Abstract
Objective
To examine whether lifelong genetically increased serum urate levels, a potent antioxidant, contribute to MS susceptibility using Mendelian randomization (MR).
Methods
This 2-sample MR study included 25 independent genetic variants strongly associated with serum urate levels in a genome-wide association study meta-analysis of 140,949 individuals. Effects on the risk of MS were assessed with summary statistics from 3 large-scale MS genetic data sets totaling 61,667 MS cases and 86,806 controls from the International MS Genetic Consortium. Multiple sensitivity analyses were performed to evaluate the assumptions of MR and remove potentially pleiotropic variants.
Results
Using inverse-variance weighted MR, we found no evidence for a causal effect of serum urate level on the risk of MS in any of the cohorts (MS1: OR 0.99 per each mg/dL unit increase in urate, 95% CI 0.89–1.08, p = 0.76; MS2: OR = 0.99, 95% CI 0.89–1.11, p = 0.90; MS3: OR = 1.00, 95% CI 0.98–1.2, p = 0.91). Pleiotropy robust MR methods yielded consistent estimates.
Conclusion
This MR study does not support a clinically relevant causal effect of serum urate levels on the risk of MS.
Urate, the anionic form of uric acid, is the final metabolite of purine metabolism in humans. Urate acts as a potent free radical scavenger and accounts for 60% of the antioxidant capacity in plasma.1 Evidence involving oxidative stress in the pathogenesis of MS2 and proposed neuroprotective effects of urate1 have motivated the investigation into the role of urate in MS. Indeed, an early population study revealed lower than predicted co-occurrence of MS and gout, a condition characterized by hyperuricemia.3 In addition, several case-control studies3–12 and 2 meta-analysis13,14 have reported lower serum urate levels in individuals with MS compared with healthy controls and other neurologic disorders. These data have led to the hypothesis that elevated serum urate may be protective against the development of MS. However, other case-controls studies have demonstrated discordant findings,15–17 with 1 prospective study suggesting that the association between lower serum urate and higher risk of MS could be due to bias from reverse causality.18 In addition, observational studies are also susceptible to residual confounding, which further limits causal inference.
Although randomized clinical trials would be ideal to provide evidence for or against a causal role for urate in MS, the few that have been performed have focused on disease outcomes among patients with MS rather than testing the proposed preventive effects.19–21 The latter question has population-wide implications, but such a trial would require prohibitively large sample sizes followed for an extended period of time and is probably not feasible. Therefore, the role of urate in the risk of developing MS remains unclear.22
In the absence of such experimental evidence, Mendelian randomization (MR) can provide an alternative approach for causal inference. MR uses genetic associations to investigate the causal effect of a risk factor on an outcome.23 This approach greatly reduces the likelihood of residual confounding because these genetic variants are randomly assigned at conception and relatively independent of socioeconomic and lifestyle characteristics that could bias transitional observational studies.24 The fact that genotypes are not modifiable by disease onset also limits reverse causality.23 Serum urate levels show a large heritable component estimated at 40%–60%.25 As such, several genetic loci have been robustly associated with serum levels, including variants in urate transporters.26 This makes MR ideally suited to resolve questions of causality involving serum urate levels, as recently shown in Parkinson disease.27
In this article, we aimed to assess whether genetic predisposition toward lifelong higher serum urate levels alters the risk of developing MS. For this analysis, we undertook a 2-sample MR approach in 3 large genetic studies of MS susceptibly with up to 61,667 MS cases and 86,806 controls.
