The role of Fas in germinal center (GC) homeostasis is controversial. Razzaghi et al. show that Fas is a strong cell-intrinsic regulator of the GC and that its loss defines an aggressive subtype of GC-derived lymphoma.
Abstract
Fas is highly expressed on germinal center (GC) B cells, and mutations of FAS have been reported in diffuse large B cell lymphoma (DLBCL). Although GC-derived DLBCL has better overall outcomes than other DLBCL types, some cases are refractory, and the molecular basis for this is often unknown. We show that Fas is a strong cell-intrinsic regulator of GC B cells that promotes cell death in the light zone, likely via T follicular helper (Tfh) cell–derived Fas ligand. In the absence of Fas, GCs were more clonally diverse due to an accumulation of cells that did not demonstrably bind antigen. FAS alterations occurred most commonly in GC-derived DLBCL, were associated with inferior outcomes and an enrichment of Tfh cells, and co-occurred with deficiency in HVEM and PD-L1 that regulate the Tfh–B cell interaction. This work shows that Fas is critically required for GC homeostasis and suggests that loss of Tfh-mediated counterselection in the GC contributes to lethality in GC-derived lymphoma.
Graphical Abstract
Introduction
Germinal centers (GCs) are the principal sites of antibody affinity maturation (Victora and Nussenzweig, 2012). Current models of the GC reaction suggest that, following immunization, selection of B cell clones expressing high-affinity B cell receptors (BCRs) occurs via iterative cycling of GC B cells between the light zone (LZ) and the dark zone (DZ). In the LZ, GC B cells acquire antigen from follicular dendritic cells via their BCR and present antigen to T cells. GC B cells that present sufficient antigen to T cells are thought to be positively selected and enter the DZ, where they proliferate and undergo somatic hypermutation of their BCR mediated by activation-induced cytidine deaminase (AID). Mutations and other genetic alterations arising from aberrant AID activity during this process can promote malignancy.
Diffuse large B cell lymphoma (DLBCL), the most common type of aggressive non-Hodgkin lymphoma, is heterogeneous and has been divided into three subtypes based on gene expression signatures (Young et al., 2019). Approximately 50% of DLBCL cases are classified as GC B cell–like (GCB)–DLBCL based on similarities to GC B cells, whereas the remainder of cases have gene expression similar to activated B cells (activated B cell–like [ABC]–DLBCL; ∼35%) or cannot be categorized into either group (unclassified; ∼15%). More recent work has sought to define subtypes of DLBCL on the basis of the landscape of specific genetic alterations in a given tumor (Chapuy et al., 2018; Schmitz et al., 2018; Wright et al., 2020). These studies have revealed a greater heterogeneity within DLBCL than previously appreciated. Two somewhat overlapping genetic classification systems have been proposed: LymphGen, consisting of EZB, ST2, MCD, A53, BN2, and N1 subtypes (Wright et al., 2020); and C1–C5 (Chapuy et al., 2018). The DLBCL genetic subtypes EZB and C3 are characterized by the presence of EZH2 gain-of-function mutations and alterations in BCL2, and ST2 and C4 are characterized by alterations in SGK1 and TET2 and are primarily composed of GCB-DLBCL cases but carry distinct clinical outcomes to immunochemotherapy. MCD and C5 are characterized by gain-of-function mutations in MYD88 and CD79B, and N1, characterized by NOTCH1 mutations, is almost exclusively composed of ABC-DLBCL cases. In contrast, A53 and C2 are characterized by TP53 mutations and aneuploidy, and BN2 and C1 are characterized by translocations of BCL6 and NOTCH2 mutations and are comprised of a mixture of all three gene expression classes of DLBCL. Distinct microenvironmental signatures have been associated with each subtype, and these signatures may contribute distinct therapeutic vulnerabilities (Wright et al., 2020). Overall, GCB-DLBCL and the EZB genetic subtype carry a more favorable prognosis than ABC-DLBCL and its closely related genetic subtype MCD. Within GCB-DLBCL and EZB, the presence of an MYC gene expression signature is associated with poor outcomes (GCB-DLBCL double-hit signature [DHIT+] and EZB-MYC+, respectively; Ennishi et al., 2019; Sha et al., 2019; Wright et al., 2020). However, some cases of GC-derived DLBCL are refractory to therapy even in the absence of an MYC signature. The molecular basis for the clinical aggressiveness of these cases is unknown.
Recent work has suggested that early activation of B cells and entry into the GC is a stochastic process that can allow for activation of cells expressing BCRs with low to negligible affinity to the immunizing antigen (Di Niro et al., 2015; Kuraoka et al., 2016; Tas et al., 2016). Early GCs exhibit high clonal diversity, where it is estimated that between 50 and 200 clones can seed an individual GC early after immunization (Tas et al., 2016). Following immunization, at least half of GC B cells express BCRs that fail to bind antigen in both native and denatured forms (Kuraoka et al., 2016). The fraction of these cells expressing BCRs that do not demonstrably bind antigen decreases at later time points during the GC response (Frank et al., 2015; Kuraoka et al., 2016). It has also been shown that, under certain conditions, B cells expressing a BCR of defined specificity can be induced to enter a GC reaction following immunization with an unrelated antigen (Silver et al., 2018). In contrast to ABC-DLBCL and the related genetic subtype, MCD, where there is selection of stereotypic BCRs that use specific heavy-chain variable (Vh) segments that confer autoreactivity or reactivity to self-antigens, no such Vh enrichment is seen across samples of GCB-DLBCL or EZB (Wright et al., 2020; Young et al., 2015). It is not clear whether this increased Vh diversity in GC-derived lymphomas is due to perturbations of normal selection in the GC.
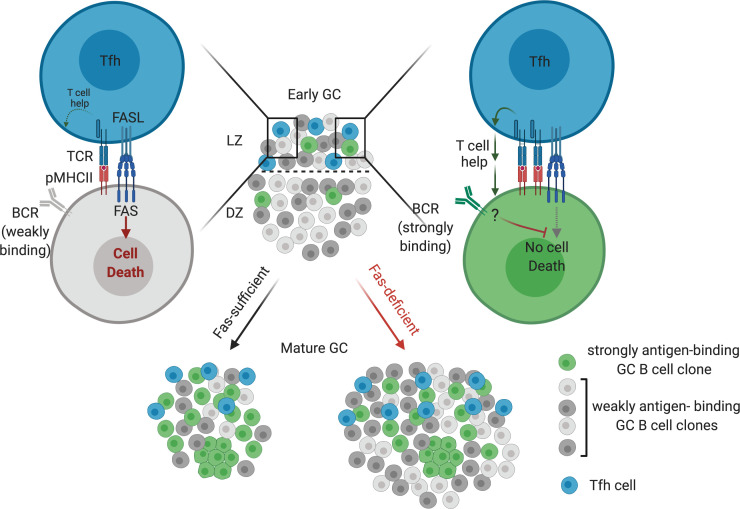
GC B cells are highly proliferative, yet the size of an individual GC remains relatively constant for several weeks after initiation, suggesting that there is a high degree of ongoing GC B cell death during a GC reaction (Victora and Nussenzweig, 2012). Recent work has estimated that approximately half of GC B cells will undergo apoptosis in 5–6 h (Mayer et al., 2017). Apoptosis occurs in both the LZ and DZ, and there are distinct signals that lead to B cell death in each zone (Mayer et al., 2017). In the DZ, B cells that have acquired deleterious mutations in their antibody genes via AID undergo apoptosis (Mayer et al., 2017; Stewart et al., 2018). In the LZ, it is currently thought that the primary mechanism by which B cells die is due to neglect from a lack of T cell help (Mayer et al., 2017). However, whether the death of GC B cells that do not demonstrably bind antigen is due solely to lack of productive interactions with T cells in the LZ or if there are signaling mechanisms that actively select against these cells remains unclear.
Fas is a member of the TNF receptor superfamily and contains an intracellular death domain (DD). Following ligation of Fas by membrane-bound Fas ligand (FasL), the adapter molecule Fas-associated protein with DD and caspase-8 are recruited to the membrane to initiate apoptosis (Nagata, 2018). Fas is highly expressed by GC B cells and can induce apoptosis of activated lymphocytes (Allen, 2015). However, how and when Fas might promote cell death during a B cell response is controversial. Several studies have shown that Fas deficiency does not result in increased numbers of antigen-specific GC B cells but can lead to aberrant GC output at late time points following immunization (Butt et al., 2015; Smith et al., 1995; Takahashi et al., 2001). Another study showed that loss of Fas in B cells results in the development of a systemic lymphoproliferative disorder and autoimmunity accompanied by increased GC B cells (Hao et al., 2008). To reconcile these findings—that there are increased GC B cells in some Fas-deficient settings, despite no clear evidence of cell-intrinsic suppression of GC B cell survival—it has been proposed that Fas does not cell-intrinsically regulate GC B cell survival in vivo and that the increase in GC B cells in some Fas-deficient settings occurs as a consequence of systemic autoimmunity (Allen, 2015). There is no clear consensus on the role of Fas in the GC reaction, and Fas is currently not thought to be a key regulator of GC selection.
In this study, we found that Fas expression was frequently lost in an animal model of GC-derived lymphoma that forms initially in the mesenteric LN (mLN), leading us to evaluate the role of Fas in polyclonal GC responses. We found that Fas deficiency provided a strong cell-intrinsic survival advantage in the GC of mLNs and in immunized lymphoid tissues. The accumulation of Fas-deficient GC B cells was due to decreased cell death in the LZ. FasL expression by a SLAM-associated protein (SAP)–dependent cell type, likely T follicular helper (Tfh) cells, was necessary to suppress GC B cell accumulation. In the absence of Fas, GCs were more clonally diverse due to persistence of clones bearing BCRs that could not demonstrably bind antigen. Genetic alterations in FAS were most commonly found in the GC-derived genetic subtype of DLBCL, EZB. EZB tumors harboring FAS mutations had inferior survival and gene signatures suggesting an altered tumor microenvironment with increased Tfh cells. Additionally, GC-derived tumors with Fas mutations were enriched for mutations in ligands that negatively regulate Tfh cell help, such as TNFRSF14 (encoding herpesvirus entry mediator [HVEM]) and CD274 (encoding PD-L1). Finally, in EZB tumors that were FAS or HVEM deficient, we found increased diversity of Vh usage across samples. Our work provides evidence for a Fas-dependent mechanism of GC B cell counterselection that limits the fraction of cells that do not demonstrably bind antigen and suggests that loss of Tfh-mediated counterselection in the GC contributes to lethality in a distinct subtype of GC-derived lymphoma.
Results
Fas is required to constrain accumulation of GC B cells in vivo in a tissue-specific manner
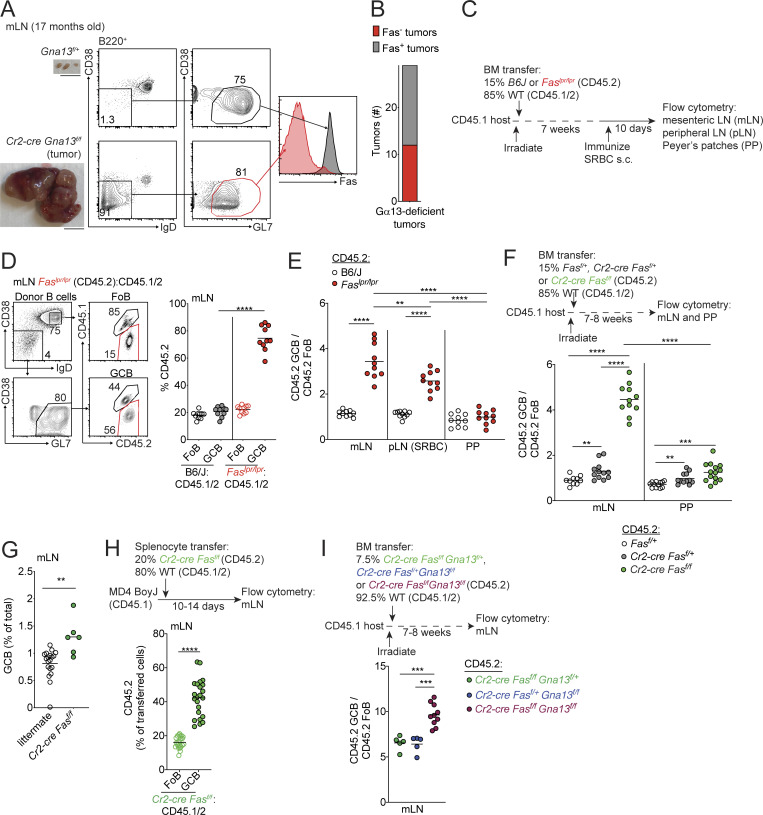
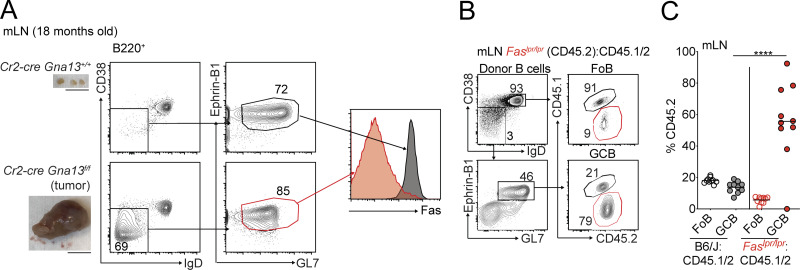
Approximately one half of animals aged ≥1 yr with deficiency in the tumor suppressor Gα13 (Gna13) in B cells or the Gα13-coupled receptor, S1pr2, develop GC-derived lymphomas that form initially in the mLN (Green et al., 2011; Muppidi et al., 2014). We immunophenotyped tumors from animals lacking Gα13 in B cells by flow cytometry and were surprised to find that ∼40% of these tumors lost surface expression of Fas in all or part of their GC B cells (n = 29; Fig. 1, A and B). Despite the loss of Fas, tumor cells expressed other markers of GC B cells, such as GL7 and the recently described GC marker Ephrin-B1 (Fig. S1 A; Laidlaw et al., 2017; Lu et al., 2017). The high frequency of Fas deficiency spurred our further investigation into the potential role of Fas in GC homeostasis.
Figure 1.
