Zhong et al. show that the SLAM family of receptors is critical for T cell–dependent humoral immunity due to its ability to enhance expression of pro-survival effectors such as Bcl-2 and BCR in germinal center B cells.
Abstract
Expression of the signaling lymphocytic activation molecule (SLAM)–associated protein (SAP) is critical for the germinal center (GC) reaction and T cell–dependent antibody production. However, when SAP is expressed normally, the role of the associated SLAM family receptors (SFRs) in these processes is nebulous. Herein, we established that in the presence of SAP, SFRs suppressed the expansion of the GC reaction but facilitated the generation of antigen-specific B cells and antibodies. SFRs favored the generation of antigen-reactive B cells and antibodies by boosting expression of pro-survival effectors, such as the B cell antigen receptor (BCR) and Bcl-2, in activated GC B cells. The effects of SFRs on the GC reaction and T cell–dependent antibody production necessitated expression of multiple SFRs, both in T cells and in B cells. Hence, while in the presence of SAP, SFRs inhibit the GC reaction, they are critical for the induction of T cell–mediated humoral immunity by enhancing expression of pro-survival effectors in GC B cells.
Introduction
Antibody production by B cells plays a key role in protection against microbial pathogens upon natural infection or vaccination (Cyster and Allen, 2019; DeFranco, 2016; Ise and Kurosaki, 2019; Shlomchik et al., 2019). It also participates in the pathophysiology of autoimmune diseases such as systemic lupus erythematosus and rheumatoid arthritis. Long-lasting “humoral” immunity is initiated by the germinal center (GC) reaction, during which a subset of antigen-specific T cells, known as T follicular helper cells (Tfh cells), triggers intense proliferation and differentiation of a subpopulation of B cells bearing B cell antigen receptors (BCRs) recognizing the same antigen. Following their activation by Tfh cells, GC B cells divide approximately every 4 h. As they proliferate, GC B cells undergo somatic hypermutation (SHM) and class switch recombination (CSR) of BCR-encoding genes, leading to the generation of B cells having BCRs of different isotypes and potentially having higher affinity for the antigen. GC B cells bearing low-affinity BCRs die and are eliminated by macrophages. However, a few GC B cells acquire BCRs of sufficiently high affinity and survive this process. These cells then exit the GC reaction and differentiate into memory B cells or antibody-secreting plasma cells, which provide long-lasting humoral immunity.
Given the crucial roles of T cell–dependent antibody production in health and disease, much effort has been directed at understanding the molecular pathways controlling this process (DeFranco, 2016; Ise and Kurosaki, 2019; Shlomchik et al., 2019). Among the proposed regulators of the GC reaction and T cell–dependent antibody production are the signaling lymphocytic activation molecule (SLAM) family receptors (SFRs). SFRs are a group of six hematopoietic cell–specific receptors termed SLAMF1, SLAMF5, SLAMF6, Ly-9, 2B4, and SLAMF7 (Cannons et al., 2011; Ma and Deenick, 2011; Shachar et al., 2019; Wu and Veillette, 2016). They are abundantly expressed on Tfh cells and GC B cells. They operate as homotypic receptors (i.e., self-ligands), with the exception of 2B4, which recognizes CD48 as ligand. Depending on the context, SFRs can be activating or inhibitory, due to their dual capacity to associate with activating effectors such as the SLAM–associated protein (SAP) family adaptors or inhibitory molecules such as Src homology 2 (SH2) domain–containing protein tyrosine phosphatase 1 (SHP-1) and SH2 domain–containing inositol phosphatase 1 (SHIP-1). Humans lacking the adaptor SAP suffer from a rare primary immunodeficiency, X-linked lymphoproliferative disease. Loss of SAP in X-linked lymphoproliferative patients or genetically engineered SAP-deficient mice results in altered SFR functions that cause multiple immune defects, including a severe defect in the GC reaction and T cell–dependent antibody production (Cannons et al., 2011; Ma and Deenick, 2011; Shachar et al., 2019; Wu and Veillette, 2016).
Genetic evidence has indirectly implicated SFRs in antibody production and antibody-mediated autoimmune diseases (Cannons et al., 2011; Ma and Deenick, 2011; Shachar et al., 2019; Wu and Veillette, 2016). First, as mentioned above, loss of SAP severely compromises the GC reaction and T cell–dependent antibody production. Studies of mice lacking both SAP and SFRs showed that this phenotype is due to conversion of SFRs into superinhibitory receptors in Tfh cells, which normally express SAP (Chen et al., 2017b; Kageyama et al., 2012). Second, polymorphisms in the genes encoding SLAMF6, 2B4, or Ly-9 have been linked to autoimmune diseases such as systemic lupus erythematosus and rheumatoid arthritis in humans or mice (Cunninghame Graham et al., 2008; Kumar et al., 2006; Suzuki et al., 2008; Wandstrat et al., 2004). However, despite this correlative evidence, analyses of mice lacking one or more SFRs, but expressing SAP, have not consistently revealed alterations in the GC reaction and T cell–dependent antibody production (Barak et al., 2020; Cannons et al., 2010; Chen et al., 2017b; Hu et al., 2016; Huang et al., 2016; Kageyama et al., 2012; Luo et al., 2019; Ma and Deenick, 2011; Veillette et al., 2008; Wandstrat et al., 2004; Wang et al., 2015; Yusuf et al., 2010). In particular, a short-term analysis of a mouse strain lacking all SFRs and expressing SAP concluded that SFRs were not required for a normal GC reaction (Chen et al., 2017b), although generation of antigen-specific B cells and antibody production were not evaluated in this study.
Hence, in the presence of SAP, whether SFRs promote, inhibit, or have no impact on the GC reaction and T cell–dependent antibody production, in particular in the context of long-lasting humoral immunity, is unclear. The possible effects of SFRs on the generation or maintenance of antigen-specific GC B cells, memory B cells, and plasma cells, if any, have not been assessed. Likewise, the cellular and molecular mechanisms by which SFRs potentially regulate T cell–mediated humoral immunity are not clarified. Lastly, the relevance of these potential effects for protection against pathogens or vaccination is not known.
To address these issues, we conducted detailed short- and long-term analyses of the GC reaction and T cell–dependent antibody production in a variety of mouse strains lacking SFRs but expressing SAP. Our studies showed that SFRs were required for T cell–dependent antibody production and protective immunity against a pathogen. Although SFRs inhibited expansion of the GC reaction, they paradoxically facilitated the generation of specific antigen-reactive GC B cells, memory B cells, and plasma cells. The latter effects were accompanied by greater expression of pro-survival effectors, in particular BCR and Bcl-2, in activated GC B cells. Accordingly, transgenic overexpression of Bcl-2 improved the humoral immunity defect seen in SFR-deficient mice. The involvement of SFRs in T cell–dependent humoral immunity required expression on T cells and B cells and multiple members of the SLAM family.
Results
Normal B cell development but decreased baseline antibody levels in mice lacking all SFRs
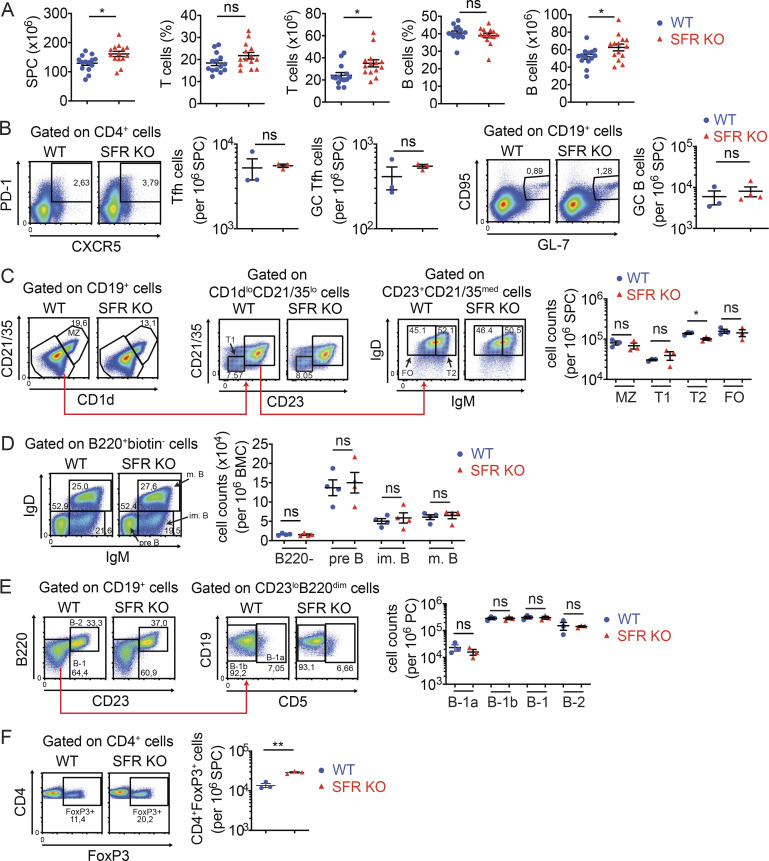
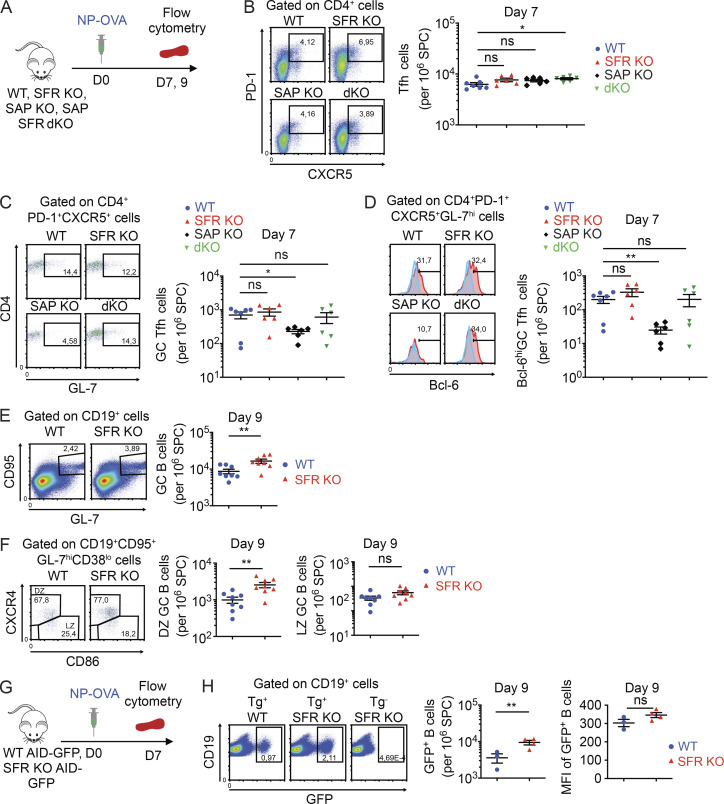
To clarify the role of SFRs in T cell–dependent antibody production, mice engineered to lack the entire Slam locus, which encodes all six SFRs in addition to CD48 (the ligand of 2B4), were studied (Chen et al., 2017a; Guo et al., 2016; Lu et al., 2019). In the absence of any immunization, SFR KO mice had little or no alterations in B cell development and subsets in bone marrow, spleen, or peritoneal cavity in comparison to WT littermates (Fig. 1, A–E). In addition, the numbers of Tfh cells (defined as CD4+PD-1+CXCR5+ cells), GC Tfh cells (CD4+PD-1+CXCR5+GL-7+), and GC B cells (CD19+CD95+GL-7+) were normal (Fig. 1 B). The only difference in B cell development was a small decrease of T2 transitional B cells (Fig. 1 C). SFR KO mice had slightly increased total splenocyte numbers, in comparison to WT mice, due to an increase in T cells and, to a lesser extent, B cells (Fig. 1 A). There was also an augmentation in the numbers of regulatory T cells (T reg cells; Fig. 1 F). The basis for these observations is not known.
Figure 1.
Normal B cell development in nonimmunized SFR KO mice. (A) Total splenocyte counts, as well as numbers and proportions of T cells and B cells, were determined in WT (n = 14) and SFR KO (n = 14) mice (8–10 wk old). Results are pooled from four independent experiments. (B) Same as A, except that Tfh cells, GC Tfh cells, and GC B cells were enumerated (WT, n = 3; SFR KO, n = 3 for Tfh cells and n = 4 for GC B cells; 14–20 wk old). (C) Same as A, except that MZ, transitional 1 (T1), transitional 2 (T2), and follicular (FO) B cells were analyzed (WT, n = 3; SFR KO, n = 3). (D) B cell development was analyzed in bone marrow of WT and SFR KO mice (8–10 wk old). The indicated B cell populations, in addition to B220− B cells, were enumerated (WT, n = 4; SFR KO, n = 4). im, immature; m, mature. (E) B-1a and B-1b B cells were enumerated in the peritoneal cavity of WT and SFR KO mice (16–20 wk old; WT, n = 3; SFR KO: n = 3). (F) T reg cells were enumerated in spleen of WT and SFR KO mice (16–20 wk old; WT, n = 3; SFR KO, n = 3). Results are from one experiment (B–F). Each symbol represents a mouse. *, P < 0.05 (two-tailed Student’s t test). Data are presented as mean ± SEM. For all panels of all figures, representative dot plots, contour plots, or histograms are shown on the left, whereas data for multiple independent mice are depicted on the right. Specific populations are boxed or highlighted by horizontal lines. Percentages are shown in the boxes or above the lines. ns, not significant; SPC, spleen cells.
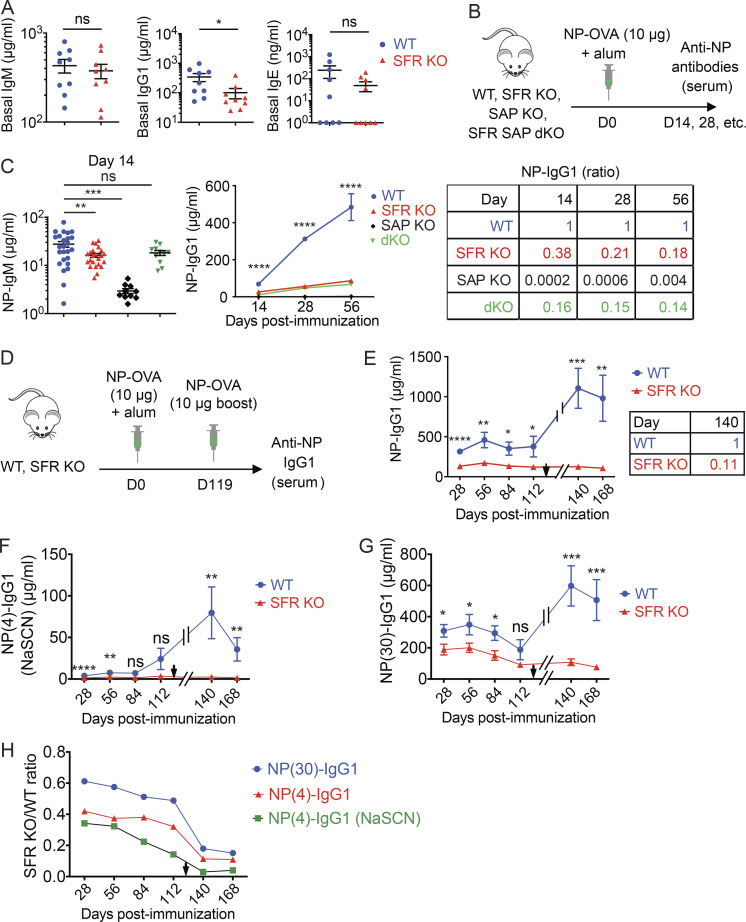
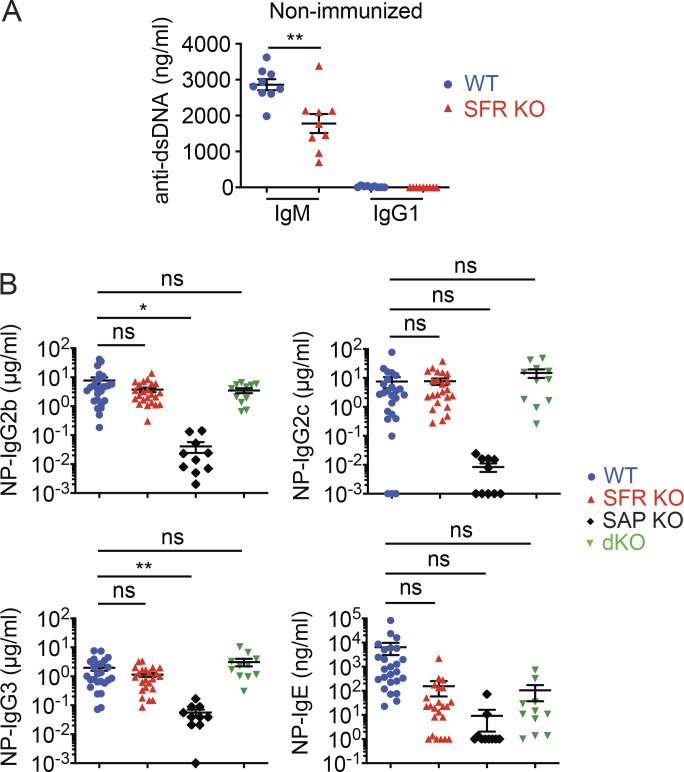
Nonetheless, nonimmunized SFR KO mice had lower levels of IgG1 (reduced by ∼70%) in serum than WT mice (Fig. 2 A). They also had lower amounts of IgE, and perhaps IgM, although the differences did not reach statistical significance, presumably because of wide variations in the abundance of the antibodies between mice. Lower levels of anti–double-stranded DNA (anti-dsDNA) IgM were also noted (Fig. S1 A).
