Abstract
Plant pathogens deliver virulence effectors into plant cells to modulate plant immunity and facilitate infection. Although species‐specific virulence effector screening approaches have been developed for several pathogens, these assays do not apply to pathogens that cannot be cultured and/or transformed outside of their hosts. Here, we established a rapid and parallel screening assay, called the virus‐induced virulence effector (VIVE) assay, to identify putative effectors in various plant pathogens, including unculturable pathogens, using a virus‐based expression vector. The VIVE assay uses the potato virus X (PVX) vector to transiently express candidate effector genes of various bacterial and fungal pathogens into Nicotiana benthamiana leaves. Using the VIVE assay, we successfully identified Avh148 as a potential virulence effector of Phytophthora sojae. Plants infected with PVX carrying Avh148 showed strong viral symptoms and high‐level Avh148 and viral RNA accumulation. Analysis of P. sojae Avh148 deletion mutants and soybean hairy roots overexpressing Avh148 revealed that Avh148 is required for full pathogen virulence. In addition, the VIVE assay was optimized in N. benthamiana plants at different developmental stages across a range of Agrobacterium cell densities. Overall, we identified six novel virulence effectors from seven pathogens, thus demonstrating the broad effectiveness of the VIVE assay in plant pathology research.
Keywords: Nicotiana benthamiana, plant pathogens, PVX, screening assay, virulence effector
The VIVE assay uses the potato virus X vector to transiently express candidate effectors in Nicotiana benthamiana leaves, and could be used for rapid identification of putative virulence effectors in various plant pathogens.

Plant pathogens pose a serious and continuous threat to global agricultural production and food security (Varden et al., 2017). Pathogens, including bacteria, fungi, oomycetes, viruses, nematodes, as well as feeding insects, subvert key host processes by suppressing host innate immunity and manipulating host nutrient supply, leading to disease. In most cases, pathogens achieve this goal by delivering virulence effector proteins into host cells, where the effectors attack the host innate immune system through various mechanisms, such as modifying signalling transduction pathways and gene transcription, altering intracellular transport and cytoskeleton stability, affecting autophagosome formation and vesicle trafficking, and triggering RNA silencing (Qiao et al., 2013, 2015; Toruno et al., 2016; Lee et al., 2019). Thus, it is important to identify the virulence genes of various pathogens and to understand their role in pathogenicity. Whole‐genome sequences of a number of pathogens have recently become available, and a multitude of effectors have been predicted. However, given their complex nature and the relatively lengthy procedures required for their functional analysis, the exact biological functions of these effectors remain unknown (Wang et al., 2019). Approaches such as gene knockout, RNA interference (RNAi)‐mediated gene knockdown, and CRISPR/Cas9‐mediated genome editing have been applied to certain pathogens amenable to genetic transformation for the identification of virulence factors; however, these approaches are rather inefficient. More importantly, it is impossible to genetically transform unculturable pathogens such as “Candidatus Liberibacter asiaticus”, the causal agent of the most severe citrus disease, huanglongbing (Clark et al., 2018), and pathogenic root‐knot nematodes (Meloidogyne spp.) (Castagnone‐Sereno et al., 2013). Thus, a robust and general screening assay is needed for the identification of virulence factors of unculturable pathogens, which will advance our understanding of pathogenesis and plant pathogen evolution.
Agrobacterium‐mediated transformation of Nicotiana benthamiana leaves has been widely used as a transient gene expression assay for gene function analysis as it is a rapid and straightforward approach (Wydro et al., 2006). Moreover, several research groups have developed transient expression assays using virus‐based expression vectors. In these assays, genes of interest of various pathogens are expressed in N. benthamiana leaves using tobacco rattle virus (TRV) and potato virus X (PVX) expression vectors (Zhang et al., 2019a; Li et al., 2020). Over the past three decades, remarkable progress has been made in the development of plant viral vectors owing to their ease of manipulation (Hefferon, 2017). PVX, a potexvirus, is a filamentous rod‐shaped virus harbouring a single‐stranded RNA genome that is capped at the 5′ end and polyadenylated at the 3′ end, and encodes five open reading frames (ORFs) including RNA‐dependent RNA polymerase (RdRP), coat protein (CP), and a triple gene block, whose products are involved in virus movement (Huisman et al., 1988). In the earliest implementation of the PVX vector system, the full‐length cDNA of PVX (P2C2S) was cloned into plasmids under the control of the T7 promoter, with the aim of transcribing PVX cDNA in vitro and manually inoculating the resultant transcripts into N. benthamiana leaves (Chapman et al., 1992). In later versions, the PVX cDNA was cloned into binary vectors (pGR106 and pGR107) under the control of the cauliflower mosaic virus (CaMV) 35S promoter for enhanced expression, and a shorter CP promoter sequence was included in the PVX cDNA to drive the expression of foreign DNA and the CP gene. The foreign DNA could be inserted into the vector between the modified promoter and the 5′ end of the CP. Then, Agrobacterium tumefaciens transformed with binary vectors was manually infiltrated into N. benthamiana leaves (Jones et al., 1999). This biologically active PVX construct was originally designed to exploit the CP promoter to drive high expression of the foreign DNA in inoculated or systemically infected leaves (Chapman et al., 1992). Recently, the PVX‐based expression vector was modified to facilitate stable and long‐term expression of the gene of interest and to simultaneously express two genes via high‐throughput cloning (Dickmeis et al., 2014; Wang et al., 2014). PVX‐based vectors have been the most widely used vectors for investigating gene expression, gene silencing, and cell death in plants (Toth et al., 2001; Lacomme and Chapman, 2008; Wang et al., 2011). Kong et al. showed that expression of the rice stripe virus (RSV) disease‐specific protein (SP) gene using the PVX vector induced visible symptoms in N. benthamiana plants, indicating the role of RSV SP in enhancing PVX symptoms (Kong et al., 2014). More recently, the PVX‐based expression system was employed for tumour immunotherapy and for the development of influenza vaccine and antigens in animal models (Denis et al., 2008; Mardanova et al., 2015; Shukla et al., 2015; Esfandiari et al., 2016; Hefferon, 2017). Given that the PVX‐based system is efficient and induces high‐level expression of foreign genes, we explored its potential to identify pathogenicity determinants or virulence effectors of plant pathogens.