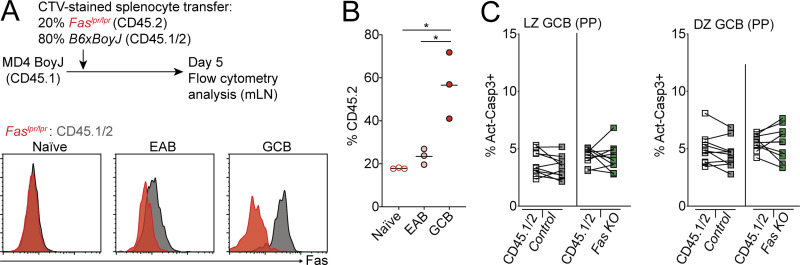
Fas is required to constrain survival of GC B cells in vivo in a tissue-specific manner. (A) Fas expression on GC B cells from an mLN tumor from a 17-mo-old Cr2-cre Gna13f/f animal or littermate control. Gross appearance of the mLN is shown in images on the left. Scale bar, 1 cm. (B) Frequency of loss of Fas expression on GC B cell tumors from animals aged 12–20 mo with B cell–specific Gα13 deficiency (n = 29). (C) Experimental scheme for data in D and E. (D) Percentages of CD45.2 FoB cells and GC B cells (GCB) in mLNs of mixed BM chimeras generated with a mixture of 85% WT (CD45.1/2) and 15% CD45.2 BM that was B6/J or Faslpr/lpr, assessed by FACS. Example gating strategy for FoB cells and GC B cells is shown on the left in D. (E) Ratio of frequency of CD45.2 GC B cells to CD45.2 FoB cells in B6J or Faslpr/lpr mixed BM chimeras in mLNs, SRBC-immunized pLNs, and PPs. Data in D and E are pooled from two independent experiments with five mice per group. (F) Ratio of frequency of CD45.2 GC B cells to CD45.2 FoB cells in mLNs and PPs of mixed BM chimeras generated with 85% WT (CD45.1/2) and 15% CD45.2 BM that was Fasf/+, Cr2-cre Fasf/+, or Cr2-cre Fasf/f. Data are pooled from two independent experiments with five to seven mice per group. (G) Frequency of GC B cells among total cells in mLNs from littermate control or Cr2-cre Fasf/f animals. Data are pooled from four independent experiments with one to four mice per group. (H) Frequency of CD45.2 cells among FoB cells or GC B cells derived from donor cells in mLNs of MD4 BoyJ mice that were given a mixture of splenocytes that were 80% WT (CD45.1/2) and 20% Cr2-cre Fasf/f (CD45.2) 10–14 d before analysis. Data are pooled from five independent experiments with four to six mice per group. (I) Ratio of frequency of CD45.2 GC B cells to CD45.2 FoB cells from mLNs of mixed chimeras generated with a mixture of 92.5% WT (CD45.1/2) and 7.5% CD45.2 BM that was Cr2-cre Fasf/f Gna13f/+, Cr2-cre Fasf/+Gna13f/f, or Cr2-cre Fasf/fGna13f/f, assessed by FACS. Data are pooled from two independent experiments with 5–10 mice per group total. ****, P < 0.0001, paired two-tailed Student’s t test for data in H. For all other data, **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, unpaired two-tailed Student’s t test.
Figure S1.
Fas is required to constrain survival of GC B cells in vivo. (A) Fas expression on Ephrin-B1+ GC B cells from an mLN tumor from an 18-mo-old Cr2-cre Gna13f/f animal or littermate control. Gross appearance of the mLN is shown in images on the left. Scale bar, 1 cm. (B and C) Example of gating scheme for Ephrin-B1+ GC B cells (B) and percentages of CD45.2 FoB cells and Ephrin-B1+ GC B cells (C) in mLNs of mixed BM chimeras generated as in Fig. 1 C, assessed by FACS. Data are from 10 mice per group. ****, P < 0.0001, unpaired two-tailed Student’s t test.
Fas is highly expressed on GC B cells, and ligation of Fas promotes apoptosis in a variety of cell types. To determine whether Fas plays a cell-intrinsic role in GC B cell selection during a polyclonal GC response, we generated mixed bone marrow (BM) chimeras by reconstituting irradiated CD45.1 hosts with BM from either Fas-sufficient (B6J) or Fas-deficient (Faslpr/lpr) mice, each expressing CD45.2, mixed with BM from WT CD45.1/2 animals at an ∼15:85 ratio (Fig. 1 C). Animals were immunized subcutaneously with the T-dependent antigen sheep RBCs (SRBCs) 7 wk after reconstitution, and frequencies of CD45.2 cells were assessed 10 d later in follicular B (FoB) cells and GC B cells in peripheral LNs (pLNs), mLNs, and Peyer’s patches (PPs). Subcutaneous immunization does not affect chronic GC responses in mLNs and PPs. Because Fas is often used to identify GC B cells by flow cytometry, we developed an alternative gating strategy in Fas-deficient mixed chimeras, where we defined GC B cells as B220+IgDloCD38loGL7hi (Fig. 1 D). In mLNs of Faslpr/lpr mixed chimeras, there was a 3.5-fold increase in Fas-deficient GC B cells compared with naive FoB cells. We also found similar GC expansions when samples were stained with Ephrin-B1 to define GC B cells (Fig. S1, B and C). In immunized pLNs, there was a 2.5-fold increase in representation of Fas-deficient GC B cells compared with FoB cells at day 10 following immunization (Fig. 1 E). Surprisingly, Fas deficiency conferred little to no competitive advantage in PP GCs (Fig. 1 E).
A fraction of T cells in the Faslpr/lpr chimera system are deficient in Fas, so it was possible that the expansion of Fas-deficient GC B cells was due in part to the presence of Fas-deficient T cells, which in some circumstances have been reported to up-regulate FasL (Alabyev et al., 2008). To rule out this possibility, we crossed animals carrying a floxed allele of Fas (Hao et al., 2004) with Cr2-cre (CD21-cre) animals to generate mice with a mature B cell–specific deletion of Fas. We then used these animals to generate mixed BM chimeras. In Cr2-cre Fasf/f mixed chimeras, there was a fourfold accumulation Fas-deficient GC B cells in mLNs but only a 1.3-fold expansion of Fas-deficient cells in PP GCs (Fig. 1 F). Fas heterozygosity promoted a small GC expansion in mLNs and PPs (Fig. 1 F). An expansion in mLN GC B cells was also observed in Cr2-cre Fasf/f mice relative to littermate controls (Fig. 1 G). Thus, Fas is a strong cell-intrinsic regulator of the GC reaction.
The absence of Fas in B cells can lead to the development of a systemic lymphoproliferative disorder, characterized by increased activated T cells and B cells, that is accompanied by autoimmunity (Hao et al., 2008). To reconcile this earlier finding with more recent data suggesting that Fas does not limit GC size (Butt et al., 2015), it has been hypothesized that the increase in GC size in mice lacking Fas in B cells is a consequence of systemic autoimmunity (Allen, 2015). To exclude the possibility that systemic lymphoproliferation and/or autoimmunity was promoting a cell-intrinsic expansion of Fas-deficient cells in the GC, we transferred polyclonal Cr2-cre Fasf/f splenocytes (CD45.2) mixed with WT splenocytes (CD45.1/2) into CD45.1+ MD4 Ig-transgenic recipients (MD4 BoyJ), where the vast majority of B cells express a BCR specific for hen egg lysozyme. This transfer approach allows participation of transferred polyclonal cells in GCs of mucosal tissues (Reboldi et al., 2016). At 10–14 d after transfer, there was a cell-intrinsic expansion of Fas-deficient GC B cells in mLNs (Fig. 1 H). These data indicate that Fas prevents accumulation of GC B cells via a cell-intrinsic mechanism in polyclonal B cell responses that is not a consequence of systemic autoimmunity or lymphoproliferation.
Given the frequent loss of Fas expression in tumors arising in animals lacking Gα13 in B cells, we then asked whether loss of Fas and Gα13 could synergize to promote exaggerated GC responses in mLNs. We intercrossed Fasf/f, Gna13f/f, and Cr2-cre animals to generate animals lacking Fas and Gα13 in mature B cells. We reconstituted irradiated hosts with mixed BM from double-knockout or single-knockout animals that were CD45.2 and WT CD45.1/2 animals at a 7.5:92.5 ratio. We found that combined loss of Gα13 and Fas promoted stronger outgrowth of GC B cells in the mLN than loss of Gα13 or Fas alone (Fig. 1 I). These data suggest that Fas and Gα13 likely act in different pathways and that loss of Fas-mediated regulation of the GC in mLNs might contribute to the development of GC-derived malignancy at this site.
Fas-dependent deletion occurs in the GC LZ
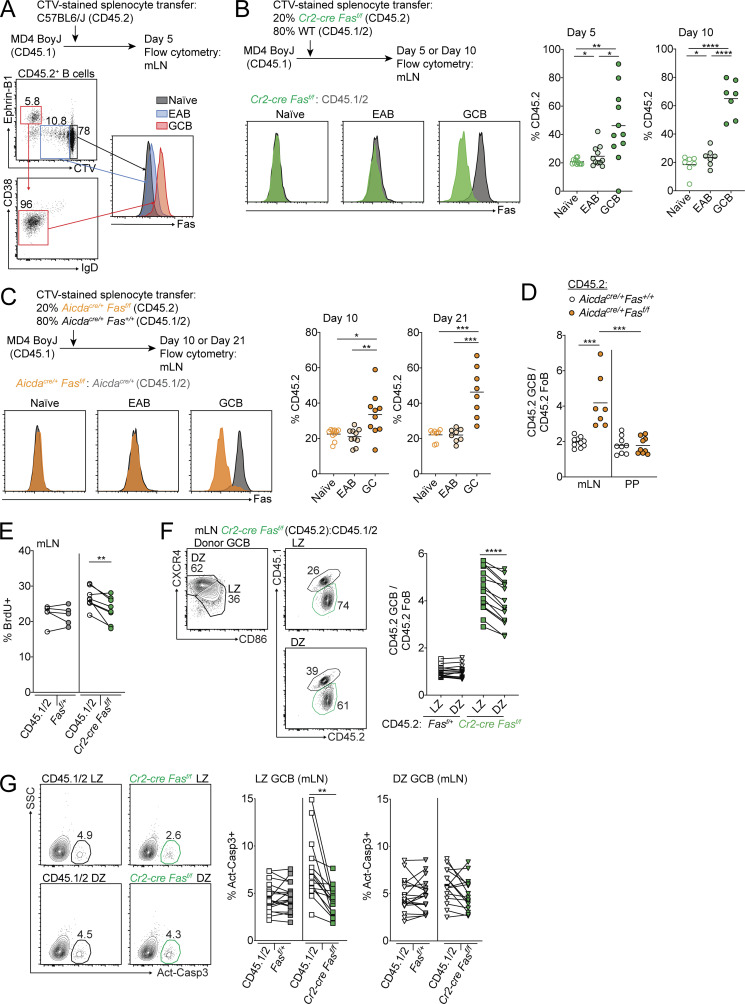
In systems using cells with a defined BCR, Fas is up-regulated early after B cell activation before entry into the GC and is expressed at even higher levels upon differentiation into GC B cells (Schwickert et al., 2011). Fas-mediated deletion of activated B cells has been reported to occur outside of GCs when B cells transgenically express a BCR that recognizes IgG2a (William et al., 2002). To determine if Fas up-regulation occurs in early activated B (EAB) cells in polyclonal responses, we transferred WT CD45.2 splenocytes that were labeled with the membrane dye CellTrace Violet (CTV) into MD4 BoyJ recipients and analyzed Fas expression on transferred cells 5 d after transfer. In this system, Fas was up-regulated in EAB cells that had diluted but still had detectable CTV (CTVlo) and did not express the GC marker Ephrin-B1 (Fig. 2 A).
Figure 2.
Fas-dependent deletion occurs in the GC LZ. (A) Fas expression on CD45.2+ naive B cells (gray), EAB cells (blue), and GC B cells (red) in mLNs of MD4 BoyJ mice that were given CTV-labeled CD45.2+ splenocytes 5 d before analysis. Naive B cells were defined as CD45.2+ B cells that had not diluted CTV; EAB cells were defined as CD45.2+ B cells that had diluted CTV but did not express the GC marker Ephrin-B1; and GC B cells were defined as CD45.2+ B cells that were Ephrin-B1+CTV−IgDloCD38lo. Data are representative of two independent experiments. (B) MD4 BoyJ mice were given a mixture of CTV-labeled splenocytes that were 20% Cr2-cre Fasf/f and 80% WT (CD45.1/2). Left panel shows Fas expression among transferred naive, EAB, or GC B cells that were Cr2-cre Fasf/f (green) or WT (CD45.1/2; gray) in mLNs 5 d after transfer. Middle and right panels show frequency of Cr2-cre Fasf/f (CD45.2+) among transferred cells that were naive, EAB, or GC B cells in mLNs 5 or 10 d after transfer. Data are pooled from two independent experiments for day 5 and three independent experiments for day 10 with three or four mice per experiment. (C) MD4 BoyJ mice were given a mixture of CTV-labeled splenocytes that were 20% Aicdacre/+Fasf/f and 80% Aicdacre/+Fas+/+ (CD45.1/2). Left panel shows Fas expression among transferred naive, EAB, or GC B cells that were Aicdacre/+Fasf/f (orange) or Aicdacre/+Fas+/+ (CD45.1/2; gray) in mLNs 10 d after transfer. Middle and right panels show the frequency of Aicdacre/+Fasf/f (CD45.2+) among transferred cells that were naive, EAB, or GC B cells in mLNs 10 or 21 d after transfer. Data are pooled from four independent experiments with two to four mice per time point. (D) Ratio of frequency of CD45.2 GC B cells to CD45.2 FoB cells in mLNs and PPs of mixed BM chimeras generated with 85% WT (CD45.1/2) and 15% CD45.2 BM that was Aicdacre/+Fas+/+ or Aicdacre/+Fasf/f. Data are from one experiment representative of two with 10 and 9 mice per group. (E) Intracellular FACS for BrdU incorporation in GC B cells from mLNs of Fasf/+ or Cr2-cre Fasf/f mixed BM chimeras generated as in Fig. 1 F that were treated i.p. with BrdU 30 min before sacrifice. Data are from five and eight mice of each type from one experiment representative of two independent experiments. (F) Ratio of frequency of Fasf/+ or Cr2-cre Fasf/f (CD45.2+) cells in LZ or DZ GC B cells relative to FoB cells in mLNs of mixed BM chimeras generated as in Fig. 1 F. Left panel shows example of gating strategy for LZ and DZ GC B cells. (G) Intracellular FACS for active caspase-3 in LZ or DZ GC B cells from mLNs of Cr2-cre Fasf/f mixed BM chimeras generated as in Fig. 1 F analyzed directly ex vivo. Left panel shows an example of the gating strategy for active caspase-3+ cells. Data in F and G are pooled from seven experiments with two or three mice per group per experiment. ***, P < 0.001, unpaired two-tailed Student’s t test for data in D. For all other data, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, paired two-tailed Student’s t test. SSC, side scatter.