Figure 2.
SFRs are required for antibody production. (A) Baseline IgM, IgG1, and IgE in nonimmunized WT littermate (n = 9) or SFRs KO (n = 9) mice (14–20 wk old) were measured by ELISA. Results are from one experiment. (B and C) WT (n = 25), SFR KO (n = 23), SAP KO (n = 10), or SFR SAP dKO (n = 11) mice were immunized with NP-OVA (10 µg) plus alum, and serum was obtained at the indicated times to measure anti-NP-specific antibodies. The protocol (B) and data for IgM (at day 14; C, left) and IgG1 over time (C, middle) are shown. The ratios of IgG1 in mutant mice over WT mice at different times are shown in (C, right). Results are pooled from four experiments. (D–H) WT (n = 9) and SFR KO mice (n = 13) were immunized as in B. In addition, at week 17 after immunization, mice were boosted by i.p. injection of a second dose of NP-OVA (10 µg) in the absence of adjuvant. Antibody production was measured at the indicated time points. Results are from one experiment. Protocol (D) and levels of anti-NP IgG1 (E, left) are shown. The time of the boost is indicated by a black arrowhead. The ratios of IgG1 in mutant mice over WT mice at day 140 are shown in (E, right). (F) The ELISA depicted in E was repeated in the presence of 1 M NaSCN. (G) Same as E, except that NP(30) was used as capture antigen. (H) The ratios of the mean titers of antibodies in SFR KO mice over WT mice over time are shown. Each symbol represents a mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-tailed Student’s t test). Data are shown as mean ± SEM. D, day; ns, not significant.
Figure S1.
Reduced T cell–dependent antibody production in SFR KO mice. Related to Fig. 2. (A) Anti-dsDNA antibody titers were measured in nonimmunized mice. The mice are same as those in Fig. 2 A (WT, n = 9; SFR KO, n = 9). Data are from one experiment. (B) Same as Fig. 2, B and C, except that anti-NP IgG2b, IgG2c, IgG3, and IgE were measured (WT, n = 25; SFR KO, n = 23; SAP KO, n = 10; SFR SAP dKO, n = 11). Data are pooled from four experiments. Each symbol represents one mouse. *, P < 0.05; **, P < 0.01 (two-tailed Student’s t test). Data are shown as mean ± SEM. D, day; ns, not significant.
Therefore, SFR KO mice had little or no defects in B cell development and B cell subsets in the absence of immunization. However, they showed a decrease in baseline levels of IgG1 and anti-dsDNA IgM.
SFR deficiency compromises antibody response to a model T cell–dependent antigen
Next, SFR KO mice were immunized with the T cell–dependent antigen 4-hydroxy-3-nitrophenyl (NP) coupled to OVA, in the presence of the adjuvant alum (Fig. 2 B). Mice lacking the adaptor SAP, which is required for the GC reaction and T cell–dependent antibody production, were analyzed in parallel (Cannons et al., 2011; Crotty et al., 2003; Czar et al., 2001; Ma and Deenick, 2011; Qi et al., 2008; Shachar et al., 2019; Wu and Veillette, 2016). When compared with WT mice, SFR KO mice exhibited a decrease in anti-NP IgM (by ∼40% at day 14) and IgG1 (by ∼60% at day 14), but not IgG2b, IgG2c, and IgG3 (Fig. 2 C and Fig. S1 B). Production of IgM and IgG1 are particularly dependent on the GC reaction. The compromise in anti-NP IgG1 levels became greater over time (Fig. 2 C). Indeed, compared with WT mice, SFR KO mice had a decrease in IgG1 of ∼60% at day 14 and ∼80% at day 56. An even greater reduction (by ∼90%) was noted after boost immunization (Fig. 1, D and E).
To address whether a lack of SFRs influences antibody affinity maturation, the affinity of anti-NP IgG1 was probed. The experiment detailed in Fig. 2 E was performed with NP(4) as antigen. By using NP(4) in the presence of the chaotropic agent sodium thiocyanate (NaSCN) or NP(30) as antigen, one can diminish or augment detection of higher and lower affinity antibodies, respectively (Fig. 2, F and G). By calculating the ratio of anti-NP IgG1 levels in SFR KO mice compared with WT mice with these various assay conditions, we estimated that SFR KO mice had a higher proportion of low affinity anti-NP antibodies compared with WT mice (Fig. 2 H).
Compared with SFR KO mice, SAP KO mice displayed a more severe and broader reduction of antibody levels. As reported previously (Crotty et al., 2003; Czar et al., 2001; Qi et al., 2008; Veillette et al., 2008), SAP KO mice had a >90% diminution in levels of all isotypes of anti-NP antibodies tested, namely IgM, IgG1, IgG2b, IgG2c, IgG3, and IgE (Fig. 2 C and Fig. S1 B). At day 14 after immunization, IgM and IgG1 levels were decreased by ∼90% and >99%, respectively. Furthermore, in keeping with the idea that the antibody defect in SAP KO mice is due to conversion of SFRs into superinhibitory receptors (Chen et al., 2017b; Kageyama et al., 2012), the decrease in anti-NP antibodies observed in SAP KO mice was attenuated when these mice were bred with SFR KO mice to the same levels as those seen in SFR KO mice (Fig. 2 C and Fig. S1 B).
Thus, mice lacking all SFRs displayed a selective defect in IgM and IgG1 production in response to the T cell–dependent protein antigen NP-OVA. This defect was less severe than that observed in SAP KO mice, but it became more pronounced over time. SFR KO mice also had a defect in antibody affinity maturation.
Lack of SFRs reduces protective antibody responses to vaccination and helminth infection
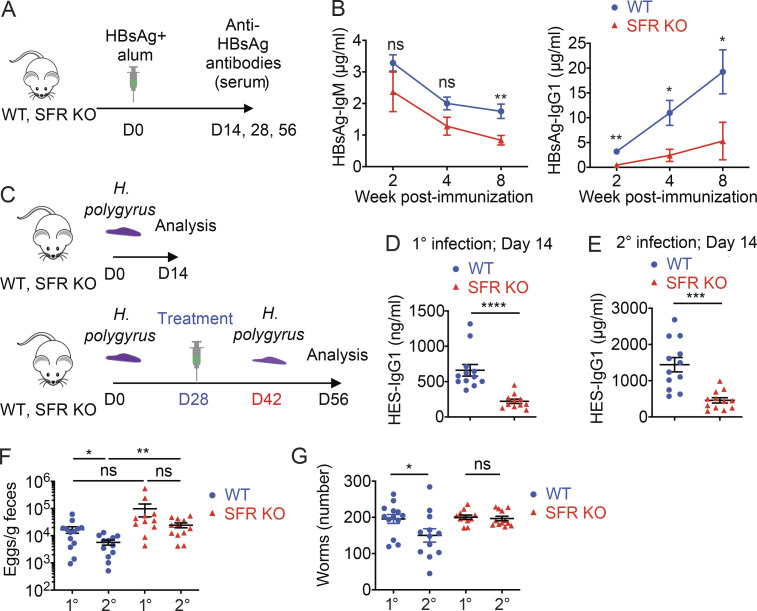
To test the impact of SFRs in more clinically relevant antibody responses, we first ascertained the production of antibodies against the hepatitis B surface antigen (HBsAg), which is used in humans for vaccination against hepatitis B virus (HBV; Fig. 3 A). Compared with WT mice, SFR KO mice displayed a ∼75% reduction of anti-HBsAg–specific IgG1 (Fig. 3 B). As was the case for NP-OVA (Fig. 2 C), the extent of the IgG1 defect in SFR KO mice became more severe over time (Fig. 3 B). A decrease in HBsAg-specific IgM (by ∼30%) was also noted, although it was statistically significant only at week 8 after immunization. Mice are not susceptible to infection by HBV, precluding us from testing protection against HBV in this model.
Figure 3.
Reduced T cell–dependent antibody production against HBsAG and H. polygyrus in SFR KO mice. (A and B) WT and SFR KO mice were immunized i.p. with HBsAg vaccine (1 µg per mouse) and alum. Anti-HBsAg antibodies in serum were monitored over time. The protocol (A) and levels of anti-HBsAg IgM and IgG1 (B) are represented (WT, n = 8; KO, n = 7). Results are from one experiment. D, day. (C–G) One group of WT (n = 13; except for ELISA, n = 12, as serum from one mouse was not available) and SFR KO (n = 11) mice was infected orally with H. polygyrus. For another group (WT, n = 12; SFR KO, n = 12), infection was treated with Pyrantel at day 28 to clear the worms, and mice were reinfected at day 42. Mice were then analyzed 2 wk after primary or secondary infection. Results are pooled from two experiments. The protocol (C) and levels of HES-specific IgG1 (D and E) are depicted. Eggs in feces were enumerated (F), and worms in small intestine were visually enumerated (G). Data are pooled from two experiments. Each symbol represents a mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-tailed Student’s t test). Data are presented as mean ± SEM. ns, not significant.
Next, we examined the ability of mice to mount a protective humoral immune response to the enteric nematode Heligmosomoides polygyrus, which requires T cell–dependent IgG1 for resistance to secondary infection (King and Mohrs, 2009). To this end, WT and SFR KO mice were orally infected with H. polygyrus larvae (Fig. 3 C). At 4 wk after infection, adult worms were eradicated by anti-helmintic treatment and mice were subsequently rechallenged with a second infection. 2 wk after secondary infection, mice were sacrificed, and eggs and adult worms were counted in feces and small intestine, respectively. Production of IgG1 against the H. polygyrus excretory–secretory (HES) antigens was also measured. For comparison, some mice were sacrificed following primary infection.
Compared with WT mice, SFR KO mice displayed decreased levels (by ∼75%) of anti-HES IgG1 after both primary and secondary infection (Fig. 3, D and E). SFR KO mice also exhibited a greater number (approximately threefold) of eggs after primary and secondary infection compared with WT mice (Fig. 3 F). However, the difference reached statistical significance only after secondary infection. More importantly, while WT mice were partially protected against secondary infection, as indicated by a decrease (by ∼50%) in worm burden, SFR KO mice failed to exhibit protection (Fig. 3 G).
Hence, the absence of SFRs compromised the IgG1 response to HBsAg. Furthermore, it led to reduced production of IgG1 and compromised protective immunity against H. polygyrus.
Expansion of the GC reaction is enhanced, rather than diminished, in SFR KO mice
Next, we analyzed the GC reaction after immunization with NP-OVA (Fig. 4 A). T cell and B cell responses in spleen were studied at day 7 or 9, which are the peak times for the T cell or B cell responses, respectively. SFR KO mice displayed a small, albeit not statistically significant, increase in the numbers of Tfh cells, GC Tfh cells, and fully polarized GC Tfh cells (Bcl-6hiCD4+PD-1+CXCR5+GL-7+) when compared with WT mice (Fig. 4, B–D). SFR KO mice also exhibited a significant (approximately twofold) increase in the numbers of GC B cells (Fig. 4 E). This increase mostly concerned the dark zone (DZ) B cells (CD19+CD95+GL-7+CD38loCXCR4hiCD86lo), which are the more proliferative subset of GC B cells (Fig. 4 F). A small increase was also seen for light zone (LZ) GC B cells (CD19+CD95+GL-7+CD38loCXCR4loCD86hi), although the difference was not statistically significant. In contrast to SFR KO mice, SAP KO mice displayed reduced numbers of GC Tfh cells and fully differentiated GC Tfh cells compared with WT mice (Fig. 4, C and D), as reported previously (Cannons et al., 2011; Ma and Deenick, 2011; Shachar et al., 2019; Wu and Veillette, 2016). These defects were corrected in SFR SAP double KO (dKO) mice.
Figure 4.
SFR KO mice display enhanced expansion of the GC reaction. (A–F) As in Fig. 2 B, except that immune cells were detected at the specified times by flow cytometry. The protocol is depicted in A. Tfh cells (B), GC Tfh cells (C), fully committed (Bcl-6hi) GC Tfh cells (D), GC B cells (E), and DZ and LZ GC B cells (F) were enumerated, as specified in Materials and methods. In D, red histograms represent anti–Bcl-6 antibodies, whereas blue histograms represent isotype control antibodies. Bcl-6hi was defined as fluorescence intensity ≥200. For B–D, WT, n = 7; SFR KO, n = 6; SAP KO, n = 6; and SFR SAP dKO, n = 6. For E and F, WT, n = 8; and SFR KO, n = 8. Data are pooled from three independent experiments. (G and H) WT (n = 3) and SFR KO (n = 4) mice were bred with AID-GFP mice, which express the GFP under the control of the AID promoter. The protocol is shown in G. GFP expression in total B cells was determined by flow cytometry (H). Both the percentage of GFP+ B cells (H, middle) and the mean fluorescence intensity (MFI) of GFP in GFP+ B cells (H, right) are depicted. Data are pooled from two independent experiments. Each symbol represents a mouse. *, P < 0.05; **, P < 0.01 (two-tailed Student’s t test). Data are shown as mean ± SEM. For all panels in all figures, representative dot plots, contour plots or histograms are shown on the left, whereas data for multiple independent mice are depicted on the right. Specific populations are boxed or highlighted by horizontal lines. Percentages are shown in the boxes or above the lines. SPC, spleen cells.
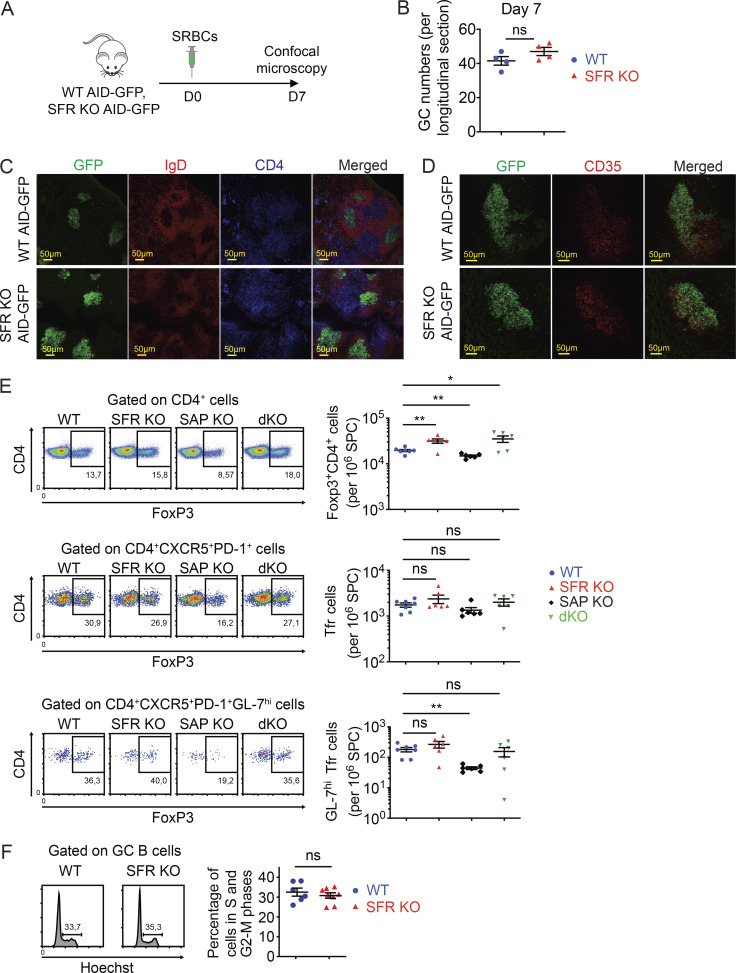
GC B cells were also enumerated using a mouse expressing an activation-induced deaminase (AID) promoter–driven GFP reporter (AID-GFP; Fig. 4 G). As AID expression is induced during the GC reaction (Casellas et al., 2016; DeFranco, 2016; Ise and Kurosaki, 2019; Shlomchik et al., 2019), GC B cells contain much greater GFP fluorescence, as compared with other B cells. SFR KO AID-GFP mice had approximately two- to threefold greater numbers of GFP+ B cells, compared with AID-GFP mice (Fig. 4 H). To examine if the increased numbers of GC B cells in SFR KO mice resulted from an augmentation in the number or structure of GCs, GC numbers and morphology were analyzed by confocal microscopy, after immunization with sheep RBCs (SRBCs), a strong T cell–dependent immunogen (Fig. S2 A). SFR KO mice had GCs of similar numbers and morphology, in comparison to WT mice (Fig. S2, B–D). The GCs of SFR KO mice were not noticeably larger than those of WT mice, although this analysis was limited by the extensive variations in the size of GCs for individual mice.
Figure S2.