We initially assessed a PVX‐based system for its ability to evaluate virulence effectors of plant pathogens, based on their ability to induce disease symptoms such as necrosis, chlorosis, dwarfing, growth retardation, and abnormal development. To critically test the system, we first separately cloned five Phytophthora sojae effectors into a binary PVX‐based expression vector (pGR106) (Figure 1a). The resulting PVX‐GFP or PVX‐Effector construct was introduced into A. tumefaciens, which was then used to inoculate the leaves of 10–14‐day‐old N. benthamiana seedlings. At 21 days postinoculation (dpi), plants infiltrated with the PVX‐Avh148 construct showed severe dwarfing, with wrinkling and necrosis on the newly emerged upper leaves, whereas those infiltrated with the other four effector constructs exhibited mild mosaic symptoms, similar to the plants infiltrated with the PVX‐GFP (Figures 1b, and S1, and Table 1). Consistently, results of northern blot analysis showed much lower accumulation of PVX genomic and subgenomic RNAs in plants infiltrated with the PVX‐GFP than in plants infiltrated with the PVX‐Avh148 (Figure 1c). Furthermore, the higher accumulation levels of Avh148 transcripts, viral RNA, and PVX CP protein were confirmed by semiquantitative reverse transcription (RT)‐PCR (Figures 1d and S2) and western blot (Figure 1e) in PVX‐Avh148 Agrobacterium‐infiltrated plants. These data suggest that Avh148 potentially functions as a virulence effector in P. sojae and enhances PVX‐induced viral symptoms in plants.
FIGURE 1.
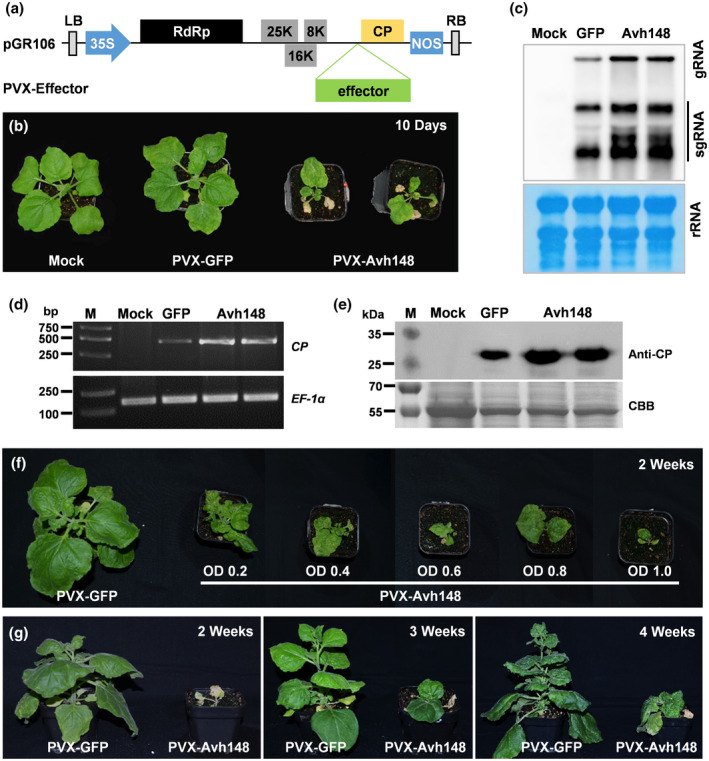
Transient expression of the Phytophthora sojae Avh148 gene in Nicotiana benthamiana leaves using the potato virus X (PVX) expression vector. (a) Schematic representation of the recombinant PVX‐Effector construct. (b) Photographs of 10‐day‐old N. benthamiana plants (n = 10) infiltrated with PVX‐Avh148, PVX‐GFP, or no construct (mock treatment; negative control) at 21 days postinfiltration (dpi). (c) RNA blot analysis of the accumulation of PVX genomic and subgenomic RNAs at 14 dpi. (d) Relative viral RNA accumulation in plants inoculated with PVX‐GFP or PVX‐Avh148. Reverse transcription PCR of the PVX coat protein (CP) gene was used to evaluate the viral RNA level. N. benthamiana EF‐1α gene was used as an internal control. (e) Western blot analysis of PVX CP in N. benthamiana leaves infiltrated with PVX‐GFP or PVX‐Avh148 using anti‐CP antibody. Equal loading was confirmed by Coomassie brilliant blue (CBB) staining. (f) Effect of Agrobacterium cell density (OD600 = 0.2–1.0) on disease development in 2‐week‐old N. benthamiana plants infiltrated with PVX‐Avh148. (g) Effect of plant age on disease development in N. benthamiana plants infiltrated with PVX‐Avh148 (OD600 = 1.2). Photographs were taken at 21 dpi. Experiments were performed twice, with similar results
TABLE 1.