Methods
Genetic variants associated with serum urate
We identified genetic variants reliably associated with serum urate levels in a large genome-wide association study meta-analysis of up to 140,949 individuals from 48 cohorts.26 Details of serum urate measurement for each of the cohorts have been described previously by the Global Urate Genetic Consortium (GUGC).26 Summary statistics were retrieved for 29 single nucleotide polymorphisms (SNPs), which were genome-wide significant in the meta-analysis. The effect of each allele on serum urate levels is presented in mg/dL and adjusted for age, sex, and study-specific covariates. To mitigate bias from population stratification,28 all effect estimates were derived from individuals of European ancestry. We calculated linkage disequilibrium (LD) between each pair of variants to ensure that they are not correlated (r2 < 0.01) as this can lead to biased MR estimates.29 We also excluded from analysis variants within the extended major histocompatibility complex (MHC) region, defined as base positions 24,000,000 to 35,000,000 on chromosome 6 (GRCh37).30 This is because the strength of the association of this region with MS and its complex LD structure renders it susceptible to horizontal pleiotropy and violation of the MR assumptions.
MS genetic data
For each genetic variant associated with serum urate levels, we obtained corresponding effect estimates on MS susceptibility using summary statistics from 3 MS genetic studies by the International MS Genetics Consortium (IMSGC) and totaling 148,473 individuals (61,667 MS cases and 86,806 controls).30–32 Participants for each data set are presented in the table. The first data set (MS1) corresponds to the discovery cohort of the latest IMSGC meta-analysis and contains more than 8 million genotyped and imputed variants.30 The 2 remaining data sets are identified by the genotyping array used for the majority of participants, namely the Immunochip32 (MS2) and Exome Chip (MS3).31 The former corresponds to the discovery cohort of IMSGC et al. 201332 and lists 161,312 markers. The latter is the combined cohort from IMSGC et al. 2018,31 which focused on low frequency and rare variants (173,746 markers).
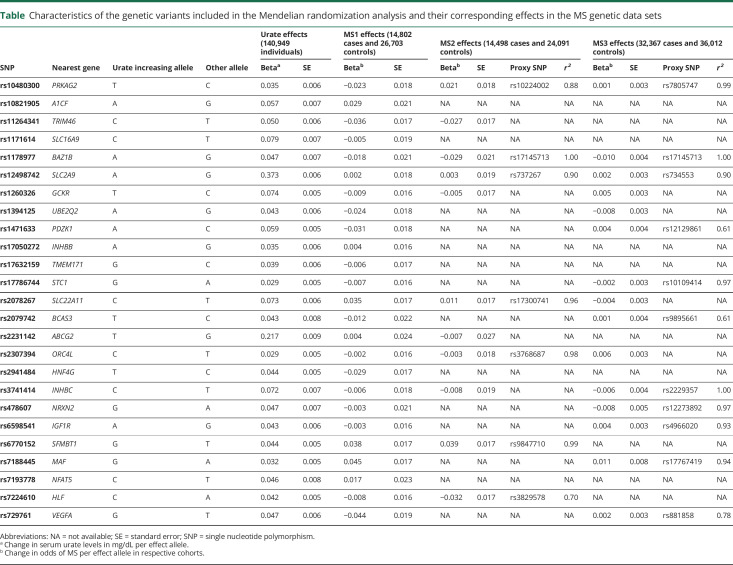
Table.
Characteristics of the genetic variants included in the Mendelian randomization analysis and their corresponding effects in the MS genetic data sets
All participants were of European ancestry. For each SNP, the effect size on MS susceptibility was aligned with respect to the urate increasing allele. All genetic data sets reported alleles on the forward strand, except the Immunochip data for which strand mismatches were reconciled. No palindromic variants were present. Participant overlap between the GUGC and IMSGC data sets was negligible and limited to controls (1.0% for MS1) or absent (MS2 and MS3), meaning that no bias should have been introduced.33 When urate-associated variants were not present in either of the MS genetic data sets, we identified a proxy SNP in LD (r2 > 0.6) using PLINK v1.934 and samples of European ancestry from 1000 Genomes phase 3.35
MR analysis
For each of the MS data sets separately, we performed an inverse-variance weighted (IVW) 2-sample MR to estimate the effect of a genetically related unit increase in plasma urate levels (in mg/dL) on the odds of MS, using previously described methods.36,37 In brief, we weighted the effect of each variant on MS susceptibility by its effect on serum urate using the Wald ratio method.37 These individual MR estimates were then combined into a summary measure using random effect meta-analysis with inverse-variance weighting. There is no participant overlap between the MS1 and MS2 cohorts, but there is at least partial overlap between the MS2 and MS3 study samples.38 Therefore, we combined the IVW MR results from the MS1 and MS2 data sets (but not MS3) in a fixed effect meta-analysis. To ensure that the overall MR results were not driven by any particular variant, we iteratively removed each SNP and recalculated the IVW MR estimate in each of the data sets.