Normal GC numbers, morphology, and cell cycle in SFR KO mice. Related to Fig. 2. (A–D) WT (n = 4) and SFR KO (n = 4) mice expressing AID-GFP were immunized i.p. with SRBCs. On day 7, spleen was removed, and cryosections were stained with either anti-IgD (B cells; red) plus anti-CD4 (CD4+ T cells; blue) antibodies (C) or biotin-anti-CD35 antibodies plus streptavidin-PE (red; D). Cells were analyzed by confocal microscopy. GC B cells were identified by detecting GFP (green). The protocol is depicted in A, and GC numbers per longitudinal section are shown in B. Representative photomicrographs of one experiment are provided in C and D. (E) The proportions of T reg cells (FoxP3+CD4+), Tfr cells (FoxP3+CD4+CXCR5+PD-1+), and GC Tfr cells (GL-7hiFoxP3+CD4+CXCR5+PD-1+) in indicated mice were analyzed by flow cytometry (WT, n = 7; SFR KO, n = 6; SAP KO, n = 6; SFR-SAP dKO, n = 6), as for Fig. 4, A–D. Data are pooled from three independent experiments. (F) Mice were immunized with NP-OVA and, on day 7, cell cycle was analyzed by staining cells for Hoechst, which detects DNA. Percentages of GC B cells showing DNA >1n (cells in S-G2-M phases) are enumerated (WT, n = 6; SFR KO, n = 8). Data are pooled from two independent experiments. Each symbol represents one mouse. *, P < 0.05; **, P < 0.01 (two-tailed Student’s t test). Data are mean ± SEM. Scale bar, 50 µM. D, day; ns, not significant.
Given that the basal numbers of T cells, including T reg cells, were slightly increased in SFR KO mice compared with WT mice, we examined if SFR deficiency also had an impact on the numbers of follicular T reg cells (Tfr cells), which can influence the GC reaction. After immunization with NP-OVA, SFR KO displayed a small (1.5-fold) and statistically nonsignificant increase in the proportions of Tfr cells compared with WT mice (Fig. S2 E).
Therefore, the defect in IgM and IgG1 caused by SFR deficiency was not due to a global compromise in the GC reaction. Rather, loss of SFRs resulted in augmented expansion of the GC reaction, resulting in increased numbers of GC B cells. Small, albeit not statistically significant, increases in GC Tfh cells and Tfr cells were also seen.
SFR KO GC B cells display increased proliferation, SHM, and isotype switching
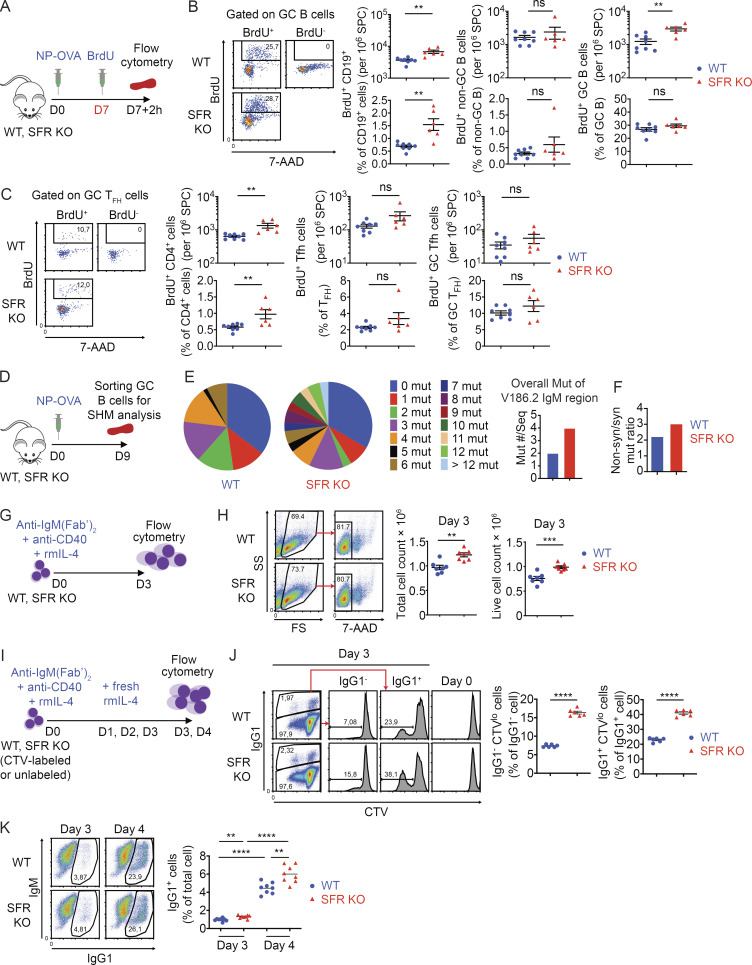
To examine if the increased expansion of the GC reaction in SFR KO mice was due to augmented cell proliferation, mice were immunized with NP-OVA, and proliferation was examined at day 7 by labeling mice with BrdU (Fig. 5 A). Compared with WT mice, SFR KO mice had augmented BrdU incorporation in total B cells (Fig. 5 B). More specifically, SFR KO mice also had higher numbers of BrdU+ GC B cells, but not non-GC B cells, per million splenocytes. When these increased numbers were corrected for the elevated numbers of total GC B cells, the percentages of BrdU+ GC B cells were only slightly increased. Staining of GC B cells with the DNA dyes 7-aminoactinomycin D (7-AAD) and Hoechst showed that the distribution of GC B cells among the various phases of the cell cycle was analogous between SFR KO and WT mice, implying that the cell cycle of SFR KO GC B cells was normal (Fig. 5 B and Fig. S2 F). An increase in BrdU incorporation was also seen in total CD4+ T cells, Tfh cells, and GC Tfh cells, although it reached statistical significance only for total CD4+ T cells (Fig. 5 C).
Figure 5.
SFR KO GC B cells display increased proliferation, SHM, and isotype switching. (A–C) As in Fig. 2 B, except that proliferation was assessed in vivo using BrdU incorporation. The protocol is outlined in A, whereas BrdU incorporation in total B cells (CD19+), non-GC B cells, GC B cells, total CD4+ cells, Tfh cells, and GC Tfh cells is depicted in B and C (WT, n = 8; SFR KO, n = 6). Data are pooled from three independent experiments. Each dot represents one mouse. (D–F) After immunization with NP-OVA, GC B cells were purified from three independent mice of each genotype. mRNA was isolated, cDNAs were synthesized, and the V186.2 region of the IgM-encoding gene was sequenced. The protocol is shown in D, and analyses of the sequencing data are shown in E and F. The proportions of cDNA sequences with the indicated numbers of mutations are depicted by pie charts (E, left). The overall average numbers of mutations per sequence are shown (E, right). The ratios of nonsynonymous over synonymous mutations are shown (F). The numbers of sequences analyzed were WT, n = 65 and SFR KO, n = 70. Data were pooled from two experiments. For statistical analysis of mutation numbers, a χ2 statistical analysis was performed on pooled data comparing the distribution of mutated sequences (i.e., the proportions of mutations with 0, 1, 2, etc. mutations per sequence) between WT and SFR KO mice. As χ2 requires that all expected values be >1.0 and at ≥20% of the expected values be >5, we pooled the more highly mutated sequences (5–10 mutations per sequence) into a bin for each genotype. The calculated P value was 0.0006 and is shown above the pie chart. (G and H) Purified naive B cells were seeded in activation medium containing anti-IgM(Fab’)2, anti-CD40 and recombinant mouse (rm) IL-4 (G). On day 3, total and live cells were enumerated (H). Data are pooled from four independent experiments. Each dot represents one mouse. (I–K) Purified naive B cells from WT (n = 2) or SFR KO (n = 2) mice were labeled or not labeled with CTV and activated in vitro as in G. The protocol is depicted (I). At day 3, CTV-labeled cells were analyzed for dividing cells (CTVlo) in IgG1− and IgG1+ subsets (J). Assays were in triplicate. At day 3 or 4, unlabeled cells were assayed for IgG1+ cells (K). Assays were in quadruplicate. Data are representative of two experiments. Each symbol represents a different well from two independent mice. Data are representative of two experiments. **, P < 0.01; ***, P < 0.001; ****, P < 0.001 (two-tailed Student’s t test). Data are presented as mean ± SEM. D, day; mut, mutations; ns, not significant; seq, cDNA sequences; syn, nonsynonymous.
To test the impact of SFR deficiency on SHM, GC B cells were purified from NP-OVA-immunized mice by cell sorting (Fig. 5 D). RNA was then used as template to sequence the VH186.2 segment of the IgM-encoding gene, which has a high SHM rate in response to NP-OVA (Motoyama et al., 1994). SFR KO GC B cells demonstrated an increased number (approximately twofold) of mutations in the VH186.2 segment, in contrast to WT B cells (Fig. 5 E). In addition, while individual cDNAs from WT B cells had at most six mutations, those from SFR KO B cells carried up to 12 mutations. We also analyzed the ratio of nonsynonymous (not maintaining the same amino acid) over synonymous (maintaining the same amino acid) mutations in the VH186.2 region. Both SFR KO and WT GC B cells displayed a preponderance of nonsynonymous mutations, compared with synonymous mutations (ratios of ∼2.2–3.7; Fig. 5 F). Very few missense or frameshift mutations were seen in either strain.
To assess the effect of loss of SFRs on CSR, naive B cells were purified and activated in vitro using anti-IgM antibodies, anti-CD40 antibodies, and IL-4, which mimic B cell stimulation during the GC reaction (Fig. 5, G and I). Activated SFR KO B cells displayed increased numbers of live B cells compared with WT B cells, as measured by cell counting or Cell Trace Violet (CTV) dilution (Fig. 5, H and J). In addition, SFR KO B cells exhibited augmented isotype switching to IgG1-positive B cells compared with WT B cells (Fig. 5 K). This was seen at day 3 and, to a greater extent, at day 4 after stimulation.
Hence, compared with WT mice, SFR KO mice showed augmented proliferation of purified naive B cells in vitro, as well as elevated numbers of GC B cells in vivo. SFR KO mice also showed increased CSR and SHM. As the chances of SHM and CSR increase with successive GC B cell divisions (Hodgkin et al., 1996), the augmented SHM and CSR were presumably related to enhanced proliferation of SFR KO GC B cells.
SFR KO mice exhibit reduced antigen-specific GC B cells, memory B cells, and plasma cells
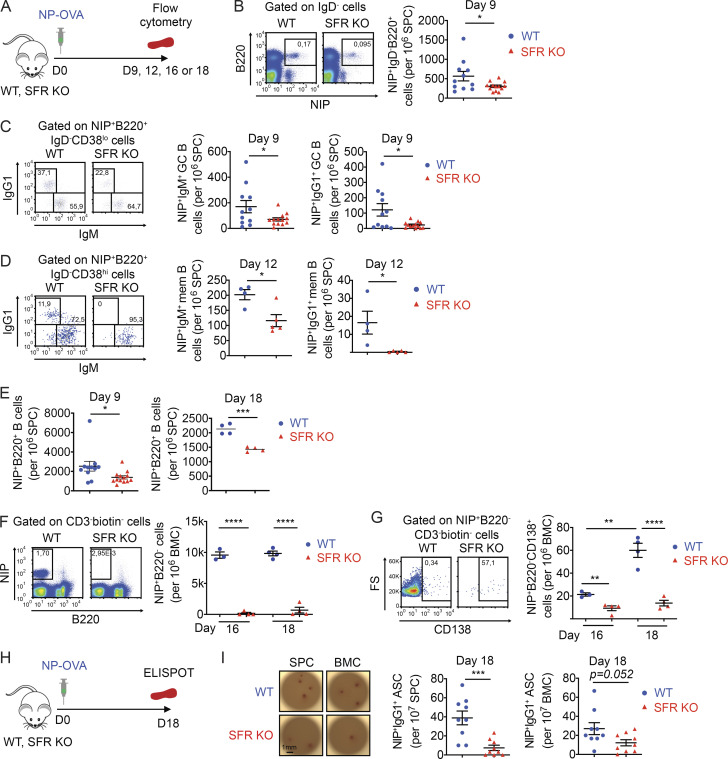
To test if the generation of antigen-specific GC B cells, memory B cells, and plasma cells was compromised in SFR KO mice, NP-specific B cells were enumerated by staining with 4-hydroxy-5-iodo-3-nitrophenyl (NIP), an analogue of NP that binds with high affinity to NP-specific BCRs (Fig. 6 A). At day 9 after immunization, SFR KO mice displayed a reduction of NIP+IgD− switched B cells (by ∼50%), NIP+IgM+ GC B cells (by ∼60%), and NIP+IgG1+ GC B cells (by ∼80%) in spleen when compared with WT mice (Fig. 6, B and C). At day 12, SFR KO mice also had decreased numbers of NIP+IgM+ memory B cells (by ∼40%) and, to a greater extent, NIP+IgG1 memory B cells (by >95%; Fig. 6 D). The extent of the defect in antigen-specific B cells did not increase between days 9 and 18, suggesting that it was stable over time in the short-term (Fig. 6 E). Moreover, at days 16 and 18, there was a marked reduction (>99%) of NIP+B220− B cells and NIP+CD138+B220− cells in the bone marrow of SFR KO mice (Fig. 6, F and G). These cells likely represent NP-specific plasmablasts and plasma cells, respectively.
Figure 6.
SFR KO mice generate fewer antigen-specific GC B cells, memory B cells, and antibody-secreting cells. (A–G) As in Fig. 2 B, except that NP-specific immune cells were detected by flow cytometry using NIP, an analogue of NP. The protocol is depicted in A. NIP+IgD− total B cells (B), NIP+IgM+ or NIP+IgG1+ GC B cells (C), NIP+IgM+ or NIP+IgG1+ memory (mem) B cells (D), and NIP+B220+ cells (E) were identified in spleen. NIP+B220− B cells (F), which represent plasma blasts and plasma cells, and NIP+CD138+B220− B cells (G), which represent plasma cells, were identified in bone marrow cells. For B, C, and E (day 9), WT, n = 11; and SFR KO, n = 13. Data are pooled from four independent experiments (B and C). For D, WT, n = 4; and SFR KO, n = 5. Data are from one experiment. For E (day 18), WT, n = 4; and KO, n = 4. Data are from four experiments (day 9) or one experiment (day 18). For F and G, WT, n = 3 for day 16 and n = 4 for day 18; and SFR KO: n = 4 for days 16 and 18. Data are from one experiment. (H and I) Mice were immunized as in Fig. 2 B, except that NIP-reactive antibody-secreting cells were enumerated in spleen and bone marrow using an ELISpot assay. The protocol is detailed in H, and results are depicted in I. Representative pictures are shown on the left. Scale bar, 1 mm. Data from multiple independent mice are shown on the right (WT, n = 9; SFR KO, n = 9). Data are pooled from three independent experiments. Each symbol represents one mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-tailed Student’s t test). Data are shown as mean ± SEM. ASC, antibody-secreting cell; BMC, bone marrow cell; D, day; FS, forward scatter.
Production of NP-specific antibody-secreting cells was also assessed using an ELISpot assay (Fig. 6 H). At day 18, SFR KO mice exhibited reduced numbers of NIP-specific IgG1-producing cells in spleen (by ∼80%) and, to a lesser extent, bone marrow (by ∼55%) in comparison to WT mice (Fig. 6 I). These cells were presumably plasma cells.
Therefore, while loss of SFRs resulted in a greater expansion of the GC reaction with more frequent SHM and CSR, it compromised the generation of antigen-specific GC B cells, memory B cells, plasmablasts, and plasma cells.
SFRs on T cells and B cells are required for normal antigen-specific B cell response
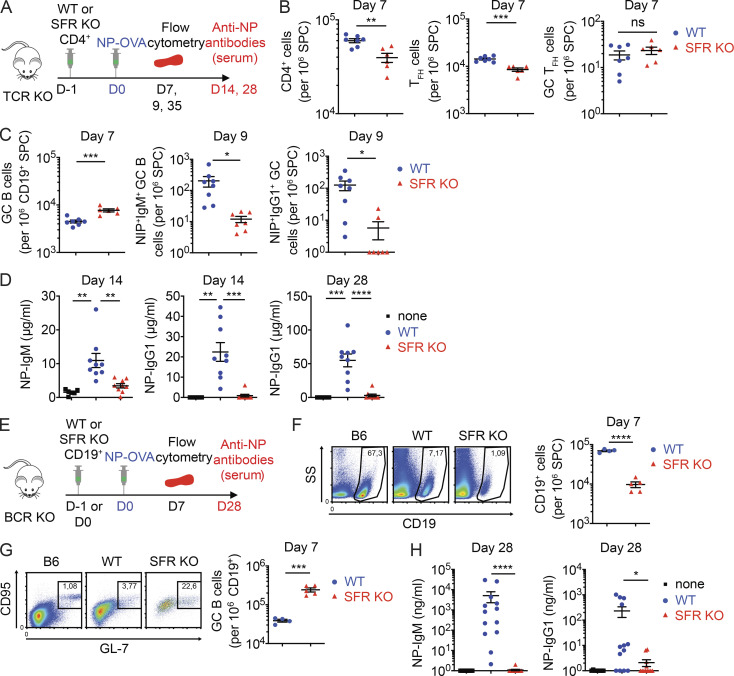
To determine if the defects in the generation of antigen-specific B cells and antibody caused by SFR deficiency occurred as a result of anomalies in T cells, B cells, or both, adoptive transfer experiments were conducted. In a first set of experiments, CD4+ T cells lacking SFRs or not were transferred into SFR-expressing T cell–deficient (TCR KO) mice before immunization of recipient mice with NP-OVA (Fig. 7 A). Compared with mice given WT CD4+ T cells, those receiving SFR KO CD4+ T cells exhibited slightly lower numbers (by ∼30%) of CD4+ T cells and Tfh cells, suggesting a slightly compromised viability of SFR KO CD4+ T cells (Fig. 7 B). However, these mice displayed little or no difference in the proportions of GC Tfh cells. More importantly, mice receiving SFR KO CD4+ T cells displayed an augmented (∼70%) proportion of GC B cells when compared with mice given WT CD4+ T cells (Fig. 7 C). Nevertheless, they had pronounced reductions in NIP+IgM+ GC B cells and NIP+IgG1+ GC B cells (by ∼95%; Fig. 7 C). They also displayed marked diminutions in the levels of NP-specific IgM (by ∼80%) and NP-specific IgG1 (by ∼99%; Fig. 7 D).