Summary of effector genes identified using the virus‐induced virulence effector (VIVE) assay
| No. | Pathogen type | Pathogen name | Effector gene | Effector function | Virulence a |
|---|---|---|---|---|---|
| 1 | Bacterium | Pseudomonas syringae | AvrPto | Known | Yes |
| 2 | AvrPtoB | Yes | |||
| 3 | “Candidatus Liberibacter asiaticus” | SDE1 | Yes | ||
| 4 | Fungus | Magnaporthe oryzae | AvrPiz‐t | Yes | |
| 5 | Ustilaginoidea virens | SCRE2 | Yes | ||
| 6 | Oomycete | Phytophthora sojae | Avh432 | Unknown | No |
| 7 | Avh447 | No | |||
| 8 | Avh266 | No | |||
| 9 | Avh45 | No | |||
| 10 | Avh148 | Yes | |||
| 11 | Bacterium | “Ca. L. asiaticus” | CLas420 | Yes | |
| 12 | CLas525 | No | |||
| 13 | CLas2250 | No | |||
| 14 | CLas1640 | Yes | |||
| 15 | Virus | Rice stripe virus | RSV‐CP | Yes | |
| 16 | RSV‐SP | No | |||
| 17 | RSV‐NSVC4 | No | |||
| 18 | Bipolaris maydis | RSV‐NS2 | No | ||
| 19 | BmPV1‐ORF1 | No | |||
| 20 | BmPV1‐ORF2 | Yes | |||
| 21 | BmPV1‐ORF3 | No | |||
| 22 | Nematode | Heterodera avenae | Ha‐1137 | No | |
| 23 | Meloidogyne incognita | NSP‐NeF | Yes | ||
| 24 | Insect | Bemisia tabaci | BTA010383 | No | |
| 25 | BTA002812 | No | |||
| 26 | BTA026759 | No | |||
| 27 | BTA021638 | Yes | |||
| 28 | BTA003413 | No |
Effector showing virulence functions using the VIVE assay.
To fully understand the role of Avh148 in the induction of viral disease symptoms, we first analysed the effect of Agrobacterium cell density on N. benthamiana plants. Leaves of 2‐week‐old N. benthamiana plants were inoculated with serial dilutions of Agrobacterium cultures expressing PVX‐GFP or PVX‐Avh148; the optical density of the cultures at 600 nm (OD600) ranged from 0.2 to 1.0. Plants infiltrated with PVX‐148 showed more severe symptoms with increasing OD600 values; at OD600 = 0.2, the systemically infected leaves exhibited smaller size and severe downward curling, whereas at higher OD600 values severe chlorotic symptoms were absent in infiltrated leaves, but necrosis was detected in the midrib of infiltrated and systemically infected leaves, eventually leading to plant death (Figure 1f).
Next, we investigated the effect of plant age on disease development. Leaves of 2‐, 3‐, and 4‐week‐old seedlings were infiltrated with Agrobacterium cultures (OD600 = 1.2) expressing PVX‐GFP or PVX‐Avh148, and symptoms were compared. Younger plants infiltrated with PVX‐Avh148 developed more severe symptoms than older plants (Figure 1g). Two‐week‐old plants infiltrated with PVX‐Avh148 showed necrosis and death (Figure 1g), which was absent in 4‐week‐old plants. Furthermore, 4‐week‐old seedlings infiltrated with PVX‐Avh148 Agrobacterium at OD600 = 1.0 showed similar symptoms at 21 dpi (dwarfing and severe downward leaf curling) as 2‐week‐old seedlings infiltrated with PVX‐Avh148 Agrobacterium at OD600 = 0.2. These results indicate that younger plants are more susceptible to the inoculum than older plants, and 2‐week‐old seedlings are ideal for this screening assay.