A fundamental assumption of MR is that the genetic variants associated with the exposure phenotype (serum urate) must not affect the risk of the outcome phenotype (MS) through independent pathways. Presence of such pathways is known as horizontal pleiotropy and can bias the MR estimates. For each of the MS data sets, we investigated the robustness of our main results to pleiotropy by applying 3 established MR sensitivity methods with complementary attributes.39 First, we formally examined for overall horizontal pleiotropy using the MR pleiotropy residual sum and outlier (MR-PRESSO) Global test.38 This method compares the observed distance of all variants to the regression line of the SNP-outcome effects on SNP-exposure effects with the expected distance under the null hypothesis of no horizontal pleiotropy. We also applied the MR-PRESSO outlier test using the imputed MS1 data set to identify and exclude from all analyses outlier and thus potentially pleiotropic variants.38 Second, we applied MR-Egger regression, a weighted linear regression of the SNP-outcome effects on the SNP-exposure effects allowing for the intercept to be estimated.40 This intercept provides a measure of average pleiotropic bias. The slope coefficient gives an unbiased estimate even in the presence of pleiotropic effects, as long as the size of these pleiotropic effects is independent of the effect of the SNP on the exposure. Third, we performed a weighted median analysis, which provides consistent estimates robust to the presence of pleiotropy in a subset (<50%) of the variants.41 In addition, we inspected funnel plots of the individual MR estimates against their precision.42 Asymmetry on this plot is indicative of directional horizontal pleiotropy.40
All statistical analyses were performed in R (version 3.6.0). The alpha level for statistical significance was set to 0.05.
Standard protocol approvals, registrations, and patient consents
The data sources used in this study (GUGC and IMSGC) obtained informed consent from all participants.26,30–32 Separate institutional review board approval was not required for this study.
Data availability
Summary level genetic data for serum urate levels and MS susceptibility are publicly available through their respective consortia. In addition, the data used to generate the main results are available in the table
Results
Selection of genetic variants
Of the 29 genome-wide significant variants for serum urate in the GUGC meta-analysis, none were in LD. We excluded a single SNP in the extended MHC region (rs1165151 in SLC17A1), leaving 28 variants. Next, we applied the MR-PRESSO outlier test using the MS1 data set and identified 3 potentially pleiotropic variants (rs642803 near OVOL1, rs653178 near ATXN2, and rs675209 near RREB1). These were excluded from all subsequent analyses. We estimated the remaining 25 variants to explain approximately 6.7% of the variance in serum urate levels, with 3.4% explained by SLC2A9 and ABCG2 alone.26 All 25 urate-associated variants were available in the MS1 data set for assessing the genetic association with MS susceptibility. Both MS2 (using Immunochip) and MS3 (using Exome Chip) studies have lower genome coverage and no imputed variants. Therefore, we were able to identify 11/25 variants in MS2, 4 directly present and 7 via proxies (median r2 = 0.96), as well as 15/25 variants in MS3, 4 directly present and 11 via proxies (median r2 = 0.94). The list of variants and proxies for each data set can be found in the table
