Abstract
Pectin is synthesized in a highly methylesterified form in the Golgi cisternae and partially de‐methylesterified in muro by pectin methylesterases (PMEs). Arabidopsis thaliana produces a local and strong induction of PME activity during the infection of the necrotrophic fungus Botrytis cinerea. AtPME17 is a putative A. thaliana PME highly induced in response to B. cinerea. Here, a fine tuning of AtPME17 expression by different defence hormones was identified. Our genetic evidence demonstrates that AtPME17 strongly contributes to the pathogen‐induced PME activity and resistance against B. cinerea by triggering jasmonic acid–ethylene‐dependent PDF1.2 expression. AtPME17 belongs to group 2 isoforms of PMEs characterized by a PME domain preceded by an N‐terminal PRO region. However, the biochemical evidence for AtPME17 as a functional PME is still lacking and the role played by its PRO region is not known. Using the Pichia pastoris expression system, we demonstrate that AtPME17 is a functional PME with activity favoured by an increase in pH. AtPME17 performs a blockwise pattern of pectin de‐methylesterification that favours the formation of egg‐box structures between homogalacturonans. Recombinant AtPME17 expression in Escherichia coli reveals that the PRO region acts as an intramolecular inhibitor of AtPME17 activity.
Keywords: Arabidopsis thaliana, Botrytis cinerea, cell wall integrity, pectin methylesterases, plant immunity
AtPME17 contributes to resistance against Botrytis cinerea by triggering jasmonic acid–ethylene‐dependent PDF1.2 expression. AtPME17 activity is inhibited by its PRO region and produces a blockwise pectin de‐methylesterification.
1. INTRODUCTION
Plant pathogens negatively affect agricultural production by reducing the plant yield and worsening the nutritional and qualitative characteristics of the harvest. To limit the damage caused by pathogen infection, crops are treated with large doses of pesticides, which cause soil and groundwater pollution. The use of crop varieties genetically resistant to necrotrophs represents a more sustainable solution but is limited by the scarcity of resistance genes to be integrated into crops. For this reason, the identification of new genetic sources to provide disease resistance for more sustainable agriculture becomes imperative. Once they have penetrated the cuticle, necrotrophs secrete different cell wall‐degrading enzymes (CWDEs) to hydrolyse cell wall (CW) polymers and facilitate access to the host's nutrients (Lionetti and Metraux, 2014; Malinovsky et al., 2014). The CW is a complex and dynamic network composed of cellulose microfibrils embedded in a matrix of hemicelluloses, pectins, and proteins. The most abundant component of pectin is homogalacturonan (HG), a linear polymer of galacturonic acid (GalA) monomers synthetized in the Golgi complex and delivered to the CW in a highly methylesterified form (Ibar and Orellana, 2007; Caffall and Mohnen, 2009; Senechal et al., 2014b).
In muro, the de‐methylesterification of HG is strictly controlled by pectin methylesterases (PMEs; EC.3.1.1.11). PMEs hydrolyse the O6‐methylester groups of the HG backbone of pectin, resulting in free carboxyl groups and releasing methanol and protons (Wu et al., 2018). PMEs are ubiquitous enzymes belonging to a large multigene family. This last evidence suggests some specificity for the single isoforms. The presence of different methylester distributions on HG in vivo indicates the existence of PME isoforms with different action patterns with respect to the removal of methyl esters (Catoire et al., 1998; Willats et al., 2001). A blockwise de‐methylesterification results in the production of adjacent free galacturonic acid units. These domains can form calcium cross‐links between HG chains, known as “egg‐box” structures, resulting in pectin stiffening (Limberg et al., 2000; Wu et al., 2018). Random de‐methylesterification results in the removal of one methyl ester group at a time from various noncontiguous residues on the HG chains, exposing HG to the activity of pectinolytic enzymes (Limberg et al., 2000). Current knowledge of the mode of de‐methylesterification of the single plant PME isoforms is scarce.
PMEs play an important role in pectin remodelling and disassembly of the CW in many physiological processes (Wormit and Usadel, 2018). Interestingly, it was discovered that plant PMEs play a critical role in multiple plant–microbe interactions and stress responses (Lionetti et al., 2012). Local and strong induction of pathogen‐induced PME (PI‐PME) activity is triggered in Arabidopsis thaliana when infected with different microbial pathogens, including fungi such as Botrytis cinerea and Alternaria brassicicola, bacteria such as Pseudomonas syringae, and viruses like turnip vein clearing virus (TVCV) (Bethke et al., 2014; Lionetti et al., 2014, 2017; Lionetti, 2015). The PI‐PME activity could favour the release and detection of de‐methylesterified oligogalacturonides (OGs) able to trigger plant immunity (Osorio et al., 2008, 2011; Ferrari et al., 2013; Kohorn et al., 2014). It is important to mention that the de‐methylesterification of pectin by PMEs generates methanol, an alarm signal (Hann et al., 2014). Exposure to methanol may result in a “priming” effect on intact tissue, setting the stage for intra‐ and interplant immunity (Dorokhov et al., 2012; Komarova et al., 2014). However, the molecular factors underlying PI‐PME activity during pathogen attack are still largely unknown and much remains to be discovered about their transcriptional and posttranscriptional regulation during pathogenesis.
In A. thaliana there are 66 PME isoforms (Pfam01095) classified in two groups on the basis of the absence (Group 1:21 isoforms) or presence (Group 2:41 isoforms) of an N‐terminal PRO region, showing structural similarities with functionally characterized PME inhibitors (PMEIs, Pfam 04,043) (Wormit and Usadel, 2018; Wu et al., 2018). Most Group 2 PMEs possess one or two conserved basic four amino acid cleavage motifs R(R/K)(R/L)L included in the linker region between the PRO region and the catalytic domain (Wolf et al., 2009). Specific subtilases (SBTs, Pfam00082), subtilisin‐like serine proteases belonging to the S8A subfamily, are involved in the cleavage of the basic tetrad motifs to release the mature PME domain from the PRO region (Schaller et al., 2018). In A. thaliana, there are 56 SBT isoforms. Some SBTs are targeted to the CW, where they take part in developmental processes and defence (Rautengarten et al., 2008; Schaller et al., 2018). In particular, AtPME17 is coexpressed with and processed by SBT3.5, at a specific single processing motif, during root development (Senechal et al., 2014a). Hitherto, however, the possible role of SBTs as posttranscriptional regulation of PMEs in plant immunity has not been investigated.
With the aim of identifying PMEs involved during B. cinerea infection, we previously demonstrated that specific PME isoform genes are up‐regulated during the A. thaliana–Botrytis interaction (Lionetti et al., 2017). Here, we focused our attention on AtPME17 (At2g45220), the isoform that showed the highest induction. We provided genetic evidence demonstrating that AtPME17 strongly contributes to PI‐PME activity and resistance against B. cinerea. Moreover, the biochemical evidence for AtPME17 as a functional PME is still lacking and the role played by its PRO region is not known. For this reason, we performed the heterologous expression of AtPME17 in Pichia pastoris and in Escherichia coli with the aim of characterizing the AtPME17 activity in terms of mode of action, pH sensitivity, and PRO region inhibitor activity. Our results shed light on AtPME17 function and regulation.
2. RESULTS
2.1. AtPME17 triggers PI‐PME activity and contributes to resistance against B. cinerea in A. thaliana
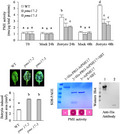
Arabidopsis uses local and strong PI‐PME activity in the fight against B. cinerea (Lionetti, 2015; Lionetti et al., 2017). However, the molecular factors triggering this response are currently unknown. To deepen this knowledge, the PI‐PME activity was compared between wild‐type (WT) plants and Arabidopsis mutants defective in immune signalling or biosynthetic pathways (Table S1) during B. cinerea infection. coi‐1, jar‐1, npr1‐1, and sid2‐2 showed a significative reduction of PI‐PME activity at 24 hr with respect to the WT (Figure S1). Later during infection, at 48 hours postinoculation (hpi), jar‐1, coi‐1, and aba2‐3 showed defective induction while a significant increase was observed in the ein2‐5 mutant with respect to control plants. These results indicate that in A. thaliana, fine and dynamic regulation of PME activity is governed by a network of signalling pathways related to jasmonic acid (JA), salicylic acid (SA), abscisic acid (ABA), and ethylene (ET). det2 and bak1‐4 were not significantly different with respect to control, indicating that brassinosteroid signalling does not participate in the modulation of this process.
The induction of the gene expression of specific PME isoforms is among the factors increasing PI‐PME activity. Among the six PME isoform genes induced in A. thaliana leaves infected with B. cinerea, AtPME17 (At2g45220) showed the highest expression level (Lionetti, 2015; Lionetti et al., 2017). To understand if the modulation of AtPME17 expression was restricted to the A. thaliana–B. cinerea pathosystem and to identify possible triggering factors, meta‐analyses of publicly available microarray data were performed. The results showed that AtPME17 expression is strongly up‐regulated in response to several pathogens with different lifestyles, including fungi, bacteria, and viruses (Figure S2). Notably, AtPME17 is affected by different pathogen‐associated molecular patterns (PAMPs) such as elf18 (Kunze et al., 2004) and flg22 (Felix et al., 1999) in seedlings, and by the effector HrpZ in leaves (Alfano et al., 1996) and does not respond to unmethylesterified and unoxidized OGs with a degree of polymerization (DP) of 9–16 in seedlings (Figure S3). AtPME17 promoter showed the presence of stress‐responsive elements as well as of elements responsive to defence hormones (Table S2). Public transcriptomes reveal that the treatment of A. thaliana plants with ABA, JA, and SA induces AtPME17 expression (Figure S4). To investigate if AtPME17 expression is controlled by immune response hormones, AtPME17 expression was compared between WT plants and ein2‐5, jar‐1, sid2‐2, and aba2‐3 mutants at 48 hpi with B. cinerea. AtPME17 induction was significantly reduced in all the mutants with respect to WT plants (Figure 1). Our results indicate that ABA, JA, SA, and ET signalling networks contribute to trigger AtPME17 expression during infection.
FIGURE 1.
