Abstract
Adaptation and efficient colonization of the phyllosphere are essential processes for the switch to an epiphytic stage in foliar bacterial pathogens. Here, we explore the interplay among light perception and global transcriptomic alterations in epiphytic populations of the hemibiotrophic pathogen Pseudomonas syringae pv. tomato DC3000 (PsPto) following contact with tomato leaves. We found that blue‐light perception by PsPto on leaf surfaces is required for optimal colonization. Blue light triggers the activation of metabolic activity and increases the transcript levels of five chemoreceptors through the function of light oxygen voltage and BphP1 photoreceptors. The inactivation of PSPTO_1008 and PSPTO_2526 chemoreceptors causes a reduction in virulence. Our results indicate that during PsPto interaction with tomato plants, light perception, chemotaxis, and virulence are highly interwoven processes.
Keywords: chemosensory proteins, light perception, Pseudomonas syringae, virulence
Blue light increases chemotaxis and metabolic activity in Pseudomonas syringae, permitting an optimal response to plant signals that facilitates bacterial plant entry.

1. INTRODUCTION
Survival in the phyllosphere is a key step in the life cycle of many phytopathogenic bacteria. Bacteria adapt to the environment on the leaf surface and eventually enter the plant tissue and cause disease (Hirano and Upper, 2000; Xin and He, 2013). The outcome of the interaction will therefore be determined by the ability to both survive in the phyllosphere and enter the plant tissues. Pseudomonas syringae pv. tomato DC3000 (PsPto) is considered a model to study plant–bacterial interactions. This foliar bacterial pathogen presents a relatively short epiphytic stage, but once inside the host, it is a highly destructive pathogen (Boureau et al., 2002; Xin and He, 2013). PsPto must perceive plant and environmental signals to elaborate a rapid and coordinated response permitting entry into the plant apoplast and establishment of infection. The study of the bacterial adaptations that govern the initial stages of interaction with the plant in the phyllosphere is essential to elucidate the switch to a pathogenic lifestyle and the occurrence of disease.
The phyllosphere is a hostile and highly dynamic environment where phytopathogenic bacteria are continuously exposed to changes in temperature, humidity, nutrient availability, CO2, and light intensity (Vorholt, 2012; Carvalho and Castillo, 2018). To achieve an optimal colonization of the leaf surface, bacteria possess specialized mechanisms involved in the recognition of plant and environmental signals. These mechanisms are commonly one‐ and two‐component systems as well as chemoreceptor‐based signalling cascades (Ulrich et al., 2005; Wuichet and Zhulin, 2010). In chemoperception, signalling is initiated by signal recognition at the ligand‐binding domain of the chemoreceptor. This recognition triggers downstream signalling involving changes in the histidine kinase CheA autokinase activity and transphosphorylation activity to the response regulator CheY, which in its phosphorylated form binds to the flagellar motor, altering its activity and causing chemotaxis (Matilla and Krell, 2018). Control of motility and chemotaxis towards plant compounds facilitates bacterial access to the apoplast through stomata or wounds on the leaf surface (Matilla and Krell, 2018). Both motility and chemotaxis during the initial stages of plant colonization are required for a subsequent successful infection (Rio‐Alvarez et al., 2014; Matilla and Krell, 2018; Santamaria‐Hernando et al., 2018). Chemoreceptors or methyl‐accepting chemotaxis proteins (MCPs) are particularly abundant in phytopathogenic bacteria as compared to bacteria living in other habitats (Matilla and Krell, 2018). However, information regarding the specific signals that trigger the chemotactic response in plant pathogens is scarce. Chemotaxis towards sugars, amino acids, and jasmonic acid in Dickeya dadantii (Antunez‐Lamas et al., 2009) as well as to malate in Ralstonia solanacearum seem to be required for virulence (Hida et al., 2015). Aerotaxis is essential in R. solanacearum for biofilm formation and full virulence (Yao and Allen, 2006). Chemotaxis towards several compounds has also been observed in different pathovars of Xanthomonas campestris (Kamoun and Kado, 1990), Xanthomonas oryzae pv. oryzae (Verma et al., 2018), and PsPto (Cuppels, 1988; Kim et al., 2007). The PsPto genome encodes 49 MCPs that are thought to stimulate five different chemosensory pathways composed of homologous signalling proteins (Figure S1a,b) encoded by gene clusters I to V (Ulrich and Zhulin, 2010). Cluster I encodes the core proteins of the chemotaxis pathway in PsPto (Parales et al., 2004). PsPto cluster III is homologous to the Pseudomonas aeruginosa wrinkly spreader (Wsp) pathway involved in the control of the bacterial second messenger cyclic diguanosine monophosphate (c‐di‐GMP) (Hickman et al., 2005). Recent work in our laboratory demonstrated a dual role for a PsPto chemoreceptor that consisted of mediating chemotaxis to specific amino acids and the regulation of virulence‐related traits like biofilm formation through the control of c‐di‐GMP levels (Cerna‐Vargas et al., 2019). This result suggests a crosstalk between different signal perception and virulence‐related mechanisms to elaborate a coordinated response that optimizes the PsPto virulence process.
Bacteria are also able to perceive changes in light quality and intensity that occur during their life in the phyllosphere by means of photosensory proteins. Several works have focused on the use of light by plants as a signal to regulate stomatal aperture and defences against the attack of pathogens (Roden and Ingle, 2009; Carvalho and Castillo, 2018; Matthews et al., 2019; Fernandez‐Milmanda et al., 2020). Recent studies have revealed that foliar bacterial pathogens can also exploit light cues to optimize their adaptation to plant surface and virulence. Green light affects the expression of virulence‐related genes and disease severity in Pseudomonas cichorii (Rajalingam and Lee, 2018). Light perception modulates adhesion, motility, and virulence in Xanthomonas axonopodis pv. citri and X. campestris pv. campestris (Kraiselburd et al., 2012; Bonomi et al., 2016), as well as swarming motility, leaf colonization, and virulence in P. syringae pv. syringae (Wu et al., 2013; McGrane and Beattie, 2017). The PsPto genome encodes a single blue‐light‐sensing light oxygen voltage (LOV)‐domain protein and two red‐light‐sensing bacteriophytochromes, BphP1 and BphP2 (Figure S2). Light perception through these photosensory proteins, especially during the initial stages of plant colonization, affects adhesion, motility, and virulence in PsPto (Moriconi et al., 2013; Rio‐Alvarez et al., 2014; Santamaria‐Hernando et al., 2018). Interestingly, we found that light functions as a global signal in PsPto. A short light treatment prior to challenge induces a genetic reprogramming that involves significant changes in subsequent leaf colonization and virulence‐related traits. These changes at the transcriptomic level were correlated with alterations in the virulence outcome depending on the wavelength of the light applied to bacteria before plant inoculation (Santamaria‐Hernando et al., 2018). These findings highlight the relevance of light as a signal in the early stages of plant colonization and prompted us to evaluate the effect of light perception on gene expression in PsPto epiphytic populations during the initial contact with the tomato leaf surface.
Our results show that flagellar motility and chemotaxis are up‐regulated during the switch to the epiphytic stage in PsPto. Moreover, in this work we provide evidence that the perception of blue light through photoreceptors regulates MCP expression in chemoheterotrophic bacteria. To our knowledge, this is the first report showing that light perception, chemotaxis, and virulence are highly interwoven processes.
2. RESULTS
2.1. Up‐regulation of motility and chemotaxis is a coordinated response during the switch of PsPto to the epiphytic stage regardless of the light conditions
Previous work carried out in our laboratory showed differential gene expression in PsPto under different light treatments in vitro, which suggested that this bacterium can exploit light cues to optimize virulence and survival on plant leaves (Santamaria‐Hernando et al., 2018). To evaluate the effect of light on gene expression during the epiphytic stage of PsPto, we carried out a global transcriptomic profiling of PsPto populations on tomato leaves under different light conditions. Bacterial cells were spray‐inoculated in both adaxial and abaxial surfaces of the leaves, and subsequently exposed to a 10‐min treatment with white, blue, or red light, or maintained in darkness. Total bacterial RNA was extracted, and microarray hybridization experiments were performed using custom‐designed microarrays (Agilent Technologies). The obtained data were compared to previous results obtained from in vitro‐cultured populations exposed to the same light treatments (Santamaria‐Hernando et al., 2018).
Comparative analyses revealed that a short contact with the leaf surface has a high impact on the PsPto transcriptome, regardless of prevailing light conditions. Transcript levels of around 30%–45% of PsPto genes were altered in populations on the leaf surface as compared to those under in vitro conditions (Table S1). About one half of the differentially expressed genes were up‐regulated (Table S1).