MR analysis
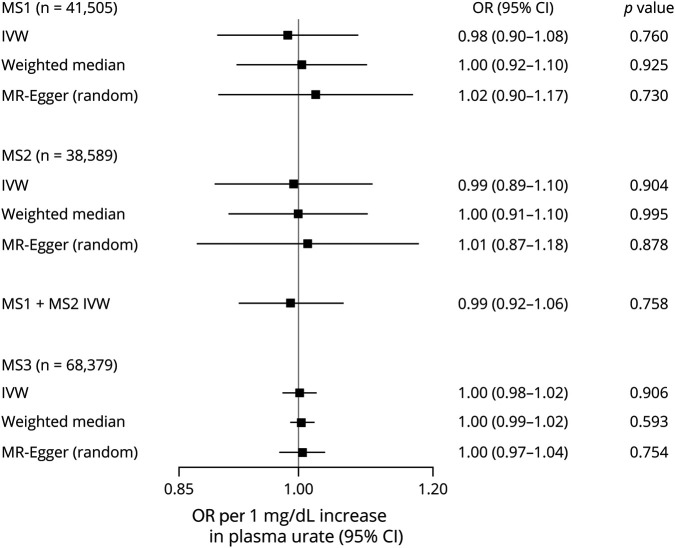
The main IVW MR analysis did not provide evidence for a causal effect of serum urate levels on odds of MS in either of the MS data sets (MS1: OR per each mg/dL unit increase in serum urate was 0.99, 95% CI 0.89–1.08; MS2: OR = 0.99, 95% CI 0.89–1.11; MS3 OR = 1.00, 95% CI 0.98–1.2). Similarly, meta-analysis of the MS2 and MS3 results in a null effect with narrow CI (OR = 0.99, 95% CI 0.92–1.06) and no heterogeneity (Q = 0.01, p = 0.91; I2 = 0%). These results remained consistent after iteratively removing each individual SNP from the IVW MR analysis (figures e-1 to e-3, links.lww.com/NXI/A349), including the strongest variants near SLC2A9 and ABCG2.
There was weak evidence of horizontal pleiotropy on the MR-PRESSO Global test (p = 0.051) using the more complete MS1 data set. Similarly, the MR-Egger intercept was centered around the null and showed no evidence of directional pleiotropy across all MS case-control studies (MS1: intercept −0.01, p = 0.41; MS2: intercept −0.003, p = 0.73; MS3: intercept −0.001, p = 0.73). The pleiotropy robust methods (weighted median and MR-Egger) yielded effect estimates consistent with the main finding across all samples (figure 1). In addition, the funnel plots for each variant were symmetric and provided no evidence of directional horizontal pleiotropy (figure 2).
Figure 1. Forest plot of the main MR estimates and sensitivity analyses for each of the MS data sets.
The combined MS1 and MS2 data set IVW MR results were obtained after fixed-effects meta-analysis. IVW = inverse-variance weighted; MR = Mendelian randomization.
Figure 2. Funnel plots of the individual MR estimates against their precision in each of the MS cohorts.
IVW = inverse-variance weighted; MR = Mendelian randomization; SE = standard error.
Discussion
In this study, we investigated the causal relationship between serum urate levels and MS susceptibility, using a 2-sample MR design in 3 MS case-control studies with a cumulative sample size of 148,473 individuals (61,667 MS cases and 86,806 controls). The main results consistently found no effect of lifelong genetically related differences in urate levels on the risk of developing MS. The narrow CIs around this null effect afforded by our large sample size provide strong evidence against any clinically relevant influence. In addition, we applied several sensitivity analyses, each with different underlying assumptions, which found no evidence of pleiotropy and further reinforce the validity of the results. This suggests that the previously reported association between serum urate and MS risk could be attributed to residual confounding, reverse causality, or both.