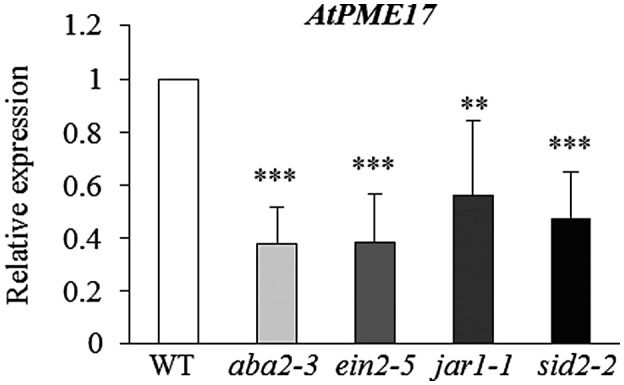
Plant immunity hormones regulate the expression of AtPME17. AtPME17 expression analysed by quantitative reverse transcription PCR at 48 hr postinoculation in Botrytis cinerea‐ or mock‐inoculated leaves of 6‐week‐old Arabidopsis thaliana wild‐type (WT) plants and defence hormone mutants. The expression levels were normalized to UBQ5 expression. The relative expression is represented as the ratio between gene expression in the mutants and WT plants. The results represent means ± SD of three independent experiments (n = 3). Asterisks indicate significant differences between mutants and WT according to Student's t test (***p < .0002; **p < .0003)
The contribution of AtPME17 to PI‐PME activity and to resistance against B. cinerea was explored through a reverse genetic approach. Two homozygous A. thaliana T‐DNA insertional mutants were isolated (Figure 2a). The T‐DNA insertions were localized in the intron for pme17‐2 (SALK_059908) and in the first exon for pme17‐3 (SM_3_25823). The abundance of AtPME17 transcripts was analysed in leaves of adult pme17 mutants infected with B. cinerea and compared to WT plants at 48 hpi. AtPME17 expression was strongly reduced in pme17‐2 and suppressed in pme17‐3 plants, indicating that pme17‐2 is a knockdown mutant and pme17‐3 is a knockout mutant (Figure 2b). The contribution of AtPME17 to the regulation of PI‐PME activity during B. cinerea infection was determined by quantifying the level of PME activity at 24 and 48 hpi using a gel diffusion assay (Lionetti, 2015). WT plants exhibited a significant 3.5‐ and 4.4‐fold induction of PME activity at 24 and 48 hpi, respectively, compared to mock‐treated plants (Figure 2c). Interestingly, pme17 mutants showed defective induction of PME activity. At 24 and 48 hpi, 2.1‐ and 2.5‐fold induction of PME activity was observed in both pme17 mutants, respectively, in comparison with the mock‐inoculated lines. The susceptibility of pme17 mutants to B. cinerea was determined by measuring the lesion size on leaves at 48 hpi. The symptoms of fungal infection were twice as high in leaves of pme17 mutants compared to WT plants (Figure 2d). A greater development of B. cinerea mycelium was observed around the inoculation sites in leaf tissues of pme17 mutants compared to WT plants (Figure 2e). These results indicate that AtPME17 contributes to the induction of PME activity and to A. thaliana resistance against B. cinerea.
FIGURE 2.
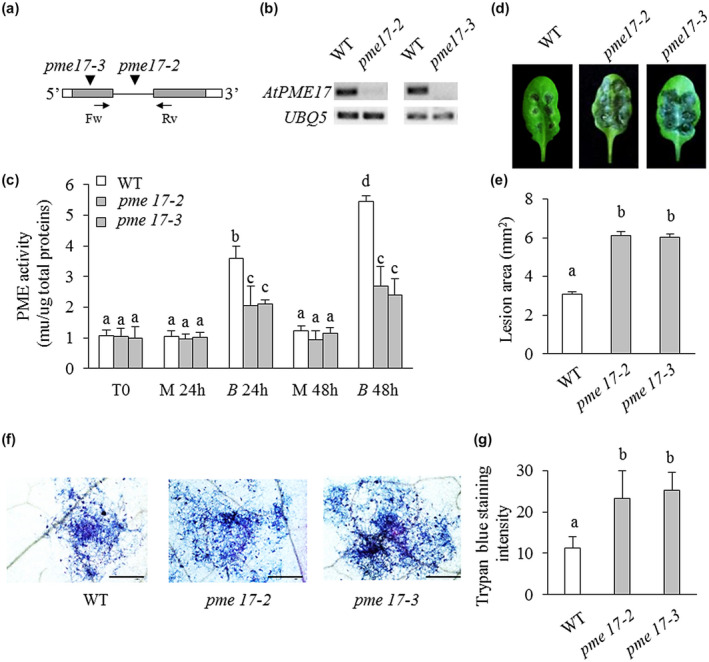
Arabidopsis pme17 mutants are defective in the induction of pectin methylesterase (PME) activity and are more susceptible to Botrytis cinerea (a) Schematic representation of AtPME17 gene structure. The localization of T‐DNA and transposon insertions in the genomic DNA sequences are shown (inverted black triangles). 5′‐UTR and 3′‐UTR are represented in white, exons in grey bars and introns as a black line. The localization of primers pairs Fw/Rv used for reverse transcription PCR are shown with black arrows (b) The expression of AtPME17 was analysed by semiquantitative reverse transcription PCR using cDNA from leaves of 4‐week‐old wild‐type (WT) and mutant plants at 48 hours postinoculation (hpi). UBQ5 was used as an internal positive control. (c) Four‐week‐old leaves of Arabidopsis thaliana WT plants and pme mutants were inoculated with B. cinerea and mock and PME activity was quantified at 0, 24, and 48 hpi. Values are means ± SD (n = 3). (d) Photographs showing lesion areas on leaves of 6‐week‐old A. thaliana WT and pme mutants. (e) Quantification of lesion areas produced by the spreading of the fungus at 48 hpi. Values are means ± SE (n ≥ 50). (f) Photomicrographs showing B. cinerea colonization revealed by trypan blue staining (scale bars = 500 μm). (g) Quantification of trypan blue intensity using ImageJ. Values are means ± SD (n = 4). Different letters indicate datasets significantly different according to analysis of variance followed by Tukey's test (p < .05)
To investigate whether the reduced resistance to B. cinerea observed in pme17 mutants could be caused by defects in the activation of pathogen‐responsive gene expression, the transcript levels of important defence genes were measured by quantitative reverse transcription PCR (RT‐qPCR) in healthy A. thaliana WT and pme17 plants and on B. cinerea infection. The expression levels of Pathogen Responsive 1 (PR1), a marker for SA (van Loon et al., 2006), and of Plant Defensin 1.2 (PDF1.2), a marker for the JA–ET signalling pathways (Penninckx et al., 1998), were analysed. Moreover, we also analysed the expression of two elicitor‐induced genes, Phytoalexin Deficient 3 (PAD3), tightly correlated with camalexin synthesis (Schuhegger et al., 2006), and RetOx, encoding a protein with homology to reticuline oxidases, a class of enzymes involved in secondary metabolism and in defence against pathogens (Dittrich and Kutchan, 1991). The pme17 mutants showed a significant reduction in the expression of PDF1.2 compared to the WT plants (Figure 3). Our results indicate that a compromised activation of PDF1.2 in the pme17 contributes to the increased susceptibility to B. cinerea. No significant differences were observed in the other genes analysed. Reactive oxygen species (ROS) production and callose deposition represent two important A. thaliana defence responses elicited against B. cinerea (Galletti et al., 2008; Pogorelko et al., 2013). A higher level of induction of both responses was detected in pme17 mutants with respect to WT, indicating that the level of elicitation correlates with the level of fungal growth observed in the genotypes. These results show that ROS and callose deposition are not compromised in pme17 mutants (Figure 4). No significant differences were observed in the growth of the vegetative rosette and no obvious changes in neutral sugars and GalA content in the leaf cell wall composition were detected between pme17 mutants with respect to WT plants (Figures S5 and S6).
FIGURE 3.
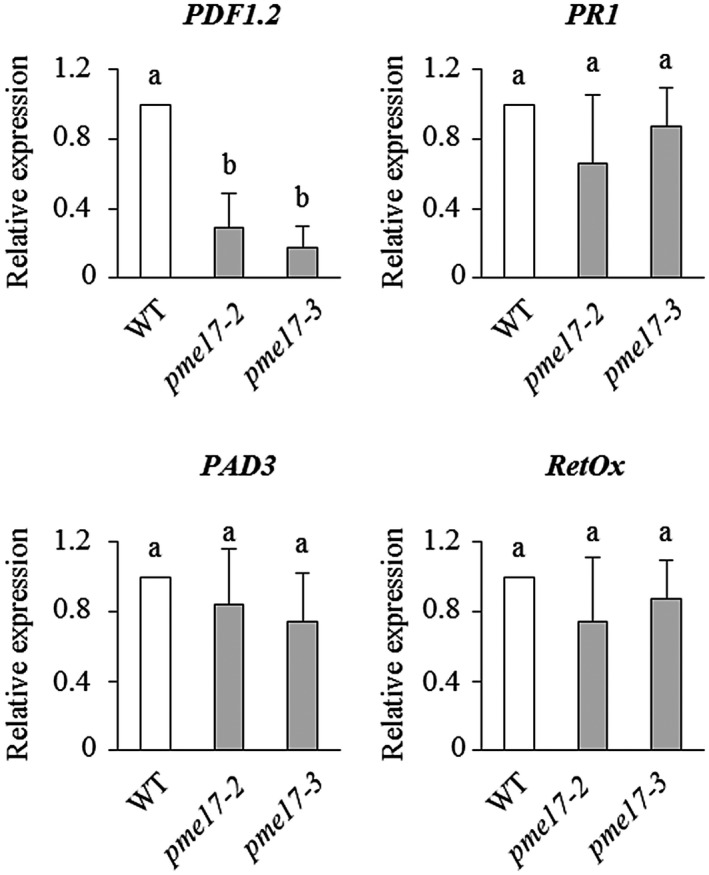
Expression of defence genes PR1, PDF1.2, PAD3, and RetOX in pme17 mutants after Botrytis cinerea induction. The expression of the selected defence genes was analysed by quantitative reverse transcription PCR at 48 hr postinoculation in B. cinerea‐ or mock‐inoculated leaves of 6‐week‐old Arabidopsis in pme17 mutants and wild‐type plants (WT). The expression levels were normalized to UBQ5 expression. The relative expression is represented as the ratio between gene expression in the mutants and WT plants. The results represent means ± SD of three independent experiments (n = 3). Different letters indicate data sets significantly different according to analysis of variance followed by Tukey's test (p < .01)
FIGURE 4.
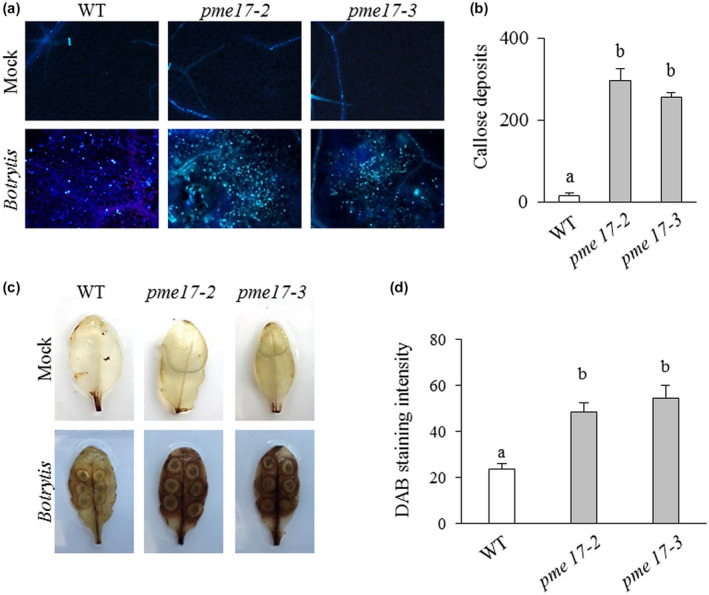
Reactive oxygen species (ROS) and callose accumulation in wild‐type (WT) and pme17 mutants in response to Botrytis cinerea. (a) Photomicrographs indicating callose deposition at the level of the lesion area produced on pme17 mutants and WT plants at 48 hr postinoculation (hpi) with B. cinerea. (b) Callose deposits were quantified per microscopic field using ImageJ. Values are means ± SD (n = 3). (c) ROS accumulation in B. cinerea and mock‐treated leaves at 48 hpi. Leaves were stained with 3,3′‐diaminobenzidine (DAB). Similar results were obtained in three independent experiments. (d) Quantification of DAB intensity using ImageJ. Values are means ± SD (n = 3). Different letters indicate datasets significantly different according to analysis of variance followed by Tukey's test (p < .05)
2.2. AtPME17 is a functional A. thaliana PME with an optimal activity at alkaline pH
We decided to produce a recombinant AtPME17 with the aim of verifying if AtPME17 is a functional enzyme and to gain insight into its possible role in the modification of pectin structures during infection. The AtPME17 gene is predicted to encode a protein of 511 amino acid residues (UniProtKB accession number O22149), which includes a putative N‐terminal signal peptide (SP) of 23 amino acids (Figure S7a). The removal of the predicted SP generates a protein of 488 amino acids (PRO‐AtPME17) with a predicted molecular mass of 53,418 Da and a theoretical isoelectric point (pI) of 9.2. The protein is formed by an N‐terminal PRO region of 148 amino acid residues with a predicted molecular mass of 16,598 Da and a theoretical pI of 6.9, and a C‐terminal catalytic PME domain (mature PME) of 307 amino acids with a predicted molecular mass of 33,125 Da and a theoretical pI of 9.62.