An analysis of the functional categories that were over‐represented among the differentially expressed genes was performed. Regardless of the light treatment, the over‐represented functional categories among the up‐regulated genes were “flagellar synthesis and motility”, “chemosensing and chemotaxis”, and to a lesser extent “polysaccharide synthesis and regulation” (Figure 1a and Table S2). Genes encoding chemosensory pathway proteins, including 40 out of the 49 PsPto MCP‐encoding genes, showed a differential expression in epiphytic populations and among them 36 were up‐regulated (Figure 2a and Table S2). Among genes encoding chemosensory pathway proteins, we observed the up‐regulation of genes belonging to the chemotaxis cluster I, cluster II, and cluster III (Table S2 and Figure S1b). Regarding cluster IV, only gene‐encoding protein PilH was up‐regulated in epiphytic populations. PsPto proteins of cluster IV are homologous to proteins that control type IV pili‐based motility and cAMP levels in P. aeruginosa (Ortega et al., 2017) and are included in the “pili synthesis and regulation” category. Interestingly, analysis of the microarray data showed that only five genes belonging to the “chemosensing and chemotaxis” category were down‐regulated in epiphytic populations (Figure 2a). Four of them correspond to MCPs that have not been previously characterized in either PsPto or any other bacteria, whereas the remaining protein is an uncharacterized CheR methyltransferase that does not belong to any of the chemosensory clusters defined in PsPto. In the “flagellar synthesis and motility” category, all genes that presented a differential expression in epiphytic populations were up‐regulated (Figure 2b).
FIGURE 1.
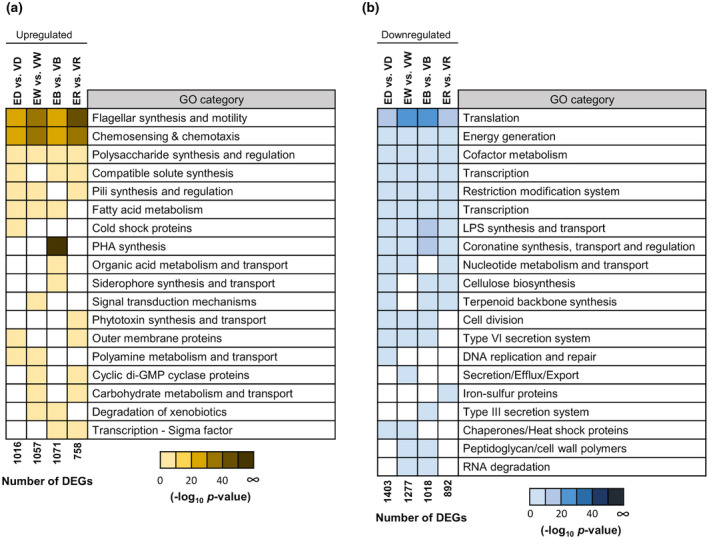
Functional categories over‐represented in light‐treated Pseudomonas syringae pv. tomato (PsPto) epiphytic populations as compared to in vitro populations. Heatmap of −log10 p values of the functional categories that are over‐represented among the differentially expressed up‐regulated (a) or down‐regulated (b) genes after a 10‐min treatment with white, blue, red light, or darkness (fold change ≥2, false discovery rate ≤ 0.01), as compared to in vitro populations subjected to the same light treatments (n = 4 biological replicates). Only functional categories that are significantly over‐represented are shown (p ≤ .01). E, epiphytic; V, in vitro; D, darkness; W, white‐light; B, blue‐light; R, red‐light; DEG, differentially expressed gene
FIGURE 2.
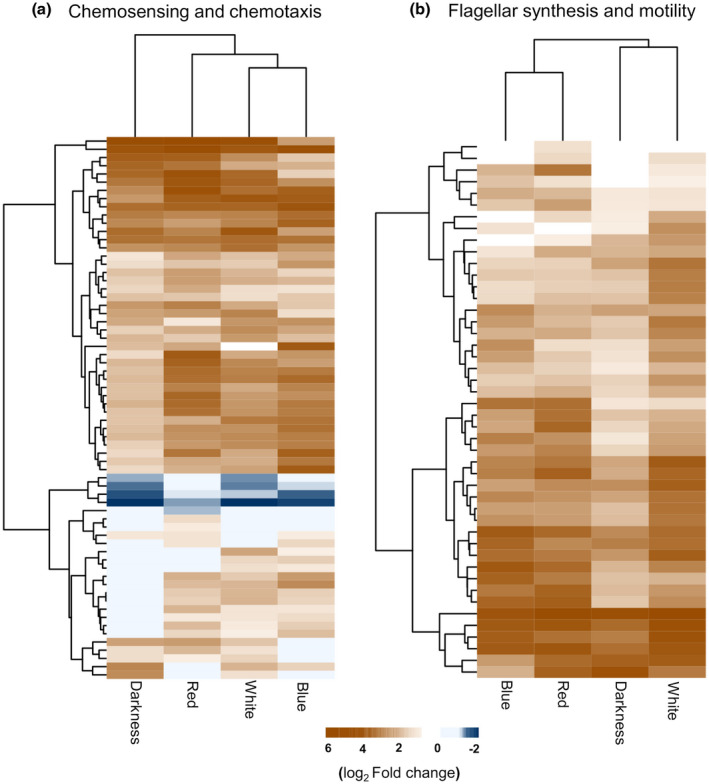
Genes related to flagellar motility and chemotaxis are up‐regulated during the switch to the epiphytic stage. Hierarchical clustering of the differentially expressed genes in chemosensing and chemotaxis (a) and flagellar synthesis and motility (b) categories in light‐treated Pseudomonas syringae pv. tomato (PsPto) epiphytic populations as compared to in vitro populations. Bar indicates log2 fold‐change in expression
Among the down‐regulated genes, we found a larger number of functional categories over‐represented than among the up‐regulated genes, regardless of light conditions (Figure 1b and Table S3). Regardless of the light treatment applied, we observed a decrease in transcript levels of genes related to translation, energy generation, cofactor metabolism, transcription, restriction‐modification systems, and lipopolysaccharide (LPS) synthesis and transport (Figure 1b). Interestingly, genes related to synthesis, transport, and regulation of the phytotoxin coronatine, a major virulence factor in PsPto, were down‐regulated in epiphytic populations regardless of the light conditions. Previous studies indicate that coronatine is produced after 3–4 hr contact with the leaf surface (Panchal et al., 2016), which suggests a more relevant role in the later stages of leaf colonization, facilitating bacterial entry to the apoplast and modulating plant defence responses.
We also observed that some other specific functional categories were over‐represented depending on the particular light treatment (Figure 1a and Table S2). Interestingly, the four genes encoding polyhydroxyalkanoate (PHA) biosynthetic proteins in PsPto were up‐regulated in blue‐light‐treated epiphytic cells. In addition, a significant number of genes involved in the metabolism and transport of diverse organic acids, and in the synthesis and transport of pyoverdine and yersiniabactin, two of the siderophores produced by PsPto, were significantly up‐regulated only in epiphytic cells after blue‐light treatment. White‐light treatment increased transcripts related to different signal transduction mechanisms while red‐light treatment up‐regulated the expression of genes involved in the biosynthesis of the biosurfactant syringafactin.
We also found specific differential gene expression depending on the light treatment among the down‐regulated genes (Figure 1b and Table S3). We observed that genes related to DNA replication and repair were only down‐regulated in dark‐treated epiphytic cells. In addition, genes for secretion/efflux/export and the type III secretion system showed a reduced expression in epiphytic populations after a white‐ or a blue‐light treatment, respectively. Transcripts for several iron‐sulphur proteins were reduced in red‐light‐treated epiphytic cells.
Overall, our results indicate that regardless of the light conditions, some bacterial traits are essentially required for adaptation to the leaf surface. Moreover, we found certain light‐responsive bacterial determinants that can modulate the general process of adaptation and colonization of the leaf.