Our findings conflict with the results of most case-control studies conducted on serum urate and MS. A previous study reported lower serum urate levels in MS when compared to noninflammatory neurologic disease controls. The same study also found that the rate of MS and gout co-occurrence was lower than expected by chance. A more recent meta-analysis of 1308 MS cases and 908 controls similarly reported lower serum urate levels in MS.14 However, there was considerable between-study heterogeneity,14 and a recent umbrella review classified the evidence for an association between serum urate levels and MS as Class IV (weak).22 Importantly, those studies used urate measurements in prevalent MS cases. Therefore, they cannot determine whether the decrease in serum levels precedes MS onset or is a consequence thereof, rendering them susceptible to bias from reverse causality. In contrast, a small prospective case-control study, which measured serum urate levels years before MS symptom onset, found no association with the risk of MS.18 The same study also observed lower urate levels as the interval between sample collection and MS symptom onset decreased.18 In combination, these findings are consistent with the interpretation that higher serum urate levels are not protective against MS, but rather a consequence of the MS disease process. This is in line with the results from our MR approach, which is robust to reverse causality. In addition, using genetic proxies of urate levels lowers the risk of confounding from diet and body weight, which were not routinely accounted for in previous observational studies.14
Indeed, serum urate levels are modulated in part by dietary purine consumption, particularly the intake of purine-rich food groups such as meat, seafood, and alcohol.43 In addition, obesity is a risk factor for the development of hyperuricemia, whereas weight loss in that group results in serum urate level reduction.43 That said, a recent study combining food frequency questionnaire and genetic data from 5 US cohorts found that dietary patterns explained only 4.3% of variation in serum urate levels, whereas common genetic variants explained 23.9% (of which 7.9% was captured by the genome-wide significant SNPs included in the present study).44
Our study did not address the separate question of whether serum urate levels influence MS disease course. This has been explored in 3 small randomized clinical trials in relapsing-remitting MS using inosine, an oral precursor of urate, which increases it serum levels.19–21 A first small study in 16 patients reported an improvement in EDSS scores at 1 year with inosine treatment compared with placebo.19 However, 2 subsequent clinical trials in 36 and 159 patients with relapsing-remitting MS, of 1- and 2-year duration, respectively, compared the combination of interferon β and inosine vs interferon β with placebo and did not found any effect on disability or other clinical and MRI metrics.20,21 These results are not directly comparable to our research findings.
Our study has some limitations. First, while the use of summary statistics in a 2-sample MR design maximized statistical power, it precluded sex-stratified analyses. However, despite women having generally lower serum urate levels, the association with MS in case-control studies was similar between sexes.14 Second, this study did not differentiate between MS subtypes. Although a small study reported higher serum urate levels in 11 primary progressive MS cases compared with other subtypes,45 others have found no difference.13 Third, the estimates reported in our study assume a linear effect between the risk factor and outcome. Last, the possibility of horizontal pleiotropy cannot be entirely excluded. Nevertheless, we undertook several sensitivity analyses, which found no evidence of pleiotropy. In addition, we excluded variants in the MHC region and those showing outlier effects using an established method.
In conclusion, we provide strong evidence against a causal effect of serum urate levels on the risk of MS. This helps inform the role of this metabolite in MS pathogenesis and indicates that approaches to increase urate levels would not be effective in preventing disease while potentially leading to adverse events.
Acknowledgment
The authors thank the IMSGC and GUGC for access to their summary statistics data.
Glossary
- GIANT
Genetic Investigation of Anthropometric Traits
- GUGC
Global Urate Genetic Consortium
- IMSGC
International Multiple Sclerosis Genetics Consortium
- IVW
inverse-variance weighted
- MHC
major histocompatibility complex
- MR
Mendelian randomization
- MR-PRESSO
Mendelian Randomization Pleiotropy RESidual Sum and Outlier
- SNP
single nucleotide polymorphism
- LD
linkage disequilibrium
Appendix. Authors
Study funding
A. Harroud is funded by the NMSS-ABF Clinician Scientist Development Award from the National Multiple Sclerosis Society (NMSS) and the Multiple Sclerosis Society of Canada (MSSC) FAN-1808-32256. J.B. Richards is supported by the Canadian Institutes of Health Research (CIHR), the Canadian Foundation for Innovation and the Fonds de Recherche du Québec—Santé (FRQS). J.B. Richards is funded by an FRQS Clinical Research Scholarship and received research support from the NMSS and the MSSC. TwinsUK is funded by the Wellcome Trust, Medical Research Council, European Union, the National Institute for Health Research (NIHR)-funded BioResource, Clinical Research Facility and Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust in partnership with King's College London.