The intron‐less region of PRO‐AtPME17 was amplified by PCR using cDNA obtained from mRNA extracted from A. thaliana leaves infected with B. cinerea. The amplified fragments were cloned into the pPICZαA vector and expressed in P. pastoris. The culture filtrate was loaded onto ion exchange CM‐sepharose column and the eluted fractions were analysed by sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS‐PAGE) and for PME activity. The chromatogram shows the presence of two peaks (peak 1 and 2) (Figure S8a). PME activity was associated with protein fractions corresponding to peak 2 (Figure S8b). SDS‐PAGE analysis of the pooled fractions corresponding to peak 2 showed the presence of a band exhibiting an apparent molecular mass of 35 kDa (Figure 5a). Pooled fractions corresponding to peak 1 showed a prevalent band of 65 kDa. The identity of the proteins corresponding to the two bands was revealed by mass spectrometry analysis (LC‐Orbitrap MS/MS) after trypsin digestion. The identified fragments of the 35 kDa band had an optimal coverage (97%) with the amino acid sequence of the mature form of AtPME17 without the PRO region (Figure S7b). The 65 kDa band of peak 1 was identified as an “unknown” protein. The absence of the PRO‐AtPME17 unprocessed form in the purified proteins expressed in P. pastoris suggests that the enzyme is cleaved by endogenous Pichia subtilases. According to this conclusion, in the culture filtrate we found the presence of protease activity, associated with an apparent molecular mass corresponding to the P. pastoris subtilase SUB2 (Figure S9) (Salamin et al., 2010). The specific activity of the purified AtPME17 mature protein was quantified as 13 ± 3 U/mg (Figure 5b).
FIGURE 5.
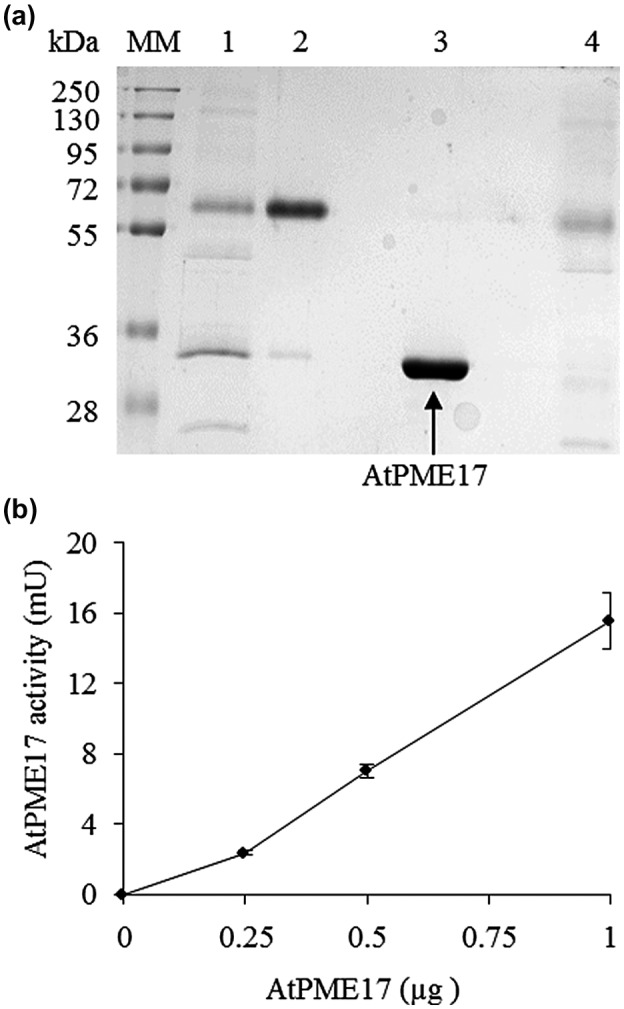
AtPME17 expressed in Pichia pastoris exhibits pectin methylesterase (PME) activity. (a) Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of AtPME17 expressed in P. pastoris and purified using CM‐sepharose chromatography. MM, molecular mass markers; lane 1, crude supernatant of transformed X33:pPICZaA/AtPME17 cells; lane 2, pooled fractions corresponding to peak 1; lane 3, pooled fractions corresponding to peak 2; lane 4, pooled fraction from flow‐through. Gel was stained with Coomassie brilliant blue after SDS‐PAGE separation. (b) Quantification of PME activity of the purified recombinant AtPME17. Values are means ± SD (n = 3)
The evidence indicates that apoplastic pH is an important factor during disease, able to influence the activity of the plant and the pathogen enzymatic machineries and CW polysaccharide rheology (Geilfus, 2017). With the aim of verifying if AtPME17 activity could be influenced by pH variation, we tested the activity of purified AtPME17 at different pHs using B. cinerea PME activity for comparison. AtPME17 activity increased with the medium alkalinization from acidic pH 5.5 up to pH 7.5. In contrast, B. cinerea PME activity was highest at the most acidic pH and decreased with medium alkalinization showing the lowest activity at pH 7.5 (Figure 6a). These results indicate that AtPME17 activity is favoured at more alkaline pH, which is unfavourable for pathogen PME activity. The thermal stability of AtPME17 was evaluated by incubating the enzyme at increasing temperatures. AtPME17 activity was stable up to 40°C, indicating a high stability at physiological temperatures. The activity decreased strongly in the range between 50 and 60°C and AtPME17 was inactivated at 70°C (Figure 6b).
FIGURE 6.
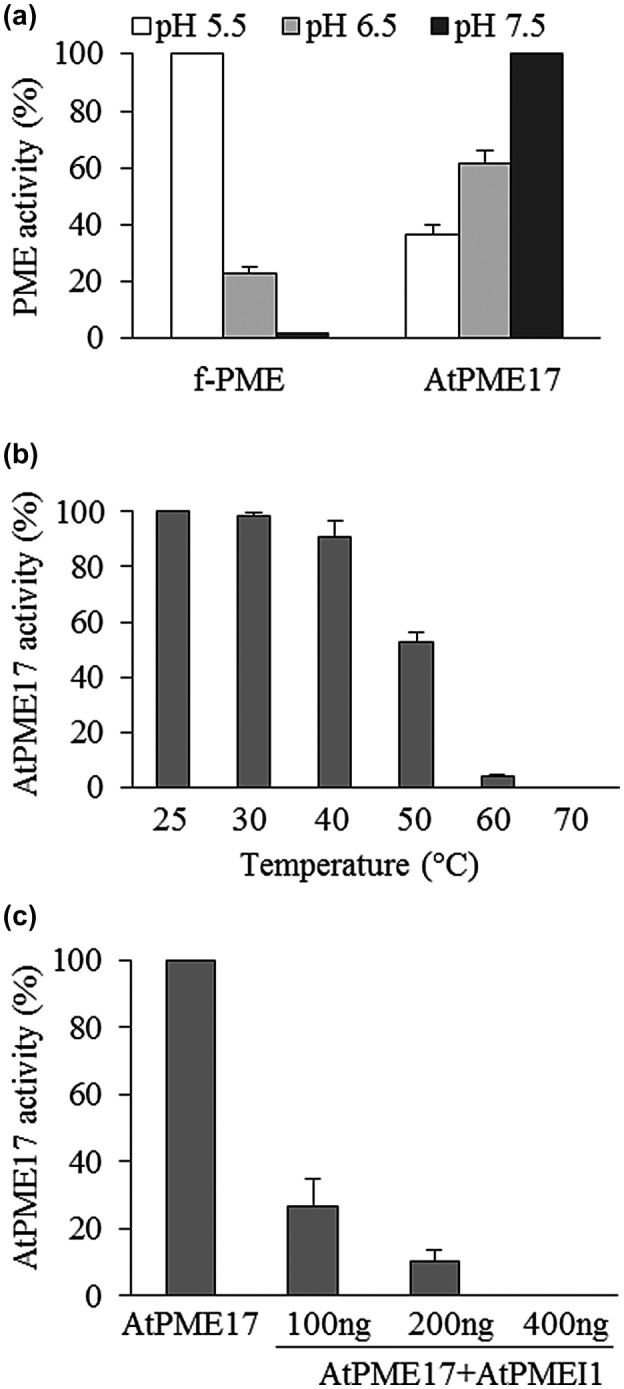
AtPME17 pectin methylesterase (PME) activity at different pHs and temperatures and treated with AtPMEI1. (a) The activity of purified AtPME17 was determined by gel diffusion assay. Botrytis cinerea PME activity (f‐PME) was also analysed. (b) Thermal stability of purified AtPME17. The activity at 25°C was set to 100% and the other values relativized. (c) Inhibition of purified recombinant AtPME17 by AtPMEI1. Values are means ± SD (n = 3)
It was previously found, by indirect evidence, that AtPMEI4 inhibits AtPME17 activity during root development (Senechal et al., 2015). We investigated if the activity of the recombinant AtPME17 is inhibited in vitro by PME inhibitors. AtPME17 was treated with the biochemically characterized AtPMEI1, already available in the laboratory (Raiola et al., 2011). AtPMEI1 inhibited AtPME17 activity with maximum inhibition at the highest PMEI concentration (c.400 ng) (Figure 6c).
2.3. AtPME17 generates HG blockwise de‐methylesterification and “egg‐box” formation
We next investigated how AtPME17 can affect the rheological properties of pectin during defence. The mode of action of purified AtPME17 was evaluated using a CaCl2 precipitation assay of ruthenium red‐labelled pectins (Dedeurwaerder et al., 2008). CaCl2 can only precipitate de‐methylesterified pectins made of consecutive GalA residues in egg‐box conformation (Hou et al., 1999). Highly methylesterified pectin (E81) was treated in parallel with a commercial orange PME, which acts in a blockwise manner (Willats et al., 2001), and with an Aspergillus aculeatus PME, known to de‐methylesterify pectin in a random manner (Kim et al., 2018) (Figure 7). Available blockwise de‐methylesterified pectin (P76), random de‐methylesterified pectin (F76), and polygalacturonic acid were also used as a comparison (Willats et al., 2001). Pectin treated with purified AtPME17 showed a 50% OD535 reduction with respect to untreated pectin, which is comparable to what was observed in the case of pectin treated with orange PME and P76, showing 54% and 51% reductions, respectively. Polygalacturonic acid showed the highest reduction due to its total de‐methylesterification. In contrast, a range of only 8%–9% OD reduction was observed in the case of pectin treated with A. aculeatus PME or F76 pectin, respectively. These results demonstrate that AtPME17 performs a blockwise pectin de‐methylesterification that leads to pectin precipitation due to calcium‐mediated egg‐box formation.