2.2. Blue light has a high impact on the regulation of gene expression in epiphytic populations
We analysed the role of white, blue, and red light in the gene expression regulation of PsPto epiphytic populations through the comparison with data of dark‐treated epiphytic cells. The analysis revealed that a 10‐min treatment of epiphytic populations with either white or red light had a small impact on PsPto transcriptome when compared to dark‐treated epiphytic cells. Only six genes under white light and 12 genes under red light were differentially expressed (≥2‐fold change; false discovery rate [FDR] ≤ 0.05) (Table S4). However, blue‐light treatment had a larger effect on gene expression because 546 genes were differentially expressed (≥2‐fold change; FDR ≤ 0.01), of which 73% were up‐regulated (Table S4). An analysis of the functional categories among the differentially expressed genes identified 12 categories that were over‐represented among genes responding to the blue‐light treatment (Figure 3 and Table S5). The analysis revealed that eight of the categories are associated with diverse metabolic‐related pathways in PsPto. We observed the up‐regulation of genes involved in the metabolism and transport of organic acids, carbohydrates, fatty acids, sulphur, and nucleotides. Genes related to metabolism and transport of γ‐amino butyric acid (GABA) and polyamines were over‐represented among induced genes. Genes encoding proteins involved in iron metabolism and transport, siderophore synthesis, and other transport processes were also induced in blue‐light‐treated epiphytic populations. Interestingly, some of these genes have been related to epiphytic fitness, assisting bacteria to cope with nutrient fluctuations and plant‐defence‐derived compounds, as well as to compete for limiting nutrients and ecological niches (Kachroo and Kachroo, 2009; Ryffel et al., 2016; Vilas et al., 2018).
FIGURE 3.
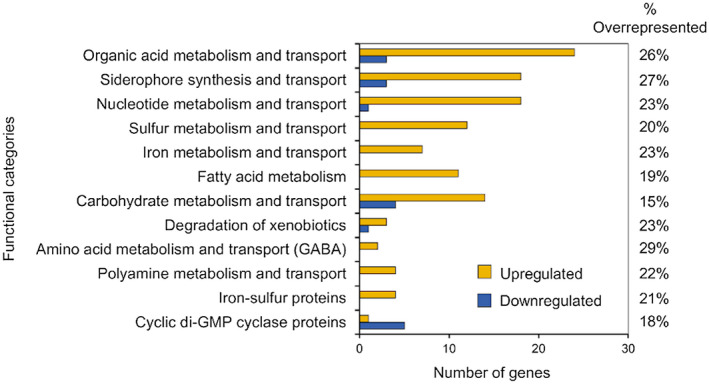
Blue light induces gene‐expression reprogramming in Pseudomonas syringa e pv. tomato (PsPto) epiphytic populations. The data shown are the functional categories that are over‐represented among the differentially expressed genes responding to a 10 min treatment with blue light (fold change ≥2, false discovery rate ≤ 0.01) in PsPto epiphytic populations as compared to dark‐treated epiphytic cells. Only functional categories that are significantly over‐represented are shown (p ≤ .05). Yellow and blue indicate the number of up‐regulated and down‐regulated genes, respectively, within each functional category. The percentage over‐represented indicates the percentage of observed genes out of the total number of genes annotated in each functional category
Taken together, these results indicate that blue light has a major role in the initial stages of leaf colonization, regulating the expression of a wide array of genes that may lead to an increased competitiveness of PsPto for nutrients and favourable sites on the leaf.
2.3. Blue light up‐regulates the expression of five genes encoding MCPs during the PsPto epiphytic stage
Chemotaxis‐driven rather than random motility is a requirement for access to nutrients and leaf invasion through stomata (Matilla and Krell, 2018). Furthermore, plant‐secreted compounds are commonly considered chemoeffectors that induce chemotaxis towards specific sites of the leaf where diffusion rates increase, or towards plant openings like stomata (Matilla and Krell, 2018). Our results showed that the “chemosensing and chemotaxis” category was over‐represented in PsPto epiphytic populations with respect to in vitro populations. Moreover, we showed that blue light had a high impact on genetic reprogramming of PsPto epiphytic cells. Considering these results, we decided to evaluate whether the expression of any gene in the “chemosensory and chemotaxis” category was specifically altered by blue‐light treatment in epiphytic populations. Our data show that the expression of five genes encoding MCPs (PSPTO_1008, PSPTO_1493, PSPTO_2526, PSPTO_4541, and PSPTO_5352) was up‐regulated in blue‐light‐treated epiphytic populations when compared to dark‐treated epiphytic cells (Table S4). Induced expression of the five genes after the 10‐min blue‐light treatment was confirmed by quantitative reverse transcription PCR (RT‐qPCR) using the RNA isolated for the microarray experiments (Figure 4). Our findings suggest that PsPto uses blue light as a source of information that drives, among other reprogramming strategies, the up‐regulation of these MCPs, which may be particularly relevant during the initial stages of bacterial adaptation to the phyllosphere.
FIGURE 4.
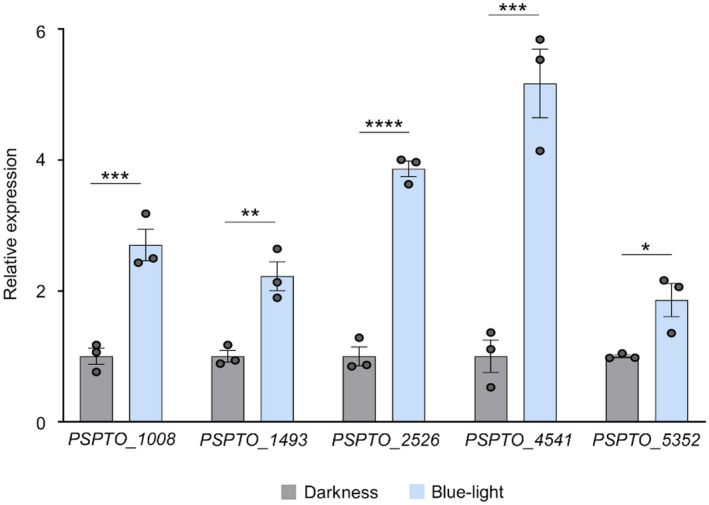
Blue light activates the expression of five methyl‐accepting chemotaxis proteins (MCPs) in Pseudomonas syringae pv. tomato (PsPto) epiphytic populations. Expression of the MCP‐encoding genes evaluated by quantitative reverse transcription PCR in PsPto epiphytic populations after 10 min treatment with blue light or darkness. Light treatment was performed just after spray‐inoculation of PsPto on tomato leaves. Bars represent relative expression changes of MCP genes in blue‐light‐treated cells with respect to dark‐treated cells. Data represent the means and standard errors of three biological replicates (n = 3). Values that are significantly different are indicated by asterisks (*p < .05; **p < .01; ***p < .005; ****p < .001)
2.4. Photoreceptors mediate changes in MCP gene transcript levels in response to blue light in epiphytic populations
Previous work in our laboratory demonstrated that PsPto has the ability to integrate different monochromatic light signals through its photosensory proteins (Santamaria‐Hernando et al., 2018). The specific up‐regulation of five genes encoding MCPs under blue light prompted us to evaluate a possible coupling between both perception systems. To this end, we analysed the expression of the genes encoding the five above‐mentioned chemoreceptors in blue‐light‐treated epiphytic populations of either PsPto‐lov mutant or PsPto‐bphP1 mutant as compared to wild‐type cells. The PsPto‐bphP1 mutant background was included because our previous data indicate that this bacteriophytochrome is also able to respond to blue light (Santamaria‐Hernando et al., 2018). Interestingly, we found that expression of the MCP‐encoding genes was reduced in the PsPto‐lov mutant (Figure 5a). Moreover, expression of the five genes was down‐regulated in the PsPto‐bphP1 mutant (Figure 5b). These results indicate that PsPto light perception is accomplished by both photosensory proteins, triggering downstream signalling.
FIGURE 5.

Function of light oxygen voltage and BphP1 photoreceptors drives the expression of the blue‐light responsive methyl‐accepting chemotaxis proteins (MCPs). Expression of the MCP‐encoding genes in epiphytic populations of the wild‐type or the photoreceptor mutants after a 10 min light treatment with blue light. Bars represent the relative expression changes of the MCP genes in the PsPto‐lov mutant (a) or the PsPto‐bphP1 mutant (b) with respect to the wild‐type strain under the same conditions. Data represent the means and standard errors of three biological replicates (n = 3). Values that are significantly different are indicated by asterisks (*p < .05; **p < .01; ***p < .005)
2.5. MCPs encoded by PSPTO_1008 and PSPTO_2526 genes are required for full virulence of PsPto in tomato plants
Chemotaxis facilitates bacterial entry into the apoplast, where some phytopathogenic bacteria like PsPto initiate the pathogenic stage. We investigated whether the inactivation of any of the MCP genes affected virulence on tomato plants. To this end, we conducted virulence assays in which leaves of tomato plants were spray‐inoculated with the wild‐type or chemoreceptor mutant strains. Data showed significant reductions in both symptom development and bacterial populations for the PsPto‐1008 and PsPto‐2526 mutant strains compared to the wild‐type strain. The reduced virulence of both mutants was restored to wild‐type levels by complementation with the corresponding gene provided in trans (Figure 6a,b). Inactivation of the other three MCPs did not cause any significant effect on virulence (Figure S3).