Disclosure
The authors report no disclosures. Go to Neurology.org/NN for full disclosures.
References
- 1.Petruzzo M, Moccia M. Time to reconsider urate: neuroprotective potential may prevail on cardiovascular risk in animal models and clinical trials. EBioMedicine 2018;37:5–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilgun-Sherki Y, Melamed E, Offen D. The role of oxidative stress in the pathogenesis of multiple sclerosis: the need for effective antioxidant therapy. J Neurol 2004;251:261–268. [DOI] [PubMed] [Google Scholar]
- 3.Hooper DC, Spitsin S, Kean RB, et al. Uric acid, a natural scavenger of peroxynitrite, in experimental allergic encephalomyelitis and multiple sclerosis. Proc Natl Acad Sci U S A 1998;95:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toncev G, Milicic B, Toncev S, Samardzic G. Serum uric acid levels in multiple sclerosis patients correlate with activity of disease and blood-brain barrier dysfunction. Eur J Neurol 2002;9:221–226. [DOI] [PubMed] [Google Scholar]
- 5.Sotgiu S, Pugliatti M, Sanna A, et al. Serum uric acid and multiple sclerosis. Neurol Sci 2002;23:183–188. [DOI] [PubMed] [Google Scholar]
- 6.Spitsin S, Hooper DC, Mikheeva T, Koprowski H. Uric acid levels in patients with multiple sclerosis: analysis in mono- and dizygotic twins. Mult Scler 2001;7:165–166. [DOI] [PubMed] [Google Scholar]
- 7.Rentzos M, Nikolaou C, Anagnostouli M, et al. Serum uric acid and multiple sclerosis. Clin Neurol Neurosurg 2006;108:527–531. [DOI] [PubMed] [Google Scholar]
- 8.Peng F, Zhang B, Zhong X, et al. Serum uric acid levels of patients with multiple sclerosis and other neurological diseases. Mult Scler 2008;14:188–196. [DOI] [PubMed] [Google Scholar]
- 9.Zamani A, Rezaei A, Khaeir F, Hooper DC. Serum and cerebrospinal fluid uric acid levels in multiple sclerosis patients. Clin Neurol Neurosurg 2008;110:642–643. [DOI] [PubMed] [Google Scholar]
- 10.Dujmovic I, Pekmezovic T, Obrenovic R, et al. Cerebrospinal fluid and serum uric acid levels in patients with multiple sclerosis. Clin Chem Lab Med 2009;47:848–853. [DOI] [PubMed] [Google Scholar]
- 11.Zoccolella S, Tortorella C, Iaffaldano P, et al. Low serum urate levels are associated to female gender in multiple sclerosis patients. PLoS One 2012;7:e40608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moccia M, Lanzillo R, Palladino R, et al. Uric acid: a potential biomarker of multiple sclerosis and of its disability. Clin Chem Lab Med 2015;53:753–759. [DOI] [PubMed] [Google Scholar]
- 13.Liu B, Shen Y, Xiao K, Tang Y, Cen L, Wei J. Serum uric acid levels in patients with multiple sclerosis: a meta-analysis. Neurol Res 2012;34:163–171. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Hu W, Wang J, Qian W, Xiao H. Low serum uric acid levels in patients with multiple sclerosis and neuromyelitis optica: an updated meta-analysis. Mult Scler Relat Disord 2016;9:17–22. [DOI] [PubMed] [Google Scholar]
- 15.Mostert JP, Ramsaransing GS, Heersema DJ, Heerings M, Wilczak N, De Keyser J. Serum uric acid levels and leukocyte nitric oxide production in multiple sclerosis patients outside relapses. J Neurol Sci 2005;231:41–44. [DOI] [PubMed] [Google Scholar]
- 16.Kastenbauer S, Kieseier BC, Becker BF. No evidence of increased oxidative degradation of urate to allantoin in the CSF and serum of patients with multiple sclerosis. J Neurol 2005;252:611–612. [DOI] [PubMed] [Google Scholar]
- 17.Amorini AM, Petzold A, Tavazzi B, et al. Increase of uric acid and purine compounds in biological fluids of multiple sclerosis patients. Clin Biochem 2009;42:1001–1006. [DOI] [PubMed] [Google Scholar]