FIGURE 7.
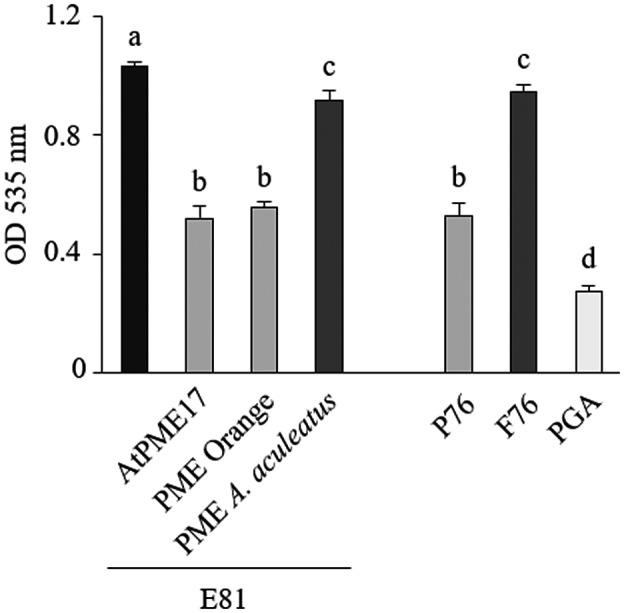
Mode of action of purified AtPME17. Measured optical densities after CaCl2 precipitation of untreated lime pectin E81 or E81 treated with AtPME17, orange pectin methylesterase (PME) or Aspergillus aculeatus PME. Lime pectins with blockwise de‐methylesterification (P76) or with random de‐methylesterification (F76) and polygalacturonic acid (PGA) were used for comparison. Values are means ± SD (n = 3). Different letters indicate datasets significantly different according to analysis of variance followed by Tukey's test (p < .05)
2.4. The PRO region is an intramolecular inhibitor of AtPME17 activity
An interesting structural similarity can be revealed between the amino acid sequences of the PRO regions, belonging to the PME isoforms induced in A. thaliana during B. cinerea infection, and the PME inhibitors contributing to A. thaliana immunity against B. cinerea (AtPMEI10 and AtPMEI11) (Figure S10). In particular, the PRO region of AtPME17 presents the conserved four cysteine residues engaged in the formation of two disulphide bridges important to stabilize the four‐helix bundle structure of PMEIs, a conserved 113Thr residue that strengthens the PME–PMEI interaction, the absence of the PKF motif in the α5 helix discriminating PME inhibitors from invertase inhibitors, and a C‐terminal hydrophobic region at the α6 helix involved in the stabilization of the four‐helical bundle structure of the inhibitors (Di Matteo et al., 2005). Moreover, secondary structure prediction reveals a prevalence of α‐helix structure in the PRO region of AtPME17, typically presented also by the PMEIs (Giovane et al., 2004). Due to the observed structural similarity of the AtPME17 PRO region with PMEI, we decided to explore a possible role for this domain in the regulation of AtPME17 activity. Aware of the experience with the P. pastoris expression system, we carried out the expression of the His‐tagged PRO‐AtPME17 (His‐PRO‐AtPME17) version in E. coli BL21, a strain that does not produce SBTs in culture (Siezen and Leunissen, 1997). After the His‐tagged protein purification, a single protein band, with an apparent molecular mass of about 55 kDa, corresponding to the His‐PRO‐AtPME17 unprocessed form, was separated by SDS‐PAGE and revealed by Coomassie blue staining and western blot analysis using anti‐His‐tag antibody (Figure 8a,b). Interestingly, the 55 kDa purified His‐PRO‐AtPME17 did not show PME activity (Figure 8c), indicating that the unprocessed form of the protein is inactive. We attempted to deprive AtPME17 of its His‐PRO region by treating the His‐PRO‐AtPME17 with SBT activity, previously isolated from P. pastoris. SDS‐PAGE analysis followed by Coomassie blue staining showed two bands of 20 and 35 kDa (Figure 8a). Western blot analysis demonstrated that the protein band of 20 kDa corresponded to the His‐PRO region (Figure 8b). The protein band of 35 kDa showed the same apparent molecular mass of the PME domain previously purified after P. pastoris expression and loaded for comparison (Figure 8a). Mass spectrometry analysis of the 20 and 35 kDa bands revealed that they corresponded to the PRO region and PME domain of PRO‐AtPME17, respectively (Figure S11). It is important to mention that the SBT‐treated His‐PRO‐AtPME17 acquired PME activity (Figure 8c). Our results clearly indicate that the PRO region acts as intramolecular inhibitor of AtPME17 enzymatic activity.
FIGURE 8.
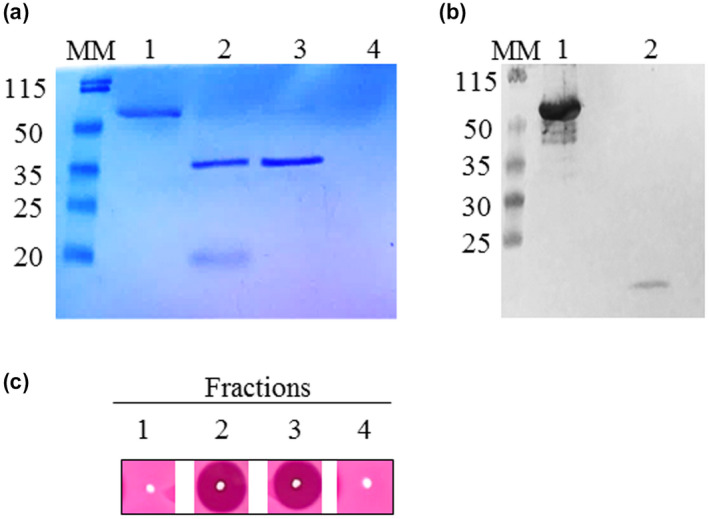
The PRO region is an intramolecular inhibitor of AtPME17 enzymatic activity. (a) Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS‐PAGE) analysis of His‐PRO‐AtPME17 expressed in Escherichia coli followed by western blot using anti‐His antibodies. Lane 1, His‐PRO‐AtPME17 form; lane 2, His‐PRO‐AtPME17 after processing by Pichia pastoris specific subtilase (SBT) activity; lane 3, recombinant AtPME17 expressed in P. pastoris; lane 4, P. pastoris culture supernatant with SBT activity cleaned on Sephadex G‐25 PD10 column. MM, molecular mass markers. (b) Western blot using anti‐His antibodies. Lane 1, His‐PRO‐AtPME17 form; lane 2, His‐PRO‐AtPME17 after processing by P. pastoris SBT activity. (c) pectin methylesterase activity revealed by gel diffusion assay in fractions loaded in SDS‐PAGE
3. DISCUSSION
Experimental evidence indicates that plants activate a local and strong PME activity in response to pathogens with different lifestyles (Hewezi et al., 2008; Bethke et al., 2014; Lionetti et al., 2014, 2017; Lionetti, 2015). Here, we studied the pathosystem A. thaliana–B. cinerea. B. cinerea is a broad‐spectrum necrotrophic fungus that causes serious pre‐ and postharvest rots in more than 200 species worldwide (Dean et al., 2012). Our results indicate that SA, JA, and ABA immune signalling pathways dynamically govern PME activity against B. cinerea. The induction of PME activity requires JA also during P. syringae infection (Bethke et al., 2014). The level of PI‐PME activity could be orchestrated by the expression of specific PME isoforms. However, this activity could be activated by SBTs or other factors, such as the apoplastic pH and is posttranscriptionally controlled by PMEIs (Lionetti et al., 2017). It is worth noting that the higher induction of PI‐PME activity revealed in the ein2‐5 mutant with respect to the WT correlates well with the lower induction of AtPME10 and AtPME11 expression observed previously in the ein2‐5 mutant with respect to the control (Lionetti et al., 2017).
The next goal was to identify the main PME isoform involved in PI‐PME activity. We focused our attention on AtPME17 (At2g45220), the PME isoform most abundantly induced on B. cinerea infection (Lionetti et al., 2017). AtPME17 expression is highly regulated in response to several pathogens and PAMPs. The analysis of ein2‐5, jar‐1, sid2‐2, and aba2‐3 mutants challenged with B. cinerea, together with meta‐analysis data and the presence of cis‐acting elements in the AtPME17 promoter, indicates that the ET, JA, ABA, and SA signalling pathways are required for AtPME17 expression. All our evidence indicates that AtPME17 expression and PI‐PME activity are regulated as part of the A. thaliana immune response to B. cinerea. Interestingly, VvPME6 (ID VIT_06s0009g02560), the orthologue of AtPME17 in Vitis vinifera, is the most induced CW‐modifying enzyme during B. cinerea penetration (Haile et al., 2017). This evidence indicates that A. thaliana and V. vinifera, and possibly other crops, induce AtPME17‐like isoforms in response to B. cinerea and other pathogens, making this PME member a molecular biomarker for microbial infection and stresses, and a promising candidate for biotechnology applications in the agronomic industry.
Prediction of subcellular location and CW proteome analysis indicated an apoplastic localization for AtPME17 (SUBA4, http://suba.live) ( Nguyen‐Kim et al., 2016; Duruflé et al., 2017). To explore the contribution of AtPME17 to A. thaliana PI‐PME activity and to resistance against B. cinerea, two T‐DNA insertional mutants of AtPME17 were isolated. The mutants were not different from WT in terms of plant vegetative growth and there were no obvious changes in neutral sugars and GalA content in the leaf cell wall of the mutants with respect to WT plants. Divergently, a reduced content of GalA in pme17‐2 mutants with respect to the WT was recently reported (Roig‐Oliver et al., 2020). This may be explained by the different growth conditions, development stage, and/or different methodology used to determine the monosaccharide composition.
Interestingly, pme17‐2 and pme17‐3 mutants showed a defective induction of PI‐PME activity and a higher susceptibility to B. cinerea with respect to WT plants. It is important to mention that both mutants showed almost 50% lower PI‐PME activity in comparison to WT plants, indicating that AtPME17 predominantly contributes to its induction. No significant differences were observed between the two mutants, indicating that the slight induction of AtPME17, still present in pme17.2 knockdown mutants, is not enough to mediate the defence responses triggered by AtPME17 in WT plants. Previous work showed an increased susceptibility of pme mutants with respect to WT plants when infected with P. syringae, a bacterial hemibiotroph (Bethke et al., 2014). However, the authors ruled out that the determinant of immunity was the total PME activity. Moreover, no significant differences were identified in terms of susceptibility to the necrotrophic fungus A. brassicicola. Our results show for the first time the functional role of AtPME17 in triggering PME activity and in resistance against a pectinolytic necrotrophic fungus such as B. cinerea.