Figure 7.
SFRs on T cells and B cells are required for proper GC reaction and T cell–dependent antibody production. (A–D) Purified CD4+ T cells from WT or SFR KO mice were injected into mice expressing SFRs but lacking all T cells (TCR KO mice). Mice were then immunized with NP-OVA and analyzed as detailed in A. Total CD4+ cells, Tfh cells, and GC Tfh cells are enumerated in B. GC B cells, NIP+IgM+ GC B cells, and NIP+IgG1+ GC B cells are enumerated in C. NP-specific IgM and IgG1 are quantified in D. TCR KO mice without injection of any CD4+ cells are represented as “none.” For B and C (day 7), mice receiving WT CD4+ T cells: n = 7; SFR KO CD4+ T cells: n = 6. For C (day 9), mice receiving WT CD4+ T cells: n = 8; and SFR KO CD4+ T cells: n = 7. Data are pooled from three (day 7) or two (day 9) independent experiments. For D, mice receiving no cells (“none”): n = 6; WT CD4+ T cells: n = 9; and SFR KO CD4+ T cells: n = 9. Data are pooled from two independent experiments. (E–H) Purified B cells from WT or SFR KO mice were injected into mice expressing SFRs but lacking all B cells (BCR KO mice). Mice were then immunized with NP-OVA and analyzed as detailed in E. Total B cells (F), GC B cells (G), and NP-specific IgM and IgG1 (H) were quantified. For F and G, mice receiving WT CD19+ B cells: n = 4; and SFR KO CD19+ B cells: n = 5. Data are from one experiment. For H, mice receiving no cells (“none”): n = 8; WT CD19+ B cells: n = 13; and SFR KO CD19+ B cells: n = 12. Data are pooled from three independent experiments. Each symbol represents one mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-tailed Student’s t test). Data are shown as mean ± SEM. D, day; ns, not significant; SPC, spleen cells; SS, side scatter.
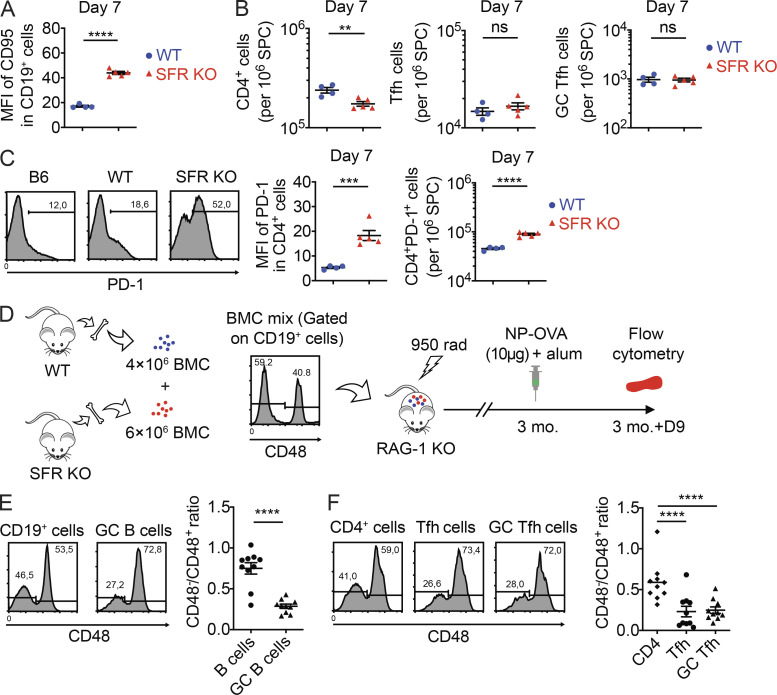
In a second set of experiments, B cells from WT or SFR KO mice were transferred into SFR-expressing B cell–deficient mice (BCR KO mice; Fig. 7 E). At day 7 after immunization with NP-OVA, mice given SFR KO B cells showed a pronounced decrease (∼85%) in the proportions of total B cells compared with mice receiving WT B cells, suggesting a serious compromise in the survival of adoptively transferred SFR KO B cells (Fig. 7 F). Despite this defect, mice provided SFR KO B cells displayed an increased proportion (by approximately sixfold) of GC B cells compared with all B cells in comparison to mice provided WT B cells (Fig. 7 G). The levels of CD95 (Fas) on SFR KO B cells were increased (Fig. S3 A). Furthermore, at day 28, mice having received SFR KO B cells had markedly decreased concentrations of NP-specific IgM and IgG1 (by ∼99%; Fig. 7 H). Although mice given SFR KO B cells did not display a reduced proportions of GC Tfh cells at day 7 after immunization (Fig. S3 B), they had an increase (approximately twofold) in the proportions of PD-1+CD4+ T cells (Fig. S3 C). Most of these PD-1+CD4+ T cells did not have high levels of CXCR5 and GL-7, implying that they were not true Tfh cells.
Figure S3.
SFR deficiency alters the GC reaction and interferes with bone marrow chimeras. Related to Fig. 7. (A–C) Same as Fig. 7, E–G, except that mean fluorescence intensity (MFI) of CD95 on total B cells was measured on day 7 (A); numbers of CD4+, Tfh cells, and GC Tfh cells were determined on day 7 (B); and MFIs of PD-1 on CD4+ cells plus numbers of PD-1+CD4+ cells were analyzed (C; WT, n = 4; SFR KO, n = 5). Data are from one experiment. (D–F) Bone marrow chimeras were created using bone marrow cells from WT (CD48+) and SFR KO (CD48−) mice. The experiment was performed as detailed in D. After reconstitution and immunization, the ratios of SFR KO to WT cell numbers in CD19+ and GC B cells (E) or CD4+, Tfh, and GC Tfh cells (F) were determined. Data are pooled from two experiments. Chimeric mice, n = 10. Each symbol represents one mouse. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (two-tailed Student’s t test). Data are shown as mean ± SEM. BMC, bone marrow cell; D, day; ns, not significant.
Given the loss of SFR KO B cells and, to a lesser extent, T cells over time in the adoptive transfer experiments, we also tested if we could circumvent this problem by making bone marrow chimeras between WT and SFR KO mice (Fig. S3 D). A greater loss of SFR KO B cells and T cells, compared with WT B cells and T cells, was still seen (Fig. S3, E and F). Although this was the case for all B cells and T cells, it was especially true for GC B cells, Tfh cells, and GC Tfh cells.
Thus, absence of SFRs on either T cells or B cells was sufficient to cause a greater expansion of the GC reaction and compromise the production of antigen-specific B cells and antibodies. SFR deficiency also appeared to compromise the survival of adoptively transferred B cells and, to a lesser degree, T cells. Likewise, in the competitive environment created by bone marrow chimeras, loss of SFRs provided a disadvantage for persistence of B cells and T cells.
SFR deficiency augments antigen receptor signal strength in GC Tfh cells
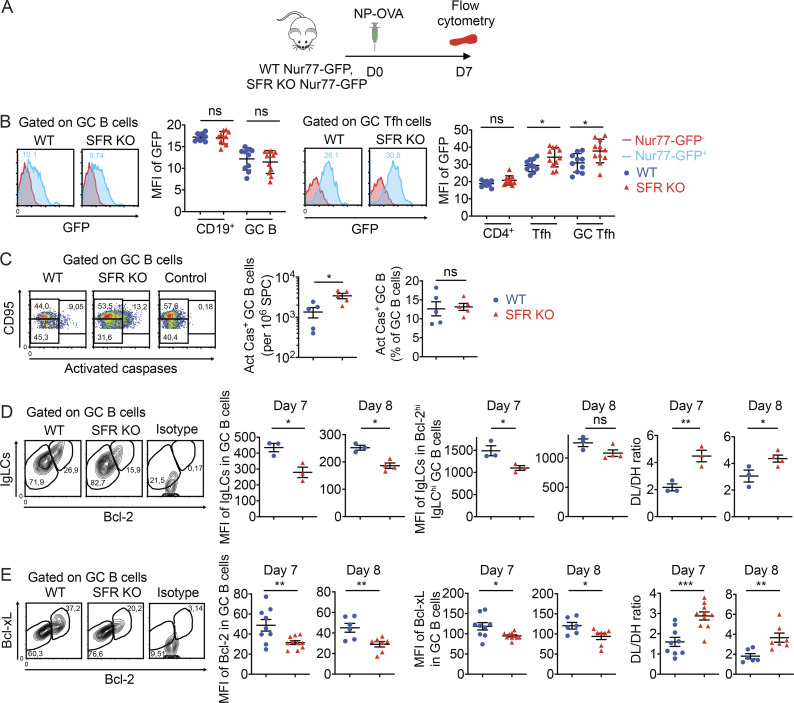
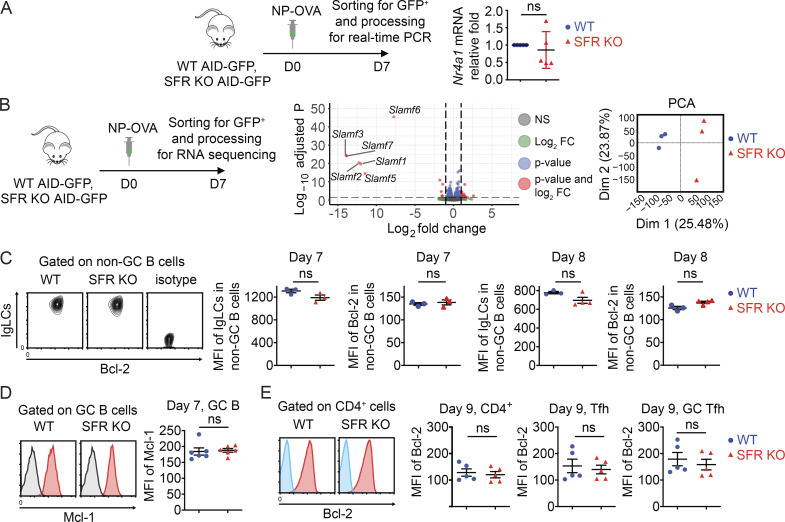
We wanted to understand how SFR deficiency led to an enhanced GC reaction while paradoxically compromising antigen-specific B cell responses. As previous work showed that SFRs inhibited TCR signal strength in invariant natural killer (NK) T cells (NKT cells; Lu et al., 2019), we first studied if SFR deficiency affected antigen receptor signaling intensity. To this end, SFR KO mice were bred with mice expressing a GFP reporter under the control of the promoter of transcription factor Nur77, which is activated in response to TCR- or BCR-triggered signals (Fig. 8 A; Holzapfel et al., 2014). No difference in Nur77-GFP was observed in GC B cells from SFR KO and WT mice (Fig. 8 B). This result was confirmed by real-time PCR (Fig. S4 A). However, an increase (∼20%) in Nur77-GFP was seen in Tfh cells and GC Tfh cells from SFR KO mice compared with WT mice (Fig. 8 B).
Figure 8.
SFRs suppress TCR signal strength in Tfh cells and promote expression of pro-survival effectors in GC B cells. (A and B) WT or SFR KO mice were bred with mice expressing GFP under the control of the Nur77-encoding promoter. Mice were then analyzed as detailed in A. GFP expression in total B cells, GC B cells, total CD4+ cells, Tfh cells, and GC Tfh cells was assessed by flow cytometry (B). Red histograms, Nur77-GFP− control mice; blue histograms, Nur77-GFP+ mice (WT, n = 11; SFR KO, n = 12). Data are pooled from three independent experiments. (C) GC B cells expressing activated caspases were detected by staining with FITC-VAD-FMK (n = 5 mice per genotype). Control was with addition of Z-VAD-FMK. Data are pooled from two independent experiments. (D and E) Mice were immunized as detailed in Fig. 2 B. Expression of IgLCs and Bcl-2 (D) and Bcl-2 and Bcl-xL (E) was determined at days 7 and 8. The mean fluorescence intensities (MFIs) of IgLCs in total GC B cells and Bcl-2hiIgLChi GC B cells were determined (D). DL/DH, ratio of the number of Bcl-2loIgLClo cells to the number of Bcl-2hiIgLChi cells. The MFIs of Bcl-2 and Bcl-xL in total GC B cells were determined (E). DL/DH, ratio of the number of Bcl-2loBcl-xLint cells to the number of Bcl-2hiBcl-xLhi cells. As control, cells were stained with isotype controls. In D, WT, n = 3; SFR KO, n = 3; data are from one experiment representative of three experiments (day 7); n = 3; SFR KO, n = 4; data are from one experiment (day 8). In E, WT, n = 9; SFR KO, n = 10 (day 7); WT, n = 6; SFR KO, n = 7; data are pooled from three (day 7) or two (day 8) independent experiments. Each symbol represents a mouse. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (two-tailed Student’s t test). Data are shown as mean ± SEM. D, day; ns, not significant.
Figure S4.
Augmented expression of Bcl-2 can rescue B cell responses in SFR KO mice. Related to Fig. 8. (A) Same as Fig. 8 B, except that abundance of Nur77-encoding RNA (Nr4a1) was determined by real-time PCR of RNA from sorted AID-GFP+ cells. Relative fold of RNA expression in SFR KO mice over that in WT mice is depicted (WT, n = 5; SFR KO, n = 5). Data are pooled from two experiments. (B) GC B cells were purified from the indicated mice. RNA was then isolated and processed for RNA sequencing. Protocol is depicted on the left. Volcano plot of gene expression differences and statistical significance between WT and SFR KO mice are shown in the middle. Log2 FC and log−10 of the P value of these changes are represented. PCA is depicted on the right. Data are from one experiment. (C) Same as Fig. 8 D, except that non-GC B cells were analyzed. For day 7, WT, n = 3; and SFR KO, n = 3. For day 8, WT, n = 3; and SFR KO, n = 4. Data are from one experiment. (D) Same as Fig. 8 D, except that expression of Mcl-1 in GC B cells was analyzed. Black line, WT cells stained with isotype control. WT, n = 6; SFR KO, n = 6. Data are pooled from two experiments. (E) Mice were immunized with NP-OVA, and on day 9, expression of Bcl-2 was detected in CD4+ T cells, Tfh cells, and GC Tfh cells. Blue line, isotype control. WT, n = 5; SFR KO, n = 5. Data are pooled from two experiments. Each symbol represents one mouse. Data are shown as mean ± SEM. D, day; FC, fold change; ns, not significant (two-tailed Student's t test); PCA, principal-component analysis.
Therefore, SFR deficiency did not result in detectably increased antigen receptor strength in GC B cells. However, an increase was detected in GC Tfh cells.
SFR-deficient GC B cells display reduced expression of anti-apoptotic proteins
Our findings suggested that while GC B cells were generated in greater numbers in SFR KO mice compared with WT mice, these cells were less successful at becoming antigen specific in SFR KO mice. To examine the possibility that SFR KO GC B cells died more frequently, apoptotic GC B cells were detected by intracellular staining for activated caspases. Compared with WT mice, SFR KO mice had increased numbers (∼2.5-fold) of GC B cells expressing activated caspases per million splenocytes, at day 7 after immunization (Fig. 8 C). When this number was corrected for the increased number of total GC B cells, the percentage of caspase+ GC B cells was only slightly increased.
We also conducted RNA-sequencing analyses using purified GC B cells from SFR KO and WT mice (Fig. S4 B). Other than differences in expression of SFR-encoding genes, there was no significant variation in the transcriptomic profile between SFR KO and WT mice, even when nonstringent filters (adjusted P value of <0.05 and |FC| >2) were used (Fig. S4 B). This observation suggested that the alterations in GC B cell biology caused by SFR deficiency were not due to transcriptional changes.