- 18.Massa J, O'Reilly E, Munger KL, Delorenze GN, Ascherio A. Serum uric acid and risk of multiple sclerosis. J Neurol 2009;256:1643–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markowitz CE, Spitsin S, Zimmerman V, et al. The treatment of multiple sclerosis with inosine. J Altern Complement Med 2009;15:619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munoz Garcia D, Midaglia L, Martinez Vilela J, et al. Associated Inosine to interferon: results of a clinical trial in multiple sclerosis. Acta Neurol Scand 2015;131:405–410. [DOI] [PubMed] [Google Scholar]
- 21.Gonsette RE, Sindic C, D'Hooghe MB, et al. Boosting endogenous neuroprotection in multiple sclerosis: the ASsociation of Inosine and Interferon beta in relapsing-remitting Multiple Sclerosis (ASIIMS) trial. Mult Scler 2010; 16: 455–462. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Meng X, Timofeeva M, et al. Serum uric acid levels and multiple health outcomes: umbrella review of evidence from observational studies, randomised controlled trials, and Mendelian randomisation studies. BMJ 2017;357:j2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. [DOI] [PubMed] [Google Scholar]
- 24.Smith GD, Lawlor DA, Harbord R, Timpson N, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med 2007;4:e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan E, Lessov-Schlaggar CN, Krasnow RE, Swan GE. Nature versus nurture in gout: a twin study. Am J Med 2012;125:499–504. [DOI] [PubMed] [Google Scholar]
- 26.Kottgen A, Albrecht E, Teumer A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 2013;45:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kia DA, Noyce AJ, White J, et al. Mendelian randomization study shows no causal relationship between circulating urate levels and Parkinson's disease. Ann Neurol 2018;84:191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet 2010;11:459–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S, Scott RA, Timpson NJ, Davey Smith G, Thompson SG, Consortium EI. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015;30:543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 2019;365:eaav7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.International Multiple Sclerosis Genetics Consortium. Low-frequency and rare-coding variation contributes to multiple sclerosis risk. Cell 2018;175:1679–1687 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Multiple Sclerosis Genetics Consortium; Beecham AH, Patsopoulos NA, Xifara DK, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013;45:1353–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol 2016; 40: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dastani Z, Hivert MF, Timpson N, et al. Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet 2012;8:e1002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–1163. [DOI] [PubMed] [Google Scholar]
- 38.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet 2018;50:693–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet 2018;27:R195–R208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torralba KD, De Jesus E, Rachabattula S. The interplay between diet, urate transporters and the risk for gout and hyperuricemia: current and future directions. Int J Rheum Dis 2012;15:499–506. [DOI] [PubMed] [Google Scholar]
- 44.Major TJ, Topless RK, Dalbeth N, Merriman TR. Evaluation of the diet wide contribution to serum urate levels: meta-analysis of population based cohorts. BMJ 2018;363:k3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piancone F, Saresella M, Marventano I, et al. Monosodium urate crystals activate the inflammasome in primary progressive multiple sclerosis. Front Immunol 2018;9:983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Summary level genetic data for serum urate levels and MS susceptibility are publicly available through their respective consortia. In addition, the data used to generate the main results are available in the table