A comparative analysis of the expression of some defence genes between WT plants and pme17 mutants was performed with the aim of identifying the molecular basis of the differential immune responses. Interestingly, a defect in the induction of PDF1.2 gene expression was identified in pme17 mutants. PDF1.2 induction requires both ET and JA pathways and is correlated with the resistance against necrotrophic pathogens, including B. cinerea (Penninckx et al., 1998). We can argue that in WT plants, AtPME17 activity participates in the activation of defence responses mediated by JA and ET, such as PDF1.2, against B. cinerea. A possibility that AtPME17 activity could trigger PDF1.2 through the generation of partially de‐methylesterified OGAs must be considered (Osorio et al., 2008). The evidence that ROS production and callose deposition, two defence responses typically induced by unmethylesterified and unoxidized OGs with DP 9 to 16 in A. thaliana (Ferrari et al., 2013), were not defective in pme17 mutants, together with the similar induction of PAD3 and RetOX in the pme17 mutants as the WT, weaken this hypothesis. However, it cannot be excluded that AtPME17 activity could modulate the OG pattern of methylesterification or the release of OGs with a different DP or structure (Voxeur et al., 2019). Moreover, AtPME17 could assist in the release of additional cell wall‐derived damage‐associated molecular patterns, like the β‐1,4 cellodextrins or the pentose‐based cell wall oligosaccharides, important in Arabidopsis immunity (Mélida et al., 2020; Souza et al., 2017). Recently, methanol was demonstrated to induce cytosolic calcium variations, membrane depolarization, and ET production in A. thaliana (Tran et al., 2018). AtPME17 could also trigger PDF1.2 through methanol release.
The biochemical and functional characterization of AtPME17 was achieved through its heterologous expression in P. pastoris. Our biochemical evidence showing that the protein expressed in P. pastoris had PME activity indicates that the AtPME17 is an A. thaliana functional isoform. Considering the effect of methyl ester distribution on cell wall degradation during pathogen infection, we next evaluated the AtPME17 mode of action. Our results show that AtPME17 performs a blockwise pattern of pectin de‐methylesterification, causing pectin precipitation due to the formation of Ca2+‐mediated egg‐box structures. AtPME17 could locally reinforce the pectin structure at the infection site to mechanically counteract the fungal penetration. It is likely that the enhanced susceptibility observed in pme17 mutants is also caused by reduced pectin stiffening. Even more intriguing is the possibility that the pectin modifications induced by AtPME17 activity could represent a trigger leading to the activation of hormone‐regulated plant immune signalling. The defective induction of JA/ET‐dependent PDF1.2 expression resulting from the lack of AtPME17 induction in pme17 mutants supports this conclusion. Consistently, treatment with pectinase induced JA production, suggesting that enzymatic pectin perturbation activates JA biosynthesis (Engelsdorf et al., 2018). Moreover, methyl and acetyl esterified OGs may modulate the JA level during B. cinerea infection (Voxeur et al., 2019). Our evidence supports the emerging role of JA in cell wall maintenance and remodelling in plant immunity (Mielke and Gasperini, 2019).
The apoplastic pH is an important factor that affects the interaction between B. cinerea and its hosts (Hua et al., 2018). Our results show that AtPME17 activity increased on increasing the pH of the medium, a condition unfavourable for B. cinerea PME activity. This result could be seen in the context of a competition between the two organisms to control the pectin and, consequently, the cell wall integrity. B. cinerea is considered an acidic fungus because it lowers the apoplastic pH in infected plant tissues through the secretion of a large amount of oxalic acid (Prusky and Yakoby, 2003). The acidification of the host tissues favours CW degradation by fungal CWDEs and contributes to the fungal colonization. Moreover, OA, a chelating agent, can sequester Ca2+ ions from the egg‐boxe structures, favouring B. cinerea PG activity in pectin degradation (Manteau et al., 2003). On the contrary, plant cells induce a rapid transient alkalinization of the apoplast during infection to counteract pathogen invasion (Aziz et al., 2003). The ability of AtPME17 to work efficiently at more alkaline pHs could indicate a molecular adaptation evolved in A. thaliana to favour a pectin‐mediated CW strengthening in a hostile extracellular environment created against B. cinerea. Another interesting hypothesis to confirm is that AtPME17 might play a role in maintaining pectin integrity. The Pathogen Recognition Receptor (PRR) Wall‐associated Kinase 1 (WAK1) and FERONIA (FER) preferentially bind to de‐methylesterified crosslinked pectins (Decreux and Messiaen, 2005; Feng et al., 2018; Guo et al., 2018; Lin et al., 2018). By producing blockwise de‐methylesterified pectin, ATPME17 could facilitate the binding of some PRRs to pectin. Intriguingly, FER, by mediating apoplast alkalization (Haruta et al., 2014), may enhance AtPME17 activity to counteract B. cinerea. Future efforts will be needed to explore this hypothesis.
Due to the observed structural similarity of the AtPME17 PRO region to PMEIs, we decided to explore the possible role of this domain in the regulation of AtPME17 activity. Our biochemical evidence indicates that the unprocessed version of PRO‐AtPME17, expressed in E. coli, did not show PME activity, while the PME domain, separated from its own PRO region, acquired PME activity. This is the first direct evidence that the PRO region is an intramolecular inhibitor of AtPME17 activity. During root development, AtPME17 is processed by AtSBT3.5 at a specific single processing motif and is probably inhibited by PMEI4 (Senechal et al., 2014a). Although a role for subtilases and PMEI in plant immunity was been reported (Lionetti et al., 2017; Figueiredo et al., 2018) the specific isoforms regulating AtPME17 activity during B. cinerea infection remain to be identified.
4. EXPERIMENTAL PROCEDURES
4.1. Plant material and growth conditions
A. thaliana Columbia‐0, WT plants or mutants were grown in a controlled environmental chamber maintained at 22°C and 70% relative humidity, with a 12hr/12hr day/night cycle (photosynthetically active radiation level of 100 μmol⋅m−2⋅s−1). Mutants isolated in this study are as follows: SALK_059908 (pme17‐2) and SM_3_25823 (pme17‐3 plants). All lines were obtained from the Nottingham Arabidopsis Stock Centre.
4.2. A. thaliana infection assay
Pathogenicity assay with B. cinerea strain SF1 was performed on leaves from 6‐week‐old A. thaliana plants inoculated with 2 × 105 conidia/ml as previously described (Lionetti et al., 2007). Plants were incubated at 24°C with a 16hr/8hr day/night photoperiod. The lesion size produced by B. cinerea was evaluated as an indicator of susceptibility to the fungus.
4.3. Gene expression analysis and mutant genotyping
B. cinerea‐infected or mock‐inoculated leaves were frozen in liquid nitrogen and homogenized with a MM301 ball mill (Retsch) for 1 min at 30 Hz, and total RNA was extracted with Isol‐RNA lysis reagent (5′‐Prime) according to the manufacturer's instructions. RNA was treated with RQ1 DNase (Promega), and first‐strand complementary DNA (cDNA) was synthesized using ImProm‐II reverse transcriptase (Promega). PCR analysis was performed in 30 μl of reaction mix containing 1 × KAPA Taq Buffer A with Mg2+ (KAPABIOSYSTEM), 10 mM of each dNTP, 10 µM of each primer, and 1 U KAPA Taq DNA polymerase (KAPABIOSYSTEM) RT‐qPCR analysis was performed using a CFX96 Real‐Time System (Bio‐Rad). One microlitre of cDNA (corresponding to 50 ng of total RNA) was amplified in 20 µl of reaction mix containing 1 × Go Taq qPCR Master Mix (Promega) and 0.5 µM of each primer. The expression levels of each gene, relative to the UBIQUITIN5 (UBQ5) gene, were determined using a modification of the Pfaffl method (Pfaffl, 2001).
For mutant genotyping, genomic DNA was extracted from rosette leaves as previously described (Edwards et al., 1991) and subjected to a PCR‐based screening using specific primers. Primer sequences are shown in Table S3.
4.4. Determination of ROS accumulation, callose deposition, and mycelium growth
A. thaliana leaves were assayed for ROS accumulation using 3,3′‐diaminobenzidine (DAB) staining as described (Reem et al., 2016). Determination of callose deposition and the detection of B. cinerea hyphae in infected plant tissues were performed as previously described (Lionetti et al., 2017). Mean grey values, expressed as DAB and trypan blue staining intensity, and callose deposits, counted in the entire image using cell counter function, were quantified using ImageJ software (https://imagej.nih.gov/ij/).
4.5. Determination of CW monosaccharide composition
Extraction of alcohol‐insoluble residue and CW monosaccharide composition was performed as previously described (Lionetti et al., 2017).
4.6. Expression of AtPME17 in P. pastoris and purification
The intronless region of AtPME17 was amplified with specific primers (Table S3) by PCR from cDNA prepared from A. thaliana infected leaves and cloned in pGEM‐T and in pPICZαA vectors. pPICZαA was used to transform P. pastoris X‐33 cells according to the Pichia EasyComp transformation kit (Invitrogen). Transformed P. pastoris cells were incubated in buffered glycerol complex medium (BMGY) at 30°C until OD600 = 2–6. Bioreactor production of AtPME17 was carried out in 1.3‐L New Brunswick BioFlo 115 fermenters (Eppendorf) following the P. pastoris fermentation process guidelines (Invitrogen) as described (Figure S12) (Couturier et al., 2018). Culture filtrate was pelleted by centrifugation at 4,000 × g for 30 min at 4°C and the supernatant assayed for PME activity. AtPME17 activity was detected in the P. pastoris supernatant after 120 hr of fermentation using methanol as the only feeding carbon source. Recombinant enzyme was secreted up to c.500 mg/L. Supernatant from the transformed P. pastoris cultures was dialysed against 25 mM MES buffer pH 6.5 and loaded into a CM‐sepharose column (Pharmacia). Retained proteins were eluted with a linear gradient of NaCl from 0 to 0.5 M in the equilibration buffer at a flow rate of 1 ml/min on a FPLC system (ÄktaTM Pure 25; GE‐Healthcare). The eluted fractions were analysed by SDS‐PAGE and for PME activity and those containing the pure protein were pooled.
4.7. Isolation of native secreted subtilisin activity from P. pastoris culture
P. pastoris X33 was grown to near‐saturation at 30°C in 10 ml of BMGY broth. Cells were harvested by centrifugation and resuspended in the same medium with 0.5% methanol as carbon source and incubated for 48 hr. After centrifugation, culture supernatant was loaded on a Sephadex G‐25 PD‐10 column (GE Healthcare) and equilibrated with 20 mM Tris–HCl pH 7.5 to remove salt and low molecular weight compounds. The high molecular weight eluted fraction was 10‐fold concentrated by Amicon Ultra 4 ml centrifugal filters. The proteolytic activity was tested as described (Salamin et al., 2010). After electrophoresis, the gels were incubated twice for 15 min at room temperature in 10 volumes of 2.5% Triton X‐100 under constant agitation. The gels were then incubated overnight with 10 mM sodium phosphate buffer, pH 7.0, and stained with 0.1% Coomassie brilliant blue R‐250 (Bio‐Rad).
4.8. Expression of His‐PRO‐AtPME17in E. coli
To express AtPME17 in E. coli DH5α cells as His‐tagged protein, the PRO‐AtPME17 was PCR‐amplified using specific primers containing EcoRI (P1) and NotI (P2) restriction enzyme sites (Table S3) and cloned into pET‐28a (+) vector digested with the same restriction enzymes. The recombinant vector was used to transform E. coli BL21 Codon‐Plus (DE3) (Agilent Technologies).