FIGURE 6.

Methyl‐accepting chemotaxis proteins (MCPs) PSPTO‐1008 and PSPTO‐2526 are required for full virulence of Pseudomonas syringae pv. tomato (PsPto) in tomato plants. (a) Virulence of PsPto wild‐type, mutants (PsPto‐1008 and PsPto‐2526), and complemented strains (PsPto‐1008‐Comp and PsPto‐2526‐Comp). Plant colonization was assessed based on bacterial population sizes in tomato leaves at 6 days postinoculation after spray inoculation of bacterial suspensions (108 cfu/ml). Shown are means and standard errors from three independent biological replicates (n = 3). In each experiment, three plants were measured by sampling five 1‐cm diameter leaf disks per plant. Values that are significantly different are indicated by asterisks (*p < .05). (b) The disease symptoms on leaves are representative of each strain. Circles in the pictures are used to highlight an area that represents the symptom expression throughout the whole leaf
3. DISCUSSION
Bacterial determinants that affect virulence have been extensively studied in phytopathogenic Pseudomonas (Xin and He, 2013; Dudnik and Dudler, 2014; Ichinose et al., 2016; Xin et al., 2018; An et al., 2020). However, only a limited number of reports explore, from a global perspective, the bacterial traits required for leaf colonization in the epiphytic stage (Yu et al., 2013; McAtee et al., 2018; Nobori et al., 2018; Helmann et al., 2019). Environmental conditions at the time of bacterial contact with the leaf surface may influence colonization success. Changing light conditions are prevalent during the interaction of plants with bacterial pathogens in nature, and little is known about the relevance of these environmental conditions during this key stage of bacterial pathogenesis. PsPto is a weak epiphytic bacterium and therefore the efficiency to deal with the phyllosphere environment would determine the output of the infection. Our analyses contemplate conditions that have not been previously included in other transcriptomic studies on epiphytic populations (Yu et al., 2013; McAtee et al., 2018; Nobori et al., 2018; Helmann et al., 2019). We determined the alterations in transcripts levels at short times and under different light conditions. Our findings illustrate that the contact with the leaf surface triggers a profound genetic reprogramming in PsPto. This drastic adaptation response is characterized by a robust activation of bacterial motility and chemotaxis genes on contact with the leaf surface, regardless of the light conditions tested. Although the relevance of this category has already been suggested in epiphytic populations (Yu et al., 2013), this early and light‐independent response has not been previously reported. However, we also found some functional categories up‐ or down‐regulated in epiphytic populations in response to a specific light treatment when compared to in vitro conditions. We propose that these determinants are not essential for the switch to the epiphytic stage but can help PsPto to modulate the adaptation process depending on the prevailing light conditions.
The main result of this work was to uncover the interwoven relationship between light, chemoperception, and virulence. Bacterial phytopathogens possess an unusually high number of chemoreceptors and there is solid experimental evidence that many of these chemoreceptors mediate chemotaxis towards compounds released from the leaf interior and from stomata, permitting plant entry and the initiation of the virulence process (Matilla and Krell, 2018). This notion is also supported by plant pathogens that are transmitted by insect bites, such as Xylella fastidiosa, which has a single chemoreceptor gene (Cursino et al., 2011). We propose a model (Figure 7) in which flagellar motility and chemotaxis are activated upon establishing contact with the leaf surface. Once on the leaf surface, blue light specifically causes important changes in transcript levels, especially in genes associated with chemotaxis and metabolism. Blue‐light exposure increases the transcript levels of five chemoreceptors. Moreover, it activates the expression of genes involved in the metabolism of essential nutrients such as organic acids, carbohydrates, and sulphur. Importantly, blue light is also a signal that was shown to open plant stomata (Zheng and Van Labeke, 2017; Matthews et al., 2019). Stomata are considered the main entry points for hemibiotrophic pathogens, and water diffusion through them can generate water films that facilitate nutrient availability (Vacher et al., 2016). The model we propose (Figure 7) envisages a functional link between the two consequences of blue‐light exposure: an increase in chemotaxis and in metabolic activity permitting an optimal response to plant signals, and a concomitant opening of the stomata that facilitates bacterial plant entry. This model is in line with recent work in which, through a mechanistic model, a light‐induced chemotactic entry of Escherichia coli into the stomata was proposed (Ranjbaran et al., 2020).
FIGURE 7.

Model illustrating the effect of blue‐light perception on Pseudomonas syringae pv. tomato epiphytic populations over tomato leaves
Our model is supported by the demonstration that the inactivation of two of these chemoreceptors reduces virulence. One of these chemoreceptors, PSPTO_1008, has a cytosolic location, contains two PAS sensor domains, and is similar to the chemoreceptors that bind oxygen (Garcia et al., 2017) and mediate aerotaxis (Garcia et al., 2016), a process that permits efficient pathogenesis in a number of species (Yao and Allen, 2007). The PSPTO_2526 chemoreceptor contains a periplasmic sensor domain that shares 52% sequence similarity with the ligand‐binding domain of the inorganic phosphate (Pi) chemoreceptor, CtpH, chemoreceptor of P. aeruginosa (Rico‐Jimenez et al., 2016). In this bacterium, inorganic phosphate is a major signal molecule controlling virulence.
Another of the blue‐light activated MCPs (PSPTO_1493) is homologous to the WspA chemoreceptor of P. aeruginosa and belongs to the Wsp chemosensory pathway. In P. aeruginosa PAO1, this pathway is involved in surface sensing, causing an increase in c‐di‐GMP levels that facilitates bacterial attachment (Lee et al., 2020). Moreover, it has been suggested that the undefined surface signal is recognized by the WspA chemoreceptor (O'Connor et al., 2012).
Our results show that the coordinated regulation of a high number of genes is required for bacterial switch to the epiphytic stage. Although our previous results showed that light had a regulatory effect on the above‐mentioned categories in PsPto in vitro populations (Santamaria‐Hernando et al., 2018), we show here that the contact with the leaf triggers a regulatory response that is upstream to the regulatory effect exerted by light. Interestingly, we have found that following contact with the leaf surface, and regardless of light conditions, all genes that encode the chemosensory cluster III are up‐regulated in PsPto (Tables S1 and S2).
When analysing changes in gene expression in epiphytic populations in response to different light conditions with respect to dark conditions, we found that during the early stages of colonization, red and white light have little effect on gene expression reprogramming, while a larger effect is observed with blue light. These results are in accordance with our previous in vitro data in which we also observed a stronger effect of blue light on gene expression regulation (Santamaria‐Hernando et al., 2018). However, the effect of blue light seems to be particularly relevant on epiphytic populations. It can therefore be hypothesized that just as plants use the information provided by blue light to regulate diverse processes, PsPto can also exploit blue light as a signal to activate metabolic‐related and other competitive traits to optimize leaf colonization.
We also found up‐regulation by blue light of the expression of genes involved in GABA metabolism, nucleotide, iron, fatty acid, and polyamine metabolism and transport, and siderophores synthesis and transport. Previous reports have shown that these compounds are key players during the confrontation of PsPto with plant immunity (Kachroo and Kachroo, 2009; McCraw et al., 2016; Ryffel et al., 2016; Nobori et al., 2018; Vilas et al., 2018). Blue light was found to be related to an increase in plant defence against P. syringae (Wu and Yang, 2010). Taken together, these results indicate that during the initial stages of leaf colonization PsPto uses blue light as a signal to adapt its metabolism to use the available nutrients that are present on the leaf surface and to cope with plant defence‐derived compounds.