During the GC reaction, the highly proliferating activated GC B cells reduce expression of several molecules, including the BCR itself and anti-apoptotic proteins like Bcl-2 and Bcl-xL, when compared with naive B cells (Hockenbery et al., 1990; Stewart et al., 2018; Tuscano et al., 1996). These changes are believed to facilitate the elimination of GC B cells having BCRs with insufficient affinity for the antigen while preserving GC B cells with high-affinity BCRs. When stained with antibodies detecting the κ and λ Ig light chains (IgLCs) of BCRs, GC B cells displayed two subpopulations of B cells, BCRhi and BCRlo (Fig. 8 D). BCRlo cells also expressed lower levels of Bcl-2. Compared with WT GC B cells, SFR KO GC B cells had a greater proportion of BCRloBcl-2lo cells. This was seen at both day 7 and day 8 after immunization. Levels of BCR and Bcl-2 were higher in non-GC B cells than in GC B cells, as reported previously (Hockenbery et al., 1990; Stewart et al., 2018; Tuscano et al., 1996), and Bcl-2 expression in non-GC B cells was not affected by loss of SFRs (Fig. S4 C).
Upon staining with antibodies recognizing Bcl-2 and Bcl-xL, GC B cells also contained Bcl-2hiBcl-xLhi and Bcl-2loBcl-xLint subpopulations (Fig. 8 E). As was the case for BCRloBcl-2lo cells, the frequency of Bcl-2loBcl-xLint cells was greater in SFR KO mice than in WT mice. Unlike Bcl-2 and Bcl-xL, the levels of Mcl-1, an anti-apoptotic protein that is increased in abundance in activated GC B cells (Vikstrom et al., 2010), were not altered in SFR KO mice when compared with WT mice (Fig. S4 D). Contrary to GC B cells, levels of Bcl-2 were not significantly reduced in SFR KO GC Tfh cells compared with WT GC Tfh cells (Fig. S4 E).
Hence, in comparison to WT B cells, SFR KO GC B cells had reduced expression of BCR and anti-apoptotic proteins Bcl-2 and Bcl-xL, but not of Mcl-1.
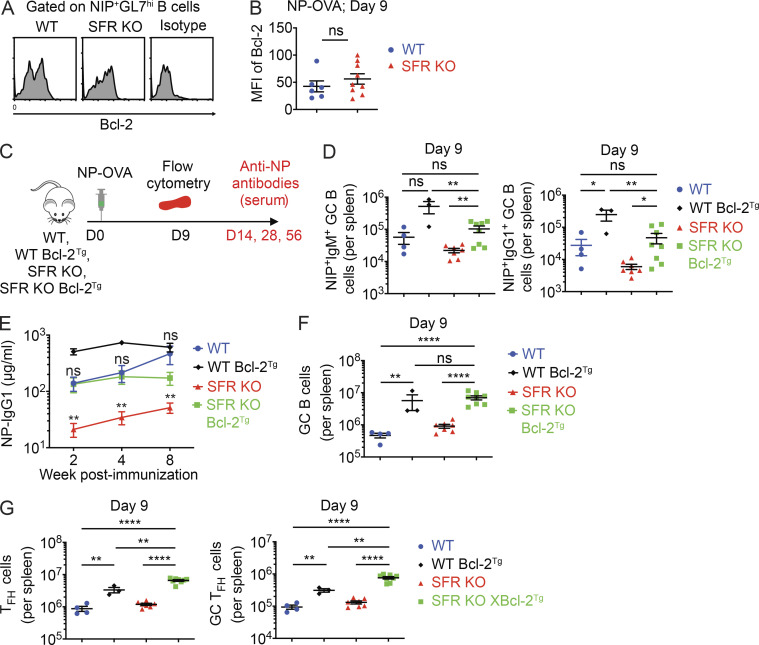
Overexpression of Bcl-2 improves antigen-specific B cell response in SFR KO mice
The results of Fig. 8 suggested that the greater loss of pro-survival factors, in particular Bcl-2, in GC B cells might be responsible for the reduced numbers of antigen-specific B cells in SFR KO mice compared with WT mice. If this were the case, one might expect that the remaining antigen-specific GC B cells developing in SFR KO mice might be “selected” for having higher levels of Bcl-2 compared with total GC B cells from these mice. In keeping with this idea, we observed that unlike total GC B cells (Fig. 8, D and E), NIP+ GC B cells from SFR KO mice had levels of Bcl-2 similar to those seen in WT mice (Fig. 9, A and B).
Figure 9.
Overexpression of Bcl-2 rescues the antibody defect in SFR KO mice. (A and B) Mice were immunized with NP-OVA, and on day 9, expression of Bcl-2 in NIP+CD19+GL7hi cells was analyzed. A sample plot is shown in A, and mean fluorescence intensities (MFIs) of Bcl-2 in multiple mice are shown in B (WT, n = 6; KO, n = 9). Data are pooled from two experiments. (C–G) WT and SFR KO mice were bred or not with mice overexpressing a transgene encoding human Bcl-2. Mice were then analyzed as outlined in C. NIP+IgM+ and NIP+IgG1+ GC B cells were enumerated in spleen (D), while anti-NP IgG1 was measured in serum (E). Numbers of total CD19+CD95+GL7hi GC B cells (F) or Tfh cells and GC Tfh cells (G) were determined. In E, statistics are provided for SFR KO versus SFR KO Bcl-2Tg and for WT versus SFR KO Bcl-2Tg. For D, F, and G, WT, n = 4; WT Bcl-2Tg, n = 3; SFR KO, n = 7; and SFR KO Bcl-2Tg, n = 8. For E, WT, n = 6; WT Bcl-2Tg, n = 6; SFR KO, n = 14; and SFR KO Bcl-2Tg, n = 9. Data were pooled from three experiments. Each symbol represents one mouse. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (two-tailed Student’s t test). Data are shown as mean ± SEM. D, day; ns, not significant; Tg, transgene.
Previous experiments using transgenic overexpression of Bcl-2 lent direct support to the notion that the reduction in Bcl-2 in normal GC B cells plays a role in the rapid death and removal of these cells (Stewart et al., 2018). Thus, to ascertain further if the exaggerated reduction of Bcl-2 expression in SFR KO GC B cells contributed to the reduced amounts of antigen-specific B cells and antibodies in SFR KO mice, SFR KO mice were bred with transgenic mice overexpressing Bcl-2 (Bcl-2 Tg mice; Fig. 9 C). These mice overexpress Bcl-2 in most cell types, including B cells.
Compared with SFR KO mice, SFR KO Bcl-2 Tg mice exhibited increased numbers of NP-specific GC B cells, either IgM+ or IgG1+, as well as augmented levels of anti-NP IgG1, upon immunization with NP-OVA (Fig. 9, D and E). An increase in GC B cell, Tfh cell, and GC Tfh cell numbers was also noted (Fig. 9, F and G). Similar effects were seen when Bcl-2 Tg mice were bred with WT mice, as expected based on published results (Vikstrom et al., 2010; Fig. 9, D–G).
Combined, these findings supported the notion that the more greatly reduced expression of Bcl-2 in GC B cells contributed to the compromised generation of antigen-specific B cells and antibodies in SFR KO mice.
Higher doses of antigen can mask the altered B cell responses caused by SFR deficiency
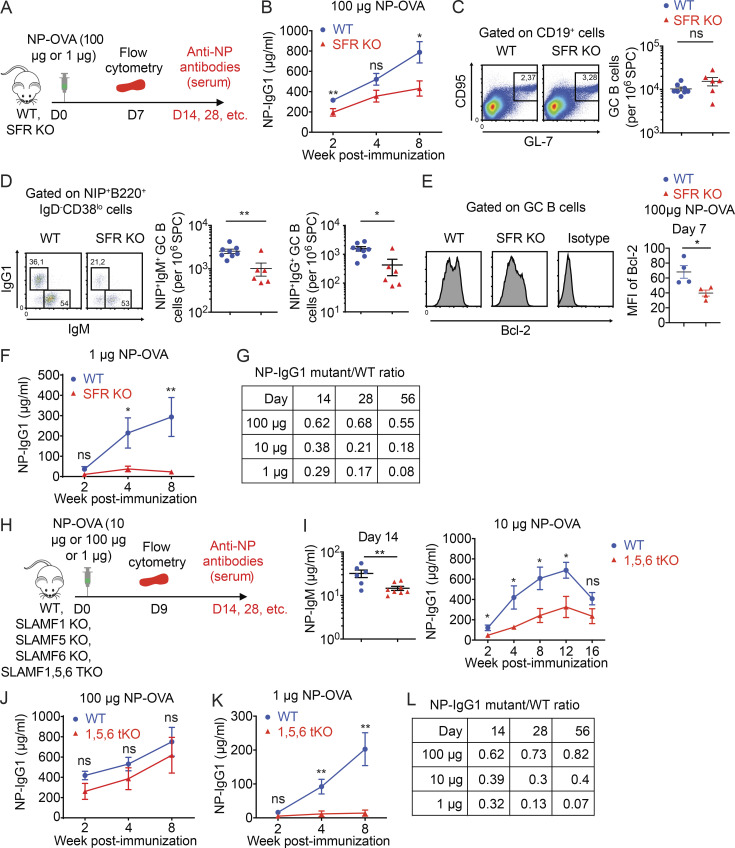
A report of another SFR KO mouse strain failed to document any significant alteration in the GC reaction, in particular in response to OVA immunization (Chen et al., 2017b). However, there were several differences between this study and ours. In the other study, OVA rather than NP-OVA was used, only short-term analyses (day 9) were conducted, antigen-specific B cells were not enumerated, and antibody production was not monitored. Moreover, mice were immunized with a much higher amount of antigen (100 µg OVA), compared with the 10 µg of NP-OVA used herein.
Given the latter distinction, we assessed if the defect in humoral immunity noted in our study could be masked by immunizing with a higher dose of antigen. To this end, mice were immunized with 100 µg NP-OVA (Fig. 10 A). Under this condition, the defect in antibody production seen in SFR KO mice, compared with WT mice, was indeed attenuated (Fig. 10 B). Moreover, the increase in GC B cell numbers caused by SFR deficiency was less marked and became statistically insignificant (Fig. 10 C). Nonetheless, a defect in the generation of NP-specific GC B cells was still seen (Fig. 10 D). Furthermore, the reduced levels of Bcl-2 in GC B cells of SFR KO mice, compared with WT mice, persisted (Fig. 10 E).
Figure 10.
The impact of SFRs on T cell–dependent humoral immunity can be masked by high doses of antigen and involves multiple SFRs. (A–G) Mice were immunized as in Fig. 2 B, except that either a higher (100 µg) or a lower (1 µg) dose of NP-OVA was used. The protocol is shown in A. For the higher dose (100 µg), production of anti-NP IgG1 (B), GC B cell numbers (C), numbers of NP-specific GC B cells (D), and Bcl-2 expression in GC B cells (E) are represented. For the lower dose (1 µg), production of anti-NP IgG1 is depicted (F). A table of the ratio of NP-IgG1 in SFR KO mice compared with WT mice is shown in (G). The data for the 10 µg dose are from Fig. 2 C. (H–K) The indicated mice were immunized as detailed for Fig. 2 B, except that various doses of NP-OVA were used. The protocol is shown in H. For SLAMF1,5,6 tKO compared with WT mice, NP-specific IgM and IgG1 were measured and depicted in I–K (I, 10 µg NP-OVA; J, 100 µg NP-OVA; K, 1 µg NP-OVA). (L) Table showing the ratio of NP-IgG1 in tKO mice compared with WT mice. For B, WT, n = 10; SFR KO, n = 14. For C and D, WT, n = 8; and SFR KO, n = 6. For E, WT, n = 4; SFR KO, n = 4; For F, WT, n = 9; and SFR KO, n = 10. For I, WT, n = 6; and SLAMF1,5,6 tKO, n = 8. For J, WT, n = 9; and SLAMF1,5,6 tKO, n = 7. For K, WT, n = 9; and SLAMF1,5,6 tKO, n = 7. Data are from one experiment (B, E, F, J, and K) or two experiments (C, D, and I). Each symbol represents one mouse. *, P < 0.05; **, P < 0.01 (two-tailed Student’s t test). Data are shown as mean ± SEM. D, day; ns, not significant.
We also tested if using a dose of antigen lower than 10 µg could further accentuate the antibody defect observed in SFR KO mice (Fig. 10 A). When 1 µg NP-OVA was used for immunization, the antibody defect seen in SFR KO mice, compared with WT mice, seemed slightly greater than when 10 µg was used (Fig. 10 F).
Thus, the ability to detect alterations in the GC reaction and antibody production in response to SFR deficiency was influenced by the dose of antigen used for immunization (Fig. 10 G). Higher doses of antigen were able to mask part of these defects.
Multiple SFRs cooperate to promote T cell–dependent antibody production
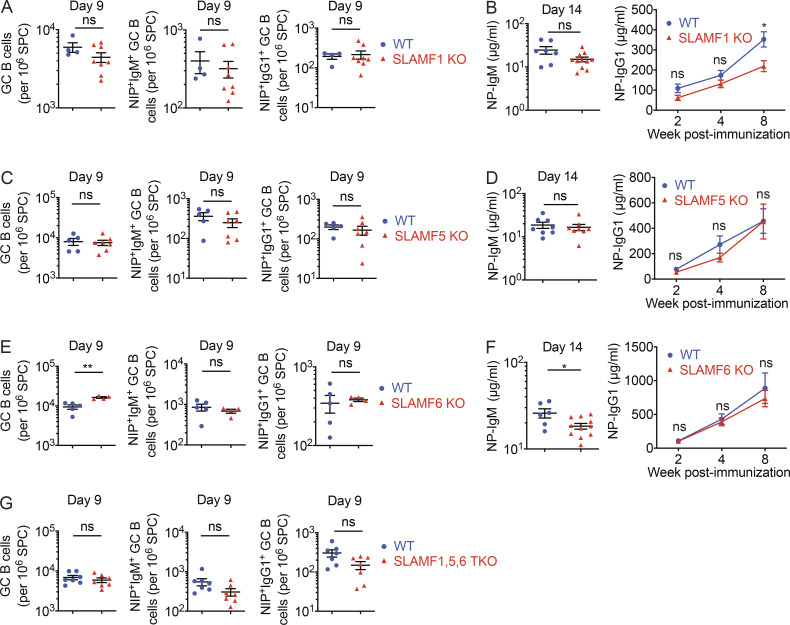
Tfh cells and GC B cells express multiple SFRs, including SLAMF1, SLAMF5, and SLAMF6 (Chen et al., 2017b; Kageyama et al., 2012). To examine whether loss of one or more of these SFRs was responsible for the B cell defects noted herein, mice individually lacking these SFRs were analyzed (Fig. 10 H). When compared with WT mice, mice lacking SLAMF1, SLAMF5, or SLAMF6 alone displayed little or no change in GC B cell numbers, NP-specific GC B cells, and NP-specific antibodies following immunization with NP-OVA (Fig. S5, A–F). The only differences were a small decrease (<50%) in NP-specific antibodies in SLAMF1 KO and SLAMF6 KO mice and a small increase in GC B cells in SLAMF6 KO mice.
Figure S5.
T cell–dependent B cell responses in mice lacking SLAMF1, SLAMF5, or SLAMF6, alone or in combination. Related to Fig. 10. (A–F) Same as Fig. 10 H, except that SLAMF1 KO (A and B), SLAMF5 KO (C and D), and SLAMF6 KO (E and F) were studied. Numbers of GC B cells, NIP+IgM+ GC B cells, and NIP+IgG1+ GC B cells were determined (A, C, and E). Levels of anti-NP IgM and IgG1 were also determined (B, D, and F). (G) Same as Fig. 10 I, except that numbers of GC B cells, NIP+IgM+ GC B cells, and NIP+IgG1+ GC B cells were assessed. For A, WT, n = 4; and SLAMF1 KO, n = 8; data are pooled from two experiments. For B, WT, n = 7; and SLAMF1 KO, n = 11. Data are from one experiment. For C, WT, n = 5; and SLAMF5 KO, n = 7. Data are pooled from two experiments. For D, WT, n = 8; and SLAMF5 KO, n = 8. Data are pooled from two experiments. For E, WT, n = 5; and SLAMF6 KO, n = 4. Data are from one experiment. For F, WT, n = 6; and SLAMF6 KO, n = 11. Data are pooled from two experiments. For G, WT, n = 7; and SLAMF1,5,6 tKO, n = 7. Data are pooled from two experiments. Each symbol represents one mouse. *, P < 0.05; **, P < 0.01 (two-tailed Student’s t test). Data are shown as mean ± SEM. ns, not significant; SPC, spleen cells.
Considering these findings, we also studied mice lacking SLAMF1, SLAMF5, and SLAMF6 (SLAMF1,5,6 triple KO [tKO] mice; Fig. 10 H). Compared with WT mice, SLAMF1,5,6 tKO mice demonstrated clearly compromised production of NP-specific IgM and IgG1 (Fig. 10 I). Whereas these defects were detectable as early as 2 wk after immunization, they were especially obvious at week 12. SLAMF1,5,6 tKO mice also seemed to display a decrease in NP-specific IgM+ GC B cells and IgG1+ GC B cells, although these differences did not reach statistical significance (Fig. S5 G). As was the case for SFR KO mice (Fig. 10 B), the antibody defect seen in SLAMF1,5,6 tKO mice was masked when animals were immunized with the higher dose (100 µg) of NP-OVA but became more prominent with the lower dose (1 µg; Fig. 10, J–L).
Therefore, loss of SLAMF1, SLAMF5, or SLAMF6 alone caused minimal defects in antibody production. However, combined loss of all three SFRs caused a noticeable, albeit partial, defect when compared with loss of all SFRs. This was especially obvious when a lower dose of antigen was used.