Overnight cultures of the transformants were grown in Luria‐Bertani medium at 37°C to OD600 = 0.6–0.8 and induced with 0.1 mM isopropyl β‐D‐1‐thiogalactopyranoside for 4 hr at 28°C. The cells were harvested by centrifugation at 10,000 × g for 15 min. The resulting pellet was resuspended in 50 mM Tris–HCl, pH 8.5, containing 1 mM phenyl methane sulfonyl fluoride. The cell suspension was subjected to ultrasonication then centrifuged at 19,000 × g at 4°C for 45 min and supernatant collected. His‐PRO‐AtPME17 was purified by immobilized metal ion affinity chromatography using HisTrap affinity column (GE Healthcare). Ten micrograms of purified His‐PRO‐AtPME17 was treated for 16 hr with 10‐fold concentrated Pichia culture supernatant (10 μl) cleaned as described above. The purified His‐tagged proteins were analysed by SDS‐PAGE and western blotting.
4.9. Protein MS/MS analysis
The excised protein bands were treated as previously described (Benedetti et al., 2018). MS and MS/MS data were acquired using Xcalibur (Thermo Scientific, version 4.1.31.9). Peptides and proteins were identified using Mascot (v. 2.6.0) with a search against a database containing the given PME modified sequence: semi‐trypsin as the enzyme, 10 ppm as the precision on the peptides, and 25 mmu on the fragments. Peptide modifications allowed during the search were as follows: carbamidomethyl (C, fixes), oxidation (M, variable), and deamidation (NQ, variable). Proline software (http://proline.profiproteomics.fr, v. 2.0) was used to perform filtering of the results (conservation of rank 1 peptides and peptide score >20).
4.10. Determination of PME activity
PME activity was isolated and the PME activity was evaluated by using apple pectin (75% methylesterification) substrate, as previously described (Lionetti, 2015). To determine the AtPME17 mode of action, lime pectins with defined degree and patterns of methylesterification were used. First, 500 µg of highly methyl esterified pectins (81%) (E81; GRINDSTED Pectin URS 1,200) were treated with 8 mU of purified recombinant AtPME17, PME from orange peel (P5400; Sigma), and a fungal PME from A. aculeatus in 500 µl of 12.5 mM citric acid, 50 mM Na2HPO4, pH 6.5. Then 250 µl of a 0.01% ruthenium red solution was added. After mixing, 250 μl of a 0.6 M CaCl2 solution was added to de‐methyl esterified and untreated E81 pectins and incubated for 15 min at room temperature. The mixture was then centrifuged at 15,000 × g for 1 min and the OD at 535 nm, the peak of absorption spectrum of ruthenium red, was measured in the supernatants. Blockwise partially de‐methylesterified, random partially de‐methylesterified pectin (P76; F76, GRINDSTED Pectin URS 1,200) and polygalacturonic acid (P3850; Sigma) were used for comparison. For the determination of recombinant AtPME17 thermal stability, AtPME17 (3.5 mU) was incubated in 20 μl of 1 M NaCl, 12.5 mM citric acid, 50 mM Na2HPO4, pH 6.5, for 10 min at each temperature tested. To assess the inhibition of AtPME17 by PMEI, AtPME17 (3.5 mU) was incubated with AtPMEI1 (100, 200, and 400 ng) for 45 min at room temperature. Recombinant AtPMEI‐1 was expressed in P. pastoris and purified to homogeneity (Raiola et al., 2004). B. cinerea PME activity was induced in the B. cinerea culture by adding 0.5% apple pectin (wt/vol; 76,282; Sigma‐Aldrich) and after 10 hr the culture filtrate was collected (Lionetti, 2015). For the determination of pH optimum, the solutions used for the preparation of the plates were buffered at different pHs.
AUTHOR CONTRIBUTIONS
D.B. and V.L. designed the research; D.D.C., M.R.F., R.M., D.P., S.G., S.K.‐J., and V.L. performed the experiments; D.D.C., M.R.F., R.M., M.L. T.G., D.B., and V.L. analysed data; D.D.C., D.B., and V.L. wrote the article.
Supporting information
FIGURE S1 Plant immunity hormones regulate the expression of total PI‐PME activity. PI‐PME activity was analysed at 0, 24, and 48 hr post B. cinerea infection (hpi) or mock‐inoculation in leaves of 6‐week‐old A. thaliana mutants defective in immune signalling or biosynthetic pathways and wild‐type (WT) plants. Values are means ± SD (n = 3). Different letters indicate data sets that are significantly different according to ANOVA followed by Tukey’s test (p < .01)
FIGURE S2 Expression pattern of AtPME17 expression in response to different pathogens. (A) Heatmaps of gene expression. Data were analysed using the Genevestigator Meta‐Analyzer Tools (Zimmermann et al., 2004). The corresponding n‐fold (log2 normalized) expressions in infected plants relative to each corresponding negative control are shown
FIGURE S3 Expression pattern of AtPME17 in response to different elicitors. Data were analysed using the Genevestigator Meta‐Analyzer Tools (Zimmermann et al., 2004). The corresponding n‐fold (log2 normalized) expressions in infected plants relative to each corresponding negative control are shown
FIGURE S4 Expression pattern of AtPME17 in response to different hormones. Data were analyzed using the Genevestigator Meta‐Analyzer Tools (Zimmermann et al., 2004). The corresponding n‐fold (log2 normalized) expressions in infected plants relative to each corresponding negative control are shown
FIGURE S5 Vegetative rosettes of 4‐week‐old pme17 mutants and WT plants
FIGURE S6 Monosaccharide composition of CW of pme17.2, pme17.3, and WT plants. The molar percentages of fucose (Fuc), rhamnose (Rha), arabinose (Ara), galactose (Gal), glucose (Glu), xylose (Xyl), mannose (Man), galacturonic acid (Gal A), and glucuronic acid (Glc A) released after 2 M TFA hydrolysis were quantified by high‐performance anion‐exchange chromatography with a pulsed amperometric detection (HPAEC‐PAD) system. The values are mean ± SD (n = 3). No significant differences were observed for each monosaccharide according to ANOVA followed by Tukey’s test (p > .05). hpi, hours postinoculation
FIGURE S7 Structural domains and amino acid sequence of AtPME17, with the peptides identified by MS underlined. (A) Structural domains of ATPME17 are highlighted with different colours: red, signal peptide; blue, PRO region with putative N‐glycosylation site in bold; orange, processing region with the putative basic processing motive RKLL; violet, PME catalytic domain. Peptides identified after trypsin digestion and analysed by liquid chromatography coupled with tandem mass spectrometry (LC‐MS/MS) are underlined. (B) Data regarding total sequence coverage of mature AtPME17 domain (percentage of the protein sequence covered by identified peptides) and sequence of identified unique peptides
FIGURE S8 AtPME17 purification using CM‐Sepharose chromatography. (A) The fractions were eluted with a gradient of 0 to 1 M NaCl. (B) Radial gel diffusion assay of eluted fractions of peak 1 and peak 2. Peak 2 fractions exhibiting PME activity (fuchsia halo) were pooled, concentrated, and used for a subsequent characterization
FIGURE S9 Native secreted subtilisin produced by P. pastoris. The samples were analysed by SDS‐PAGE with a separation gel of 12% polyacrylamide stained with Coomassie brilliant blue. To detect endoproteolytic activity of culture supernatant, gelatin at a concentration of 0.1% was added to the gel. Lane 1, culture supernatant of P. pastoris grown in the presence of methanol; Lane 2, culture supernatant cleaned on a Sephadex G‐25 PD10 column; MM, molecular mass markers
FIGURE S10 Sequence comparison of PRO regions of B. cinerea‐induced AtPMEs with B. cinerea‐induced AtPMEI10 and AtPMEI11. The biochemically characterized functional AtPMEI1 inhibitor is added for comparison. Highlight homology level: invariant residues are in black, homology level >75% in pink and homology level >50% in turquoise. The secondary structure elements as elucidated in AtPMEI1 crystal structures are indicated below the alignment. Multiple sequence alignment was performed using the DNAman software package (Lynnon Biosoft)
FIGURE S11 MS analysis of 20 and 35 kDa bands isolated by SDS‐PAGE after treatment of His‐PRO‐AtPME17 with SBTs. (A) Structural domains of His‐PRO‐AtPME17are highlighted with different colours: red, pET‐28a‐His tag sequence; blue, PRO region; yellow, processing region; violet, PME catalytic domain. Peptides identified after trypsin digestion and analysed by liquid chromatography coupled with tandem mass spectrometry (LC‐MS/MS) are underlined and peptides identified in the 20 kDa band are shown in italics. (B) Number of peptides and amino acid sequences of identified unique peptides in 20 and 35 kDa bands
FIGURE S12 Fermentation profile of P. pastoris expressing AtPME17. Time profiles of dissolved oxygen (DO), temperature, and optical density (OD) at 600 nm during the production of AtPME17 by P. pastoris in a 1.3‐L bioreactor fermentation. 1, glycerol batch phase; 2, sorbitol and methanol transition phase; 3, methanol fed‐batch phase
TABLE S1 Arabidopsis mutants defective in immune hormone signalling used in this study
TABLE S2 Cis‐acting element related to biotic stresses present in the 1000 bp promoter of AtPME17
TABLE S3 Primers used for semi‐quantitative and quantitative reverse transcript PCR, for genotyping, for PRO‐AtPME17 cloning in the pPICZαA vector, and for His‐PRO‐AtPME17 cloning in pET‐28a(+) vector
ACKNOWLEDGMENTS
We thank H.C. Buchholt (DANISCO) for kindly providing the methylated pectins. We thank Daniele Coculo for technical assistance. The work was supported by Sapienza University of Rome, grants RM11816432F244FD, RP1181642E99296A, and RM11916B7A142CF1, by LazioInnova grant ABASA CUPB81G18000770002 and by ANR “Investissement d'Avenir Infrastructures Nationales en Biologie et Santé” grant, Programme ProFI project, ANR‐10‐INBS‐08. The authors have no conflicts of interest to declare.