Signal transduction mechanisms that connect light perception to the control of motility have been more extensively studied in phototrophic prokaryotes. In these microorganisms, this process allows movement towards more favourable sites for photosynthesis and protection from harmful light intensities (Armitage and Hellingwerf, 2003; Wilde and Mullineaux, 2017). In Haloarchaea salinarum, the best‐studied example, after light perception by rhodopsin photoreceptors, a signal is transmitted to an MCP that regulates archaella rotation (Hoff et al., 1997; Armitage and Hellingwerf, 2003). Nonphototrophic bacteria can also cue into light to obtain information from the plant and the environment. Although several studies demonstrated light‐mediated alteration of flagellar motility in chemoheterotrophic bacteria (Kraiselburd et al., 2012; Wu et al., 2013; Rio‐Alvarez et al., 2014; Bonomi et al., 2016; Santamaria‐Hernando et al., 2018), the link between light perception and chemosensory pathways has not been elucidated until now. In a study carried out in Bacillus subtilis, the authors suggest that exposure to UV light triggers a chemotactic response in the bacteria that allows a movement away from its waste products (Delprato et al., 2001). Entry of Salmonella enterica to lettuce stomata is induced by light and mediated by chemotaxis (Kroupitski et al., 2009). Moreover, a recent study indicates that in E. coli the response to blue light is mediated by its five chemoreceptors (Perlova et al., 2019). E. coli presents a blue‐light sensing BLUF (blue light using flavin adenine dinucleotide, FAD) photoreceptor; however, no interaction with the chemosensory machinery has been demonstrated. Here we show that in PsPto the expression of the five blue‐light responsive MCPs is coupled to the function of the LOV and BphP1 photoreceptors. BphP1 is a bacteriophytochrome, traditionally considered a red/far red photoreceptor; however, in a previous study we demonstrated that it is also able to respond to blue light (Santamaria‐Hernando et al., 2018). Similar observations were noted for BphP1 in P. syringae B728a (Wu et al., 2013). Although further studies are required to determine the signal transduction mechanisms that link light perception to the chemosensory pathways, our study provides evidence that in PsPto blue‐light perception through photoreceptors drives the expression of specific chemoreceptors. This work constitutes, to our knowledge, the first testimony in the literature reporting a link between light‐sensing photoreceptors and chemoreceptors in phytopathogenic bacteria. Moreover, we found that during the initial phase of leaf colonization, PsPto cues into blue light as a signal to regulate the interaction with the plant. Taken together, our results illustrate that PsPto, by coordinating diverse perception mechanisms, integrates plant and environmental signals to generate a response that allows optimal colonization of the leaf surface and successful infection.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains, culture media, and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S6. PsPto derivatives were grown at 28°C in King's B (KB) medium (King et al., 1954). E. coli derivatives were grown at 37°C in lysogeny broth (Bertani, 1951). When appropriate, antibiotics were added to the medium at the following final concentrations (μg/ml): rifampicin 25, streptomycin 50, kanamycin 50.
4.2. Illumination conditions
White light was provided using Osram Lumilux L 30W/830 fluorescent tubes (Osram) at an intensity of 70 μmol⋅m−2⋅s−1. Red and blue lights were provided using GreenPower LED HF red (660 nm) and GreenPower LED HF blue (458 nm) lights (Philips), respectively, each at an intensity of 20 μmol⋅m−2⋅s−1. LED lighting was chosen because this lighting does not produce heat in the form of infrared radiation. To maintain dark conditions, plates and flasks were covered with two layers of aluminium foil.
4.3. Molecular biology techniques
Plasmid DNA was isolated using a NucleoSpin Plasmid Kit (Macherey‐Nagel). DNA restriction, dephosphorylation, ligation, competent cells transformations, and electrophoresis were performed using standard protocols (Sambrook et al., 1989) and manufacturers' instructions. DNA fragments were recovered from agarose gels using a GenJet Gel Extraction Kit (Thermo Scientific). PrimeSTAR HS DNA polymerase (Takara Bio Inc.) was used for the amplification of DNA fragments. Bacterial growth was monitored based on optical density measured at 600 nm (OD600) using a Jenway 6300 spectrophotometer.
4.4. Construction of MCP mutants
To generate the five MCP mutants, internal fragments of PSPTO_1008, PSPTO_1493, PSPTO_2526, PSPTO_4541, and PSPTO_5352 were amplified by PCR from the PsPto genome using the corresponding primers listed in Table S7. PCR‐amplified fragments were cloned into pKOSac101 (Cerna‐Vargas et al., 2019). The resulting plasmids were introduced in E. coli CC118λpir and subsequently transferred into PsPto by electroporation. Plasmid integration was confirmed using PCR.
For complementation assays, the MCP‐encoding genes and their promoter regions were PCR‐amplified using the primers listed in Table S7. PCR fragments were cloned into the broad‐host‐range plasmid pBBR1MCS‐2 (Kovach et al., 1995) using the Gibson Assembly cloning method (New England Biolabs). The resulting plasmids were verified by DNA sequencing and transferred into MCP mutants by electroporation.
4.5. Isolation of epiphytic bacteria from tomato leaves
Isolation of epiphytic bacteria was performed according to Yu et al. (2013) with some modifications. PsPto and mutant strains were grown at 28°C for 24 hr on KB agar under dark conditions. Cells were resuspended in 10 mM MgCl2 and diluted to 109 cfu/ml with all manipulations performed under dark conditions (<0.05 μmol⋅m−2⋅s−1). Three‐week‐old tomato plants were spray‐inoculated with the bacterial suspension on the adaxial and abaxial surfaces of the leaves. Subsequently, tomato plants were either subjected to a light treatment (white, red, or blue light) for 10 min or maintained in darkness. After the light treatments, 400–500 primary leaves were immediately collected and submerged in an RNA‐stabilizing solution (5 ml water‐saturated phenol [pH < 7], 95 ml ethanol, 0.9 L water). Leaves were sonicated for 20 min, removed from the solution and harvested by centrifugation at 5,000 × g for 10 min. Cells were suspended in residual supernatant and subjected to several filtering steps using 6‐μm filter paper (Whatman). Cells in the filtrate were collected by centrifugation at 5,000 × g for 5 min, the supernatant was discarded, and the pellets were stored at −80°C. Each light treatment was carried out four times to provide four biological independent replicates. The cells collected from the 400–500 leaves on each treatment served as a biological replicate.
4.6. RNA extraction
Total RNA was extracted by using TRI reagent solution (ThermoFisher Scientific) as recommended by the manufacturer. The purification was accomplished by using a High Pure RNA Isolation Kit (Roche) following the manufacturer's recommendations. RNA samples were quantified using an ND‐1000 spectrophotometer (NanoDrop Technologies, Inc.) and their quality was evaluated using a Bioanalyser 2100 (Agilent Technologies).
4.7. Microarray hybridization experiments
For each treatment, four biological replicates were independently hybridized onto custom‐designed microarrays containing the complete PsPto genome (PsPto Oligo Microarrays 8 × 15K; #042459, Agilent Technologies). Fluorescently labelled cDNA for microarray hybridization was obtained using the SuperScript Indirect cDNA Labeling System (Invitrogen Corp.). Briefly, 15 μg of total RNA was converted into cDNA using SuperScript III reverse transcriptase and random hexamers as primers and including aminoallyl‐modified nucleotides in the reaction mixture. After cDNA purification, the Cy3 fluorescent dyes (Amersham Biosciences Corp.) were coupled to the amino‐modified first‐strand cDNA, and the labelling efficiencies were assessed using an ND‐1000 spectrophotometer. Hybridization of these probes to the microarray was performed as described (One‐Colour Microarray Based Gene Expression Analysis Manual v. 6.5, Agilent Technologies). Briefly, for each hybridization 600 ng of the Cy3 probes was combined with 5 μl of 10 × blocking agent and nuclease‐free water in a 25 μl reaction, which was then combined with 25 μl of 2 × GExHybridization buffer and placed on ice, then loaded onto a microarray and allowed to hybridize at 65°C for 17 hr. The microarrays were washed in GE wash buffer 1 at room temperature for 1 min and in GE wash buffer 2 at 37°C for 1 min. Images from the Cy3 channel were equilibrated and captured with a high‐resolution microarray scanner (Agilent Technologies) and the fluorescence in each spot was quantified using Feature Extraction Software (Agilent Genomics). Background correction and normalization of expression data were performed using LIMMA (Smyth and Speed, 2003; Smyth, 2004). LIMMA is part of Bioconductor, an R language project (Ihaka and Gentleman, 1996). For local background correction and normalization, the methods “normexp” and loess in LIMMA were used, respectively (Smyth, 2004). To have similar distribution across arrays and to achieve consistency among the arrays, the log ratio values were scaled using the median‐absolute value as a scale estimator (Smyth and Speed, 2003). Differentially expressed genes were determined using methods for linear models. Each probe was tested for changes in expression using an empirical Bayes moderated t statistic (Smyth, 2004). To control the FDR, p values were corrected using the method of Benjamini and Hochberg (1995). The expected FDR was controlled to be less than 1%, thus genes were declared to be differentially expressed if they exhibited a fold‐change ≥2 and FDR ≤ 0.01. Hybridizations and statistical analyses were performed by the Genomics Facility at Centro Nacional de Biotecnología (CSIC, Madrid, Spain). Heatmaps were generated using the pheatmap R package (Kolde, 2012) using the Euclidean distance and the “complete” clustering method to cluster both rows and columns.
