.ppat.1004457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi N., Ryan C.J. Androgen hazards with COVID-19. Endocr. Relat. Canc. 2020;27:E1–E3. doi: 10.1530/ERC-20-0133. [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K.K., Chaubey G., Chen J.Y., Suravajhala P. Virus-host cell interactions and the viral life cycle: basic science to therapeutics: decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. 2020;319:C258. doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone J.H., Frigault M.J., Serling-Boyd N.J., Fernandes A.D., Harvey L., Foulkes A.S., Horick N.K., Healy B.C., Shah R., Bensaci A.M., Woolley A.E., Nikiforow S., Lin N., Sagar M., Schrager H., Huckins D.S., Axelrod M., Pincus M.D., Fleisher J., Sacks C.A., Dougan M., North C.M., Halvorsen Y.D., Thurber T.K., Dagher Z., Scherer A., Wallwork R.S., Kim A.Y., Schoenfeld S., Sen P., Neilan T.G., Perugino C.A., Unizony S.H., Collier D.S., Matza M.A., Yinh J.M., Bowman K.A., Meyerowitz E., Zafar A., Drobni Z.D., Bolster M.B., Kohler M., D'Silva K.M., Dau J., Lockwood M.M., Cubbison C., Weber B.N., Mansour M.K., Investigators B.B.T.T. Efficacy of tocilizumab in patients hospitalized with covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2028836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. Vitro and animal models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020;41:513–517. doi: 10.1016/j.tips.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. Vitro and animal models for SARS-CoV-2 research. Trends Pharmacol. Sci. 2020 doi: 10.1016/j.tips.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te Velthuis A.J., Van Den Worm S.H., Snijder E.J. The SARS-coronavirus nsp7+ nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., Psaltopoulou T., Gerotziafas G., Dimopoulos M.A. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020;95:834–847. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.F., Chan P.K. Identification of human cell line model of persistent SARS coronavirus infection and studies of the response to cytokines and chemokines. Hong Kong Med. J. 2009;6(15 Suppl):39–43. [PubMed] [Google Scholar]
- Touret F., Gilles M., Barral K., Nougairede A., van Helden J., Decroly E., de Lamballerie X., Coutard B. In vitro screening of a FDA approved chemical library reveals potential inhibitors of SARS-CoV-2 replication. Sci. Rep. 2020;10 doi: 10.1038/s41598-020-70143-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020;16:434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Eynde J.J. COVID-19: a brief overview of the discovery clinical trial. Pharmaceuticals. 2020;13:65. doi: 10.3390/ph13040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatagopalan P., Daskalova S.M., Lopez L.A., Dolezal K.A., Hogue B.G. Coronavirus envelope (E) protein remains at the site of assembly. Virology. 2015;478:75–85. doi: 10.1016/j.virol.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A., Puranik A., Anand A., Zemmour D., Yao X., Wu X., Chilaka R., Murakowski D.K., Standish K., Raghunathan B. Knowledge synthesis from 100 million biomedical documents augments the deep expression profiling of coronavirus receptors. arXiv preprint. 2020. [DOI] [PMC free article] [PubMed]
- Viswanathan T., Arya S., Chan S.-H., Qi S., Dai N., Misra A., Park J.-G., Oladunni F., Kovalskyy D., Hromas R.A. Structural basis of RNA cap modification by SARS-CoV-2. Nat. Commun. 2020;11:1–7. doi: 10.1038/s41467-020-17496-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428. doi: 10.1016/j.cell.2020.05.034. e413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., Zhang Z., Shu F., Huang W., Li Y., Zhang Z., Chen R.A., Wu Y.J., Peng S.M., Huang M., Xie W.J., Cai Q.H., Hou F.H., Chen W., Xiao L., Shen Y. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- Xu X., Dang Z. Promising inhibitor for 2019-nCoV in drug development. Cancer Transl. Med. 2020;6:17–20. doi: 10.4103/ctm.ctm_3_20. [DOI] [Google Scholar]
- Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Tan Z., Zhou L., Yang M., Peng L., Liu J., Cai J., Yang R., Han J., Huang Y. Effects of ARBs and ACEIs on virus infection, inflammatory status and clinical outcomes in COVID-19 patients with hypertension: a single center retrospective study. Hypertension. 2020;76:51–58. doi: 10.1161/HYPERTENSIONAHA.120.15143. [DOI] [PubMed] [Google Scholar]
- Yang G., Tan Z., Zhou L., Yang M., Peng L., Liu J., Cai J., Yang R., Han J., Huang Y., He S. Effects of angiotensin II receptor blockers and ACE (Angiotensin-Converting enzyme) inhibitors on virus infection, inflammatory status, and clinical outcomes in patients with COVID-19 and hypertension: a single-center retrospective study. Hypertension. 2020;76:51–58. doi: 10.1161/HYPERTENSIONAHA.120.15143. [DOI] [PubMed] [Google Scholar]
- Ye Q., Wang B., Mao J. The pathogenesis and treatment of the ‘Cytokine Storm’ in COVID-19. J. Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., Langereis M.A., van Vliet A.L., Huizinga E.G., de Groot R.J. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Zhong Z., Xie X., Yu Q., Liu J. CT scans of patients with 2019 novel coronavirus (COVID-19) Pneumonia. Theranostics. 2020;10:4606–4613. doi: 10.7150/thno.45016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J., Tang J., Ye C., Dong L. The immunology of COVID-19: is immune modulation an option for treatment? Lancet Rheumatol. 2020;2:E428–E436. doi: 10.1016/S2665-9913(20)30120-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dai S.M., Tong Q. COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression. J. Antimicrob. Chemother. 2020;75:1667–1670. doi: 10.1093/jac/dkaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;24:490–502. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]