To further evaluate the plant expression vector, two additional constructs were generated in which Avh148 was expressed under the control of the CaMV 35S promoter, pEG100‐Avh148 and pEG104‐Avh148. Unlike the plants infiltrated with Agrobacterium containing the PVX‐Avh148 plasmid, 2‐week‐old seedlings infiltrated with the pEG100, pEG100‐Avh148, or pEG104‐Avh148 constructs showed no unusual phenotypes at 21 dpi (Figure S3a). At 2 dpi, similar levels of Avh148 transcripts were detected in leaves inoculated with all expression vectors; however, at 21 dpi, Avh148 transcripts were detected only in leaves inoculated with PVX‐Avh148 and not in those inoculated with pEG100‐Avh148 or pEG104‐Avh148 (Figure S3b). These data suggest that the strong virulence phenotype of PVX‐Avh148‐inoculated plants results from the continuous and high‐level expression of Avh148. Next, we determined the effect of the stop codon of Avh148 on the virulence phenotype of PVX‐Avh148‐inoculated plants. The results indicated that removal of the stop codon did not affect the disease phenotype of PVX‐Avh148‐inoculated plants (Figure S4a). Western blot analysis showed that the CP alone, but not the Avh148‐CP fusion protein, was consistently present in all inoculated plants. The expression of both effector formats was stronger than that of PVX‐GFP at 21 dpi (Figure S4b). Taken together, these results suggest that Agrobacterium‐mediated delivery of a binary PVX‐based vector serves as an efficient tool for screening effectors that enhance viral disease symptoms in N. benthamiana.
To investigate the biological role of Avh148 in P. sojae infection, we analysed the expression pattern of Avh148 in the roots of susceptible soybean cultivar Jack at 1.5, 3, 6, 12, 16, 20, and 24 hr postinoculation (hpi). The results of quantitative reverse transcription RT‐PCR (RT‐qPCR) analysis showed that Avh148 expression was induced at the early stages of infection, with maximal expression at 3 hpi (approximately 200‐fold greater than that at 0 hpi) (Figure 2a).
FIGURE 2.
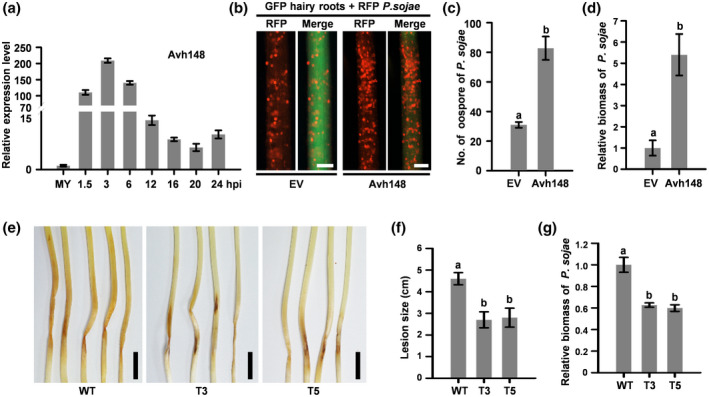
Phytophthora sojae effector Avh148 is crucial for pathogen virulence. (a) Expression profile of Avh148 in soybean hairy roots inoculated with P. sojae P6497. Total RNA was extracted from mycelia (MY) or infected soybean roots at the indicated time points, and transcript levels of Avh148 were detected by quantitative real‐time reverse transcription PCR (RT‐qPCR). P. sojae Actin gene was used as an internal control. hpi, hours postinoculation. (b) Analysis of Avh148 expression in soybean hairy roots upon P. sojae infection (n = 10). Hairy roots expressing GFP‐Avh148 or GFP were inoculated with mycelia plugs of red fluorescent protein (RFP)‐labelled P. sojae. Oospore production in infected hair roots was investigated under a fluorescence microscope, and lesion length was determined at 48 hpi. (c) Quantification of P. sojae oospores on soybean hairy roots at 48 hpi. (d) Quantification of P. sojae biomass in soybean hairy roots by quantitative PCR (qPCR). (e) Effect of Avh148 silencing in P. sojae on infection in soybean seedlings (n = 8). Disease symptoms were monitored in aetiolated hypocotyls. Photographs were taken at 7 days postinoculation (dpi). (f) Analysis of relative expression of Avh148 in P. sojae transformants by RT‐qPCR. (g) Analysis of P. sojae biomass in soybean hypocotyls by genomic DNA (gDNA)‐based qPCR. This experiment was performed twice, with similar results. Scale bars: 1 cm in (e), 0.3 mm in (e). Data in (c), (d), (f), and (g) represent mean ± SE of three independent biological replicates. Different letters indicate statistically significant differences among samples (p < .01; Duncan's multiple range test)
To determine whether Avh148 promotes P. sojae infection in planta, a construct carrying both 35S::GFP and 35S::Avh148‐3 × FLAG was introduced in Agrobacterium rhizogenes K599 and then transiently expressed in soybean hairy roots. Subsequently, soybean hairy roots were inoculated with P. sojae P6497 expressing the monomeric red fluorescent protein (mRFP) gene (Ma et al., 2017; Zhang et al., 2019b). At 48 hpi, soybean hairy roots expressing the 35S::Avh148‐3 × FLAG construct showed more oospores than those transformed with the empty vector (EV) control (Figure 2b,c). Consistently, P. sojae biomass was much higher in soybean hairy roots inoculated with Avh148‐3 × FLAG than in roots inoculated with the EV control (Figure 2d). Expression of the recombinant Avh148 protein was confirmed by western blot analysis using the anti‐FLAG antibody (Figure S5). These data suggest that Avh148 is a crucial virulence factor that contributes to plant susceptibility.