4.8. Analysis of gene representation in functional categories
Following the procedure described by Yu et al. (2013), we found that PsPto genes can be assigned to 69 functional categories (Santamaria‐Hernando et al., 2018). Using the microarray data, we computed for each functional category the number of differentially and nondifferentially expressed genes in that category, as well as the number of differentially and nondifferentially expressed genes not in that category. Genes that were enriched in a given functional category were identified by performing a hypergeometric test (p ≤ 0.01) using the R function “phyper”. The whole analysis was performed using R v. 3.3.1 (R Core Team, 2014; http://www.R‐project.org/) with ad hoc scripts (available upon request).
4.9. RT‐qPCR
Total RNA isolated as described above was converted to cDNA using a High‐Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primers were designed to amplify fragments of approximately 100 bp (Table S7) and the rpoD gene was used as an internal control for normalization (Sawada et al., 1999). Reverse transcription quantitative PCR amplifications were carried out on an ABI PRISM 7300 RT PCR System (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems). Thermal cycling conditions were as follows: one cycle at 95°C for 10 min; 50 cycles at 95°C for 15 s and 64°C for 1 min; and a final cycle at 95°C for 15 s, 60°C for 1 min, 95°C for 15 s, and 60°C for 15 s. The relative gene expression ratio was calculated using the comparative critical threshold (∆∆C t) method (Pfaffl, 2001; Rotenberg et al., 2006).
4.10. Tomato virulence assays
P. syringae pv. tomato strains were grown at 28°C for 24 hr on KB agar plates under dark conditions. Cells were resuspended in 10 mM MgCl2 and diluted to 108 cfu/ml. Three‐week‐old tomato plants (Solanum lycopersicum ‘Moneymaker’) were spray‐inoculated with the bacterial suspension. Silwet L‐77 was added to the bacterial suspensions at a final concentration of 0.02% (vol/vol). Plants were infected at subjective dawn, which was at the end of the night cycle with immediate transfer to the light. Plants were incubated in a growth chamber at 25°C at 60% relative humidity with a daily light period of 12 hr. Six days after inoculation, the leaf symptoms were recorded and bacterial populations were measured by sampling five 1‐cm diameter leaf disks per plant. We performed three independent biological replicates. In each replicate three plants were used, and five independent leaves were sampled from each plant. The infected leaf disks were washed twice with 10 mM MgCl2 prior to homogenization to eliminate the bacteria from the leaf surface. Plant material was homogenized in 10 mM MgCl2 and drop plated onto KB agar supplemented with the appropriate antibiotics. The average number of bacteria per cm2, isolated from five infected tomato leaves, was determined based on log‐transformed data.
4.11. Statistical analysis
Variables that were heteroscedastic and did not follow a normal distribution were compared using generalized linear models. The remaining variables were analysed using general linear models. All analyses were performed using the statistical software package SPSS 25.0 (SPSS Inc.) and Centurion 18 (Statgraphics Technologies, Inc.).
Supporting information
FIGURE S1 PsPto chemoreceptors and chemosensory clusters. (a) Chemoreceptor repertoire of PsPto. Ligand binding domains (LBDs) were annotated according to Pfam (https://pfam.xfam.org/). (b) Schematic representation of chemotaxis clusters found in PsPto. Genes whose expression was up‐regulated in epiphytic populations are coloured green. Adapted from Cerna‐Vargas et al. (2019) (mBio 10, e01868‐01819)
FIGURE S2 Domain organization of PsPto photosensory proteins. PAS, Per‐Arnt‐Sim domain; HATPase_c, family of structurally related ATPase domains of histidine kinase, DNA gyrase B, and HSP90; RR, response regulator; GAF, GAF domain; HisKA, histidine kinase A; HWE HK, HWE histidine kinase. Adapted from Pfam database (https://pfam.xfam.org/)
FIGURE S3 Inactivation of MCPs PSPTO_1493, PSPTO_4541, and PSPTO_5352 does not affect virulence of PsPto in tomato plants. Virulence of PsPto wild‐type and MCP mutants (PsPto‐1493, PsPto‐4541, and PsPto‐5352). Plant colonization was assessed based on bacterial population sizes in tomato leaves at 6 days postinoculation after spray inoculation of bacterial suspensions (108 cfu/ml). Shown are means and standard errors from three independent experiments (n = 3)
TABLE S1 Differentially expressed genes in PsPto epiphytic populations after the light treatments as compared to in vitro populations
TABLE S2 Overrepresented functional categories and differentially expressed genes among the up‐regulated genes in PsPto epiphytic populations after the light treatments as compared to in vitro populations
TABLE S3 Overrepresented functional categories and differentially expressed genes among the down‐regulated genes in PsPto epiphytic populations as compared to in vitro populations in the different light treatments
TABLE S4 Differentially expressed genes in light‐treated PsPto epiphytic populations as compared to dark‐treated epiphytic populations
TABLE S5 Overrepresented functional categories and differentially expressed genes in blue‐light‐treated PsPto epiphytic populations as compared to dark‐treated epiphytic populations
TABLE S6 Bacteria and plasmids used
TABLE S7 Primers used in this study
ACKNOWLEDGMENTS
This research was supported by project grant RTI2018‐095222‐B‐I00 from the Ministerio de Ciencia, Innovación y Universidades, Spain. We thank Gloria García for microarray analysis. We thank Dr Tino Krell for critical review of the manuscript and helpful advice. The authors declare no competing financial interests.
Santamaría‐Hernando S, Cerna‐Vargas JP, Martínez‐García PM, et al. Blue‐light perception by epiphytic Pseudomonas syringae drives chemoreceptor expression, enabling efficient plant infection. Molecular Plant Pathology 2020;21:1606–1619. 10.1111/mpp.13001
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in the GEO database at https://www.ncbi.nlm.nih.gov/geo/, reference number GSE152188 (token for reviewers: inadksmuppchder). Record GSE101783 (Santamaria‐Hernando et al., 2018) previously generated in our laboratory has been used for comparisons.