Fas was expressed at higher levels in GC B cells, which were Ephrin-B1+CTV− (Fig. 2 A). Therefore, we sought to determine whether Fas-mediated selection of B cells occurs in the GC. We labeled a mixture of Cr2-cre Fasf/f or Faslpr/lpr (CD45.2) and WT (CD45.1/2) splenocytes with CTV and transferred them into MD4 BoyJ mice. We assessed Fas expression on CD45.2 cells among Cr2-cre Fasf/f or Faslpr/lpr B cells that had not undergone cell division (naive), EAB cells, and GC B cells at 5 and 10 d after transfer. In contrast to Faslpr/lpr B cells, Fas expression is only partially lost in EAB cells and fully deleted in GC B cells in Cr2-cre Fasf/f B cells (Figs. 2 B and S2 A). We found that Faslpr/lpr and Cr2-cre Fasf/f cells were enriched in the GC in comparison with both naive and EAB cells and that the GC enrichment increased over the course of the immune response (Figs. 2 B and S2 B).
Figure S2.
Fas is up-regulated in EAB cells. (A and B) MD4 BoyJ mice were given a mixture of CTV-labeled splenocytes that were 20% Faslpr/lpr and 80% WT (CD45.1/2). Histograms show Fas expression among transferred naive cells, EAB cells, or GC B cells that were Faslpr/lpr (red) or WT (CD45.1/2; gray) in mLNs 5 d after transfer (A). Frequency of Faslpr/lpr (CD45.2+) among transferred cells that were naive, EAB, or GC B cells in mLNs 5 d after transfer (B). Data are from one experiment with three mice representative of two independent experiments. (C) Intracellular FACS for active caspase-3 in LZ or DZ GC B cells from PPs of Cr2-cre Fasf/f mixed BM chimeras generated as in Fig. 1 F, analyzed directly ex vivo. Data are from two independent experiments with 10 mice total per group. *, P < 0.05, paired two-tailed Student’s t test.
As an additional test of whether Fas-mediated selection occurs in the GC, we crossed animals carrying a floxed allele of Fas to animals expressing cre under the control of the AID promoter (Aicdacre). Although AID is expressed at low levels in activated B cells, Aicdacre was not able to efficiently delete Fas in EAB cells in mixed transfers (Fig. 2 C). We saw an expansion of Aicdacre/+Fasf/f GC B cells that increased over time following transfer (Fig. 2 C). As a complementary approach to test if Fas deficiency in GC B cells provides a competitive advantage, we generated Aicdacre/+Fas+/+ or Aicdacre/+Fasf/f mixed BM chimeras. Fas deficiency in GC B cells conferred a growth advantage in the GC of mLNs but not PPs (Fig. 2 D). These data suggest that Fas-mediated negative selection occurs primarily in the GC itself.
To determine whether the accumulation of Fas-deficient GC B cells was due to differences in GC B cell proliferation, we treated control or Fas-deficient mixed chimeras with the nucleoside analogue BrdU for 30 min and assessed BrdU incorporation in GC B cells by FACS (Fig. 2 E). The accumulation of Fas-deficient GC B cells in mLNs was not due to increased proliferation, as Fas-deficient GC B cells showed reduced BrdU incorporation compared with WT GC B cells. Consistent with a reduction in proliferation among Fas-deficient GC B cells compared with WT competitors, there was a modest increase among Fas-deficient GC B cells in the LZ compared with their representation in the DZ in mixed chimeras (Fig. 2 F).
To maintain GC size, high rates of proliferation are balanced by high rates of cell death that occur via distinct mechanisms in the LZ and the DZ (Mayer et al., 2017). It is currently not known whether there are active mechanisms that might promote death of LZ GC B cells (Mayer et al., 2017). Ligation of Fas in a variety of cell types can induce cell death. Therefore, we assessed cell death by staining for active caspase-3 in Fas-deficient GC B cells from mixed chimeras directly ex vivo. In Fas-deficient mixed chimeras, we found a reduction in active caspase-3 among Fas-deficient LZ cells compared with WT competitor cells but did not see a difference among DZ cells (Fig. 2 G). Importantly, in PP, where there is no outgrowth of Fas-deficient GC B cells, we did not see a difference in active caspase-3 staining in Fas-deficient GC B cells in the LZ or DZ (Fig. S2 C). These data suggest that the accumulation of Fas-deficient GC B cells in the mLN is principally due to reduced death of GC B cells in the LZ.
GC Tfh cells likely promote death of GC B cells via Fas
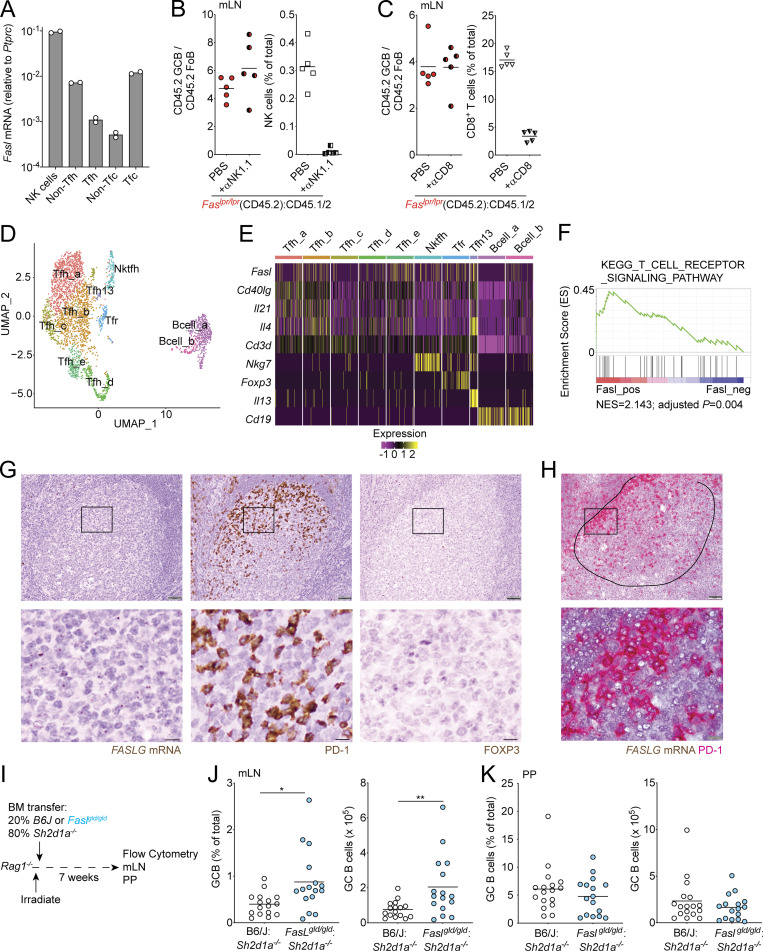
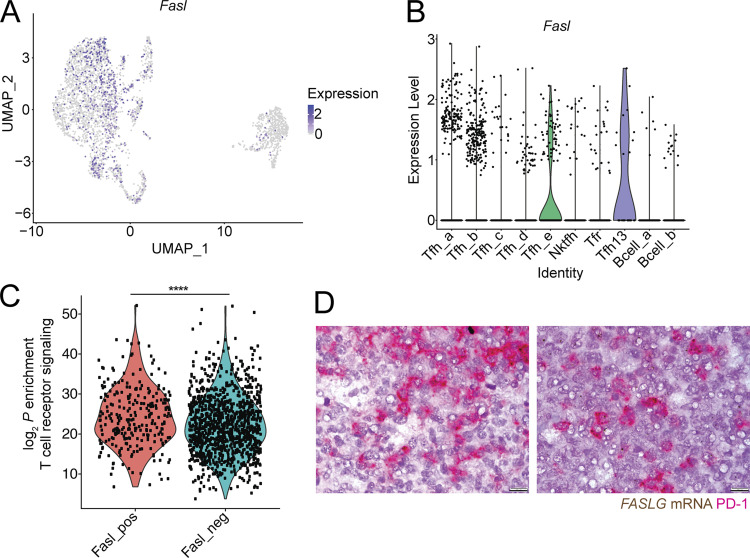
Our data suggest that Fas controls GC size by mediating the death of GC B cells primarily in the LZ. We sought to determine what cell types might be responsible for Fas-mediated death in the GC. Natural killer (NK) cells and some CD8 T cell subsets can express high levels of FasL mRNA (http://www.immgen.org/). However, NK cells are not commonly thought to be present in the B cell follicle or the GC. Although a subset of CD8+ T cells that express Cxcr5 (T follicular cytotoxic cells) have been reported to be present in the B cell follicle, it is unclear whether these cells are present in the GC itself. It has also been hypothesized that Tfh cells can kill GC B cells via Fas–FasL interactions. However, this has not been demonstrated experimentally, and bulk-sorted Tfh cells have been reported to express little Fasl mRNA (Bentebibel et al., 2011; Crotty, 2011; Weinstein et al., 2014). We found that sorted NK cells and T follicular cytotoxic cells from mLNs express Fasl mRNA. Consistent with prior findings, bulk-sorted Tfh cells did not show increased amounts of Fasl transcript compared with non-Tfh cells (Fig. 3 A). To determine if NK cells or CD8 T cells were responsible for deleting GC B cells via Fas, we treated Fas-mixed chimeras with NK-depleting or CD8 T cell–depleting antibody for 3 or 4 wk, respectively, and found that the outgrowth of Fas-deficient GC B cells in mLNs was not reduced following depletion of either NK cells or CD8+ T cells (Fig. 3, B and C). To determine whether a fraction or subset of Tfh cells expresses Fasl, we assessed Fasl expression on a per-cell basis in a publicly available single-cell RNA sequencing (scRNA-seq) dataset of sorted Tfh cells (Fig. 3, D and E; and Fig. S3; Gowthaman et al., 2019). Among the 2,455 cells that were present in Tfh clusters, 472 cells expressed detectable Fasl transcript, and these cells were present in all Tfh clusters (Fig. 3 E and Fig. S3, A and B). Among cells in Tfh clusters, Fasl expressers were more enriched for a TCR signaling gene signature than nonexpressers (Figs. 3 F and S3 C). Next, we performed RNAscope analysis of tonsillar GCs to assess FASL (FASLG) expression in situ. We found that FASLG was expressed in a distribution that was similar to PD-1+ cells (Tfh cells) in the GC (Fig. 3 G). Consistent with previous reports, Foxp3+ cells were very rare in GCs, suggesting that T follicular regulatory (Tfr) cells are unlikely to be responsible for Fas-mediated selection in the GC (Sayin et al., 2018). Co-staining showed that the large majority of FASLG-expressing cells in the GC also expressed PD-1 (Figs. 3 H and S3 D). To determine if Tfh-derived FasL could play a role in deleting GC B cells, we used Rag1−/− hosts to generate mixed chimeras where 80% of the BM was from SAP-deficient (Sh2d1a−/−) animals that are unable to generate Tfh cells and 20% was from Faslgld/gld BM (Fig. 3 I). In these animals, the only T cells capable of interacting with B cells are FasL deficient. When Tfh cells lacked FasL, we observed increased numbers of GC B cells in mLNs (Fig. 3 J). Importantly, GC B cells were not increased in PP when Tfh cells lacked FasL, consistent with our previous data showing that Fas does not limit GC B cell survival in PP (Fig. 3 K). These data suggest that Tfh cells are the likely source of FasL that mediates Fas-dependent selection of GC B cells in vivo.
Figure 3.
GC Tfh cells promote death of GC B cells via Fas. (A) Quantitative PCR for Fasl from sorted cells in indicated lymphocyte subsets from mLNs. Data are from two independent experiments. Tfc, cytotoxic T follicular cells. (B and C) Ratio of frequency of CD45.2 GC B cells to CD45.2 FoB cells or frequency of NK cells (B) or CD8 cells (C) in Faslpr/lpr mixed BM chimeras generated as in Fig. 1 C in mLNs treated with PBS or anti-NK1.1 for 3 wk (B) or with anti-CD8 antibody for 4 wk (C) before analysis. Data are from five mice per group. (D) Two-dimensional Uniform Manifold Approximation and Projection (UMAP) of the Gowthaman et al. (2019) scRNA-seq dataset of sorted Tfh cells showing 10 distinct cell clusters. (E) Heatmap of expression of Fasl, Cd40lg, Il21, Il4, Cd3d, Nkg7, Foxp3, Il13, and Cd19 among cell clusters. Five clusters of cells express Cd40lg, Il21, Il4, and Cd3d and are not enriched for Nkg7, Foxp3, Il13, or Cd19 (Tfh_a–e). One cell cluster coexpresses Cd3d and Nkg7 (Nktfh). One cluster coexpresses Cd3d and Foxp3 (Tfr). One cell cluster is enriched for Il13 and represents the recently described IL-13–producing Tfh (Tfh13) cell population (Gowthaman et al., 2019). Cd19 is enriched in two clusters of B cells that are not enriched for Cd3d (Bcell_a and Bcell_b). Cd79a and Ms4a1 are also enriched in these clusters (not shown). (F) Gene set enrichment analysis of differentially expressed genes from cells expressing Fasl versus nonexpressers in Tfh clusters (Tfh_a–e) compared with the TCR signaling pathway gene set. NES, normalized enrichment score. (G and H) RNAscope analysis for FASLG mRNA (brown) in human tonsillar sections. PD-1 and FOXP3 staining of serial sections is shown in G. Costaining for FASLG (brown) and PD-1 (red) is shown in H. Original magnification, 20× in G, 20× in top panel in H, and 100× in bottom panel in H. Circled area in H denotes GC boundary. Scale bars, 50 µm in top panels and 10 µm in bottom panels. Data in G and H are representative of at least three independent experiments. (I) Experimental scheme for data in J and K. (J and K) Frequency or number of GCB in mLNs (J) or PPs (K) of mixed BM chimeras generated by reconstituting irradiated Rag1−/− hosts with a mixture of 80% Sh2d1a−/− (SAP KO) and 20% B6/J or Faslgld/gld BM assessed by FACS. Data in J and K are pooled from three independent experiments with five to seven mice per group per experiment. *, P < 0.05; **, P < 0.01, unpaired two-tailed Student’s t test.
Figure S3.
Fasl is expressed in a fraction of Tfh. (A) Single-cell transcript levels of Fasl in the Gowthaman et al. (2019) scRNA-seq dataset of sorted Tfh illustrated in a Uniform Manifold Approximation and Projection (UMAP) plot. (B) Violin plot of Fasl expression among cell clusters. (C) Enrichment (log2 P values) of TCR signaling gene signature in Fasl-expressing (Fasl_pos) versus -nonexpressing (Fasl_neg) cells in Tfh clusters (Tfh_a–e) determined by a one-sided Fisher’s exact test. (D) Additional examples of costaining for FASLG (brown) and PD-1 (red) in human tonsillar GCs. Original magnification, 100×. Scale bar, 10 µm. ****, P < 0.0001, unpaired two-tailed Student’s t test, for data in C.