To further explore the contribution of Avh148 to the pathogenicity of P. sojae, we generated P. sojae Avh148 deletion in the wild‐type (WT) background (P6497) using the CRISPR/Cas9 system. A unique single‐guide RNA (sgRNA) targeting the Avh148 coding sequence was used to replace the Avh148 with mCherry in the deletion mutant, as described previously (Fang and Tyler, 2016) (Figure S6a). Two independent homozygous deletion mutant lines (T3 and T5) were identified by genomic DNA (gDNA)‐based PCR (Figure S6b) and confirmed by single‐colony sequencing. Compared with the WT strain, both T3 and T5 transformants showed no abnormal developmental phenotypes (Figure S6c,d). However, both Avh148 deletion mutant lines of P. sojae caused smaller lesions on soybean seedlings than the WT strain (Figure 2e,f). Furthermore, P. sojae biomass was significantly lower in soybean seedlings infected with transgenic strains T3 and T5 than in seedlings infected with the WT strain (Figure 2g). These results indicate that Avh148 is essential for the virulence of P. sojae. In summary, these observations confirmed the utility of the PVX‐based expression system as a functional screen for the identification of P. sojae effectors. Thus, we termed this screening method the virus‐induced virulence effector (VIVE) assay.
Having established and optimized the VIVE assay for the identification of P. sojae virulence effectors, we tested its applicability to other plant pathogens. Five previously known virulence effectors were chosen for this experiment, including two bacterial effectors (AvrPto and AvrPtoB from Pseudomonas syringae; SDE1 from “Ca. L. asiaticus”) and two fungal effectors (AvrPiz from Magnaporthe oryzae; SCRE2 from Ustilaginoidea virens) (Nguyen et al., 2010; Park et al., 2012; Clark et al., 2018; Fang et al., 2019). These virulence effectors were cloned into PVX vectors and introduced in A. tumefaciens, which was then used to inoculate N. benthamiana leaves to test whether these effectors can aggravate viral symptoms. All inoculated plants exhibited typical viral disease symptoms, including downward leaf curling, leaf puckering, and necrosis (Figures 3a and S7). By contrast, N. benthamiana plants expressing PVX‐GFP showed no symptoms (Figures 3a and S7). Northern blot assay revealed that the level of PVX genomic and subgenomic RNAs in plants inoculated with the known effectors was higher than in control plants at 21 dpi (Figure 3b). Additionally, RT‐qPCR was performed to verify the accumulation of PVX‐CP transcripts in plants inoculated with PVX‐AvrPtoB and PVX‐AvrPiz (Figure 3c). These results suggest that the VIVE assay can be used for the identification of virulence effectors in a wide range of plant pathogens, including unculturable pathogens.
FIGURE 3.
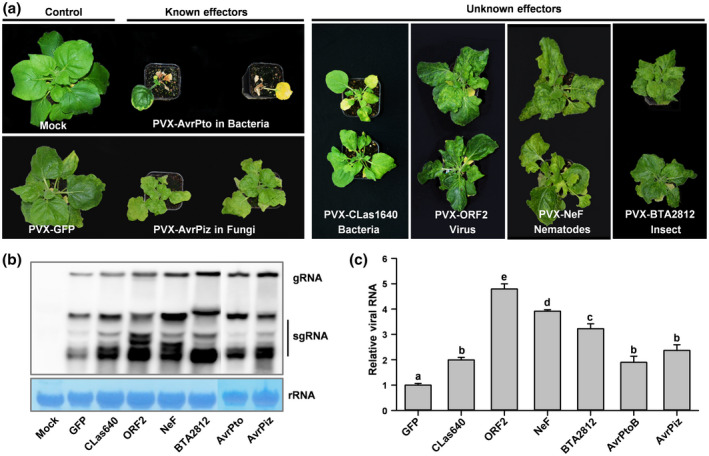
Confirmation and identification of virulence effectors in multiple plant pathogens using the virus‐induced virulence effector (VIVE) assay. (a) Analysis of two known and four novel virulence effectors from six plant pathogens in leaves of 10–14‐day‐old Nicotiana benthamiana plants (n = 10) using the PVX‐based expression system. Leaves were infiltrated with Agrobacterium (OD600 = 0.6–0.8) carrying the PVX‐Effector construct, and photographs were taken at 21 days postinfiltration (dpi). (b) RNA blot analysis of the accumulation of PVX genomic and subgenomic RNAs in N. benthamiana plants at 21 dpi. Total RNA was extracted from infected N. benthamiana leaves at 21 dpi. Wild‐type N. benthamiana plants (mock) were used as a negative control. (c) Relative viral RNA accumulation in plants inoculated with the PVX‐GFP or PVX‐Avh148 construct. Analysis of the PVX CP gene by quantitative reverse transcription PCR (RT‐qPCR) was used to determine the viral RNA level. Data represent mean ± SE of three independent experiments. The EF‐1α gene was used as an internal control. Each experiment was performed twice, with similar results. Different letters indicate statistically significant differences among samples (p < .01; Duncan's multiple range test)