Del Corpo D, Fullone MR, Miele R, et al. AtPME17 is a functional Arabidopsis thaliana pectin methylesterase regulated by its PRO region that triggers PME activity in the resistance to Botrytis cinerea . Molecular Plant Pathology 2020;00:1–14. 10.1111/mpp.13002
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Alfano, J.R. , Bauer, D.W. , Milos, T.M. and Collmer, A. (1996) Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non‐polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Molecular Microbiology, 19, 715–728. [DOI] [PubMed] [Google Scholar]
- Aziz, A. , Poinssot, B. , Daire, X. , Adrian, M. , Bézier, A. , Lambert, B. et al. (2003) Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola . Molecular Plant‐Microbe Interactions, 16, 1118–1128. [DOI] [PubMed] [Google Scholar]
- Benedetti, M. , Verrascina, I. , Pontiggia, D. , Locci, F. , Mattei, B. , De Lorenzo, G. et al. (2018) Four Arabidopsis berberine bridge enzyme‐like proteins are specific oxidases that inactivate the elicitor‐active oligogalacturonides. The Plant Journal, 94, 260–273. [DOI] [PubMed] [Google Scholar]
- Bethke, G. , Grundman, R.E. , Sreekanta, S. , Truman, W. , Katagiri, F. and Glazebrook, J. (2014) Arabidopsis PECTIN METHYLESTERASES contribute to immunity against Pseudomonas syringae . Plant Physiology, 164, 1093–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffall, K.H. and Mohnen, D. (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydrate Research, 344, 1879–1900. [DOI] [PubMed] [Google Scholar]
- Catoire, L. , Pierron, M. , Morvan, C. , du Penhoat, C.H. and Goldberg, R. (1998) Investigation of the action patterns of pectinmethylesterase isoforms through kinetic analyses and NMR spectroscopy. Implications in cell wall expansion. Journal of Biological Chemistry, 273, 33150–33156. [DOI] [PubMed] [Google Scholar]
- Couturier, M. , Ladevèze, S. , Sulzenbacher, G. et al. (2018) Lytic xylan oxidases from wood‐decay fungi unlock biomass degradation. Nature Chemical Biology, 14, 306–310. [DOI] [PubMed] [Google Scholar]
- Dean, R. , van Kan, J.A.L. , Pretorius, Z.A. et al. (2012) The top 10 fungal pathogens in molecular plant pathology. Molecular Plant Pathology, 13, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decreux, A. and Messiaen, J. (2005) Wall‐associated kinase WAK1 interacts with cell wall pectins in a calcium‐induced conformation. Plant and Cell Physiology, 46, 268–278. [DOI] [PubMed] [Google Scholar]
- Dedeurwaerder, S. , Menu‐Bouaouiche, L. , Mareck, A. , Lerouge, P. and Guerineau, F. (2008) Activity of an atypical Arabidopsis thaliana pectin methylesterase. Planta, 229, 311–321. [DOI] [PubMed] [Google Scholar]
- Di Matteo, A. , Giovane, A. , Raiola, A. , Camardella, L. , Bonivento, D. , Lorenzo, G.D. et al. (2005) Structural basis for the interaction between pectin methylesterase and a specific inhibitor protein. The Plant Cell, 17, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich, H. and Kutchan, T.M. (1991) Molecular cloning, expression, and induction of berberine bridge enzyme, an enzyme essential to the formation of benzophenanthridine alkaloids in the response of plants to pathogenic attack. Proceedings of the National Academy of Sciences of the United States of America, 88, 9969–9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov, Y.L. , Komarova, T.V. , Petrunia, I.V. , Frolova, O.Y. , Pozdyshev, D.V. and Gleba, Y.Y. (2012) Airborne signals from a wounded leaf facilitate viral spreading and induce antibacterial resistance in neighboring plants. PLoS Pathogens, 8, e1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duruflé H., San Clemente H., Balliau T., Zivy M., Dunand C., Jamet E. (2017) Cell wall proteome analysis of Arabidopsis thaliana mature stems. PROTEOMICS, 17, 1600449. [DOI] [PubMed] [Google Scholar]
- Edwards K., Johnstone C., Thompson C. (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research, 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsdorf, T. , Gigli‐Bisceglia, N. , Veerabagu, M. , McKenna, J.F. , Vaahtera, L. , Augstein, F. et al. (2018) The plant cell wall integrity maintenance and immune signaling systems cooperate to control stress responses in Arabidopsis thaliana . Science Signalling, 11, eaao3070. [DOI] [PubMed] [Google Scholar]
- Felix, G. , Duran, J.D. , Volko, S. and Boller, T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. The Plant Journal, 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Feng, W. , Kita, D. , Peaucelle, A. , Cartwright, H.N. , Doan, V. , Duan, Q. et al. (2018) The FERONIA receptor kinase maintains cell‐wall integrity during salt stress through Ca2+ signaling. Current Biology, 28, 666–675.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S., Savatin, D. , Sicilia, F. , Gramegna, G. , Cervone, F. & De Lorenzo, G. (2013) Oligogalacturonides: plant damage‐associated molecular patterns and regulators of growth and development. Frontiers in Plant Science, 4, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo, J. , Silva, M.S. and Figueiredo, A. (2018) Subtilisin‐like proteases in plant defence: the past, the present and beyond. Molecular Plant Pathology, 19, 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletti, R. , Denoux, C. , Gambetta, S. , Dewdney, J. , Ausubel, F.M. , De Lorenzo, G. et al. (2008) The AtrbohD‐mediated oxidative burst elicited by oligogalacturonides in Arabidopsis is dispensable for the activation of defense responses effective against Botrytis cinerea . Plant Physiology, 148, 1695–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geilfus, C.‐M. (2017) The pH of the apoplast: dynamic factor with functional impact under stress. Molecular Plant, 10, 1371–1386. [DOI] [PubMed] [Google Scholar]
- Giovane, A. , Servillo, L. , Balestrieri, C. , Raiola, A. , D’Avino, R. , Tamburrini, M. et al. (2004) Pectin methylesterase inhibitor. Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics, 1696, 245–252. [DOI] [PubMed] [Google Scholar]
- Guo, H. , Nolan, T.M. , Song, G. , Liu, S. , Xie, Z. , Chen, J. et al. (2018) FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana . Current Biology, 28, 3316–3324.e6. [DOI] [PubMed] [Google Scholar]
- Haile, Z.M. , Pilati, S. , Sonego, P. , Malacarne, G. , Vrhovsek, U. , Engelen, K. et al. (2017) Molecular analysis of the early interaction between the grapevine flower and Botrytis cinerea reveals that prompt activation of specific host pathways leads to fungus quiescence. Plant, Cell and Environment, 40, 1409–1428. [DOI] [PubMed] [Google Scholar]
- Hann C.T., Bequette C.J., Dombrowski J.E., Stratmann J.W. (2014) Methanol and ethanol modulate responses to danger‐ and microbe‐associated molecular patterns. Frontiers in Plant Science, 5, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta, M. , Sabat, G. , Stecker, K. , Minkoff, B.B. and Sussman, M.R. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science, 343, 408–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi, T. , Howe, P. , Maier, T.R. , Hussey, R.S. , Mitchum, M.G. , Davis, E.L. et al. (2008) Cellulose binding protein from the parasitic nematode Heterodera schachtii interacts with Arabidopsis pectin methylesterase: cooperative cell wall modification during parasitism. The Plant Cell, 20, 3080–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, W.‐C. , Chang, W.‐H. and Jiang, C.‐M. (1999) Qualitative distinction of carboxyl group distributions in pectins with ruthenium red. Botanical Bulletin of Academia Sinica, 40, 115–119. [Google Scholar]
- Hua, L. , Yong, C. , Zhanquan, Z. , Boqiang, L. , Guozheng, Q. and Shiping, T. (2018) Pathogenic mechanisms and control strategies of Botrytis cinerea causing post‐harvest decay in fruits and vegetables. Food Quality and Safety, 2, 111–119. [Google Scholar]
- Ibar, C. and Orellana, A. (2007) The import of S‐adenosylmethionine into the golgi apparatus is required for the methylation of homogalacturonan. Plant Physiology, 145, 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. , Cameron, R.G. , Williams, M.A.K. and Luzio, G.A. (2018) Structural and functional effects of manipulating the degree of methylesterification in a model homogalacturonan with a pseudo‐random fungal pectin methylesterase followed by a processive methylesterase. Food Hydrocolloids, 77, 879–886. [Google Scholar]
- Kohorn, B.D. , Kohorn, S.L. , Saba, N.J. and Martinez, V.M. (2014) Requirement for pectin methyl esterase and preference for fragmented over native pectins for wall‐associated kinase‐activated, EDS1/PAD4‐dependent stress response in Arabidopsis. Journal of Biological Chemistry, 289, 18978–18986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova T.V., Sheshukova E.V., Dorokhov Y.L. (2014) Cell wall methanol as a signal in plant immunity. Frontiers in Plant Science, 5, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze, G. , Zipfel, C. , Robatzek, S. , Niehaus, K. , Boller, T. and Felix, G. (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. The Plant Cell, 16, 3496–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limberg, G. , Korner, R. , Buchholt, H.C. , Christensen, T.M. , Roepstorff, P. and Mikkelsen, J.D. (2000) Analysis of different de‐esterification mechanisms for pectin by enzymatic fingerprinting using endopectin lyase and endopolygalacturonase II from A. niger . Carbohydrate Research, 327, 293–307. [DOI] [PubMed] [Google Scholar]
- Lin, W. , Tang, W. , Anderson, C.T. and Yang, Z. (2018) FERONIA’s sensing of cell wall pectin activates ROP GTPase signaling in Arabidopsis. bioRxiv, 269647 [Preprint]. [Google Scholar]
- Lionetti, V. (2015) PECTOPLATE: the simultaneous phenotyping of pectin methylesterases, pectinases, and oligogalacturonides in plants during biotic stresses. Frontiers in Plant Science, 6, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti, V. and Metraux, J.P. (2014) Plant cell wall in pathogenesis, parasitism and symbiosis. Frontiers in Plant Science, 5, 612 10.3389/fpls.2014.00612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti, V. , Cervone, F. and Bellincampi, D. (2012) Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. Journal of Plant Physiology, 169, 1623–1630. [DOI] [PubMed] [Google Scholar]
- Lionetti V., Fabri E., De Caroli M., Hansen A.R., Willats W.G.T., et al. (2017) Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis . Plant Physiology, 173, 1844–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti, V. , Raiola, A. , Camardella, L. , Giovane, A. , Obel, N. , Pauly, M. et al. (2007) Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea . Plant Physiology, 143, 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti, V. , Raiola, A. , Cervone, F. and Bellincampi, D. (2014) Transgenic expression of pectin methylesterase inhibitors limits tobamovirus spread in tobacco and Arabidopsis. Molecular Plant Pathology, 15, 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M.J. (2006) Significance of inducible defense‐related proteins in infected plants. Annual Review of Phytopathology, 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Malinovsky, F.G. , Fangel, J.U. and Willats, W.G.T. (2014) The role of the cell wall in plant immunity. Frontiers in Plant Science, 5, 178 10.3389/fpls.2014.00178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteau, S. , Abouna, S. , Lambert, B. and Legendre, L. (2003) Differential regulation by ambient pH of putative virulence factor secretion by the phytopathogenic fungus Botrytis cinerea . FEMS Microbiology Ecology, 43, 359–366. [DOI] [PubMed] [Google Scholar]
- Mélida H., Bacete L., Ruprecht C., Rebaque D., del Hierro I., López G., (2020) Arabinoxylan‐oligosaccharides act as damage associated molecular patterns in plants regulating disease resistance. Frontiers in Plant Science, 11, 1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke, S. and Gasperini, D. (2019) Interplay between plant cell walls and jasmonate production. Plant and Cell Physiology, 60, 2629–2637. [DOI] [PubMed] [Google Scholar]
- Nguyen‐Kim, H. , San Clemente, H. , Balliau, T. , Zivy, M. , Dunand, C. , Albenne, C. et al. (2016) Arabidopsis thaliana root cell wall proteomics: increasing the proteome coverage using a combinatorial peptide ligand library and description of unexpected Hyp in peroxidase amino acid sequences. Proteomics, 16, 491–503. [DOI] [PubMed] [Google Scholar]