REFERENCES
- An, S.Q. , Potnis, N. , Dow, M. , Vorholter, F.J. , He, Y.Q. , Becker, A. et al (2020) Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas . FEMS Microbiology Reviews, 44, 1–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunez‐Lamas, M. , Cabrera, E. , Lopez‐Solanilla, E. , Solano, R. , Gonzalez‐Melendi, P. , Chico, J.M. et al (2009) Bacterial chemoattraction towards jasmonate plays a role in the entry of Dickeya dadantii through wounded tissues. Molecular Microbiology, 74, 662–671. [DOI] [PubMed] [Google Scholar]
- Armitage, J.P. and Hellingwerf, K.J. (2003) Light‐induced behavioral responses (“phototaxis”) in prokaryotes. Photosynthesis Research, 76, 145–155. [DOI] [PubMed] [Google Scholar]
- Benjamini, Y. and Hochberg, Y. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B, 57, 289–300. [Google Scholar]
- Bertani, G. (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli . Journal of Bacteriology, 62, 293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomi, H.R. , Toum, L. , Sycz, G. , Sieira, R. , Toscani, A.M. , Gudesblat, G.E. et al (2016) Xanthomonas campestris attenuates virulence by sensing light through a bacteriophytochrome photoreceptor. EMBO Reports, 17, 1565–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boureau, T. , Routtu, J. , Roine, E. , Taira, S. and Romantschuk, M. (2002) Localization of hrpA‐induced Pseudomonas syringae pv. tomato DC3000 in infected tomato leaves. Molecular Plant Pathology, 3, 451–460. [DOI] [PubMed] [Google Scholar]
- Carvalho, S.D. and Castillo, J.A. (2018) Influence of light on plant–phyllosphere interaction. Frontiers in Plant Science, 9, 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerna‐Vargas, J.P. , Santamaria‐Hernando, S. , Matilla, M.A. , Rodriguez‐Herva, J.J. , Daddaoua, A. , Rodriguez‐Palenzuela, P. et al (2019) Chemoperception of specific amino acids controls phytopathogenicity in Pseudomonas syringae pv. tomato . mBio, 10, e01868–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppels, D.A. (1988) Chemotaxis by Pseudomonas syringae pv. tomato . Applied and Environmental Microbiology, 54, 629–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cursino, L. , Galvani, C.D. , Athinuwat, D. , Zaini, P.A. , Li, Y. , de La Fuente, L. et al (2011) Identification of an operon, Pil‐Chp, that controls twitching motility and virulence in Xylella fastidiosa . Molecular Plant‐Microbe Interactions, 24, 1198–1206. [DOI] [PubMed] [Google Scholar]
- Delprato, A.M. , Samadani, A. , Kudrolli, A. and Tsimring, L.S. (2001) Swarming ring patterns in bacterial colonies exposed to ultraviolet radiation. Physical Review Letters, 87, 158102. [DOI] [PubMed] [Google Scholar]
- Dudnik, A. and Dudler, R. (2014) Virulence determinants of Pseudomonas syringae strains isolated from grasses in the context of a small type III effector repertoire. BMC Microbiology, 14, 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Milmanda, G.L. , Crocco, C.D. , Reichelt, M. , Mazza, C.A. , Köllner, T.G. , Zhang, T. et al (2020) A light‐dependent molecular link between competition cues and defence responses in plants. Nature Plants, 6, 223–230. [DOI] [PubMed] [Google Scholar]
- Garcia, D. , Orillard, E. , Johnson, M.S. and Watts, K.J. (2017) Gas sensing and signaling in the PAS‐Heme domain of the Pseudomonas aeruginosa Aer2 receptor. Journal of Bacteriology, 199, e00003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, D. , Watts, K.J. , Johnson, M.S. and Taylor, B.L. (2016) Delineating PAS‐HAMP interaction surfaces and signalling‐associated changes in the aerotaxis receptor Aer. Molecular Microbiology, 100, 156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann, T.C. , Deutschbauer, A.M. and Lindow, S.E. (2019) Genome‐wide identification of Pseudomonas syringae genes required for fitness during colonization of the leaf surface and apoplast. Proceedings of the National Academy of Sciences of the United States of America, 116, 18900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman, J.W. , Tifrea, D.F. and Harwood, C.S. (2005) A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proceedings of the National Academy of Sciences of the United States of America, 102, 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida, A. , Oku, S. , Kawasaki, T. and Nakashimada, Y. (2015) Identification of the mcpA and mcpM genes, encoding methyl‐accepting proteins involved in amino acid and l‐malate chemotaxis, and involvement of McpM‐mediated chemotaxis in plant infection by Ralstonia pseudosolanacearum (formerly Ralstonia solanacearum phylotypes I and III). Applied and Environmental Microbiology, 81, 7420–7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano, S.S. and Upper, C.D. (2000) Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae a pathogen, ice nucleus, and epiphyte. Microbiology and Molecular Biology Reviews, 64, 624–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff, W.D. , Jung, K.H. and Spudich, J.L. (1997) Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annual Review of Biophysics and Biomolecular Structure, 26, 223–258. [DOI] [PubMed] [Google Scholar]
- Ichinose, Y. , Sawada, T. , Matsui, H. , Yamamoto, M. , Toyoda, K. , Noutoshi, Y. et al (2016) Motility‐mediated regulation of virulence in Pseudomonas syringae . Physiological and Molecular Plant Pathology, 95, 50–54. [Google Scholar]
- Ihaka, R. and Gentleman, R. (1996) A language for data analysis and graphics. Journal of Computational and Graphical Statistics, 5, 299–314. [Google Scholar]
- Kachroo, A. and Kachroo, P. (2009) Fatty acid‐derived signals in plant defense. Annual Review of Phytopathology, 47, 153–176. [DOI] [PubMed] [Google Scholar]
- Kamoun, S. and Kado, C.I. (1990) A plant‐inducible gene of Xanthomonas campestris pv. campestris encodes an exocellular component required for growth in the host and hypersensitivity on nonhosts. Journal of Bacteriology, 172, 5165–5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.E. , Shitashiro, M. , Kuroda, A. , Takiguchi, N. and Kato, J. (2007) Ethylene chemotaxis in Pseudomonas aeruginosa and other Pseudomonas species. Microbes and Environments, 22, 186–189. [Google Scholar]
- King, E.O. , Ward, M.K. and Raney, D.E. (1954) Two simple media for the demonstration of pyocyanin and fluorescin. Journal of Laboratory and Clinical Medicine, 44, 301–307. [PubMed] [Google Scholar]
- Kolde, R. (2012) Pheatmap: pretty heatmaps. R Package Version, 61, 1–7. [Google Scholar]
- Kovach, M.E. , Elzer, P.H. , Hill, D.S. , Robertson, G.T. , Farris, M.A. , Roop, R.M. et al (1995) Four new derivatives of the broad‐host‐range cloning vector pBBR1MCS, carrying different antibiotic‐resistance cassettes. Gene, 166, 175–176. [DOI] [PubMed] [Google Scholar]
- Kraiselburd, I. , Alet, A.I. , Tondo, M.L. , Petrocelli, S. , Daurelio, L.D. , Monzon, J. et al (2012) A LOV protein modulates the physiological attributes of Xanthomonas axonopodis pv. citri relevant for host plant colonization. PLoS One, 7, e38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroupitski, Y. , Golberg, D. , Belausov, E. , Pinto, R. , Swartzberg, D. , Granot, D. et al (2009) Internalization of Salmonella enterica in leaves is induced by light and involves chemotaxis and penetration through open stomata. Applied and Environmental Microbiology, 75, 6076–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.K. , Vachier, J. , de Anda, J. , Zhao, K. , Baker, A.E. , Bennett, R.R. et al (2020) Social cooperativity of bacteria during reversible surface attachment in young biofilms: a quantitative comparison of Pseudomonas aeruginosa PA14 and PAO1. mBio, 11, e02644–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla, M.A. and Krell, T. (2018) The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiology Reviews, 42, 40–67. [DOI] [PubMed] [Google Scholar]
- Matthews, J.S.A. , Vialet‐Chabrand, S. and Lawson, T. (2019) Role of blue and red light in stomatal dynamic behaviour. Journal of Experimental Botany, 7, 2253–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAtee, P.A. , Brian, L. , Curran, B. , van der Linden, O. , Nieuwenhuizen, N.J. , Chen, X. et al (2018) Re‐programming of Pseudomonas syringae pv. actinidiae gene expression during early stages of infection of kiwifruit. BMC Genomics, 19, 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCraw, S.L. , Park, D.H. , Jones, R. , Bentley, M.A. , Rico, A. , Ratcliffe, R.G. et al (2016) GABA (gamma‐Aminobutyric Acid) uptake via the GABA permease GabP represses virulence gene expression in Pseudomonas syringae pv. tomato DC3000. Molecular Plant‐Microbe Interactions, 29, 938–949. [DOI] [PubMed] [Google Scholar]
- McGrane, R. and Beattie, G.A. (2017) Pseudomonas syringae pv. syringae B728a regulates multiple stages of plant colonization via the bacteriophytochrome BphP1. mBio, 8, e01178–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriconi, V. , Sellaro, R. , Ayub, N. , Soto, G. , Rugnone, M. and Shah, R. (2013) LOV‐domain photoreceptor, encoded in a genomic island, attenuates the virulence of Pseudomonas syringae in light‐exposed Arabidopsis leaves. The Plant Journal, 76, 322–331. [DOI] [PubMed] [Google Scholar]