Fas is required to select against cells that do not bind antigen in immunized GCs
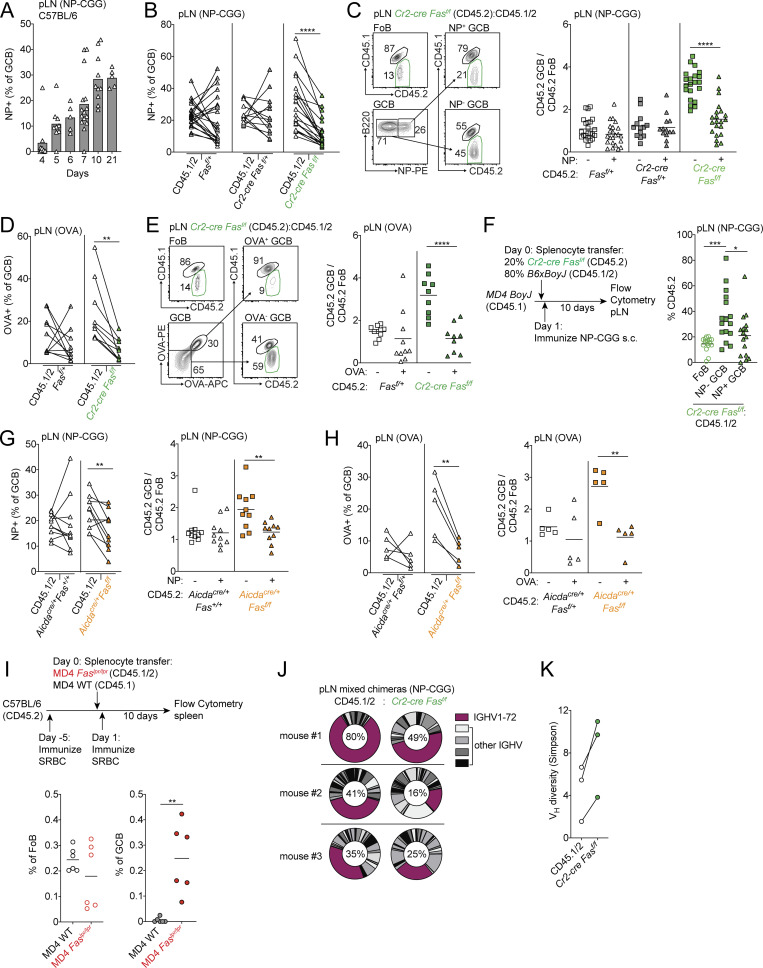
In polyclonal systems, B cells expressing BCRs with a wide range of affinity for the immunizing antigen can enter nascent GCs (Kuraoka et al., 2016; Tas et al., 2016). A significant fraction of GC B cells express BCR with no measurable affinity to the immunizing antigen in native or denatured forms (Kuraoka et al., 2016). During the course of the GC response, the frequency of cells that do not demonstrably bind antigen in the GC decreases. It has been proposed that the frequency of cells that bind antigen increases over time as a result of these cells receiving increased T cell help and consequently undergoing clonal bursts (Mesin et al., 2016). However, whether there are mechanisms that actively delete cells that do not demonstrably bind antigen during the course of a GC response is unclear. We immunized WT C57BL/6 mice with 4-hydroxy-3-nitrophenyl acetyl conjugated to chicken gamma globulin (NP-CGG) in alum and analyzed the frequency of NP-binding cells among GC B cells in nascent GCs and at later time points. In WT animals, the frequency of NP-binding cells among total GC B cells increased steadily over the course of immunization (Fig. 4 A). Expansion of clones that have been positively selected as a result of acquisition of antigen via BCR likely explains part of this increase.
Figure 4.
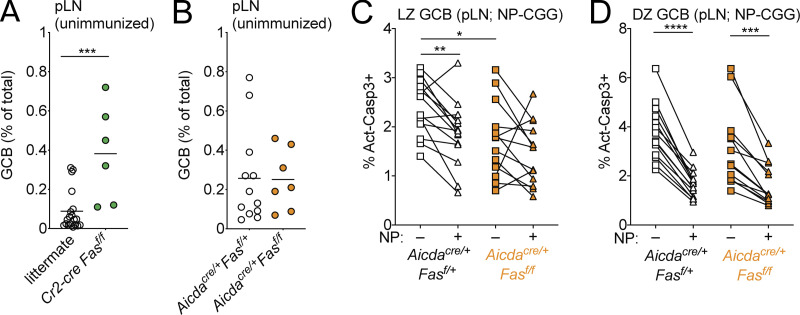
Fas is required to select against cells that do not strongly bind antigen in immunized GCs. (A) Frequency of NP-binding GC B cells among total GC B cells in pLNs of WT mice at indicated time points following immunizations with NP-CGG in alum. Data are pooled from four independent experiments with 5–10 mice per time point. (B) Frequency of NP binding in WT (CD45.1/2) GC B cells or CD45.2 GC B cells that were Fasf/+, Cr2-cre Fasf/+, or Cr2-cre Fasf/f in pLNs of mixed BM chimeras generated as in Fig. 1 F 10 d following s.c. immunization with NP-CGG in alum. (C) Ratio of frequency of CD45.2 non–NP-binding or NP-binding GC B cells to CD45.2 FoB cells in mixed chimeras from B. Example gating strategy for NP-specific GC B cells is shown on the left. Data in B and C are pooled from two independent experiments with six or seven mice per group per experiment. (D) Frequency of OVA binding in WT (CD45.1/2) GC B cells or CD45.2 GC B cells that were Fasf/+ or Cr2-cre Fasf/f in pLNs of mixed BM chimeras generated as in Fig. 1 F on day 21 following s.c. immunization with OVA in alum on days 0, 2, and 4. (E) Ratio of frequency of CD45.2 non–OVA-binding or OVA-binding GC B cells to CD45.2 FoB cells in mixed chimeras from D. Example gating strategy for OVA-specific GC B cells is shown on the left. Data in D and E are pooled from two experiments with four or five mice per group per experiment. (F) Frequency of CD45.2 cells among FoB cells or non–NP-binding GC B cells or NP-binding GC B cells derived from donor cells in pLNs of MD4 BoyJ mice that were given a mixture of splenocytes that were 80% WT (CD45.1/2) and 20% Cr2-cre Fasf/f, immunized with NP-CGG in alum s.c. 1 d after transfer, and analyzed 10–14 d later. Data are pooled from five independent experiment with three to five mice per experiment. Only mice with >5% of total GC B cells that bound NP were included in this analysis. (G and H) Frequency of antigen binding in WT (CD45.1/2) GC B cells or CD45.2 GC B cells that were Aicdacre/+Fas+/+ or Aicdacre/+Fasf/f in pLNs of mixed BM chimeras generated as in Fig. 2 D and immunized as in B and D, respectively (left panels), or the ratio of frequency of CD45.2 non–antigen-binding or antigen-binding GC B cells to CD45.2 FoB cells in mixed chimeras (right panels). Data in G and H are from 10 and 5 mice per group, respectively. (I) CD45.2 C57BL/6 mice were immunized with SRBC 5 d before receiving a mixture of CD45.1 MD4 WT or CD45.1/2 MD4 Faslpr/lpr splenocytes. Recipients were immunized with SRBC 1 d after transfer and analyzed 10 d later. Shown is the frequency of CD45.1 MD4 WT or CD45.1/2 MD4 Faslpr/lpr cells among all FoB cells or all GC B cells (GCB). Data are pooled from two independent experiments with six mice in total. (J) Vh usage from heavy-chain repertoire sequencing of sorted day 10 pLNs from WT CD45.1/2 or Cr2-cre Fasf/f GCB cells from mixed chimeras generated as in Fig. 1 F that were immunized s.c. with NP-CGG. The percentage of reads with IGHV1-72 among total reads is shown. (K) Simpson’s diversity of Vh usage from repertoire sequencing in J. Data in J and K are from three mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, paired two-tailed Student’s t test.
B cells expressing BCRs that fail to demonstrably bind immunizing antigen can be activated and recruited to early GCs, but there appears to be selection against these cells during GC reactions (Frank et al., 2015; Kuraoka et al., 2016; Tas et al., 2016). Earlier work demonstrated that overexpression of anti-apoptotic Bcl2 family members in B cells led to impaired negative selection against non–hapten-binding cells, which are likely to include cells that do not bind or only poorly bind antigen (Takahashi et al., 1999). To test whether Fas could play a role in selection against non–hapten-binding GC B cells in polyclonal B cell responses, we immunized Cr2-cre Fasf/f or control mixed BM chimeras with NP-CGG in alum and assessed the frequency of NP-binding cells at day 10 following immunization. In the absence of Fas in mature B cells, the frequency of NP-binding GC B cells was reduced (Fig. 4 B). Outgrowths of Fas-deficient GC B cells following immunization were restricted to cells that failed to bind NP (Fig. 4 C). To assess whether these findings were extendable beyond immune responses to haptens, we immunized Cr2-cre Fasf/f or control mixed BM chimeras with OVA in alum and stained GC B cells with OVA tetramers 21 d after immunization (Kim et al., 2019). We found that the frequency of OVA-binding Fas-deficient GC B cells was reduced compared with internal controls and that outgrowths of Fas-deficient GC B cells were restricted to cells that could not demonstrably bind OVA (Fig. 4, D and E).
Cr2-cre Fasf/f animals develop increased GCs in peripheral tissue in the absence of immunization relative to littermate controls, whereas Aicdacre/+Fasf/f do not (Fig. S4, A and B). Therefore, it was possible that the outgrowth of Fas-deficient GC B cells that failed to bind NP following immunization were derived from preexisting spontaneous GCs that form when mature B cells lack Fas. To exclude this possibility, we first transferred CD45.2 Cr2-cre Fasf/f and CD45.1/2 WT splenocytes into CD45.1 MD4 Ig-transgenic hosts and immunized animals with NP-CGG subcutaneously 1 d later. We analyzed animals 10–14 d after immunization and again found that the accumulation of Fas-deficient GC B cells was restricted to those cells that did not bind NP (Fig. 4 F). Second, we immunized Aicdacre/+ Fasf/f or control mixed chimeras with NP-CGG or OVA and assessed the frequency of antigen-binding cells 10 or 21 d following immunization, respectively. Again, the frequency of antigen-specific cells was reduced in Fas-deficient GC B cells, and outgrowths of Fas-deficient GC B cells were restricted to non–antigen-binding cells (Fig. 4, G and H). We next assessed active caspase-3 in non–antigen-binding and antigen-binding GC B cells from control or Aicdacre/+ Fasf/f animals directly ex vivo. There was reduced cell death in antigen-binding LZ GC B cells compared with non–antigen-binding cells in control but not Fas-deficient animals (Fig. S4 C). In contrast, in the DZ, cell death was reduced in antigen-binding DZ GC B cells in both control and Fas-deficient animals (Fig. S4 D).
Figure S4.
Frequency of GC B cells in pLNs of unimmunized Fas-deficient mice. (A and B) Frequency of GC B cells among total cells in unimmunized pLNs from littermate control or Cr2-cre Fasf/f animals (A) or Aicdacre/+Fasf/+ or Aicdacre/+Fasf/f animals (B). Data are from four and three independent experiments, respectively, with one to four mice per group. (C and D) Intracellular FACS for active caspase-3 in non–NP-binding or NP-binding LZ or DZ GC B cells from pLNs of Aicdacre/+Fasf/+ or Aicdacre/+Fasf/f animals 12 d following s.c. immunization with NP-CGG in alum. Data are from three independent experiments with three to six animals per group. ***, P < 0.001, unpaired two-tailed Student’s t test, for data in A. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, paired two-tailed Student’s t test, for data in C and D comparing non–NP-binding with NP-binding cells. *, P < 0.05, unpaired two-tailed Student’s t test, for data in C comparing non–NP-binding cell control and Aicdacre/+Fasf/f animals.
We then asked whether Fas deficiency could promote the persistence of GC B cells of defined specificity in animals that had been immunized with an unrelated antigen. We immunized WT CD45.2 animals with SRBCs and 5 d later transferred a mixture of splenocytes from CD45.1/2 MD4 Ig-transgenic Faslpr/lpr and CD45.1 MD4 Ig-transgenic Fas+/+ animals. The next day, recipients were immunized again with SRBCs and analyzed 10 d later. We found that MD4 Faslpr/lpr cells were able to enter and persist in GCs of recipient animals, whereas MD4 WT cells were not (Fig. 4 I). It has previously been shown in a BCR transgenic system that “rogue” non–antigen-binding GC B cells arise from antigen-binding precursors through somatic hypermutation in the absence of Fas late in the GC response (Butt et al., 2015). To determine if, in polyclonal systems, the increase in non–antigen-binding cells was due to a similar mechanism or, alternatively, due to a failure to delete non–antigen-binding clones that are often found in early GCs, we performed heavy-chain repertoire sequencing of sorted WT CD45.1/2 or Cr2-cre Fasf/f GC B cells from pLNs of NP-CGG–immunized mixed chimeras and assessed V gene usage. Among Fas-deficient GC B cells, we found reduced use of the canonical NP-binding variable segment IGHV1-72 and increased diversity of V genes compared with internal controls in all three animals that were tested (Fig. 4, J and K). These data demonstrate that Fas promotes negative selection of GC B cells that do not demonstrably bind antigen in polyclonal immune responses and is an important mechanism that allows for accumulation of antigen-binding GC B cells during the course of an immune response.