- Osorio, S. , Castillejo, C. , Quesada, M.A. , Medina‐Escobar, N. , Brownsey, G.J. , Suau, R. et al. (2008) Partial demethylation of oligogalacturonides by pectin methyl esterase 1 is required for eliciting defence responses in wild strawberry (Fragaria vesca). The Plant Journal, 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Osorio, S. , Bombarely, A. , Giavalisco, P. , Usadel, B. , Stephens, C. , Aragüez, I. et al. (2011) Demethylation of oligogalacturonides by FaPE1 in the fruits of the wild strawberry Fragaria vesca triggers metabolic and transcriptional changes associated with defence and development of the fruit. Journal of Experimental Botany, 62, 2855–2873. [DOI] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A. , Thomma, B.P.H.J. , Buchala, A. , Métraux, J.P. and Broekaert, W.F. (1998) Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. The Plant Cell, 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001) A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Research, 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelko, G. , Lionetti, V. , Fursova, O. , Sundaram, R.M. , Qi, M.S. , Whitham, S.A. et al. (2013) Arabidopsis and Brachypodium distachyon transgenic plants expressing Aspergillus nidulans acetylesterases have decreased degree of polysaccharide acetylation and increased resistance to pathogens. Plant Physiology, 162, 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusky, D. and Yakoby, N. (2003) Pathogenic fungi: leading or led by ambient pH? Molecular Plant Pathology, 4, 509–516. [DOI] [PubMed] [Google Scholar]
- Raiola A., Camardella L., Giovane A., Mattei B., De Lorenzo G., Cervone F., et al. (2004) Two Arabidopsis thaliana genes encode functional pectin methylesterase inhibitors1. FEBS Letters, 557, 199–203. [DOI] [PubMed] [Google Scholar]
- Raiola, A. , Lionetti, V. , Elmaghraby, I. , Immerzeel, P. , Mellerowicz, E.J. , Salvi, G. et al. (2011) Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Molecular Plant‐Microbe Interactions, 24, 432–440. [DOI] [PubMed] [Google Scholar]
- Reem, N. , Pogorelko, G. , Lionetti, V. , Chambers, L. , Held, M.A. , Bellincampi, D. et al. (2016) Decreased polysaccharide feruloylation compromises plant cell wall integrity and increases susceptibility to necrotrophic fungal pathogens. Frontiers in Plant Science, 7, 630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C., Usadel B., Neumetzler L., Hartmann J., Büssis D., Altmann T. (2008) A subtilisin‐like serine protease essential for mucilage release from Arabidopsis seed coats. The Plant Journal, 54, 466–480. [DOI] [PubMed] [Google Scholar]
- Roig‐Oliver M., Rayon C., Roulard R., Fournet F., Bota J., Flexas J. (2020) Reduced photosynthesis in Arabidopsis thaliana atpme17.2 and atpae11.1 mutants is associated to altered cell wall composition. Physiologia Plantarum. 10.1111/ppl.13186. [DOI] [PubMed] [Google Scholar]
- Salamin, K. , Sriranganadane, D. , Léchenne, B. , Jousson, O. and Monod, M. (2010) Secretion of an endogenous subtilisin by Pichia pastoris strains GS115 and KM71. Applied and Environmental Microbiology, 76, 4269–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, A. , Stintzi, A. , Rivas, S. , Serrano, I. , Chichkova, N.V. , Vartapetian, A.B. et al. (2018) From structure to function—A family portrait of plant subtilases. New Phytologist, 218, 901–915. [DOI] [PubMed] [Google Scholar]
- Schuhegger, R. , Nafisi, M. , Mansourova, M. , Petersen, B.L. , Olsen, C.E. , Svatoš, A. et al. (2006) CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiology, 141, 1248–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal, F. , Graff, L. , Surcouf, O. , Marcelo, P. , Rayon, C. , Bouton, S. et al. (2014a) Arabidopsis PECTIN METHYLESTERASE17 is co‐expressed with and processed by SBT3.5, a subtilisin‐like serine protease. Annals of Botany, 114, 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal, F. , Wattier, C. , Rusterucci, C. and Pelloux, J. (2014b) Homogalacturonan‐modifying enzymes: structure, expression, and roles in plants. Journal of Experimental Botany, 65, 5125–5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senechal, F. , Mareck, A. , Marcelo, P. , Lerouge, P. and Pelloux, J. (2015) Arabidopsis PME17 activity can be controlled by pectin methylesterase inhibitor4. Plant Signaling & Behavior, 10, e983351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen, R.J. and Leunissen, J.A.M. (1997) Subtilases: the superfamily of subtilisin‐like serine proteases. Protein Science, 6, 501–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza, C. A. , Li, S. , Lin, A.Z. , Boutrot, F. , Grossmann, G. , Zipfel, C. and Somerville, S.C. (2017) Cellulose‐derived oligomers act as damage‐associated molecular patterns and trigger defense‐like responses. Plant Physiology, 173, 2383–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, D. , Dauphin, A. , Meimoun, P. , Kadono, T. , Nguyen, H.T. , Arbelet‐Bonnin, D. et al. (2018) Methanol induces cytosolic calcium variations, membrane depolarization and ethylene production in Arabidopsis and tobacco. Annals of Botany, 122, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur, A. , Habrylo, O. , Guénin, S. , Miart, F. , Soulié, M.C. , Rihouey, C. et al. (2019) Oligogalacturonide production upon Arabidopsis thaliana–Botrytis cinerea interaction. Proceedings of the National Academy of Sciences of the United States of America, 116, 19743–19752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats, W.G. , Orfila, C. , Limberg, G. , Buchholt, H.C. , van Alebeek, G.J.W. , Voragen, A.G. et al. (2001) Modulation of the degree and pattern of methyl esterification of pectic homogalacturonan in plant cell walls: implications for pectin methyl esterase action, matrix properties and cell adhesion. Journal of Biological Chemistry, 276, 19404–19413. [DOI] [PubMed] [Google Scholar]
- Wolf, S. , Rausch, T. and Greiner, S. (2009) The N‐terminal pro region mediates retention of unprocessed type‐I PME in the Golgi apparatus. The Plant Journal, 58, 361–375. [DOI] [PubMed] [Google Scholar]
- Wormit A., Usadel B. (2018) The multifaceted role of pectin methylesterase inhibitors (PMEIs). International Journal of Molecular Sciences, 19, 2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.‐C., Bulgakov V.P., Jinn T.‐L. (2018) Pectin methylesterases: cell wall remodeling proteins are required for plant response to heat stress. Frontiers in Plant Science, 9, 1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Plant immunity hormones regulate the expression of total PI‐PME activity. PI‐PME activity was analysed at 0, 24, and 48 hr post B. cinerea infection (hpi) or mock‐inoculation in leaves of 6‐week‐old A. thaliana mutants defective in immune signalling or biosynthetic pathways and wild‐type (WT) plants. Values are means ± SD (n = 3). Different letters indicate data sets that are significantly different according to ANOVA followed by Tukey’s test (p < .01)
FIGURE S2 Expression pattern of AtPME17 expression in response to different pathogens. (A) Heatmaps of gene expression. Data were analysed using the Genevestigator Meta‐Analyzer Tools (Zimmermann et al., 2004). The corresponding n‐fold (log2 normalized) expressions in infected plants relative to each corresponding negative control are shown
FIGURE S3 Expression pattern of AtPME17 in response to different elicitors. Data were analysed using the Genevestigator Meta‐Analyzer Tools (Zimmermann et al., 2004). The corresponding n‐fold (log2 normalized) expressions in infected plants relative to each corresponding negative control are shown
FIGURE S4 Expression pattern of AtPME17 in response to different hormones. Data were analyzed using the Genevestigator Meta‐Analyzer Tools (Zimmermann et al., 2004). The corresponding n‐fold (log2 normalized) expressions in infected plants relative to each corresponding negative control are shown
FIGURE S5 Vegetative rosettes of 4‐week‐old pme17 mutants and WT plants
FIGURE S6 Monosaccharide composition of CW of pme17.2, pme17.3, and WT plants. The molar percentages of fucose (Fuc), rhamnose (Rha), arabinose (Ara), galactose (Gal), glucose (Glu), xylose (Xyl), mannose (Man), galacturonic acid (Gal A), and glucuronic acid (Glc A) released after 2 M TFA hydrolysis were quantified by high‐performance anion‐exchange chromatography with a pulsed amperometric detection (HPAEC‐PAD) system. The values are mean ± SD (n = 3). No significant differences were observed for each monosaccharide according to ANOVA followed by Tukey’s test (p > .05). hpi, hours postinoculation
FIGURE S7 Structural domains and amino acid sequence of AtPME17, with the peptides identified by MS underlined. (A) Structural domains of ATPME17 are highlighted with different colours: red, signal peptide; blue, PRO region with putative N‐glycosylation site in bold; orange, processing region with the putative basic processing motive RKLL; violet, PME catalytic domain. Peptides identified after trypsin digestion and analysed by liquid chromatography coupled with tandem mass spectrometry (LC‐MS/MS) are underlined. (B) Data regarding total sequence coverage of mature AtPME17 domain (percentage of the protein sequence covered by identified peptides) and sequence of identified unique peptides
FIGURE S8 AtPME17 purification using CM‐Sepharose chromatography. (A) The fractions were eluted with a gradient of 0 to 1 M NaCl. (B) Radial gel diffusion assay of eluted fractions of peak 1 and peak 2. Peak 2 fractions exhibiting PME activity (fuchsia halo) were pooled, concentrated, and used for a subsequent characterization
FIGURE S9 Native secreted subtilisin produced by P. pastoris. The samples were analysed by SDS‐PAGE with a separation gel of 12% polyacrylamide stained with Coomassie brilliant blue. To detect endoproteolytic activity of culture supernatant, gelatin at a concentration of 0.1% was added to the gel. Lane 1, culture supernatant of P. pastoris grown in the presence of methanol; Lane 2, culture supernatant cleaned on a Sephadex G‐25 PD10 column; MM, molecular mass markers
FIGURE S10 Sequence comparison of PRO regions of B. cinerea‐induced AtPMEs with B. cinerea‐induced AtPMEI10 and AtPMEI11. The biochemically characterized functional AtPMEI1 inhibitor is added for comparison. Highlight homology level: invariant residues are in black, homology level >75% in pink and homology level >50% in turquoise. The secondary structure elements as elucidated in AtPMEI1 crystal structures are indicated below the alignment. Multiple sequence alignment was performed using the DNAman software package (Lynnon Biosoft)
FIGURE S11 MS analysis of 20 and 35 kDa bands isolated by SDS‐PAGE after treatment of His‐PRO‐AtPME17 with SBTs. (A) Structural domains of His‐PRO‐AtPME17are highlighted with different colours: red, pET‐28a‐His tag sequence; blue, PRO region; yellow, processing region; violet, PME catalytic domain. Peptides identified after trypsin digestion and analysed by liquid chromatography coupled with tandem mass spectrometry (LC‐MS/MS) are underlined and peptides identified in the 20 kDa band are shown in italics. (B) Number of peptides and amino acid sequences of identified unique peptides in 20 and 35 kDa bands
FIGURE S12 Fermentation profile of P. pastoris expressing AtPME17. Time profiles of dissolved oxygen (DO), temperature, and optical density (OD) at 600 nm during the production of AtPME17 by P. pastoris in a 1.3‐L bioreactor fermentation. 1, glycerol batch phase; 2, sorbitol and methanol transition phase; 3, methanol fed‐batch phase
TABLE S1 Arabidopsis mutants defective in immune hormone signalling used in this study
TABLE S2 Cis‐acting element related to biotic stresses present in the 1000 bp promoter of AtPME17
TABLE S3 Primers used for semi‐quantitative and quantitative reverse transcript PCR, for genotyping, for PRO‐AtPME17 cloning in the pPICZαA vector, and for His‐PRO‐AtPME17 cloning in pET‐28a(+) vector
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