Discussion
Herein, we found that mice lacking all SFRs displayed a marked defect in antibody production in response to T cell–dependent antigens, including NP-OVA, HBsAg, and the helminth H. polygyrus. This defect concerned specifically IgM and IgG1, the isotypes most dependent on the GC reaction. The deficit in antibody production was progressive over time and was accentuated after boost immunization. It was accompanied by a defect in affinity maturation. SFR KO mice also had decreased clearance of H. polygyrus in vivo, implying that the antibody defect was physiologically impactful. Altogether, these observations firmly established that SFRs are key regulators of T cell–dependent humoral immunity.
Other groups previously examined the impact of a deficiency in one or more SFRs on T cell–dependent humoral immunity using mouse models but in general saw little or no defects (Chen et al., 2017b; Hu et al., 2016; Huang et al., 2016; Wang et al., 2015; Yusuf et al., 2010). However, there were several differences between these studies and ours. First, the earlier studies solely included short-term analyses (day 7 or 9 after immunization). We observed that the antibody defect of SFR KO mice was progressive over time and best seen at later time points. Second, antigen-specific B cells were not analyzed in the previous studies. We noted that antigen-specific B cells, but not total GC B cells, were prominently reduced in SFR KO mice. Third, the other studies uniformly used much higher doses of antigen (100 µg compared with 10 µg in our case for NP-OVA). We found that higher doses of antigen masked the impact of SFR deficiency on the GC reaction and antibody production. Fourth, there may also be as-yet-unappreciated effects due to differences in mouse genetic background or microbiome.
Despite having a marked defect in antibody production, SFR KO mice displayed an augmentation in the number of GC B cells during the GC reaction compared with WT mice. As the number of anatomical GCs in spleen was not obviously increased, this augmentation was likely caused, at least in part, by a greater expansion of individual GCs, although the marked variation in the size and shape of individual GCs makes a definitive determination regarding this matter difficult. SFR KO mice also had increased B cell proliferation, as measured by BrdU incorporation and in vitro proliferation. There was also an increased frequency of SHM in vivo and isotype switching to IgG1 in vitro. SHM and isotype switching occur more frequently when B cell proliferation is augmented (Cyster and Allen, 2019; DeFranco, 2016; Ise and Kurosaki, 2019; Shlomchik et al., 2019).
Adoptive transfer experiments showed that loss of SFRs on T cells or B cells was sufficient to cause a reduction in antibody levels, as well as a greater expansion of the GC reaction. These findings were consistent with the notion that SFRs controlled antibody production largely by influencing the GC reaction. In all likelihood, SFRs on Tfh cells were engaging SFRs on GC B cells and vice versa. This mutual triggering led to bidirectional signaling in Tfh cells and GC B cells that restrained the GC reaction but also promoted the generation of antigen-specific B cells. However, it is likely that part of the effect of SFRs on antibody production also occurred in a B cell–intrinsic manner, independently of T cells. In support of this possibility, purified SFR KO B cells had increased proliferation and isotype switching in vitro when compared with WT B cells.
As was the case for conventional T cells, the numbers of Tfr cells were slightly increased in SFR KO mice compared with WT mice. This finding raised the possibility that augmented Tfr cell–dependent functions might contribute to the altered generation of antigen-specific B cells and antibodies observed in SFR KO mice. Although future studies are needed to examine this idea, it seems unlikely to be the case for several reasons. First, the increase in Tfr cell numbers was rather small (1.5-fold). Second, expansion of the GC reaction was enhanced, rather than diminished, in SFR KO mice. Third, adoptive transfer of SFR KO B cells in mice where T cells expressed SFRs re-created the defects in generation of antigen-specific B cells and antibodies.
Using a Nur77-GFP reporter mouse, we determined that TCR signal strength was enhanced in GC Tfh cells of SFR KO mice compared with WT mice. The magnitude of this increase was similar to that previously noted in SFR KO invariant NKT cells (iNKT cells) compared with WT iNKT cells (Lu et al., 2019). This increase in activation of SFR KO GC Tfh cells was likely responsible, at least in part, for the greater activation of SFR KO GC B cells. However, it is plausible that activation of SFR KO GC B cells was also enhanced in part due a B cell–intrinsic effect, as proliferation of purified SFR KO B cells was augmented upon activation in vitro. Nevertheless, we did not observe an increase in Nur77-GFP or Nur77-encoding RNA in SFR KO GC B cells. Therefore, SFRs were presumably inhibiting other pathways in GC B cells, either other downstream effectors of BCR signaling or BCR-independent pathways.
The lack of significant transcriptional changes in SFR KO GC B cells, compared with WT GC B cells, may seem surprising. It presumably indicates that the changes in the biology of GC B cells in SFR KO mice are likely to be due to posttranscriptional alterations. Such a feature may make sense, as transcriptional changes leading to altered steady-state RNA levels are typically slow. For rapid changes to occur, as needed during the GC reaction, it may be more efficient to regulate proteins at the posttranslational level, through processes such as phosphorylation, ubiquitination, internalization, degradation, or change in plasma membrane distribution.
Given the low abundance of GC Tfh cells and GC B cells in vivo, it is difficult to conduct biochemical studies to identify the immediate downstream effectors of SFRs in these cells. However, in other systems such as iNKT cells and NK cells, SFR-mediated inhibition was mediated by SHP-1 or SHIP-1 (Dong et al., 2012; Guo et al., 2016; Lu et al., 2019). Thus, similar SFR effectors were possibly at play in GC Tfh cells and GC B cells. Interestingly, a previous report showed that SHP-1 and SHIP-1 were hyperphosphorylated and presumably more active in GC B cells compared with naive B cells (Khalil et al., 2012). Moreover, selective elimination of SHP-1 expression in B cells led to severely compromised generation of antigen-specific GC B cells, in a manner analogous to SFR deficiency as reported herein (Khalil et al., 2012). Thus, SFRs may play a role in the recruitment and activation of SHP-1 and, perhaps, SHIP-1 in GC B cells.
Despite the increased expansion of their GC reaction, SFR KO mice exhibited a decrease in the numbers of antigen-specific B cells, including GC B cells, memory B cells, and plasma cells. GC B cells from SFR KO mice also had compromised expression of BCR, Bcl-2, and Bcl-xL. Although expression of these pro-survival effectors is normally down-regulated in activated GC B cells compared with naive B cells, such a down-regulation was accentuated in SFR KO mice compared with WT mice. This effect was likely detrimental to the survival of antigen-specific SFR KO GC B cells. Support for this notion was adduced by the observation that the reduced numbers of antigen-specific GC B cells that were able to develop in SFR KO mice had normal levels of Bcl-2 compared with WT mice. Moreover, transgenic overexpression of Bcl-2 was able to rescue the number of antigen-specific GC B cells and the antibody levels in SFR KO mice. While the interpretation of the latter experiment is limited by the fact that similar effects were seen when Bcl-2 transgenic mice were bred with WT mice, as expected, the finding was nonetheless supportive of our hypothesis. Clearly, SFR KO GC B cells with higher levels of Bcl-2 had a greater capacity to react with an immunizing antigen.
The alterations in the GC reaction and antibody production seen in SFR KO mice significantly differed from those noted in SAP KO mice. First, unlike SFR KO mice, SAP KO mice displayed a near-complete absence of GC Tfh cells, GC B cells, and anatomical GCs (Cannons et al., 2010; Chen et al., 2017b; Kageyama et al., 2012; Qi et al., 2008; Veillette et al., 2008; this report). Second, compared with SFR KO mice, SAP KO mice had a much more severe defect in antibody production that affected all isotypes of IgG. Third, breeding of SAP KO mice with SFR KO mice indicated that the more severe defects observed in SAP KO mice were not due to a loss, but rather to a change, in the function of SFRs, which become superinhibitory in the absence of SAP (Chen et al., 2017b; Kageyama et al., 2012; this report). As Tfh cells, but not GC B cells, express SAP (Veillette et al., 2008), loss of SAP presumably prevented the GC reaction by causing SFRs to abrogate TCR signaling in Tfh cells.
Our analyses of mouse strains lacking combinations of SFRs indicated that the coordinated efforts of multiple members of the SLAM family were required to restrain the expansion of the GC reaction and promote T cell–dependent antibody production. These included SLAMF1, SLAMF5, and SLAMF6, although other members of the family were also likely implicated. Along these lines, Tfh cells and GC B cells express two other SFRs, SLAMF7 and Ly-9, in addition to CD48, a distant relative of SFRs (Chen et al., 2017b). All these receptors were absent in our SFR KO mouse strain. While defects in antibody production have not been reported in mice lacking SLAMF7, Ly-9, or CD48, we presume that one or a combination of these other receptors contributed to the regulation of the GC reaction and antibody production.
Put together, our findings suggest the following model. During the initial stimulation of Tfh cells by antigen and APCs, SFRs suppress TCR signal strength in Tfh cells in order to prevent excessive activation of Tfh cells. This inhibitory effect in turn limits the activation of GC B cells. The reduced activation of GC B cells then attenuates the normal loss of pro-survival effectors such as BCR, Bcl-2, and Bcl-xL in these cells, until such a time that the hypermutated BCRs become expressed in sufficient quantities to allow survival and selection of GC B cells. Thus, we postulate that by controlling the GC reaction, SFRs augment the generation of antigen-specific B cells and antibodies. In addition to their effects on the GC reaction, SFRs are likely to act via a B cell–intrinsic effect, which promotes the survival of antigen-specific B cells. Although future studies will be needed to dissect this role, another study linked SLAMF5 to a B cell–intrinsic survival mechanism in leukemic B cells (Barak et al., 2020).
In combination with previous findings (Khalil et al., 2012; Luo et al., 2019; Shlomchik et al., 2019; Stewart et al., 2018), our data highlight the importance of maintaining low levels of signaling in GC B cells. This may seem paradoxical, given that GC B cells are highly proliferative and undergo very active processes such as SHM and CSR. Nonetheless, we posit that such an inhibitory environment has a dual purpose. On the one hand, it is critical to control selection of antigen-specific GC B cells, as proposed elsewhere (Khalil et al., 2012; Luo et al., 2019; Shlomchik et al., 2019; Stewart et al., 2018). Only GC B cells expressing BCRs of sufficiently high affinity are able to generate enough activating signals to survive. On the other hand, as alluded to here, this inhibitory environment allows GC B cells to lose to a lesser extent the expression of pro-survival molecules. Hence, as a consequence, GC B cells become less susceptible of dying while awaiting acquisition of their newly created high-affinity BCR.
Materials and methods
Mice
Mice lacking all SFRs (SFR KO), SAP KO mice, SFR SAP dKO mice, SLAMF1,5,6 tKO mice, and Slamf6 bacterial artificial chromosome (BAC) transgenic mice have been described elsewhere (Chen et al., 2017a; Guo et al., 2016; Lu et al., 2019). SFR KO mice were backcrossed to C57BL/6 (B6) mice for up to 20 generations. Independently of the extent of backcrossing, SFR KO mice displayed a stable phenotype. Slamf1 gene–disrupted mice were generated by CRISPR-Cas9–mediated genome editing using the plasmid pSpCas9 (BB; formerly pX330; Addgene) and the guide RNA sequence 5′-GCCTGTTCCCGAGGATCTTC-3′. DNA was injected into fertilized oocytes of C57BL/6J mice. C57BL/6.Cg-Slamf1em1Av mice bearing an 11-bp deletion and C57BL/6.Cg-Slamf1em2Av bearing a 7-bp deletion in exon 2 of Slamf1 were identified and bred to homozygosity. Either deletion resulted in a frameshift in the second coding exon and caused loss of SLAMF1 expression by flow cytometry. The following mice were obtained from the Jackson Laboratory: RAG-1 KO mice, TCR αβ/γδ-deficient (TCR KO) mice, Igµ-deficient (BCR KO) mice, SLAMF5 KO (Slamf5−/−) mice, SLAMF6 KO (Slamf6−/−) mice, and Nur77-GFP transgenic mice (C57BL/6-Tg(Nr4a1-EGFP/cre)820Khog/J). AID-GFP reporter mice were obtained from Dr. R. Casellas (National Institutes of Health, Bethesda, MD; Crouch et al., 2007). H2-K-BCL-2 transgenic mice, which ubiquitously overexpress human Bcl-2, were provided by Dr. T. Möröy (Institut de recherches cliniques de Montréal, Montréal, Canada; Kondo et al., 1997). All mice were maintained in the B6 background and kept in a specific pathogen–free environment. Males and females between 3 and 6 mo of age were used for experiments. Littermates were used, except for the experiments comparing Bcl-2 Tg mice with SFR KO Bcl-2 Tg mice and comparing AID-GFP mice with SFR KO AID-GFP mice. All studies were performed in a nonblinded fashion. Animal experimentation was approved by the Animal Care Committees of the Institut de recherches cliniques de Montréal or McGill University and performed as defined by the Canadian Council of Animal Care.
Immunization with T cell–dependent antigens
Mice were injected i.p. with 200 µl of a suspension containing 50% volume of immunogen solution and 50% volume of alum (Thermo Fisher Scientific; catalog no. 77161). The immunogens were: NP(18)-OVA (Biosearch Technologies; catalog no. N-5051-100) and recombinant hepatitis surface antigen (HBsAg; ProSpec; catalog no. HBS-872-3). The dose of immunogen was 10 µg per mouse, unless otherwise indicated. For immunohistology of GCs in spleen, SRBCs (Innovative Research; IC10-0210-15ML) were washed and resuspended in PBS and then injected i.p. at 250 × 106 cells per mouse (in 0.2 ml).
H. polygyrus infection
Infections were performed as described (King and Mohrs, 2009). In brief, mice were infected by gavage with 200 L3-stage H. polygyrus bakeri larvae diluted in sterile water. At week 4 after infection, some mice were treated orally with the anti-helminthic antibiotic Pyrantel (10 mg/ml) to clear the primary infection. For this group, mice were reinfected at week 6 after primary infection with the same dose of H. polygyrus bakeri. At day 14 after primary or secondary infection, mice were bled to measure HES antigen–specific antibodies. Moreover, mice were sacrificed and adult worms were counted in small intestine. Eggs were also enumerated in feces.
Antibody production assays
Antibody levels in serum were measured by ELISA, as described previously (Zhong and Veillette, 2013a; Zhong and Veillette, 2013b). To determine total IgM, IgG1, and IgE in nonimmunized mice, 100 µl unlabeled goat anti-mouse IgH+L (Southern Biotechnology; catalog no. 1010–01) or unlabeled rat anti-mouse IgE (BD Bioscience; catalog no. 553413, clone R35-72) was coated at 2 µg/ml on the ELISA plate overnight. To detect antigen-specific antibodies, 100 µl antigen dissolved in PBS (2 µg/ml) was coated. The coating antigens were NP(4)-BSA (Biosearch Technology; catalog no. N-5050-10), NP(30)-BSA (Biosearch Technology; catalog no. N-5050H-10), HBsAg (ProSpec; catalog no. hbs-872, lot no. 717PHBSAG23) and HES (prepared in Dr. I. King’s laboratory). The plate was washed with PBS containing 0.05% Tween 20, followed by blocking for 1.5 h with 1% BSA dissolved in PBS. For all ELISAs, serum at different dilutions was then added in duplicates to the wells and incubated at room temperature for 2 h. After thoroughly washing, 100 µl of HRP-conjugated secondary antibody against mouse IgM, IgG1, IgG2b, IgG2c, or IgG3 (Southern Biotechnology; catalog no. 1020–05, 1070–05, 1090–05, 1079–05, 1100–05, respectively) was added and incubated at room temperature for 1 h. After more washes, wells were incubated with 100 µl HRP substrate (R&D Systems; catalog no. DY999) at room temperature for 20 min, and the reaction was stopped by adding 50 µl of 2N H2SO4. The OD450 and OD540 were read with an ELISA reader. A standard curve was done for each ELISA plate. For IgE, the wells were incubated with biotin-rat anti-mouse IgE (BD Bioscience; catalog no. 553419, clone 35–118), followed by HRP-conjugated streptavidin and substrate. To assess affinity maturation, ELISA were performed either with NP(4) in the presence of 1 M NaSCN (Fluka Biochemica; catalog no. 80518) or NP(30) as capture antigens. For the assays using NaSCN, serial dilutions of serum were first incubated for 2 h in NP(4)-coated wells. Then, 150 µl of 1 M NaSCN solution was added for 15 min to the wells. The wells were then washed, and assays were subsequently performed according to standard procedures. To detect anti-dsDNA antibodies, plates were coated with 100 µl salmon sperm DNA (10 µg/ml in 0.02 mol/liter carbonate buffer, pH 9.6; Sigma-Aldrich; catalog no. D-1626).