We then tested the utility of the VIVE assay for the identification of novel virulence effectors from six plant pathogens. Instead of culturable species such as P. sojae, we focused on plant pathogens that cannot be cultured or genetically transformed, including “Ca. L. asiaticus”, cereal cyst nematode (Heterodera avenae), root‐knot nematode (Meloidogyne incognita), silverleaf whitefly (Bemisia tabaci), RSV, and Bipolaris maydis (fungal pathogen of southern corn leaf blight). A total of 18 effector genes belonging to these six plant pathogens were cloned into the pGR106 expression vector (Table 1). The resulting constructs were introduced into A. tumefaciens, which was then used to inoculate the leaves of 2‐week‐old N. benthamiana plants via agroinfiltration. A total of six putative virulence effectors were identified: NeF in M. incognita, BTA021638 in B. tabaci, CLas420 and CLas1640 in “Ca. L. asiaticus”, viral proteins CP and ORF2 in RSV, and Bipolaris maydis partitivirus 1 (BmPV1) in B. maydis. N. benthamiana plants infiltrated with Agrobacterium carrying the PVX vector harbouring each of these six proteins separately showed distinct and discernibly severe viral disease symptoms. Among all plants, those infiltrated with the PVX‐NeF showed the most prominent viral disease phenotypes (Figure 3a and Table 1). Northern blot analysis revealed that levels of PVX genomic and subgenomic RNAs were much higher in leaves infiltrated with these four effector genes than in leaves infiltrated with PVX‐GFP at 21 dpi (Figure 3b). Overall, our results revealed that the VIVE assay is a reliable and powerful tool for the identification of effector proteins of pathogens that function as potential virulence factors in plants.
Many mammalian and plant pathogens secrete virulence proteins into the host cell via diverse secretion systems. These proteins attack the host innate immune system, modify the cytoskeleton and cellular membranes, alter vesicle trafficking, and facilitate nutrient acquisition, eventually causing infection and disease (He et al., 2004). In this study, we successfully identified six potential virulence effectors in bacteria, filamentous pathogens, viruses, nematodes, and insects using the VIVE assay (Figures 1 and 3). The identification of such a wide range of virulence factors suggests that VIVE is a general and robust tool that can be used to obtain preliminary biological information related to pathogenic phenotypes.
Despite its wide applicability, it is possible that a small subset of host‐specific virulence factors is not compatible with the VIVE assay. One possibility is that plant genes are silenced via virus‐induced gene silencing (VIGS) if the effector gene shows relatively high sequence similarity with a plant gene, leading to false positives (Takken et al., 2000). In addition, the development of severe systemic symptoms in N. benthamiana leaves infiltrated with the PVX‐Effector construct indicates that the effector gene interacts synergistically with the virus in causing infection, with a severe pathogenic response, and effectors that fail to interact with the virus may be less likely to be detected. Although true synergism between two viruses has been reported in plants (Ogwok et al., 2010), synergism between an effector protein and a virus has not been investigated to date. On the other hand, disease symptoms also might be associated with small RNA (sRNA) movement between plants and pathogens. Recent studies have shown that recovery from viral disease involves sRNA‐mediated posttranscriptional gene silencing (PTGS), and components of the transcriptional gene silencing (TGS) pathway facilitate the movement of the virus from the infection site to upper leaves, leading to sRNA‐mediated resistance in distal tissues (Baulcombe, 2004; Korner et al., 2018). Additionally, we cannot rule out the possibility that abnormal phenotypes of N. benthamiana plants are caused by the loss of function of specific development‐related genes or proteins, which might be targeted by specific effector proteins. Thus, the precise mechanism of the exacerbation of PVX disease symptoms by an effector protein remains largely unknown.
The power of the VIVE approach relies on several factors, most importantly the cell density of Agrobacterium cultures and the age of N. benthamiana plants. A previous study showed that virus titre and plant age are critical for the successful infection of plants and replication of the virus; young plants infected with a high virus concentration exhibit severely stunted growth (Ogwok et al., 2010). Consistent with this result, our data showed that 10–14‐day‐old N. benthamiana seedlings inoculated with Agrobacterium (OD600 = 0.6–0.8) were ideal for the screening of virulence effectors (Figure 1b–e). However, high cell density can be used to screen weak virulence effectors. Additionally, we were unable to examine the effect of other environmental factors, such as temperature, on plant infection; these factors greatly affect plant–virus interactions and infection (Chellappan et al., 2005).
In summary, our data suggest that the VIVE assay serves as a general, straightforward, and unbiased method of obtaining rudimentary information about the virulence determinants of various plant pathogens. The identification of crucial virulence effectors using the VIVE assay will broaden our understanding of the molecular basis of pathogenesis and plant resistance in fundamentally diverse pathosystems.