FAS alterations define a distinct subtype of lethal GC-derived DLBCL
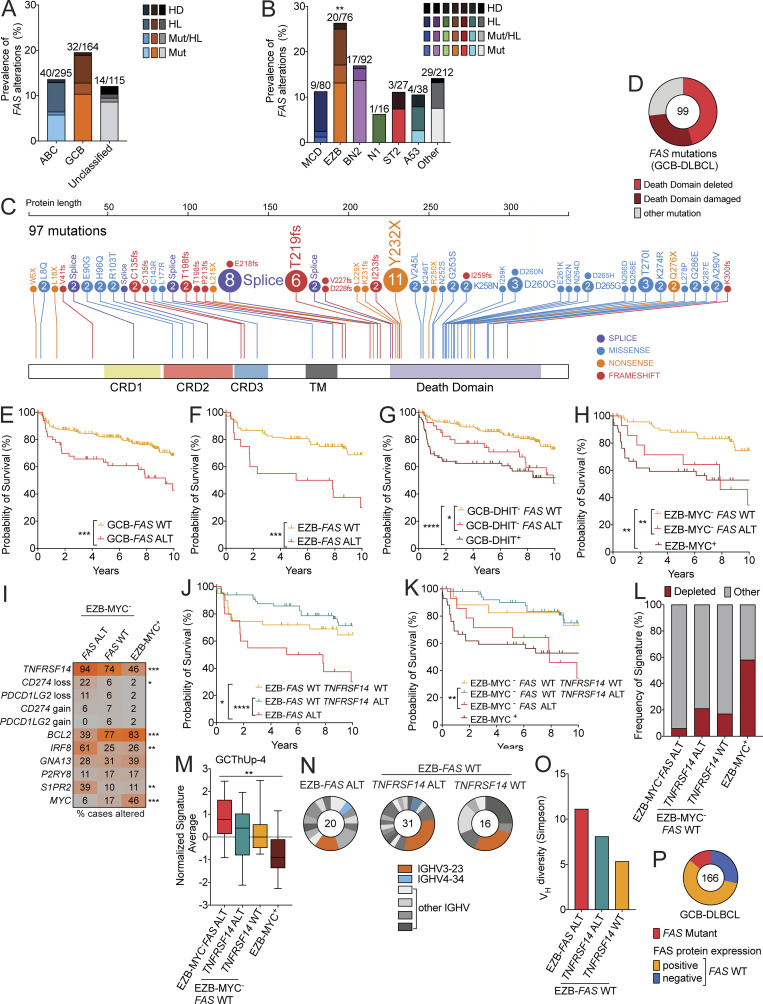
In a cohort of 574 DLBCL cases in which the mutational landscape, copy number alterations, and gene expression have been comprehensively characterized (National Cancer Institute [NCI] cohort; Schmitz et al., 2018; Wright et al., 2020), alterations in FAS were seen more often in GCB-DLBCL than in ABC-DLBCL or unclassified DLBCL cases (Fig. 5 A). Among genetic subtypes of DLBCL, FAS mutations, heterozygous loss, and homozygous deletions (alterations [ALTs]) were most enriched among EZB cases (26% of cases; Fig. 5 B). To more comprehensively characterize the landscape of FAS mutations in GCB-DLBCL, we analyzed exome sequencing data from this and other published studies (Arthur et al., 2018; Chapuy et al., 2018; Ennishi et al., 2019; Morin et al., 2013; Scherer et al., 2016; Schmitz et al., 2018) and found that Fas was mutated more frequently in GCB-DLBCL than other subtypes (Fig. S5 A; Arthur et al., 2018; Chapuy et al., 2018; Ennishi et al., 2019; Morin et al., 2013; Scherer et al., 2016; Schmitz et al., 2018). Overall, 99 FAS mutations were found in 90 cases of GCB-DLBCL. 45 mutations were predicted to express a surface protein that completely lacked a DD. An additional 28 cases had DD mutations that were predicted to be structurally damaging by PolyPhen-2 analysis (Adzhubei et al., 2010; Fig. 5, C and D; and Table S1). The distribution of FAS mutations in GCB-DLBCL was strikingly similar to the distribution of FAS mutations in the autoimmune lymphoproliferative syndrome (ALPS). In a well-described cohort of ALPS patients, of the 125 FAS mutations reported, 52 were predicted to express a protein that was expressed on the surface that completely lacked a DD, and an additional 30 cases had missense mutations in the DD that perturb FAS function (Price et al., 2014). Due to the trimeric structure of Fas oligomers before ligation, a single allele of FAS with a disrupted or deleted DD, as frequently occurs in both GCB-DLBCL and ALPS, functions as a dominant negative (Siegel et al., 2000).
Figure 5.
FAS alterations define a distinct subtype of lethal GC-derived DLBCL. (A) Frequency of FAS mutations (Mut), heterozygous loss (HL), or homozygous deletion (HD) in GCB-DLBCL, ABC-DLBCL, and unclassified cases reported by Schmitz et al. (2018). (B) Frequency of FAS Mut, HL, or HD in genetic subtypes of DLBCL. (C) Comprehensive analysis of nonsynonymous coding mutations of FAS in GCB-DLBCL from published cohorts. Cysteine-rich domains (CRD) 1, 2, and 3; transmembrane (TM) domain; and DD are indicated. (D) Frequency of mutations leading to a FAS protein that is expressed on the surface lacking a DD or mutations predicted to disrupt DD structure. (E and F) Overall survival of GCB-DLBCL (E) or EZB (F) in cases with or without a FAS mut, HL, or HD (ALT). (G and H) Overall survival of GCB-DLBCL (G) or EZB (H) lacking a double-hit signature (DHIT– or MYC–, respectively) with or without a FAS alteration. (I) Frequency of alterations of selected genes in EZB-MYC– FAS ALT, EZB-MYC– FAS WT, or EZB-MYC+ cases. (J and K) Overall survival of EZB (J) or EZB-MYC– (K) that were FAS ALT or FAS WT TNFRSF14 ALT. (L and M) Frequency of LME-depleted signature (L) or Tfh signature (M) in EZB-MYC– FAS ALT, EZB-MYC– FAS WT TNFRSF14 ALT, EZB-MYC– FAS WT TNFRSF14 WT, or EZB-MYC+ cases. (N) Vh usage in EZB-FAS ALT, EZB-FAS WT TNFRSF14 ALT, or EZB-FAS WT TNFRSF14 WT. The number of cases with identified Vh segments is shown. IGHV3-23 is the most commonly used V gene in the normal human B cell repertoire. IGHV4-34 encodes a self-reactive BCR that is enriched in several non–GC-derived lymphomas. (O) Simpson’s diversity of Vh usage across EZB samples that were FAS ALT, FAS WT TNFRSF14 ALT, or FAS WT TNFRSF14 WT. (P) FAS protein expression assessed on a tissue microarray including GCB-DLBCL. The total number of cases is indicated. **, P < 0.01 Fisher’s exact test of EZB compared with all others in B. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****,P < 0.0001, log-rank test, for data in E, F, G, H, J, and K. *, P < 0.05; **, P < 0.01; ***, P < 0.001, χ2 test, for data in I. **, P < 0.01, one-way ANOVA, for data in M.
Figure S5.
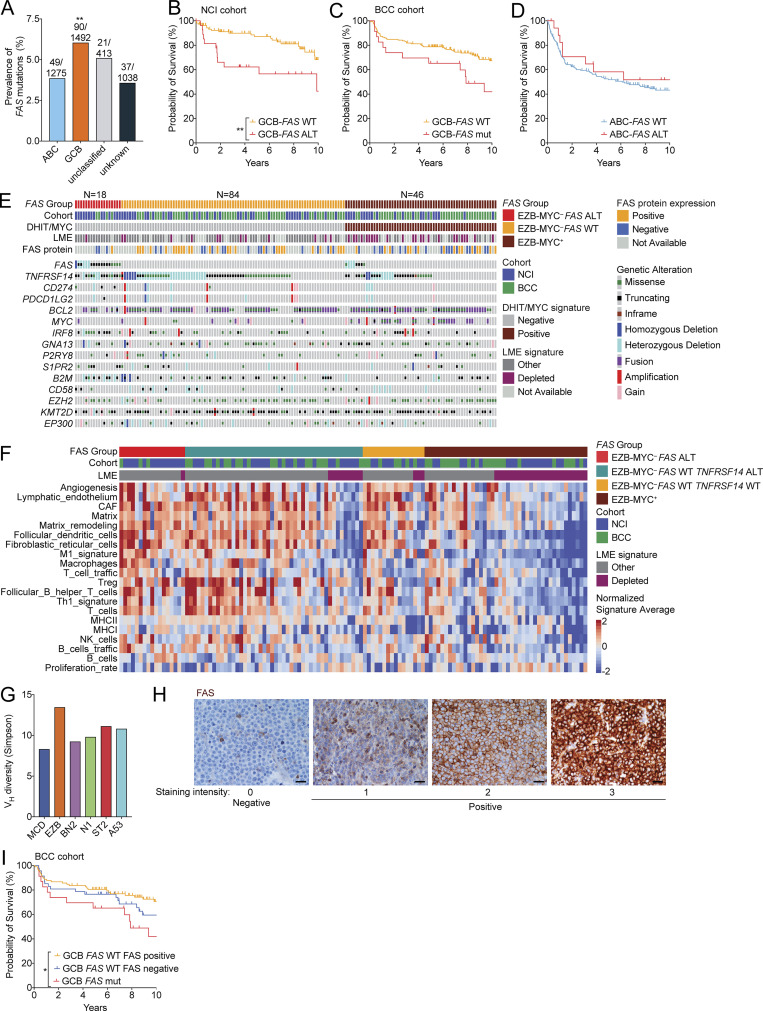
Landscape of genetic alterations in FAS mutant EZB DLBCL. (A) Frequency of FAS mutations in GCB-DLBCL, ABC-DLBCL, and unclassified and unknown cases across all published cohorts. (B and C) Overall survival of GCB-DLBCL with or without a FAS copy number alteration (CNA) or mutation from the NCI (A) or BCC (B) cohorts. FAS CNA information was not available in the BCC cohort. (D) Overall survival of ABC-DLBCL with or without a FAS mutation. (E) Landscape of genetic alterations in selected genes, LME signatures, or FAS protein expression assessed in a tissue microarray in EZB-MYC– FAS ALT, EZB-MYC– FAS WT, or EZB-MYC+ cases. (F) Heatmap of gene signatures used to assess LME signatures in EZB-MYC– FAS ALT, FAS WT TNFRSF14 ALT, FAS WT TNFRSF14 WT, or EZB-MYC+ cases. (G) Simpson’s diversity of Vh usage across genetic subtypes of DLBCL. (H) Representative FAS immunohistochemistry in GCB-DLBCL. Scale bar, 50 µm. (I) Overall survival of FAS WT GCB-DLBCL cases with or without FAS protein expression in the BCC cohort. **, P < 0.01, χ2 test of GCB-DLBCL compared with all others in A. *, P < 0.05; **, P < 0.01, log-rank test, for data in B and I.
We assessed the overall survival of patients within GCB-DLBCL or EZB with or without FAS ALTs in a combined cohort of patients from two centers who had been classified into gene expression and genetic subtypes with paired gene expression data (NCI and British Columbia Cancer [BCC] cohorts; Ennishi et al., 2019, 2020; Schmitz et al., 2018; Wright et al., 2020). In both GCB-DLBCL and EZB, loss of FAS was associated with worse outcomes compared with patients with WT FAS (Fig. 5, E and F; Fig. S5, B and C; and Table S2). In contrast, FAS ALTs were not associated with inferior outcomes in ABC-DLBCL (Fig. S5 D). To exclude the possibility that an enrichment for an MYC signature in FAS alteration cases was accounting for this difference in survival, we assessed overall survival of FAS-deficient cases among GCB-DLBCL DHIT− and EZB-MYC− cases and again found that loss of FAS was associated with a worse overall survival (Fig. 5, G and H).
We then compared the landscape of genetic alterations in EZB-MYC− FAS ALT, EZB-MYC– FAS WT, and EZB-MYC+ cases (Fig. 5 I and Fig. S5 E). Interestingly, we found that alterations in TNFRSF14 (encoding HVEM) were highly enriched among EZB-MYC− FAS mutant cases compared with EZB-MYC− FAS WT and EZB-MYC+ cases. HVEM, a tumor suppressor in both GCB-DLBCL and follicular lymphoma, has recently been shown to negatively regulate the GC by reducing the amount of CD40L delivered by Tfh cells to GC B cells (Mintz et al., 2019). HVEM deficiency in mouse models more strongly promotes expansion of non–hapten-binding GC B cells (Mintz et al., 2019). Importantly, combined loss of FAS and HVEM, as occurs in 17 of 18 EZB-MYC− FAS ALT cases, was associated with worse overall survival compared with loss of HVEM alone independent of an MYC signature (Fig. 5, J and K). In addition to alterations in TNFRSF14, loss-of-function mutations in CD274 (encoding PD-L1) were more frequent in EZB-MYC− FAS ALT (3 of 18 cases) compared with FAS WT (1 of 84 cases; Figs. 5 I and S5 E). An additional EZB-MYC− FAS ALT case harbored a loss-of-function mutation in PDCD1LG2 (encoding PD-L2). Ligation of PD-1 on Tfh cells by PD-L1 on GC B cells has also been shown to negatively regulate T cell help and enforce selection of hapten-binding GC B cells (Shi et al., 2018). A significant enrichment of TNFRSF14 (P < 0.0001) and CD274 (P = 0.0014) mutations in FAS mutant cases was also seen in GCB-DLBCL across published DLBCL cohorts (Table S3). IRF8 mutations, which frequently target the C-terminus of the protein and may result in altered function, were also enriched in EZB-MYC− FAS ALT cases. IRF8 activity has been reported to promote transcription of FAS, suggesting that loss of FAS may be one mechanism that enables GC-derived tumors to tolerate higher levels of IRF8 activity (Yang et al., 2007). BCL2 alterations, in contrast, were enriched among EZB-MYC− FAS WT cases.
Given the enrichment of loss-of-function alterations in multiple additional negative regulators of the Tfh–GC B cell interaction in EZB-MYC− FAS ALT cases, we then asked if there were changes in the cellular composition of these tumors compared with FAS WT or EZB-MYC+ cases. We assessed microenvironmental gene expression signatures in RNA sequencing datasets from EZB-MYC− with or without alterations in FAS, TNFRSF14, or EZB-MYC+ cases. Recent work has identified a subgroup of tumors with signatures correlated with a microenvironment depleted of immune and stromal cells and enriched for malignant cells (lymphoma microenvironment [LME] depleted) that is associated with a poor prognosis independent of DLBCL subtype (Cerchietti et al., 2019). Surprisingly, despite the poor prognosis of EZB-MYC− FAS ALT cases, only one case had the LME-depleted signature, suggesting that cues in the microenvironment might be more important for driving lymphoma progression in this subset of tumors than others (Figs. 5 L and S5 F). Consistent with these data, we found that there was an enrichment of a Tfh cell gene signature in EZB-MYC− FAS ALT cases (Fig. 5 M). We assessed the diversity of Vh usage across samples of genetic subtypes of DLBCL. EZB had higher Vh diversity than other genetic subtypes (Fig. S5 G). Within EZB, cases with alterations in FAS (n = 20) or cases with alterations in TNFRSF14 that were FAS WT (n = 31) had greater Vh diversity across samples compared with cases that were WT for both (n = 16) in the limited number of tumors for which Vh usage was available (Fig. 5, N and O). Taken together, these data show that FAS genetic alterations in GC-derived lymphomas are associated with a worse prognosis and co-occur with loss of other negative regulators of the Tfh–GC B cell interaction that may contribute to loss of counterselection.
Given the high frequency of loss of Fas expression in tumors from the mouse (Fig. 1, A and B), we asked whether FAS expression could be lost in GC-derived tumors via nongenetic mechanisms. We assessed FAS protein expression in a DLBCL tissue microarray and found that FAS protein expression was lost in 33% of FAS WT GCB-DLBCL and 44% of FAS WT EZB (Fig. 5 P and Fig. S5, E and H). However, loss of FAS expression in FAS WT cases was not associated with a statistically significant difference in survival or a distinct mutational landscape (Fig. S5, E and I). Finally, to determine how often nongenetic mechanisms contributed to loss of Fas expression in Gα13-deficient tumors in the mouse, we performed Sanger sequencing of Fas from cDNA from Fas-deficient tumors. Of the nine tumors analyzed, four had Fas coding mutations that were predicted to be structurally disruptive (Table S4). We were not able to identify a Fas coding mutation in cDNA or genomic DNA from the remaining five tumors. Therefore, FAS is lost via genetic and nongenetic mechanisms in GC-derived tumors in both humans and mice.