Cell isolation and purification
For splenocyte isolation, single-cell suspensions were obtained and centrifuged at 1,500 rpm at 4°C for 10 min. After adding 1 ml red blood cell lysis buffer (Sigma-Aldrich; R7757) to the cell pellet for 5 min at room temperature, 10 ml ice-cold PBS containing 2% FBS was added, and cells were centrifuged for 10 min at 1,500 rpm (4°C). Cell pellets were washed again to remove residual lysis buffer. Finally, cell pellets were resuspended in 10 ml ice-cold PBS with 2% FBS and filtered through a 40-µm cell strainer. For purification of CD4+ T cells or B cells, splenocytes were prepared in 4 ml ice-cold complete medium and transferred to a 15-ml tube containing 4 ml Lympholyte (Cedarlane; catalog no. CL5030). After centrifugation at 2,600 rpm for 15 min (room temperature), the upper layer of lymphocytes was carefully collected and transferred to a new tube. After washing twice with complete medium, the cells were resuspended in 1 ml and processed for further purification according to the manufacturer’s protocols. For isolation of bone marrow cells, tibia and femur were dissected. Cavities were then flushed with 5 ml ice-cold PBS containing 2% FBS. After centrifugation at 4°C, cell pellets were treated with red cell lysis buffer, as described for splenocytes. Then, cells were resuspended in 5 ml PBS containing 2% FBS and passed through a cell strainer. For isolation of peritoneal cells, 5 ml ice-cold PBS supplemented with 2% FBS and 1 mM EDTA was injected into the peritoneal cavity of euthanized mice. The abdomen was then gently massaged for 2 min to dislodge attached cells. Peritoneal cells were collected using a syringe. Finally, the skin was cut, and the remaining liquid in the cavity was collected. Cells were then centrifuged at 1,500 rpm for 10 min (4°C). The cell pellets were resuspended in 5 ml PBS with 2% FBS and passed through a cell strainer.
Flow cytometry
Spleen or bone marrow cells were harvested and treated with red cell lysis buffer at room temperature for 5 min to deplete RBCs. After washing and passing through a cell strainer, cells were counted and aliquoted and then blocked with anti-CD16/32 mAb 2.4G2 hybridoma supernatant. Typically, 5 × 106 cells were used for staining, with the exception of staining for NIP+ GC B cells, when 7 × 106 cells were used. In all cases, cells were stained with 7-AAD (BioLegend; catalog no. 420404) to exclude dead cells from the analyses.
For identification of T cell subsets, splenocytes were stained with anti-CD44-PE, anti-CD62L-PE-Cy7, anti-CD8-Pacific Blue, and anti-CD4-APC. Subpopulations of T cells (CD4+, CD8+, naive [CD62LhiCD44−], central memory [CD62LhiCD44+], and effector-memory [CD62L−CD44+]) were then identified. For Tfh cell subsets, splenocytes were stained with anti-CD4-PE-Cy7, anti-PD-1-PE, anti-CXCR5-biotin plus streptavidin-APC and anti-GL-7-Pacific Blue. Tfh cells were identified as CD4+PD-1+CXCR5+, while GC Tfh cells were identified as CD4+PD-1+CXCR5+GL-7+. For identification of fully polarized GC Tfh cells, splenocytes were stained with anti-CD4-PE-Cy7, anti-GL7-FITC, anti-PD-1-PE, anti-CXCR5-biotin plus streptavidin-eFluor450, and anti-Bcl-6-Alexa647. Fully polarized GC Tfh cells were identified as Bcl-6hiCD4+PD-1+CXCR5+GL-7+. Bcl-6hi was defined as fluorescence intensity of ≥200. For identification of T reg cells, splenocytes were stained with anti-CD4-FITC and anti-FoxP3-eFluor660. T reg cells were identified as CD4+FoxP3+. For identification of Tfr cells, splenocytes were stained with anti-GL-7-FITC, anti-PD-1-PE, anti-CD4-PE-Cy7, anti-CXCR5-biotin + streptavidin-eFluor450, and anti-FoxP3-eFluor660. Tfr cells were identified as CD4+PD-1+CXCR5+FoxP3+, and GC Tfr cells were CD4+PD-1+CXCR5+GL-7hi FoxP3+.
For identification of B cell subsets in spleen, splenocytes were stained with anti-CD19-PE-Cy7, anti-CD95-Alexa647, and anti-GL-7-Pacific Blue. Total B cells were identified as CD19+, whereas GC B cells were identified as CD19+CD95+GL-7hi. For identification of marginal zone (MZ), transitional 1, transitional 2, and follicular B cells, splenocytes were stained with anti-CD19-PE-Cy7, anti-IgM-eFluor450, anti-IgD-APC-eFluor780, anti-CD21/35-FITC, anti-CD23-PE, and anti-CD1d-APC. MZ B cells were CD19+CD1dhiCD21/35hi, T1 B cells were CD19+CD1dloCD21/35loCD23−CD21lo, T2 B cells were CD19+CD23+CD21/35medIgDhiIgMhi, and follicular B cells were CD19+CD23+CD21/35medIgDhiIgMlo.
For identification of B cell subsets in bone marrow, cells were stained with anti-B220-PE-Cy7, anti-GL-7-FITC, anti-IgM-eFluor450, anti-IgD-APC-eFluor780, and a cocktail of biotinylated antibodies (anti-CD3, anti-Thy1.2, anti-Gr-1, anti-F4/80, anti-Ter119, anti-DX5, and anti-NK1.1) plus streptavidin-APC. The biotin−B220+ population was analyzed to identify pre–B cells (B220+IgM−IgD−), immature B cells (B220+IgM+IgD−), and mature B cells (B220+IgM+IgD+).
For the peritoneal cavity, cells were stained with anti-B220-FITC, anti-CD19-PE-Cy7, anti- CD23-PE, anti-IgM-eFluor450, anti-IgD-Alexa647, and anti-CD5-eFluor780. B-1 cells were CD19+B220dimCD23lo, B-2 cells were CD19+B220hiCD23hi, B-1a cells were CD19+B220dimCD23loCD5+, and B-1b B cells were CD19+B220dimCD23loCD5−.
For identification of DZ and LZ GC B cells, splenocytes were stained with anti-B220-PE-Cy7, anti-CD95-FITC, anti-GL-7-Pacific Blue, anti-CD38-PE, anti-CXCR4-APC, and anti-CD86-APC-Cy7. DZ GC B cells were CD19+CD95+GL-7+CD38loCXCR4hiCD86lo, while LZ GC B cells were CD19+CD95+GL-7+CD38loCXCR4loCD86hi.
To detect NP-specific B cells, splenocytes were stained with anti-B220-PE-Cy7, anti-IgM-eFluor 450, anti-IgG1-FITC, anti-IgD-APC eFluor780 and anti-CD38-PE in the presence of NIP(5)-BSA-Alexa647. NP-specific GC B cells were NIP+IgD−IgM+B220+CD38lo or NIP+IgD−IgG1+B220+CD38lo. NP-specific memory B cells were NIP+IgD−IgM+B220+CD38hi or NIP+IgD−IgG1+B220+CD38hi. For plasma cells, spleen or bone marrow cells were stained with NIP(5)-BSA-Alexa647, anti-B220-PE-Cy7, anti-CD138-BV421, anti-CD3-FITC, and biotinylated antibodies against Thy1.2, Gr-1, F4/80, Ter119, DX5, and NK1.1 plus streptavidin-PE as above. The biotin−CD3− population was analyzed for expression of B220 and NIP binding. Plasmablasts and plasma cells were NIP+B220−. The NIP+B220− population was further analyzed for expression of CD138. Plasma cells were NIP+B220−CD138+.
To analyze expression of BCR, Bcl-2, Bcl-xL, and Mcl-1 in GC B cells and non-GC B cells, splenocytes were stained with anti-CD19-PE-Cy7, anti-CD95-biotin + streptavidin-eFluor450, and anti-GL-7-FITC in the presence of anti-Ig kappa-PE + Ig lambda-PE, anti-Bcl-2-Alexa647, anti-Bcl-xL-PE, and anti-Mcl-1-AF647, alone or in combination. Isotype controls were used. GC B cells and non-GC B cells were gated as CD19+CD95+GL-7hi and CD19+CD95−GL-7lo, respectively. Expression of BCR, Bcl-2, Bcl-xL, and Mcl-1 was analyzed.
To determine Bcl-2 expression in antigen-specific GC B cells, splenocytes were stained with anti-CD19-PE-Cy7, anti-GL7-Pacific Blue, NIP(5)-BSA-Alexa647, and anti-Bcl-2-PE. Antigen-specific GC B cells were identified as NIP+CD19+GL7hi.
To detect activated caspases, spleen cells were incubated for 1 h at 37°C in culture medium supplemented with FITC-VAD-FMK (BioVision; catalog no. K180-100). Cells were then washed twice, resuspended in 100 µl PBS containing 1% BSA supplemented with 0.2 µg mAb 2.4G2 (BD Biosciences; catalog no. 553142), and incubated on ice for 20 min. Cells were then stained with anti-CD19-PE-Cy7, anti-GL-7-Pacific Blue, and anti-CD95-Alexa647 to identify GC B cells.
For cell cycle analysis, spleen cells were incubated for 30 min at 37°C in culture medium containing Hoechst 33342 (1 µg/ml; Thermo Fisher Scientific; catalog no. 62249). They were then processed for flow cytometry.
For BrdU labeling, a kit from BioLegend (catalog no. 370704) was used. On day 7 after immunization, mice were injected i.p. with 1 mg BrdU (in 0.1 ml). After 2 h, mice were sacrificed and spleen cells were stained with markers for GC B cells (CD19-PE-Cy7, CD95-Alexa647, and GL-7-BV421) or GC Tfh cells (CD4-PE-Cy7, PD-1-PE, CXCR5-APC, and GL7-BV421). Cells were then fixed and processed for staining with anti-BrdU antibodies and 7-AAD, as detailed by the manufacturer.
Antibodies and conjugates
The following antibodies or reagents were from eBioscience: biotin-anti-mouse CD3 (clone 145-2C11), PE-Cy7-anti-B220 (clone RA3-6B2), biotin-anti-mouse CD49b (clone DX5), eFluor780-anti-mouse CD5 (clone 53–7.3), PE-anti-mouse CD38 (clone 90), biotin-anti-mouse CD90.2 (clone 30-H12), biotin-anti-mouse CD93 (clone AA4.1), biotin-anti-mouse NK1.1 (clone PK136), biotin-anti-mouse Ly6G (clone RB6-8C5), biotin-anti-mouse F4/80 (clone BM8), biotin-anti-mouse TER119 (clone TER119), eFluor450-anti-mouse IgM (clone eB121-15F9), APC-eFluor780-anti-mouse IgD (clone 11-26C), PE-anti-mouse PD-1 (clone J43), FITC-anti-mouse CD21/35 (clone 4E3), PE-anti-mouse CD23 (clone B3B4), APC-anti-mouse CD1d (clone 1B1), APC-anti-mouse CXCR4 (clone 2B11), eFluor660-anti-FoxP3 (clone FJK-16s), APC-streptavidin, and eFluor450-streptavidin.
The following antibodies were from BD Bioscience: FITC-anti–T and B cell activation antigen (clone GL-7), BV421-anti–T and B cell activation antigen (clone GL-7), biotin-anti-mouse CXCR5 (clone 2G8), Alexa647-anti-Bcl-6 (clone K112-91), FITC-anti-mouse IgG1 (clone A85-1), biotin-anti-mouse IgG1 (clone A85-1), BV421-anti-mouse CD138 (clone 281–2), Alexa647-anti-mouse CD95 (clone Jo2), FITC-anti-mouse CD95 (clone Jo2), and biotin-anti-mouse CD95 (clone Jo2).
The following antibodies were from BioLegend: Pacific blue-anti–T and B cell activation antigen (clone GL-7), PE-Cy7-anti-mouse CD19 (clone 6D5), PE-anti-mouse CD4 (clone GK1.5), PE-Cy7-anti-mouse CD4 (clone GK1.5), Alexa647-anti-Bcl-2 (clone BCL/10C4), PE-anti-Bcl-2 (clone BCL/10C4), Alexa647-mouse IgG1 isotype control (clone MOPC-21), PE-anti-mouse Ig kappa chain (clone RMK-45), PE-anti-mouse Ig lambda chain (clone RML-42), PE-rat IgG1 isotype control (clone RTK2071), APC-anti-mouse Ki-67 (clone 16A8), APC-rat IgG2a isotype control (clone RTK2758), PE-anti-mouse CD44 (clone IM7), PE-Cy7-anti-mouse CD62L (clone MEL-14), Pacific blue anti-mouse CD8 (clone 53–6.7), APC anti-mouse CD4 (clone GK1.5), FITC anti-B220 (clone RA3-6B2), FITC anti-mouse CD3 (clone 145-2C11), APC-Cy7 anti-mouse CD86 (clone GL-1), Alexa647-anti-mouse IgD (clone 11-26C.2a), and PE-streptavidin.
The following antibodies were from Cell Signaling: PE-anti-Bcl-xL (clone 54H6), PE-rabbit IgG isotype control (clone DA1E), Alexa647-anti-Mcl-1 (clone D2W9E), and Alexa647-rabbit IgG isotype control (clone DA1E).
NIP(5)-BSA (Biosearch Technology; catalog no. N-5040-10) was conjugated to Alexa Fluor 647 in the Veillette laboratory using Alexa Fluor 647 protein labeling kit (Thermo Fisher Scientific; catalog no. A20173).
ELISpot
Multiscreen IP filter 96-well plates (Millipore; catalog no. MSIPS4W10) were coated overnight with 100 µl NIP(5)-BSA (Biosearch Technologist; catalog no. N-5040-10; 2 µg/ml) and then blocked with RPMI-1640 medium supplemented with 10% FBS. Spleen or bone marrow cells (5 × 105 in 100 µl complete medium per well) were seeded into 12 independent wells for each mouse and incubated overnight in a tissue culture incubator. Then, the wells were thoroughly washed and incubated for 2 h at room temperature with HRP-conjugated anti-mouse IgG1 (Southern Biotech; catalog no. 1070–05). After washing, 100 µl 3-amino-9-ethylcarbazole substrate (BD Biosciences; catalog no. 551951) was added to each well and incubated for 20 min. Spots representing NP-specific IgG1-secreting ASCs were counted visually in all 12 wells using a microscope. Representative photographs were taken and are shown. The total numbers of dots for each mouse were calculated.
Morphological analyses of GCs
Spleens from mice immunized with SRBCs were fixed for 10–12 h at 4°C in periodate-lysine-paraformaldehyde–containing fixative medium and then embedded in Optimal Cutting Temperature compound. 12-µm-thick cryosections were subsequently stained with PE-anti-CD4 (BioLegend; catalog no. 100408, clone GK1.5; 1:200) and Alexa647-anti-IgD (BioLegend; catalog no. 405708, clone 11-26C.2a; 1:200). For staining follicular dendritic cells, sections were incubated with biotin-anti-mouse CD35 (BD Bioscience; catalog no. 553816, clone 8C12; 1:200) for 1 h at room temperature and stained with PE-streptavidin (BioLegend; catalog no. 405203; 1:200) for 1 h at room temperature. Stained sections were covered with a coverslip plus antifade reagent (Thermo Fisher Scientific; catalog no. P36934) and observed with a confocal microscope (Zeiss; LSM710).
Adoptive transfer of T cells or B cells
CD4+ and CD19+ cells were purified by negative selection from splenocytes of nonimmunized mice using purification kits from Stem Cell Technology (catalog nos. 19852 and 19754, respectively). For T cells, 5 × 106 CD4+ cells (in 0.2 ml PBS) were injected i.v. into TCR KO mice. For B cells, 20 × 106 CD19+ cells were injected i.v. into BCR KO mice. Within 24 h of adoptive transfer, recipient mice were injected i.p. with NP(18)-OVA with alum as detailed above. Cell purity was routinely >95%.
Bone marrow chimeras
RAG-1 KO mice (2–3 mo old) were irradiated with 950 rads and then immediately injected i.v. with 10 × 106 bone marrow cells (4 × 106 WT + 6 × 106 SFR KO). After 3 mo, mice were immunized. On day 9 after immunization with NP-OVA, mice were sacrificed, and expression of CD48 was analyzed on GC B cells and GC Tfh cells. As CD48 is expressed at high levels on all B cells and T cells from WT mice, but not from SFR KO mice, it was used as marker to distinguish WT and SFR KO cells in the chimeras.
IgM VH186.2 SHM analysis
Nine days after immunization with NP-OVA, splenocytes were harvested and stained with anti-CD19, anti-CD95, and anti-GL-7. GC B cells (CD19+CD95+GL-7hi) were then sorted using a FACSAria Cell Sorter. RNA was extracted, and cDNAs were synthesized by RT-PCR, as described elsewhere (Zahn et al., 2013). cDNAs encoded by the VH186.2 segment of the IgM-encoding gene were then amplified by PCR using the oligonucleotides 5′-TCTTCTTGGCAGCAACAG-3′ and 5′-CCAGATTCTTATCAGACAGGG-3′. The purified PCR products were cloned into pGEM T-Easy vector according to the manufacturer’s instructions and transformed into competent Escherichia coli DH5α. Plasmid DNA was obtained from single colonies and subjected to IgM VH186.2 sequence analysis using IMGT/V-quest (http://imgt.org).