Supporting information
ACKNOWLEDGMENTS
We thank Professors Youjun Zhang (Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences [CAAS]), Huan Peng (Institute of Plant Protection, CAAS), and Songbai Zhang (Yangtze University) for providing the genome sequences of insect, nematode, and virus, respectively. We also thank Professor Fei Yan (Ningbo University) for providing anti‐coat protein antibody. This work was supported by grants from the National Key Research and Development Program of China (2018YFD0201500), the Thousand Talents Program for Young Professionals of China, and the Science and Technology Commission of Shanghai Municipality (18DZ2260500).
Shi J, Zhu Y, Li M, et al. Establishment of a novel virus‐induced virulence effector assay for the identification of virulence effectors of plant pathogens using a PVX‐based expression vector. Molecular Plant Pathology 2020;21:1654–1661. 10.1111/mpp.13000
Jinxia Shi, Yuanhong Zhu, and Ming Li contributed equally to this work.
[Correction added on 11 November 2020 after first publication: Figure S7 and Table S2 have been updated in this version.]
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. [DOI] [PubMed] [Google Scholar]
- Castagnone‐Sereno, P. , Danchin, E.G. , Perfus‐Barbeoch, L. and Abad, P. (2013) Diversity and evolution of root‐knot nematodes, genus Meloidogyne: new insights from the genomic era. Annual Review of Phytopathology, 51, 203–220. [Google Scholar]
- Chapman, S. , Kavanagh, T. and Baulcombe, D. C. (1992) Potato virus X as a vector for gene expression in plants. The Plant Journal, 2, 549–557. [DOI] [PubMed] [Google Scholar]
- Chellappan, P. , Vanitharani, R. , Ogbe, F. and Fauquet, C.M. (2005) Effect of temperature on geminivirus‐induced RNA silencing in plants. Plant Physiology, 138, 1828–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K. , Franco, J.Y. , Schwizer, S. , Pang, Z. , Hawara, E. , Liebrand, T.W.H. et al. (2018) An effector from the huanglongbing‐associated pathogen targets citrus proteases. Nature Communications, 9, 1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis, J. , Acosta‐Ramirez, E. , Zhao, Y. , Hamelin, M.E. , Koukavica, I. , Baz, M. et al. (2008) Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine, 26, 3395–3403. [DOI] [PubMed] [Google Scholar]
- Dickmeis, C. , Fischer, R. and Commandeur, U. (2014) Potato virus X‐based expression vectors are stabilized for long‐term production of proteins and larger inserts. Biotechnology Journal, 9, 1369–1379. [DOI] [PubMed] [Google Scholar]
- Esfandiari, N. , Arzanani, M.K. , Soleimani, M. , Kohi‐Habibi, M. and Svendsen, W.E. (2016) A new application of plant virus nanoparticles as drug delivery in breast cancer. Tumour Biology, 37, 1229–1236. [DOI] [PubMed] [Google Scholar]
- Fang, A. , Gao, H. , Zhang, N. , Zheng, X. , Qiu, S. , Li, Y. et al. (2019) A novel effector gene SCRE2 contributes to full virulence of Ustilaginoidea virens to rice. Frontiers in Microbiology, 10, 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Y. and Tyler, B.M. (2016) Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Molecular Plant Pathology, 17, 127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, S.Y. , Nomura, K. and Whittam, T.S. (2004) Type III protein secretion mechanism in mammalian and plant pathogens. Biochimica et Biophysica Acta, 1694, 181–206. [DOI] [PubMed] [Google Scholar]
- Hefferon, K. (2017) Plant virus expression vectors: a powerhouse for global health. Biomedicines, 5, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman, M.J. , Linthorst, H.J. , Bol, J.F. and Cornelissen, J.C. (1988) The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus‐stranded RNA viruses. Journal of General Virology, 69, 1789–1798. [DOI] [PubMed] [Google Scholar]
- Jones, L. , Hamilton, A.J. , Voinnet, O. , Thomas, C.L. , Maule, A.J. and Baulcombe, D.C. (1999) RNA–DNA interactions and DNA methylation in post‐transcriptional gene silencing. The Plant Cell, 11, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, L. , Wu, J. , Lu, L. , Xu, Y. and Zhou, X. (2014) Interaction between Rice stripe virus disease‐specific protein and host PsbP enhances virus symptoms. Molecular Plant, 7, 691–708. [DOI] [PubMed] [Google Scholar]
- Korner, C.J. , Pitzalis, N. , Pena, E.J. , Erhardt, M. , Vazquez, F. and Heinlein, M. (2018) Crosstalk between PTGS and TGS pathways in natural antiviral immunity and disease recovery. Nature Plants, 4, 157–164. [DOI] [PubMed] [Google Scholar]
- Lacomme, C. and Chapman, S. (2008) Use of potato virus X (PVX)‐based vectors for gene expression and virus‐induced gene silencing (VIGS). Current Protocols in Microbiology, 8, 1–16. [DOI] [PubMed] [Google Scholar]