Discussion
Although GC B cells express high amounts of the death receptor Fas, its function in supporting GC homeostasis has been controversial. In this study, we showed that Fas is a critical regulator of GC selection that strongly suppresses accumulation of GC B cells in a cell-intrinsic manner. Fas promoted death of GC B cells in the LZ, which was likely due to Tfh-derived FasL. Fas suppressed clonal diversity in the GC by promoting the elimination of B cell clones that did not strongly bind antigen to support counterselection. Genetic alterations in FAS occurred most often in GCB-DLBCL and in the related genetic subtype of DLBCL, EZB. FAS alterations co-occurred with loss of HVEM and PD-1 ligands and was associated with increased lethality and an altered microenvironment with evidence of increased Tfh cells. Finally, lymphomas with genetic alterations in FAS or HVEM showed greater Vh diversity across samples, raising the possibility that these tumors are derived from GC B cells that do not strongly bind antigen.
Several studies focusing on antigen-binding cells or using B cells with a defined specificity have shown no increase in GC size in the absence of Fas (Butt et al., 2015; Smith et al., 1995; Takahashi et al., 2001), whereas increased total GC B cells were observed in animals lacking Fas in B cells that also showed systemic autoimmunity and lymphoproliferation (Hao et al., 2008). Increased GC size in this latter study was thought to occur as a result of systemic autoimmunity rather than a GC B cell–intrinsic function of Fas (Allen, 2015). Our study addresses this issue by showing that in polyclonal settings, Fas prevents accumulation of cells that do not demonstrably bind antigen but has little effect on cells that do bind antigen. We also show that in mixed transfer settings, a strong cell-intrinsic expansion of Fas-deficient GC B cells still occurs in the absence of autoimmunity or systemic lymphoproliferation.
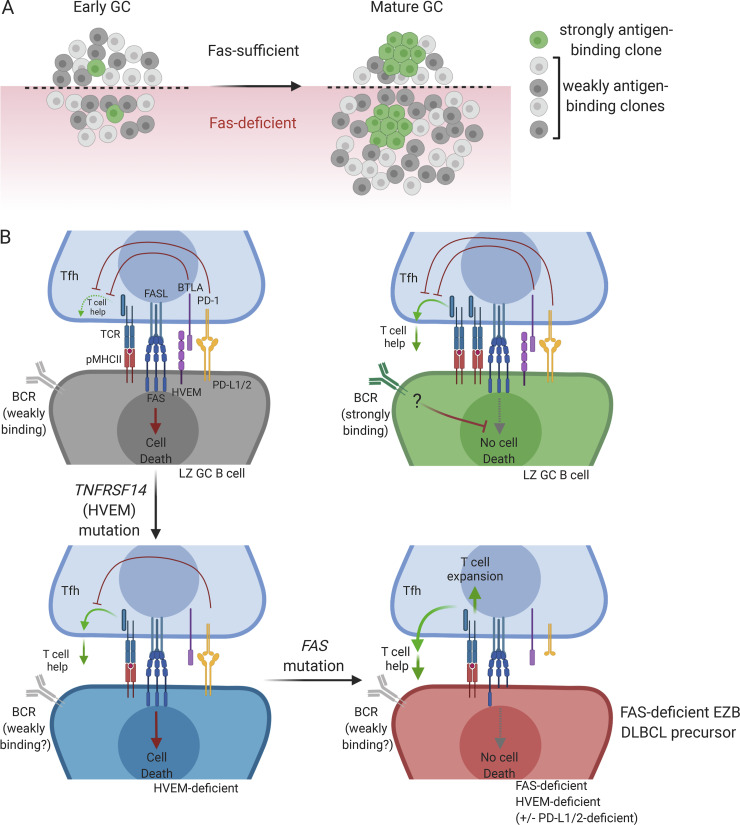
In the absence of Fas in monoclonal systems, rogue non–antigen-binding clones arise from B cells with a defined antigen specificity late during the GC reaction (Butt et al., 2015). However, recent work in polyclonal systems has estimated that individual GCs can be seeded by tens to hundreds of B cell clones, many of which express BCRs that do not demonstrably bind immunizing antigen (Kuraoka et al., 2016; Tas et al., 2016). The frequency of cells expressing BCR that do not demonstrably bind antigen decreases over time (Frank et al., 2015; Kuraoka et al., 2016). Rather than principally functioning to eliminate rogue GC B cells in systems with a defined antigen specificity, our data suggest that in polyclonal systems, Fas-mediated death is an important mechanism to delete the large number of clones that are recruited to the GC that do not strongly bind antigen (Fig. 6 A). BCR stimulation can protect activated B cells from Fas-mediated death (Rathmell et al., 1996; Rothstein et al., 1995). Further work is needed to establish whether increased BCR signaling protects GC clones that strongly bind antigen from FAS-mediated death in vivo (Fig. 6 B). In GC-derived lymphoma, loss of FAS may limit the ability of the GC to focus the reaction toward clones that bind particular antigens and away from clones that do not bind antigens strongly, some of which may acquire deleterious mutations in other lymphoma-associated loci.
Figure 6.
Loss of multiple negative regulators of the T cell–GC B cell integration in FAS mutant EZB DLBCL. (A) Early GCs are seeded by many B cell clones with negligible affinity for the immunizing antigen. Over time, these weakly antigen-binding clones are deleted in a Fas-dependent manner but accumulate in the absence of Fas. (B) GC B cells that do not strongly bind antigen are counterselected via FAS-mediated cell death and diminished T cell help as a result of inhibition of T cell help by HVEM/BTLA and PD-L1/PD-1 interactions (upper left panel). GC B cells that strongly bind antigen are protected from Fas-mediated death, possibly as a result of BCR signaling (upper right panel). In the absence of HVEM, the inhibition of T cell help is weakened, thereby allowing for some expansion of weakly antigen-binding B cell clones, but these cells can still be killed via Fas (lower left panel). With loss of FAS and HVEM (and, in some cases, PD-L1/2), as occurs in FAS-deficient EZB, there is positive feedback between weakly antigen-binding GC B cells and Tfh, promoting expansion of both cell types (lower right panel).
Our data suggest that Tfh cells are responsible for deleting GC B cells via Fas in the LZ. A subset of CD8+ T cells are present in the B cell follicle, and, in a spontaneous mouse model of non–GC B cell–derived lymphoma that forms in the absence of T cells, CD8+ T cells have been reported to kill lymphoma cells in a partially FasL-dependent manner in vivo (Afshar-Sterle et al., 2014). However, these cells are not reported to be SAP dependent, and we found that the competitive advantage of Fas-deficient GC B cells was not dependent on the presence of CD8+ T cells. NK cells have been reported to suppress GC size during viral infection (Rydyznski et al., 2018). However, NK cells have not been reported to be present in the GC, and we did not find that the competitive advantage of Fas-deficient GC B cells was dependent on the presence of NK cells. Importantly, Tfh cells have previously been shown to have FasL-mediated cytotoxic potential in vivo (Kotov et al., 2018), and analysis of single-cell transcriptomic data showed that Fasl was expressed in a significant fraction of Tfh cells that were enriched for a TCR signaling signature. Our data show that Tfh cells are the primary FASLG-expressing cell in the GC in situ. Additionally, we found an accumulation of mLN GC B cells when FasL was absent in SAP-dependent cells, strongly suggesting that GC Tfh-derived FasL is responsible for Fas-dependent death in the LZ. It will be important in future studies to determine when and how expression of FasL is controlled in GC Tfh cells and to determine what role specific subsets of Tfh cells, such as Tfr, might play in Fas-mediated death of GC B cells.
Fas deficiency promoted a strong cell-intrinsic survival advantage in the GC in mLNs and immunized pLNs but not PP. Further studies are necessary to determine whether this might be due to an intrinsic resistance to Fas-mediated death in PP GC B cells or a decrease in FasL availability in this organ. The lack of survival advantage in PP could be explained by the increased presence of microbially derived antigens or other gut-derived cues in this location, which might act directly on PP GC B cells via BCR or innate immune receptors to promote resistance to Fas-mediated death. Further work is also needed to determine whether the lack of Fas-mediated negative selection in this organ is important for maintaining clonal diversity over long periods of time, as clones can persist in mouse PP GCs for at least 1 yr (Le Gallou et al., 2018). Maintenance of clonal diversity in PP GCs may play a role in the repertoire diversification that has been proposed to occur at this site (Reboldi and Cyster, 2016).
In the DZ, GC B cells that acquire structurally disruptive mutations to their BCR undergo apoptosis. In the LZ, it was previously hypothesized that the primary reason GC B cells undergo death is due to neglect, and those clones that die have failed to productively interact with T cells. Our data suggest that Fas-mediated negative selection by Tfh cells in the LZ is an important mechanism that induces the death of GC B cells. Fas-deficient GC B cells are biased to the LZ phenotype and undergo less proliferation than WT competitors, suggesting that cells that are normally targeted for Fas-mediated deletion are less competitive than those not targeted. Further work is needed to address the nature of interactions that lead to deletion via Fas. It remains to be determined whether FasL is present in all interactions and whether some GC B cells are resistant to Fas-mediated death as a result of other inputs or if FasL is only present in some interactions as a result of feedback between GC B cells and Tfh cells. The enrichment of a TCR signaling gene signature among Fasl-expressing Tfh cells suggests that the former possibility is more likely. Similar to the accumulation of non–antigen-binding cells in the absence of Fas, increased frequency of non–hapten-binding GC B cells has been reported in the absence of coinhibitory interactions from GC B cells to Tfh cells via PD-L1/PD-1 and HVEM/B and T lymphocyte-associated protein (BTLA). HVEM and PD-L1 limit positive selection of weakly antigen-binding cells by limiting the amount of help GC B cells receive from T cells (Mintz et al., 2019; Shi et al., 2018). In contrast, recent work has shown that expression of an oncogenic gain-of-function Ezh2 mutation in mouse GCs results in restricted clonality (Béguelin et al., 2020). Restricted clonality in this setting might be occurring as a result of ongoing signaling through Fas/FasL, HVEM/BTLA, and PD-L1/PD-1.
We found that nongenetic loss of FAS was very frequent in GC-derived DLBCL but was not associated with the distinct genetic landscape and clinical outcome of tumors carrying a genetic alterations in FAS. These data suggest that nongenetic loss of FAS may occur at a later stage of tumor development due to new selective pressures introduced as a result of a disrupted microenvironment.
The increased overall frequency of HVEM deficiency in EZB tumors compared with genetic loss of FAS and the presence of TNFRSF14 alterations in nearly all FAS altered cases suggests that TNFRSF14 mutations or deletions occur earlier than FAS alterations during lymphomagenesis. We speculate that survival of weakly antigen-binding cells is initially favored with loss of HVEM due to increased CD40L from Tfh cells. However, Tfh-mediated deletion of these cells can still occur via FAS (Fig. 6 B). We propose that when FAS is genetically lost, there is both increased T cell help as a result of loss of HVEM and/or PD-1 ligand inputs to T cells and a loss of Tfh-mediated apoptosis. Expansion of malignant FAS- and HVEM-deficient GC B cells would then support expansion of Tfh cells (Baumjohann et al., 2013). Although we cannot exclude the possibility that the inferior survival of patients with FAS-altered lymphomas is due to resistance to killing by CD8+ T cells or NK cells, our data support the hypothesis that, in the absence of FAS, HVEM, and/or PD-L1/2, Tfh cells play a supportive role in the survival of malignant B cells that enhances their pathogenicity. GC-derived DLBCL cases containing mutations in negative regulators of the Tfh–GC B cell interaction might derive greater benefit from therapies that interfere with this interaction rather than from therapies directed at promoting T cell activation, such as checkpoint blockade.
Materials and methods
Animals
Adult B6-Ly5.1/Cr (B6.SJL-PtprcaPepcb/BoyCrCrl; stock no. 564) and C57BL/6 (C57BL/6NCrl; stock no. 556) mice at least 6 wk of age were obtained from Charles River Frederick Research Model Facility. B6J (C57BL/6J; stock no. 000664), Faslpr/lpr (B6.MRL-Faslpr/J; stock no. 000482), Cr2-cre (B6.Cg-Tg(Cr2-cre)3Cgn/J; stock no. 006368), Fasf/f (C57BL/6-Fastm1Cgn/J; stock no. 007895), MD4 (C57BL/6-Tg(IghelMD4)4Ccg/J; stock no. 002595), Faslgld/gld (B6Smn.C3-Faslgld/J; stock no. 001021), Sh2d1a−/− (B6.129S6-Sh2d1atm1Pls/J; stock no. 025754), Rag1−/− (B6.129S7-Rag1tm1Mom/J; stock no. 002216), and Aicdacre/cre (B6.129P2-Aicdatm1(cre)Mnz/J; stock no. 007770) mice were on a B6 background and were from The Jackson Laboratory. Gna13f/f mice were backcrossed to B6 for nine generations and were from S. Coughlin (University of California, San Francisco, San Francisco, CA). MD4 mice were intercrossed with BoyJ (B6.SJL-Ptprca Pepcb/BoyJ; stock no. 002014) to generate MD4 BoyJ animals. Spontaneous lymphomas were assessed in Cr2-cre Gna13f/f or Mb1-cre Gna13f/f aged between 12 and 20 mo.
Treatments
BM chimeras were made using B6-Ly5.1/Cr mice from Charles River or Rag1−/− mice as hosts. B6-Ly5.1/Cr hosts were lethally irradiated with 900 rads, and Rag1−/− mice with 600 rads, both in split doses. Hosts were later reconstituted by tail vein injection with at least 3 × 106 BM cells from the indicated donors. Mice were analyzed at least 7 wk after reconstitution. Splenocyte transfers were made by using MD4 BoyJ mice as hosts. Approximately 2.5 × 107 total splenocytes from the indicated donors were transferred by tail vein injection. In some experiments, splenocytes were labeled with CTV (Invitrogen) according to the manufacturer’s protocol before transfer. CD8+ T cells were depleted in chimeras by weekly doses of 250 µg anti-CD8 (YTS-169.4; Bio X Cell) injected i.p. starting 4 wk before analysis. NK cells were depleted in chimeras by treating mice with 200 µg anti-NK1.1 (PK136; Bio X Cell) injected i.p. every 5 d starting 3 wk before analysis. Mice were immunized with NP-CGG (BioSearch Technologies) precipitated in Imject Alum (Thermo Fisher Scientific; 25 µg s.c. per LN), SRBCs (Colorado Serum Company) at the indicated time points, or OVA (Sigma-Aldrich) 10 µg, 20 µg, and 40 µg s.c. per LN in flank or footpad on days 0, 2, and 4, respectively. Mice were housed in a specific pathogen–free environment, and all mouse experiments were approved by the NCI Animal Care and Use Committee (ACUC) and were performed in accordance with NCI ACUC guidelines and under approved protocols.