RNA sequencing and real-time PCR
After immunization of AID-GFP and SFR KO AID-GFP mice with NP-OVA, GFP+ cells were sorted, and RNA was obtained. RNA-sequencing library preparation was performed using the Illumina TruSeq Stranded mRNA Kit, according to manufacturer’s instructions, and sequenced with the Illumina HiSeq 2000 Sequencer. Read quality was confirmed using FastQC v0.11.8 before alignment using STAR v2.5.0 on the mouse GRCm38 v98 reference genome. Differential expression analysis was performed with DESeq2 v1.22.2 from the raw alignment counts calculated with FeatureCounts v1.5. Differentially expressed genes were defined as genes with an adjusted P value of <0.05 and log2 fold change of ≥1.0. Principal-component analysis was applied using the FactoMineR R package to r-log transformed normalized count outputted by DESeq2 (Love et al., 2014; https://www.jstatsoft.org/article/view/v025i01). Levels of Nur77-encoding RNA (Nr4a1) were determined using real-time PCR, as described (Lu et al., 2019).
In vitro B cell proliferation and isotype switching
Naive B cells were purified by negative selection using anti-CD43 antibodies, which remove T cells, NK cells, and activated B cells (MACS beads; Miltenyi Biotec; catalog no. 130–049-801). Cell purity was >95%. To measure growth rate, 0.5 × 106 cells were seeded in 24-well plates with 1 ml complete RPMI-1640 medium supplemented with the following B cell activation cocktail: goat F(ab′)2 fragments of anti-mouse IgM (0.2 µg/ml; Jackson ImmunoResearch Labs; catalog no. 115–006-075), rat anti-mouse CD40 (3 µg/ml; Thermo Fisher Scientific; catalog no. 16–0401-86), and recombinant mouse IL-4 (5 ng/ml; PeproTech; catalog no. 214–14). Multiple independent wells were used for each mouse. Cell counts were then determined daily by harvesting and counting cells in one well for each mouse. Dead and live cells were identified by 7-AAD staining and flow cytometry. To determine proliferation and isotype switching, 4 × 106 cells were labeled or not labeled with CTV (Thermo Fisher Scientific; catalog no. C34557), and seeded in 6-well plates with 4 ml of activation medium as detailed for the growth rate assays. Each day, 1–2 ml culture medium was removed from each well, and fresh medium supplemented with IL-4 at 10 ng/ml was added. Cells were then analyzed for CTV dilution and expression of surface IgG1 and IgM using flow cytometry.
Accession number
The GEO accession number for the RNA-sequencing data is GSE147990.
Statistical analyses
Data were analyzed with Prism 8.2.1. Statistics were obtained using Student’s t tests (two sided) for all experiments, except for the SHM analysis, which used the χ2 test. P values of <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 describes the levels of anti-dsDNA antibodies and various isotypes of anti-NP Ig in SFR KO mice. Fig. S2 shows the morphology and numbers of GCs in SFR KO mice immunized with SRBCs, as well as the numbers of Tfr cells in SFR KO mice immunized with NP-OVA. Fig. S3 depicts transfer of SFR KO B cells in B cell–deficient mice and bone marrow chimeras between WT and SFR KO mice. Fig. S4 shows the expression of Nur77-encoding RNA, RNA-sequencing analyses, Mcl-1 expression in B cells, and Bcl-2 expression in T cells for SFR KO mice. Fig. S5 represents the numbers of antigen-specific B cells in mice lacking one or three SFRs.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (grants MT-14429, MOP-82906, and FDN-143338 to A. Veillette, grant PJT-166028 to J.M. Di Noia, and grant FRN-153048 to I.L. King). It was also supported by the National Natural Science Foundation of China (grant 81771754 to L. Dong). Y. Zhu is supported by a scholarship from the China Scholarship Council. J.M. Di Noia is a Chercheur-boursier de mérite from the Fonds de Recherche Santé de Québec. A. Veillette holds the Canada Research Chair on Signaling in the Immune System.
Author contributions: Conceptualization: M.-C. Zhong, J.M. Di Noia, I.L. King, and A. Veillette. Formal analysis: M.-C. Zhong, A. Zahn, J.M. Di Noia, D. Karo-Atar, I.L. King, and A. Veillette. Funding acquisition: J.M. Di Noia, I.L. King, and A. Veillette. Investigation: M.-C. Zhong, Y. Zhu, A. Zahn, and D. Karo-Atar. Methodology: M.-C. Zhong, Y. Lu, A. Zahn, and D. Karo-Atar. Project administration: A. Veillette. Resources: M.-C. Zhong, Y. Lu, and J. Qian. Supervision: L. Dong, J.M. Di Noia, I.L. King, and A. Veillette. Validation: Y. Zhu. Visualization: M.-C. Zhong, J. Qian, A. Zahn, and D. Karo-Atar. Writing (original draft): M.-C. Zhong and A. Veillette. Writing (review and editing): all authors.
References
- Barak, A.F., Lewinsky H., Perpinial M., Huber V., Radomir L., Kramer M.P., Sever L., Wolf Y., Shapiro M., Herishanu Y., et al. 2020. Bone marrow dendritic cells support the survival of chronic lymphocytic leukemia cells in a CD84 dependent manner. Oncogene. 39:1997–2008. 10.1038/s41388-019-1121-y [DOI] [PubMed] [Google Scholar]
- Cannons, J.L., Qi H., Lu K.T., Dutta M., Gomez-Rodriguez J., Cheng J., Wakeland E.K., Germain R.N., and Schwartzberg P.L.. 2010. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. 32:253–265. 10.1016/j.immuni.2010.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons, J.L., Tangye S.G., and Schwartzberg P.L.. 2011. SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 29:665–705. 10.1146/annurev-immunol-030409-101302 [DOI] [PubMed] [Google Scholar]
- Casellas, R., Basu U., Yewdell W.T., Chaudhuri J., Robbiani D.F., and Di Noia J.M.. 2016. Mutations, kataegis and translocations in B cells: understanding AID promiscuous activity. Nat. Rev. Immunol. 16:164–176. 10.1038/nri.2016.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., Zhong M.C., Guo H., Davidson D., Mishel S., Lu Y., Rhee I., Pérez-Quintero L.A., Zhang S., Cruz-Munoz M.E., et al. 2017a. SLAMF7 is critical for phagocytosis of haematopoietic tumour cells via Mac-1 integrin. Nature. 544:493–497. 10.1038/nature22076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S., Cai C., Li Z., Liu G., Wang Y., Blonska M., Li D., Du J., Lin X., Yang M., and Dong Z.. 2017b. Dissection of SAP-dependent and SAP-independent SLAM family signaling in NKT cell development and humoral immunity. J. Exp. Med. 214:475–489. 10.1084/jem.20161312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty, S., Kersh E.N., Cannons J., Schwartzberg P.L., and Ahmed R.. 2003. SAP is required for generating long-term humoral immunity. Nature. 421:282–287. 10.1038/nature01318 [DOI] [PubMed] [Google Scholar]
- Crouch, E.E., Li Z., Takizawa M., Fichtner-Feigl S., Gourzi P., Montaño C., Feigenbaum L., Wilson P., Janz S., Papavasiliou F.N., and Casellas R.. 2007. Regulation of AID expression in the immune response. J. Exp. Med. 204:1145–1156. 10.1084/jem.20061952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunninghame Graham, D.S., Vyse T.J., Fortin P.R., Montpetit A., Cai Y.C., Lim S., McKenzie T., Farwell L., Rhodes B., Chad L., et al. CaNIOS GenES Investigators . 2008. Association of LY9 in UK and Canadian SLE families. Genes Immun. 9:93–102. 10.1038/sj.gene.6364453 [DOI] [PubMed] [Google Scholar]
- Cyster, J.G., and Allen C.D.C.. 2019. B Cell Responses: Cell Interaction Dynamics and Decisions. Cell. 177:524–540. 10.1016/j.cell.2019.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czar, M.J., Kersh E.N., Mijares L.A., Lanier G., Lewis J., Yap G., Chen A., Sher A., Duckett C.S., Ahmed R., and Schwartzberg P.L.. 2001. Altered lymphocyte responses and cytokine production in mice deficient in the X-linked lymphoproliferative disease gene SH2D1A/DSHP/SAP. Proc. Natl. Acad. Sci. USA. 98:7449–7454. 10.1073/pnas.131193098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco, A.L. 2016. The germinal center antibody response in health and disease. F1000 Res. 5:5. 10.12688/f1000research.7717.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, Z., Davidson D., Pérez-Quintero L.A., Kurosaki T., Swat W., and Veillette A.. 2012. The adaptor SAP controls NK cell activation by regulating the enzymes Vav-1 and SHIP-1 and by enhancing conjugates with target cells. Immunity. 36:974–985. 10.1016/j.immuni.2012.03.023 [DOI] [PubMed] [Google Scholar]
- Guo, H., Cranert S.A., Lu Y., Zhong M.C., Zhang S., Chen J., Li R., Mahl S.E., Wu N., Davidson D., et al. 2016. Deletion of Slam locus in mice reveals inhibitory role of SLAM family in NK cell responses regulated by cytokines and LFA-1. J. Exp. Med. 213:2187–2207. 10.1084/jem.20160552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockenbery, D., Nuñez G., Milliman C., Schreiber R.D., and Korsmeyer S.J.. 1990. Bcl-2 is an inner mitochondrial membrane protein that blocks programmed cell death. Nature. 348:334–336. 10.1038/348334a0 [DOI] [PubMed] [Google Scholar]
- Hodgkin, P.D., Lee J.H., and Lyons A.B.. 1996. B cell differentiation and isotype switching is related to division cycle number. J. Exp. Med. 184:277–281. 10.1084/jem.184.1.277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzapfel, K.L., Tyznik A.J., Kronenberg M., and Hogquist K.A.. 2014. Antigen-dependent versus -independent activation of invariant NKT cells during infection. J. Immunol. 192:5490–5498. 10.4049/jimmunol.1400722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J.K., Crampton J.C., Locci M., and Crotty S.. 2016. CRISPR-Mediated Slamf1Δ/Δ Slamf5Δ/Δ Slamf6Δ/Δ Triple Gene Disruption Reveals NKT Cell Defects but Not T Follicular Helper Cell Defects. PLoS One. 11:e0156074. 10.1371/journal.pone.0156074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Gomez-Rodriguez J., Preite S., Garrett L.J., Harper U.L., and Schwartzberg P.L.. 2016. CRISPR-Mediated Triple Knockout of SLAMF1, SLAMF5 and SLAMF6 Supports Positive Signaling Roles in NKT Cell Development. PLoS One. 11:e0156072. 10.1371/journal.pone.0156072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise, W., and Kurosaki T.. 2019. Plasma cell differentiation during the germinal center reaction. Immunol. Rev. 288:64–74. 10.1111/imr.12751 [DOI] [PubMed] [Google Scholar]
- Kageyama, R., Cannons J.L., Zhao F., Yusuf I., Lao C., Locci M., Schwartzberg P.L., and Crotty S.. 2012. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 36:986–1002. 10.1016/j.immuni.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, A.M., Cambier J.C., and Shlomchik M.J.. 2012. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 336:1178–1181. 10.1126/science.1213368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, I.L., and Mohrs M.. 2009. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J. Exp. Med. 206:1001–1007. 10.1084/jem.20090313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, M., Akashi K., Domen J., Sugamura K., and Weissman I.L.. 1997. Bcl-2 rescues T lymphopoiesis, but not B or NK cell development, in common gamma chain-deficient mice. Immunity. 7:155–162. 10.1016/S1074-7613(00)80518-X [DOI] [PubMed] [Google Scholar]
- Kumar, K.R., Li L., Yan M., Bhaskarabhatla M., Mobley A.B., Nguyen C., Mooney J.M., Schatzle J.D., Wakeland E.K., and Mohan C.. 2006. Regulation of B cell tolerance by the lupus susceptibility gene Ly108. Science. 312:1665–1669. 10.1126/science.1125893 [DOI] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y., Zhong M.C., Qian J., Calderon V., Cruz Tleugabulova M., Mallevaey T., and Veillette A.. 2019. SLAM receptors foster iNKT cell development by reducing TCR signal strength after positive selection. Nat. Immunol. 20:447–457. 10.1038/s41590-019-0334-0 [DOI] [PubMed] [Google Scholar]
- Luo, W., Hawse W., Conter L., Trivedi N., Weisel F., Wikenheiser D., Cattley R.T., and Shlomchik M.J.. 2019. The AKT kinase signaling network is rewired by PTEN to control proximal BCR signaling in germinal center B cells. Nat. Immunol. 20:736–746. 10.1038/s41590-019-0376-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C.S., and Deenick E.K.. 2011. The role of SAP and SLAM family molecules in the humoral immune response. Ann. N. Y. Acad. Sci. 1217:32–44. 10.1111/j.1749-6632.2010.05824.x [DOI] [PubMed] [Google Scholar]
- Motoyama, N., Miwa T., Suzuki Y., Okada H., and Azuma T.. 1994. Comparison of somatic mutation frequency among immunoglobulin genes. J. Exp. Med. 179:395–403. 10.1084/jem.179.2.395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, H., Cannons J.L., Klauschen F., Schwartzberg P.L., and Germain R.N.. 2008. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 455:764–769. 10.1038/nature07345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shachar, I., Barak A., Lewinsky H., Sever L., and Radomir L.. 2019. SLAMF receptors on normal and malignant B cells. Clin. Immunol. 204:23–30. 10.1016/j.clim.2018.10.020 [DOI] [PubMed] [Google Scholar]
- Shlomchik, M.J., Luo W., and Weisel F.. 2019. Linking signaling and selection in the germinal center. Immunol. Rev. 288:49–63. 10.1111/imr.12744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, I., Radtke D., Phillips B., McGowan S.J., and Bannard O.. 2018. Germinal Center B Cells Replace Their Antigen Receptors in Dark Zones and Fail Light Zone Entry when Immunoglobulin Gene Mutations are Damaging. Immunity. 49:477–489.e7. 10.1016/j.immuni.2018.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, A., Yamada R., Kochi Y., Sawada T., Okada Y., Matsuda K., Kamatani Y., Mori M., Shimane K., Hirabayashi Y., et al. 2008. Functional SNPs in CD244 increase the risk of rheumatoid arthritis in a Japanese population. Nat. Genet. 40:1224–1229. 10.1038/ng.205 [DOI] [PubMed] [Google Scholar]
- Tuscano, J.M., Druey K.M., Riva A., Pena J., Thompson C.B., and Kehrl J.H.. 1996. Bcl-x rather than Bcl-2 mediates CD40-dependent centrocyte survival in the germinal center. Blood. 88:1359–1364. 10.1182/blood.V88.4.1359.bloodjournal8841359 [DOI] [PubMed] [Google Scholar]
- Veillette, A., Zhang S., Shi X., Dong Z., Davidson D., and Zhong M.C.. 2008. SAP expression in T cells, not in B cells, is required for humoral immunity. Proc. Natl. Acad. Sci. USA. 105:1273–1278. 10.1073/pnas.0710698105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom, I., Carotta S., Lüthje K., Peperzak V., Jost P.J., Glaser S., Busslinger M., Bouillet P., Strasser A., Nutt S.L., and Tarlinton D.M.. 2010. Mcl-1 is essential for germinal center formation and B cell memory. Science. 330:1095–1099. 10.1126/science.1191793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandstrat, A.E., Nguyen C., Limaye N., Chan A.Y., Subramanian S., Tian X.H., Yim Y.S., Pertsemlidis A., Garner H.R. Jr., Morel L., and Wakeland E.K.. 2004. Association of extensive polymorphisms in the SLAM/CD2 gene cluster with murine lupus. Immunity. 21:769–780. 10.1016/j.immuni.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Wang, N., Halibozek P.J., Yigit B., Zhao H., O’Keeffe M.S., Sage P., Sharpe A., and Terhorst C.. 2015. Negative Regulation of Humoral Immunity Due to Interplay between the SLAMF1, SLAMF5, and SLAMF6 Receptors. Front. Immunol. 6:158. 10.3389/fimmu.2015.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, N., and Veillette A.. 2016. SLAM family receptors in normal immunity and immune pathologies. Curr. Opin. Immunol. 38:45–51. 10.1016/j.coi.2015.11.003 [DOI] [PubMed] [Google Scholar]
- Yusuf, I., Kageyama R., Monticelli L., Johnston R.J., Ditoro D., Hansen K., Barnett B., and Crotty S.. 2010. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol. 185:190–202. 10.4049/jimmunol.0903505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn, A., Daugan M., Safavi S., Godin D., Cheong C., Lamarre A., and Di Noia J.M.. 2013. Separation of function between isotype switching and affinity maturation in vivo during acute immune responses and circulating autoantibodies in UNG-deficient mice. J. Immunol. 190:5949–5960. 10.4049/jimmunol.1202711 [DOI] [PubMed] [Google Scholar]
- Zhong, M.C., and Veillette A.. 2013a. The adaptor molecule signaling lymphocytic activation molecule (SLAM)-associated protein (SAP) is essential in mechanisms involving the Fyn tyrosine kinase for induction and progression of collagen-induced arthritis. J. Biol. Chem. 288:31423–31436. 10.1074/jbc.M113.473736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, M.C., and Veillette A.. 2013b. Critical role of SAP in progression and reactivation but not maintenance of T cell-dependent humoral immunity. Mol. Cell. Biol. 33:1223–1232. 10.1128/MCB.01591-12 [DOI] [PMC free article] [PubMed] [Google Scholar]