- Lee, A.H. , Bastedo, D.P. , Youn, J.Y. , Lo, T. , Middleton, M.A. , Kireeva, I. et al. (2019) Identifying Pseudomonas syringae type III secreted effector function via a yeast genomic screen. G3 (Bethesda), 9, 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Ying, X. , Shang, L. , Redfern, B. , Kypraios, N. , Xie, X. et al. (2020) Heterologous expression of CLIBASIA_03915/CLIBASIA_04250 by tobacco mosaic virus resulted in phloem necrosis in the senescent leaves of Nicotiana benthamiana . International Journal of Molecular Sciences, 21, 1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, Z. , Zhu, L. , Song, T. , Wang, Y. , Zhang, Q. , Xia, Y. et al. (2017) A paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science, 355, 710–714. [DOI] [PubMed] [Google Scholar]
- Mardanova, E.S. , Kotlyarov, R.Y. , Kuprianov, V.V. , Stepanova, L.A. , Tsybalova, L.M. , Lomonosoff, G.P. et al. (2015) Rapid high‐yield expression of a candidate influenza vaccine based on the ectodomain of M2 protein linked to flagellin in plants using viral vectors. BMC Biotechnology, 15, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, H.P. , Yeam, I. , Angot, A. and Martin, G.B. (2010) Two virulence determinants of type III effector AvrPto are functionally conserved in diverse Pseudomonas syringae pathovars. New Phytologist, 187, 969–982. [DOI] [PubMed] [Google Scholar]
- Ogwok, E. , Patil, B.L. , Alicai, T. and Fauquet, C.M. (2010) Transmission studies with Cassava brown streak Uganda virus (Potyviridae: Ipomovirus) and its interaction with abiotic and biotic factors in Nicotiana benthamiana . Journal of Virological Methods, 169, 296–304. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. et al. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen‐associated molecular pattern‐triggered immunity in rice. The Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y. , Liu, L. , Xiong, Q. , Flores, C. , Wong, J. , Shi, J. et al. (2013) Oomycete pathogens encode RNA silencing suppressors. Nature Genetics, 45, 330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, Y. , Shi, J. , Zhai, Y. , Hou, Y. and Ma, W. (2015) Phytophthora effector targets a novel component of small RNA pathway in plants to promote infection. Proceedings of the National Academy of Sciences of the United States of America, 112, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. , DiFranco, N.A. , Wen, A.M. , Commandeur, U. and Steinmetz, N.F. (2015) To target or not to target: active vs. passive tumor homing of filamentous nanoparticles based on potato virus X. Cellular and Molecular Bioengineering, 8, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken F.L.W., Luderer R., Gabriels S.H.E.J., Westerink N., Lu R., de Wit P.J.G.M., et al (2000) A functional cloning strategy, based on a binary PVX‐expression vector, to isolate HR‐inducing cDNAs of plant pathogens. The Plant Journal, 24, 275–283. [DOI] [PubMed] [Google Scholar]
- Toruno, T.Y. , Stergiopoulos, I. and Coaker, G. (2016) Plant‐pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annual Review of Phytopathology, 54, 419–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, R.L. , Chapman, S. , Carr, F. and Santa Cruz, S. (2001) A novel strategy for the expression of foreign genes from plant virus vectors. FEBS Letters, 489, 215–219. [DOI] [PubMed] [Google Scholar]
- Varden, F.A. , De la Concepcion, J.C. , Maidment, J.H. and Banfield, M.J. (2017) Taking the stage: effectors in the spotlight. Current Opinion in Plant Biology, 38, 25–33. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Han, C. , Ferreira, A.O. , Yu, X. , Ye, W. , Tripathy, S. et al. (2011) Transcriptional programming and functional interactions within the Phytophthora sojae RXLR effector repertoire. The Plant Cell, 23, 2064–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Cong, Q.Q. , Lan, Y.F. , Geng, C. , Li, X.D. , Liang, Y.C. et al. (2014) Development of new potato virus X‐based vectors for gene over‐expression and gene silencing assay. Virus Research, 191, 62–69. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Tyler, B.M. and Wang, Y. (2019) Defense and counterdefense during plant–pathogenic oomycete infection. Annual Review of Microbiology, 73, 667–696. [DOI] [PubMed] [Google Scholar]
- Wydro, M. , Kozubek, E. and Lehmann, P. (2006) Optimization of transient Agrobacterium‐mediated gene expression system in leaves of Nicotiana benthamiana . Acta Biochimica Polonica, 53, 289–298. [PubMed] [Google Scholar]
- Zhang, C. , Wang, X. , Liu, X. , Fan, Y. , Zhang, Y. , Zhou, X. et al. (2019a) A novel “Candidatus Liberibacter asiaticus”‐encoded Sec‐dependent secretory protein suppresses programmed cell death in Nicotiana benthamiana . International Journal of Molecular Sciences, 20, 5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P. , Jia, Y. , Shi, J. , Chen, C. , Ye, W. , Wang, Y. et al. (2019b) The WY domain in the Phytophthora effector PSR1 is required for infection and RNA silencing suppression activity. New Phytologist, 223, 839–852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author upon reasonable request.