Flow cytometry and cell sorting
pLN, spleen, mLN, and PP cell suspensions were generated by mashing the organs through 70-mm cell strainers in RPMI containing 2% (vol/vol) FBS, antibiotics (50 IU/ml penicillin and 50 mg/ml streptomycin; Cellgro), and 10 mM Hepes, pH 7.2 (Cellgro). Cells were stained as indicated with the following antibodies and dyes: BV786- or BUV395-conjugated anti-B220 (RA3-6B2; BD Biosciences); FITC- or APC-conjugated anti-CD4 (RM4-5; BioLegend); Pacific blue– or PerCP Cy5.5–conjugated GL7 (GL-7; BioLegend); BV650-conjugated anti-IgD (11-26c.2a; BioLegend); PerCP-Cy5.5-, PE-Cy7–, or APC-conjugated anti-CD38 (90; BioLegend); PE-Cy7– or PE-conjugated anti-Fas (Jo2; BD Biosciences); FITC- or Alexa Fluor 700–conjugated anti-CD45.2 (104; BioLegend); PerCP-Cy5.5–conjugated anti-CD45.1 (A20; BioLegend); BV786-conjugated anti-CD86 (GL-1; BioLegend); PE-, APC-, or Alexa Fluor 488–conjugated or biotinylated anti-CXCR4 (2B11; eBioscience), BV605 conjugated to streptavidin (BD Biosciences); biotinylated anti–Ephrin-B1 (R&D Systems); BV605-conjugated CXCR5 (L138D7; BioLegend); Pacific blue–conjugated PD-1 (RMP1-14; BioLegend); PE-conjugated CD44 (IM7; BioLegend); FITC-conjugated CD8 (53-6.7; BioLegend); Alexa Fluor 647–conjugated NKp46 (29A1.4; BioLegend); FITC-conjugated NK1.1 (PK136; BioLegend); PE-conjugated IgMa (MA-69; BioLegend); Alexa Fluor 647–conjugated active caspase-3 (C92-605; BD Biosciences); and NP-PE (BioSearch Technologies). OVA tetramers were generated by biotinylating OVA (Sigma-Aldrich) with the EZ-Link Sulfo-NHS-LC-Biotinylation Kit (Pierce) per the manufacturer’s protocols, and biotinylated OVA was labeled with streptavidin-PE or streptavidin-APC (ProZyme).
For BrdU incorporation experiments, animals were given 2.5 mg of BrdU in a single i.p. injection and sacrificed 30 min later. Staining was performed using the FITC BrdU Flow Kit (BD PharMingen) according to the manufacturer’s instructions. To stain for intracellular antigens, cells were first stained for surface markers and then fixed and permeabilized using the BD Cytofix/Cytoperm Kit per the manufacturer’s instructions. For anti–active caspase-3 staining, cells were maintained on ice during harvesting and processing with prechilled cell strainers to minimize cell death ex vivo. Cells were stained for with Alexa Fluor 647–conjugated anti–active caspase-3 (C92-605; BD Biosciences) following fixation and permeabilization according to the manufacturer’s instructions. Flow cytometry was performed on a Cytoflex LX device (Beckman Coulter). Cells were sorted on a BD FACS Aria Fusion sorter and were sorted directly into TRIzol LS reagent (Life Technologies).
RNA isolation, RT-PCR, heavy-chain repertoire sequencing, and Fas sequencing
Cells from mLNs or pLNs were sorted directly into TRIzol LS reagent (Life Technologies), and RNA was extracted according to the manufacturer’s protocol. Real-time PCR was performed with SYBR Green PCR Mix (Roche) and an ABI Prism 7500 sequence detection system (Applied Biosystems). The following primers were used: Fasl forward: 5′-TTAACAGGGAACCCCCACTC-3′, Fasl reverse: 5′-GGCTGGTTGTTGCAAGACTG-3′; Ptprc forward: 5′-TTCCAAGAGGAAGGAGCCCA-3′, Ptprc reverse: 5′-ACAAGGCACAGAACAACCCT-3′. For assessment of Vh gene usage, RNA from 3,000 to 25,000 sorted GC B cells was sent on dry ice to iRepertoire, Inc. cDNA synthesis, PCR amplification of heavy-chain repertoire, and analysis on an Illumina MiSeq Nano were performed with proprietary reagents by iRepertoire, Inc. 28,000–231,000 reads were obtained for each sample. Simpson’s diversity (D) of V genes present in >0.1% of reads was calculated using the formula
where p is the proportion of reads for an individual V gene among total reads, and S is the number of V genes used in >0.1% of reads. Coding regions for Fas were amplified by PCR using primers listed in Table S5 from cDNA or genomic DNA isolated from snap-frozen bulk tumor cell pellets. Sanger sequencing of PCR products was done bidirectionally. Sequence electropherograms were manually reviewed.
RNAscope in situ hybridization
The RNAscope 2.5 HD Assay-BROWN (Advanced Cell Diagnostics) was used according to the manufacturer’s instructions to perform in situ hybridization of Hs-FASLG (449051; ACDBio) on formalin-fixed, paraffin-embedded tonsil samples. POLR2A (310451; ACDBio) mRNA labeling was used as a positive control for the RNA quality in samples. For serial sections, PD-1 (NAT105; Roche) staining was performed on automated Leica BOND-MAX using BOND Polymer Refine Detection, and FOXP3 (236A/E7; Abcam) staining was performed on a Benchmark ULTRA (Roche) using an ultraView DAB Detection Kit. For costaining of FASLG mRNA and PD-1, directly after the 3,3′-diaminobenzidine incubation step of RNAscope, samples were rinsed in distilled water, and heat-induced antigen reretrieval was performed by steaming the slides in low-pH buffer (Dako Retrieval Solution; Dako) for 30 min. The slides were rinsed in Tris buffer solution, then immersed in 3% Tris–goat serum to block nonspecific binding. A 1:50 dilution of PD-1 mouse antibody (NAT105; Abcam) in DAKO antibody diluent (Dako) was applied and incubated for 2 h at room temperature. Slides were rinsed in Tris buffer, and the detection was performed on an automated immunostainer (Benchmark ULTRA; Roche) using a ultraView Universal AP Red Detection Kit with amplifier. Slides were rinsed in tap water, counterstained with hematoxylin, rinsed in tap water again, and dehydrated in graded alcohols and xylenes (3 × 5 min) before coverslipping.
scRNA-seq analysis
A publicly available scRNA-seq dataset of sorted Tfh cells (Gowthaman et al., 2019) was reanalyzed to assess Fasl expression using Seurat version 3.0 (Stuart et al., 2019). We generated rank files based on the fold change of mRNA expression between Fasl-expressing and -nonexpressing Tfh cells. The KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY gene set was downloaded from https://www.gsea-msigdb.org/gsea/msigdb/cards/KEGG_T_CELL_RECEPTOR_SIGNALING_PATHWAY. Gene set enrichment analysis was performed with GSEA_4.0.2 using the default setting (number of permutations = 1,000). T cell signaling enrichment was determined by calculating P values of Fisher’s exact test of the TCR signaling gene set in each cell.
DLBCL analysis
FAS alterations in gene expression and genetic subtypes of DLBCL were assessed in the NCI cohort (Schmitz et al., 2018; Wright et al., 2020). Primary data for this cohort are available through the National Institutes of Health Database of Genotypes and Phenotypes (accession nos. phs001444, phs001184, and phs000178). FAS, TNFRSF14, and CD274 mutations in GCB-DLBCL were characterized using data curated from 150 published studies that have reported somatic mutations in lymphoma. The reported mutations (n > 400,000) from >10,000 individuals were harmonized using TransVar (https://bioinformatics.mdanderson.org/transvar/) and subsequently annotated using VarScan (http://varscan.sourceforge.net/). All mutations present at a prevalence of >0.0001 among human donors were identified among exosomes from nonmalignant blood samples (http://gnomad.broadinstitute.org/) and removed from the harmonized dataset, as were variants present in the National Center for Biotechnology Information Single Nucleotide Polymorphism Database. Known cancer hotspot mutations (https://www.cancerhotspots.org/#/home) were retained. A schematic of FAS mutation locations was generated with ProteinPaint (https://pecan.stjude.cloud/proteinpaint). Overall survival and the landscape of genetic alterations in EZB cases were assessed in the NCI cohort and the BCC cohort (Ennishi et al., 2019, 2020). The depleted LME signature was recently described (Cerchietti et al., 2019). Briefly, an unsupervised Louvain clustering technique (Blondel et al., 2008) was applied to >4,500 DLBCL samples from the public data in the space of 25 LME- and tumor cell–related gene signatures. Four clusters with different biology and clinical outcomes were identified. The depleted LME cluster represents the DLBCL cluster with the lowest nontumor cell content and aggressive behavior. For the Tfh signature (GCThUp-4; GC_T_helper_up4x_Chtanova; Chtanova et al., 2004), digital gene expression signal values of the signature genes were averaged to provide a signature average value for each sample in the NCI cohort (Schmitz et al., 2018). Assembly of Vh from RNA-seq data in the NCI cohort has been reported previously (Wright et al., 2020). For immunohistochemical analysis of FAS expression in DLBCL, a 4-µm slide of a tissue microarray containing samples from the BCC cohort was assessed using a rabbit monoclonal antibody directed at the C-terminus of FAS (EPR5700; Abcam). Samples were scored as negative if <10% of cells expressed FAS. Most FAS-positive cases expressed FAS in >70% of cells. Clinical samples in the NCI and BCC cohorts were studied in accordance with the ethics and principles of the Declaration of Helsinki and under NCI Institutional Review Board/University of British Columbia–BCC Research Ethics Board–approved protocols, respectively. Informed consent was obtained from all patients or given an institutional review board or research ethics board waiver as archived tissue submitted for consultation to the Hematopathology Section.
Statistical analysis
Prism software (GraphPad Software) was used for all statistical analysis. Data were analyzed with a two-sample unpaired (or paired, where indicated) Student’s t test or Fisher’s exact test, or log-rank (Mantel-Cox) test or one-way ANOVA test, where indicated. P values were considered significant when ≤0.05.
Online supplemental material
Fig. S1 shows that Fas constrains Ephrin-B1+ GC B cells. Fig. S2 shows that Fas is up-regulated in EAB cells. Fig. S3 shows additional data related to FasL expression in Tfh cells. Fig. S4 shows the frequency of GC B cells in pLNs of unimmunized Fas-deficient mice. Fig. S5 shows additional data regarding genetic alterations associated with FAS mutations in DLBCL. Table S1 lists FAS mutations in GCB-DLBCL. Table S2 shows data from DLBCL cases. Table S3 shows co-occurrence of FAS mutations with mutations of TNFRSF14 or CD274.Table S4 shows analysis of Fas in mouse tumors. Table S5 shows primers used for amplification and sequencing of Fas from mouse tumors.
Supplementary Material
shows FAS mutations in GCB-DLBCL.
lists data from primary DLBCL cases.
displays the co-occurrence of FAS mutations with mutations of TNFRSF14 or CD274 across published DLBCL datasets.
shows the sequencing of Fas in Fas-negative mouse tumors.
lists the primers used for amplification and sequencing of Fas from mouse tumors.
Acknowledgments
We thank L. Staudt for assistance with DLBCL genomic analyses and critical discussions; J. Cyster for providing resources and guidance in early stages of this project and for comments on the manuscript; M. Mintz, A. Reboldi, P. Schwartzberg, and R. Young for helpful discussions and providing comments on the manuscript; O. Bannard for advice on active caspase-3 staining; and M. Fontana for advice on B cell tetramer generation. Cell sorting was performed in the Center for Cancer Research Flow Cytometry Core Facility. The graphical abstract and model figure were created with BioRender.com.
This research was supported by the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, and NCI.
Author contributions: R. Razzaghi designed and performed experiments, analyzed and interpreted data, and wrote the manuscript. S. Agarwal designed and performed experiments, analyzed and interpreted data, and edited the manuscript. N. Kotlov, O. Plotnikova, and K. Nomie analyzed and interpreted data and edited the manuscript. D.W. Huang and G.W. Wright analyzed and interpreted data. M. Li and M. Yamadi performed experiments. K. Takata and D.W. Scott performed experiments and analyzed data. C. Yao and J.J. O’Shea analyzed data. G.A. Smith and S. Pittaluga performed experiments and analyzed data. J.D. Phelan analyzed data and edited the manuscript. J.R. Muppidi designed and performed experiments, analyzed and interpreted data, and wrote the manuscript.