- Nobori, T. , Velasquez, A.C. , Wu, J. , Kvitko, B.H. , Kremer, J.M. and Wang, Y. (2018) Transcriptome landscape of a bacterial pathogen under plant immunity. Proceedings of the National Academy of Sciences of the United States of America, 115, E3055–E3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor, J.R. , Kuwada, N.J. , Huangyutitham, V. , Wiggins, P.A. and Harwood, C.S. (2012) Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory‐like system leading to c‐di‐GMP production. Molecular Microbiology, 86, 720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega, D.R. , Fleetwood, A.D. , Krell, T. , Harwood, C.S. , Jensen, G.J. and Zhulin, I.B. (2017) Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa . Proceedings of the National Academy of Sciences of the United States of America, 114, 12809–12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal, S. , Roy, D. , Chitrakar, R. , Price, L. , Breitbach, Z.S. , Armstrong, D.W. et al (2016) Coronatine facilitates Pseudomonas syringae infection of Arabidopsis leaves at night. Frontiers in Plant Science, 7, 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales, R.E. , Ferrandez, A. and Harwood, C.S. (2004) Chemotaxis in pseudomonads In: Ramos J.L. (Ed.) Pseudomonas Volume I: Genomics, Life Style and Molecular Architecture. New York, NY: Kluwer Academic/Plenum Publishers, pp. 793–815. [Google Scholar]
- Perlova, T. , Gruebele, M. and Chemla, Y.R. (2019) Blue light is a universal signal for Escherichia coli chemoreceptors. Journal of Bacteriology, 201, e00762–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research, 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam, N. and Lee, Y.H. (2018) Effects of green light on the gene expression and virulence of the plant pathogen Pseudomonas cichorii JBC1. European Journal of Plant Pathology, 150, 223–236. [Google Scholar]
- Ranjbaran, M. , Solhtalab, M. and Datta, A.K. (2020) Mechanistic modeling of light‐induced chemotactic infiltration of bacteria into leaf stomata. PLoS Computational Biology, 16, e1007841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico‐Jimenez, M. , Reyes‐Darias, J.A. , Ortega, A. , Diez Pena, A.I. , Morel, B. and Krell, T. (2016) Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa . Scientific Reports, 6, 28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio‐Alvarez, I. , Rodriguez‐Herva, J.J. , Martinez, P.M. , Gonzalez‐Melendi, P. , Garcia‐Casado, G. , Rodriguez‐Palenzuela, P. et al (2014) Light regulates motility, attachment and virulence in the plant pathogen Pseudomonas syringae pv. tomato DC3000. Environmental Microbiology, 16, 2072–2085. [DOI] [PubMed] [Google Scholar]
- Roden, L.C. and Ingle, R.A. (2009) Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant–pathogen interactions. The Plant Cell, 21, 2546–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg, D. , Thompson, T.S. , German, T.L. and Willis, D.K. (2006) Methods for effective real‐time RT‐PCR analysis of virus‐induced gene silencing. Journal of Virological Methods, 138, 49–59. [DOI] [PubMed] [Google Scholar]
- Ryffel, F. , Helfrich, E.J.N. , Kiefer, P. , Peyriga, L. , Portais, J.C. , Piel, J. et al (2016) Metabolic footprint of epiphytic bacteria on Arabidopsis thaliana leaves. The ISME Journal, 10, 632–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Frits, E.F. and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edition. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Santamaria‐Hernando, S. , Rodriguez‐Herva, J.J. , Martinez‐Garcia, P.M. , Rio‐Alvarez, I. , Gonzalez‐Melendi, P. , Zamorano, J. et al (2018) Pseudomonas syringae pv. tomato exploits light signals to optimize virulence and colonization of leaves. Environmental Microbiology, 20, 4261–4280. [DOI] [PubMed] [Google Scholar]
- Sawada, H. , Suzuki, F. , Matsuda, I. and Saitou, N. (1999) Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. Journal of Molecular Evolution, 49, 627–644. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. (2004) Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology, 3, 1–25. [DOI] [PubMed] [Google Scholar]
- Smyth, G.K. and Speed, T. (2003) Normalization of cDNA microarray data. Methods, 31, 265–273. [DOI] [PubMed] [Google Scholar]
- Ulrich, L.E. , Koonin, E.V. and Zhulin, I.B. (2005) One‐component systems dominate signal transduction in prokaryotes. Trends in Microbiology, 13, 52–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich, L.E. and Zhulin, I.B. (2010) The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Research, 38, D401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher, C. , Hampe, A. , Porté, A.J. , Sauer, U. , Compant, S. and Morris, C.E. (2016) The phyllosphere: microbial jungle at the plant–climate interface. Annual Review of Ecology, Evolution, and Systematics, 47, 1–24. [Google Scholar]
- Verma, R. , Samal, B. and Chatterjee, S. (2018) Xanthomonas oryzae pv. oryzae chemotaxis components and chemoreceptor Mcp2 are involved in the sensing of constituents of xylem sap and contribute to the regulation of virulence‐associated functions and entry into rice. Molecular Plant Pathology, 19, 2397–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilas, J.M. , Romero, F.M. , Rossi, F.R. , Marina, M. , Maiale, S.J. , Calzadilla, P.I. et al (2018) Modulation of plant and bacterial polyamine metabolism during the compatible interaction between tomato and Pseudomonas syringae . Journal of Plant Physiology, 231, 281–290. [DOI] [PubMed] [Google Scholar]
- Vorholt, J.A. (2012) Microbial life in the phyllosphere. Nature Reviews Microbiology, 10, 828–840. [DOI] [PubMed] [Google Scholar]
- Wilde, A. and Mullineaux, C.W. (2017) Light‐controlled motility in prokaryotes and the problem of directional light perception. FEMS Microbiology Reviews, 41, 900–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. , McGrane, R.S. and Beattie, G.A. (2013) Light regulation of swarming motility in Pseudomonas syringae integrates signaling pathways mediated by a bacteriophytochrome and a LOV protein. mBio, 4, e00334–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, L. and Yang, H.Q. (2010) CRYPTOCHROME 1 is implicated in promoting R protein‐mediated plant resistance to Pseudomonas syringae in Arabidopsis . Molecular Plant, 3, 539–548. [DOI] [PubMed] [Google Scholar]
- Wuichet, K. and Zhulin, I.B. (2010) Origins and diversification of a complex signal transduction system in prokaryotes. Science Signaling, 3, ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, X.F. and He, S.Y. (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annual Review of Phytopathology, 51, 473–498. [DOI] [PubMed] [Google Scholar]
- Xin, X.F. , Kvitko, B. and He, S.Y. (2018) Pseudomonas syringae: what it takes to be a pathogen. Nature Reviews Microbiology, 16, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2006) Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum . Journal of Bacteriology, 188, 3697–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J. and Allen, C. (2007) The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. Journal of Bacteriology, 189, 6415–6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Lund, S.P. , Scott, R.A. , Greenwald, J.W. , Records, A.H. , Nettleton, D. et al (2013) Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proceedings of the National Academy of Sciences of the United States of America, 110, E425–E434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, L. and Van Labeke, M.C. (2017) Long‐term effects of red‐ and blue‐light emitting diodes on leaf anatomy and photosynthetic efficiency of three ornamental pot plants. Frontiers in Plant Science, 8, 917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 PsPto chemoreceptors and chemosensory clusters. (a) Chemoreceptor repertoire of PsPto. Ligand binding domains (LBDs) were annotated according to Pfam (https://pfam.xfam.org/). (b) Schematic representation of chemotaxis clusters found in PsPto. Genes whose expression was up‐regulated in epiphytic populations are coloured green. Adapted from Cerna‐Vargas et al. (2019) (mBio 10, e01868‐01819)
FIGURE S2 Domain organization of PsPto photosensory proteins. PAS, Per‐Arnt‐Sim domain; HATPase_c, family of structurally related ATPase domains of histidine kinase, DNA gyrase B, and HSP90; RR, response regulator; GAF, GAF domain; HisKA, histidine kinase A; HWE HK, HWE histidine kinase. Adapted from Pfam database (https://pfam.xfam.org/)
FIGURE S3 Inactivation of MCPs PSPTO_1493, PSPTO_4541, and PSPTO_5352 does not affect virulence of PsPto in tomato plants. Virulence of PsPto wild‐type and MCP mutants (PsPto‐1493, PsPto‐4541, and PsPto‐5352). Plant colonization was assessed based on bacterial population sizes in tomato leaves at 6 days postinoculation after spray inoculation of bacterial suspensions (108 cfu/ml). Shown are means and standard errors from three independent experiments (n = 3)
TABLE S1 Differentially expressed genes in PsPto epiphytic populations after the light treatments as compared to in vitro populations
TABLE S2 Overrepresented functional categories and differentially expressed genes among the up‐regulated genes in PsPto epiphytic populations after the light treatments as compared to in vitro populations
TABLE S3 Overrepresented functional categories and differentially expressed genes among the down‐regulated genes in PsPto epiphytic populations as compared to in vitro populations in the different light treatments
TABLE S4 Differentially expressed genes in light‐treated PsPto epiphytic populations as compared to dark‐treated epiphytic populations
TABLE S5 Overrepresented functional categories and differentially expressed genes in blue‐light‐treated PsPto epiphytic populations as compared to dark‐treated epiphytic populations
TABLE S6 Bacteria and plasmids used
TABLE S7 Primers used in this study
Data Availability Statement
The data that support the findings of this study are openly available in the GEO database at https://www.ncbi.nlm.nih.gov/geo/, reference number GSE152188 (token for reviewers: inadksmuppchder). Record GSE101783 (Santamaria‐Hernando et al., 2018) previously generated in our laboratory has been used for comparisons.