References
- Adzhubei, I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., and Sunyaev S.R.. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar-Sterle, S., Zotos D., Bernard N.J., Scherger A.K., Rödling L., Alsop A.E., Walker J., Masson F., Belz G.T., Corcoran L.M., et al. 2014. Fas ligand-mediated immune surveillance by T cells is essential for the control of spontaneous B cell lymphomas. Nat. Med. 20:283–290. 10.1038/nm.3442 [DOI] [PubMed] [Google Scholar]
- Alabyev, B., Vuyyuru R., and Manser T.. 2008. Influence of Fas on the regulation of the response of an anti-nuclear antigen B cell clonotype to foreign antigen. Int. Immunol. 20:1279–1287. 10.1093/intimm/dxn087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, C.D. 2015. Germinal center quality control: death by Fas. Immunity. 42:783–785. 10.1016/j.immuni.2015.05.005 [DOI] [PubMed] [Google Scholar]
- Arthur, S.E., Jiang A., Grande B.M., Alcaide M., Cojocaru R., Rushton C.K., Mottok A., Hilton L.K., Lat P.K., Zhao E.Y., et al. 2018. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat. Commun. 9:4001. 10.1038/s41467-018-06354-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann, D., Preite S., Reboldi A., Ronchi F., Ansel K.M., Lanzavecchia A., and Sallusto F.. 2013. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 38:596–605. 10.1016/j.immuni.2012.11.020 [DOI] [PubMed] [Google Scholar]
- Béguelin, W., Teater M., Meydan C., Hoehn K.B., Phillip J.M., Soshnev A.A., Venturutti L., Rivas M.A., Calvo-Fernández M.T., Gutierrez J., et al. 2020. Mutant EZH2 induces a pre-malignant lymphoma niche by reprogramming the immune response. Cancer Cell. 37:655–673.e11. 10.1016/j.ccell.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentebibel, S.E., Schmitt N., Banchereau J., and Ueno H.. 2011. Human tonsil B-cell lymphoma 6 (BCL6)-expressing CD4+ T-cell subset specialized for B-cell help outside germinal centers. Proc. Natl. Acad. Sci. USA. 108:E488–E497. 10.1073/pnas.1100898108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel, V.D., Guillaume J.-L., Lambiotte R., and Lefebvre E.. 2008. Fast unfolding of communities in large networks. J. Stat. Mech. 108:P10008. 10.1088/1742-5468/2008/10/P10008 [DOI] [Google Scholar]
- Butt, D., Chan T.D., Bourne K., Hermes J.R., Nguyen A., Statham A., O’Reilly L.A., Strasser A., Price S., Schofield P., et al. 2015. FAS inactivation releases unconventional germinal center B cells that escape antigen control and drive IgE and autoantibody production. Immunity. 42:890–902. 10.1016/j.immuni.2015.04.010 [DOI] [PubMed] [Google Scholar]
- Cerchietti, L., Inghirami G., Kotlov N., Svelkolkin V., Bagaev A., Frenkel F., Revuelta M.V., Phillip J.M., Cacciapuoti M.T., Rutherford S.C., et al. 2019. Microenvironmental signatures reveal biological subtypes of diffuse large B-cell lymphoma (DLBCL) distinct from tumor cell molecular profiling [abstract]. Blood. 134(Supplement_1):656. 10.1182/blood-2019-12888931416815 [DOI] [Google Scholar]
- Chapuy, B., Stewart C., Dunford A.J., Kim J., Kamburov A., Redd R.A., Lawrence M.S., Roemer M.G.M., Li A.J., Ziepert M., et al. 2018. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 24:679–690. 10.1038/s41591-018-0016-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova, T., Tangye S.G., Newton R., Frank N., Hodge M.R., Rolph M.S., and Mackay C.R.. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173:68–78. 10.4049/jimmunol.173.1.68 [DOI] [PubMed] [Google Scholar]
- Crotty, S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Di Niro, R., Lee S.J., Vander Heiden J.A., Elsner R.A., Trivedi N., Bannock J.M., Gupta N.T., Kleinstein S.H., Vigneault F., Gilbert T.J., et al. 2015. Salmonella infection drives promiscuous B cell activation followed by extrafollicular affinity maturation. Immunity. 43:120–131. 10.1016/j.immuni.2015.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennishi, D., Jiang A., Boyle M., Collinge B., Grande B.M., Ben-Neriah S., Rushton C., Tang J., Thomas N., Slack G.W., et al. 2019. Double-hit gene expression signature defines a distinct subgroup of germinal center B-cell-like diffuse large B-cell lymphoma. J. Clin. Oncol. 37:190–201. 10.1200/JCO.18.01583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennishi, D., Healy S., Bashashati A., Saberi S., Hother C., Mottok A., Chan F.C., Chong L., Abraham L., Kridel R., et al. 2020. TMEM30A loss-of-function mutations drive lymphomagenesis and confer therapeutically exploitable vulnerability in B-cell lymphoma. Nat. Med. 26:577–588. 10.1038/s41591-020-0757-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, G.M., Angeletti D., Ince W.L., Gibbs J.S., Khurana S., Wheatley A.K., Max E.E., McDermott A.B., Golding H., Stevens J., et al. 2015. A simple flow-cytometric method measuring B cell surface immunoglobulin avidity enables characterization of affinity maturation to influenza A virus. MBio. 6:e01156-15. 10.1128/mBio.01156-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowthaman, U., Chen J.S., Zhang B., Flynn W.F., Lu Y., Song W., Joseph J., Gertie J.A., Xu L., Collet M.A., et al. 2019. Identification of a T follicular helper cell subset that drives anaphylactic IgE. Science. 365:eaaw6433. 10.1126/science.aaw6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, J.A., Suzuki K., Cho B., Willison L.D., Palmer D., Allen C.D., Schmidt T.H., Xu Y., Proia R.L., Coughlin S.R., and Cyster J.G.. 2011. The sphingosine 1-phosphate receptor S1P2 maintains the homeostasis of germinal center B cells and promotes niche confinement. Nat. Immunol. 12:672–680. 10.1038/ni.2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Z., Hampel B., Yagita H., and Rajewsky K.. 2004. T cell-specific ablation of Fas leads to Fas ligand-mediated lymphocyte depletion and inflammatory pulmonary fibrosis. J. Exp. Med. 199:1355–1365. 10.1084/jem.20032196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Z., Duncan G.S., Seagal J., Su Y.W., Hong C., Haight J., Chen N.J., Elia A., Wakeham A., Li W.Y., et al. 2008. Fas receptor expression in germinal-center B cells is essential for T and B lymphocyte homeostasis. Immunity. 29:615–627. 10.1016/j.immuni.2008.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, C.C., Baccarella A.M., Bayat A., Pepper M., and Fontana M.F.. 2019. FCRL5+ memory B cells exhibit robust recall responses. Cell Rep. 27:1446–1460.e4. 10.1016/j.celrep.2019.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov, D.I., Kotov J.A., Goldberg M.F., and Jenkins M.K.. 2018. Many Th cell subsets have Fas ligand-dependent cytotoxic potential. J. Immunol. 200:2004–2012. 10.4049/jimmunol.1700420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraoka, M., Schmidt A.G., Nojima T., Feng F., Watanabe A., Kitamura D., Harrison S.C., Kepler T.B., and Kelsoe G.. 2016. Complex antigens drive permissive clonal selection in germinal centers. Immunity. 44:542–552. 10.1016/j.immuni.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw, B.J., Schmidt T.H., Green J.A., Allen C.D., Okada T., and Cyster J.G.. 2017. The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. J. Exp. Med. 214:639–649. 10.1084/jem.20161461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gallou, S., Zhou Z., Thai L.H., Fritzen R., de Los Aires A.V., Mégret J., Yu P., Kitamura D., Bille E., Tros F., et al. 2018. A splenic IgM memory subset with antibacterial specificities is sustained from persistent mucosal responses. J. Exp. Med. 215:2035–2053. 10.1084/jem.20180977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, P., Shih C., and Qi H.. 2017. Ephrin B1-mediated repulsion and signaling control germinal center T cell territoriality and function. Science. 356:eaai9264. 10.1126/science.aai9264 [DOI] [PubMed] [Google Scholar]
- Mayer, C.T., Gazumyan A., Kara E.E., Gitlin A.D., Golijanin J., Viant C., Pai J., Oliveira T.Y., Wang Q., Escolano A., et al. 2017. The microanatomic segregation of selection by apoptosis in the germinal center. Science. 358:eaao2602. 10.1126/science.aao2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin, L., Ersching J., and Victora G.D.. 2016. Germinal center B cell dynamics. Immunity. 45:471–482. 10.1016/j.immuni.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz, M.A., Felce J.H., Chou M.Y., Mayya V., Xu Y., Shui J.W., An J., Li Z., Marson A., Okada T., et al. 2019. The HVEM-BTLA axis restrains T cell help to germinal center B cells and functions as a cell-extrinsic suppressor in lymphomagenesis. Immunity. 51:310–323.e7. 10.1016/j.immuni.2019.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin, R.D., Mungall K., Pleasance E., Mungall A.J., Goya R., Huff R.D., Scott D.W., Ding J., Roth A., Chiu R., et al. 2013. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood. 122:1256–1265. 10.1182/blood-2013-02-483727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muppidi, J.R., Schmitz R., Green J.A., Xiao W., Larsen A.B., Braun S.E., An J., Xu Y., Rosenwald A., Ott G., et al. 2014. Loss of signalling via Gα13 in germinal centre B-cell-derived lymphoma. Nature. 516:254–258. 10.1038/nature13765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata, S. 2018. Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 36:489–517. 10.1146/annurev-immunol-042617-053010 [DOI] [PubMed] [Google Scholar]
- Price, S., Shaw P.A., Seitz A., Joshi G., Davis J., Niemela J.E., Perkins K., Hornung R.L., Folio L., Rosenberg P.S., et al. 2014. Natural history of autoimmune lymphoproliferative syndrome associated with FAS gene mutations. Blood. 123:1989–1999. 10.1182/blood-2013-10-535393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathmell, J.C., Townsend S.E., Xu J.C., Flavell R.A., and Goodnow C.C.. 1996. Expansion or elimination of B cells in vivo: dual roles for CD40- and Fas (CD95)-ligands modulated by the B cell antigen receptor. Cell. 87:319–329. 10.1016/S0092-8674(00)81349-5 [DOI] [PubMed] [Google Scholar]
- Reboldi, A., and Cyster J.G.. 2016. Peyer’s patches: organizing B-cell responses at the intestinal frontier. Immunol. Rev. 271:230–245. 10.1111/imr.12400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi, A., Arnon T.I., Rodda L.B., Atakilit A., Sheppard D., and Cyster J.G.. 2016. IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science. 352:aaf4822. 10.1126/science.aaf4822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein, T.L., Wang J.K., Panka D.J., Foote L.C., Wang Z., Stanger B., Cui H., Ju S.T., and Marshak-Rothstein A.. 1995. Protection against Fas-dependent Th1-mediated apoptosis by antigen receptor engagement in B cells. Nature. 374:163–165. 10.1038/374163a0 [DOI] [PubMed] [Google Scholar]
- Rydyznski, C.E., Cranert S.A., Zhou J.Q., Xu H., Kleinstein S.H., Singh H., and Waggoner S.N.. 2018. Affinity maturation is impaired by natural killer cell suppression of germinal centers. Cell Rep. 24:3367–3373.e4. 10.1016/j.celrep.2018.08.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayin, I., Radtke A.J., Vella L.A., Jin W., Wherry E.J., Buggert M., Betts M.R., Herati R.S., Germain R.N., and Canaday D.H.. 2018. Spatial distribution and function of T follicular regulatory cells in human lymph nodes. J. Exp. Med. 215:1531–1542. 10.1084/jem.20171940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer, F., Kurtz D.M., Newman A.M., Stehr H., Craig A.F., Esfahani M.S., Lovejoy A.F., Chabon J.J., Klass D.M., Liu C.L., et al. 2016. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci. Transl. Med. 8:364ra155. 10.1126/scitranslmed.aai8545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz, R., Wright G.W., Huang D.W., Johnson C.A., Phelan J.D., Wang J.Q., Roulland S., Kasbekar M., Young R.M., Shaffer A.L., et al. 2018. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 378:1396–1407. 10.1056/NEJMoa1801445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert, T.A., Victora G.D., Fooksman D.R., Kamphorst A.O., Mugnier M.R., Gitlin A.D., Dustin M.L., and Nussenzweig M.C.. 2011. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J. Exp. Med. 208:1243–1252. 10.1084/jem.20102477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, C., Barrans S., Cucco F., Bentley M.A., Care M.A., Cummin T., Kennedy H., Thompson J.S., Uddin R., Worrillow L., et al. 2019. Molecular high-grade B-cell lymphoma: defining a poor-risk group that requires different approaches to therapy. J. Clin. Oncol. 37:202–212. 10.1200/JCO.18.01314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J., Hou S., Fang Q., Liu X., Liu X., and Qi H.. 2018. PD-1 controls follicular T helper cell positioning and function. Immunity. 49:264–274.e4. 10.1016/j.immuni.2018.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel, R.M., Frederiksen J.K., Zacharias D.A., Chan F.K., Johnson M., Lynch D., Tsien R.Y., and Lenardo M.J.. 2000. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 288:2354–2357. 10.1126/science.288.5475.2354 [DOI] [PubMed] [Google Scholar]
- Silver, J., Zuo T., Chaudhary N., Kumari R., Tong P., Giguere S., Granato A., Donthula R., Devereaux C., and Wesemann D.R.. 2018. Stochasticity enables BCR-independent germinal center initiation and antibody affinity maturation. J. Exp. Med. 215:77–90. 10.1084/jem.20171022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, K.G., Nossal G.J., and Tarlinton D.M.. 1995. FAS is highly expressed in the germinal center but is not required for regulation of the B-cell response to antigen. Proc. Natl. Acad. Sci. USA. 92:11628–11632. 10.1073/pnas.92.25.11628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, I., Radtke D., Phillips B., McGowan S.J., and Bannard O.. 2018. Germinal center B cells replace their antigen receptors in dark zones and fail light zone entry when immunoglobulin gene mutations are damaging. Immunity. 49:477–489.e7. 10.1016/j.immuni.2018.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M. III, Hao Y., Stoeckius M., Smibert P., and Satija R.. 2019. Comprehensive integration of single-cell data. Cell. 177:1888–1902.e21. 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Cerasoli D.M., Dal Porto J.M., Shimoda M., Freund R., Fang W., Telander D.G., Malvey E.N., Mueller D.L., Behrens T.W., and Kelsoe G.. 1999. Relaxed negative selection in germinal centers and impaired affinity maturation in bcl-xL transgenic mice. J. Exp. Med. 190:399–410. 10.1084/jem.190.3.399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., Ohta H., and Takemori T.. 2001. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity. 14:181–192. 10.1016/S1074-7613(01)00100-5 [DOI] [PubMed] [Google Scholar]
- Tas, J.M., Mesin L., Pasqual G., Targ S., Jacobsen J.T., Mano Y.M., Chen C.S., Weill J.C., Reynaud C.A., Browne E.P., et al. 2016. Visualizing antibody affinity maturation in germinal centers. Science. 351:1048–1054. 10.1126/science.aad3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora, G.D., and Nussenzweig M.C.. 2012. Germinal centers. Annu. Rev. Immunol. 30:429–457. 10.1146/annurev-immunol-020711-075032 [DOI] [PubMed] [Google Scholar]
- Weinstein, J.S., Lezon-Geyda K., Maksimova Y., Craft S., Zhang Y., Su M., Schulz V.P., Craft J., and Gallagher P.G.. 2014. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood. 124:3719–3729. 10.1182/blood-2014-06-582700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- William, J., Euler C., Christensen S., and Shlomchik M.J.. 2002. Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science. 297:2066–2070. 10.1126/science.1073924 [DOI] [PubMed] [Google Scholar]
- Wright, G.W., Huang D.W., Phelan J.D., Coulibaly Z.A., Roulland S., Young R.M., Wang J.Q., Schmitz R., Morin R.D., Tang J., et al. 2020. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 37:551–568.e14. 10.1016/j.ccell.2020.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, D., Thangaraju M., Browning D.D., Dong Z., Korchin B., Lev D.C., Ganapathy V., and Liu K.. 2007. IFN regulatory factor 8 mediates apoptosis in nonhemopoietic tumor cells via regulation of Fas expression. J. Immunol. 179:4775–4782. 10.4049/jimmunol.179.7.4775 [DOI] [PubMed] [Google Scholar]
- Young, R.M., Wu T., Schmitz R., Dawood M., Xiao W., Phelan J.D., Xu W., Menard L., Meffre E., Chan W.C., et al. 2015. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc. Natl. Acad. Sci. USA. 112:13447–13454. 10.1073/pnas.1514944112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, R.M., Phelan J.D., Shaffer A.L., Wright G.W., Huang D.W., Schmitz R., Johnson C., Oellerich T., Wilson W., and Staudt L.M.. 2019. Taming the heterogeneity of aggressive lymphomas for precision therapy. Annu. Rev. Cancer Biol. 3:429–455. 10.1146/annurev-cancerbio-030518-055734 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows FAS mutations in GCB-DLBCL.
lists data from primary DLBCL cases.
displays the co-occurrence of FAS mutations with mutations of TNFRSF14 or CD274 across published DLBCL datasets.
shows the sequencing of Fas in Fas-negative mouse tumors.
lists the primers used for amplification and sequencing of Fas from mouse tumors.