Abstract
Single-cell RNA sequencing (scRNA-seq) is a powerful approach for reconstructing cellular differentiation trajectories. However, inferring both the state and direction of differentiation is challenging. Here, we demonstrate a simple, yet robust, determinant of developmental potential—the number of expressed genes per cell—and leverage this measure of transcriptional diversity to develop a computational framework (CytoTRACE) for predicting differentiation states from scRNA-seq data. When applied to diverse tissue types and organisms, CytoTRACE outperformed previous methods and nearly 19,000 annotated gene sets for resolving 52 experimentally determined developmental trajectories. Additionally, it facilitated the identification of quiescent stem cells and revealed genes that contribute to breast tumorigenesis. This study thus establishes a key RNA-based feature of developmental potential and a platform for delineation of cellular hierarchies.
In multicellular organisms, tissues are hierarchically organized into distinct cell types and cellular states with intrinsic differences in function and developmental potential (1). Common methods for studying cellular differentiation hierarchies, such as lineage tracing and functional transplantation assays, have revealed detailed roadmaps of cellular ontogeny at scales ranging from tissues and organs to entire model organisms (2–4). While powerful, these technologies, cannot be applied to human tissues in vivo and generally require prior knowledge of cell type-specific genetic markers (2). These limitations have made it difficult to study the developmental organization of primary human tissues under physiological and pathological conditions.
Single-cell RNA-sequencing (scRNA-seq) has emerged as a promising approach to study cellular differentiation trajectories at high resolution in primary tissue specimens (5). Although a large number of computational methods for predicting lineage trajectories have been described, they generally rely upon (i) a priori knowledge of the starting point (and thus, direction) of the inferred biological process (6, 7) and (ii) the presence of intermediate cell states to reconstruct the trajectory (8, 9). These requirements can be challenging to satisfy in certain contexts such as human cancer development (10). Moreover, with existing in silico approaches, it is difficult to distinguish quiescent (noncycling) adult stem cells that have long-term regenerative potential from more specialized cells. While gene expression-based models can potentially overcome these limitations (e.g., transcriptional entropy (11–13), pluripotency-associated gene sets (14), and machine learning strategies (15)), their utility across diverse developmental systems and single-cell sequencing technologies is still unclear.
Here, we systematically evaluated RNA-based features, including nearly 19,000 annotated gene sets, to identify factors that accurately predict cellular differentiation status independently of tissue type, species, and platform. We then leveraged our findings to develop an unsupervised framework for predicting relative differentiation states from single-cell transcriptomes. We validated our approach through comparison to leading methods and explored its utility for identifying key genes associated with stem cells and differentiation in both healthy tissues and human cancer.
Results
RNA-based correlates of single-cell differentiation states
Our initial goal was to identify robust, RNA-based determinants of developmental potential without the need for a priori knowledge of developmental direction or intermediate cell states marking cell fate transitions. We evaluated ~19,000 potential correlates of cell potency in scRNA-seq data, including all available gene sets in the Molecular Signatures Database (n = 17,810) (16), 896 gene sets covering transcription factor binding sites from ENCODE (17) and ChEA (18), an mRNA-expression-derived stemness index (mRNAsi) (15), and three computational techniques that infer stemness as a measure of transcriptional entropy (StemID, SCENT, SLICE (11–13)). We also explored the utility of “gene counts,” or the number of detectably expressed genes per cell. Although anecdotally observed to correlate with differentiation status in a limited number of settings (alveolar development in mouse and thrombocyte development in zebrafish (19, 20)), the reliability of this association, and whether it reflects a general property of cellular ontogeny, are unknown.
To assess these RNA-based features, we compiled a training cohort consisting of nine gold standard scRNA-seq datasets with experimentally-confirmed differentiation trajectories. These datasets were selected to prioritize commonly used benchmarking datasets from earlier studies and to ensure a broad sampling of developmental states from the mammalian zygote to terminally differentiated cells (table S1). Overall, the training cohort encompassed 3174 single cells spanning 49 phenotypes, six biological systems, and three scRNA-seq platforms (fig. S1A and table S1). To determine performance, we used Spearman correlation to compare each RNA-based feature, averaged by phenotype, against known differentiation states (Fig. 1A). We then averaged the results across the nine training datasets to yield a final score and rank for every feature (table S2).
Fig. 1. RNA-based determinants of developmental potential.
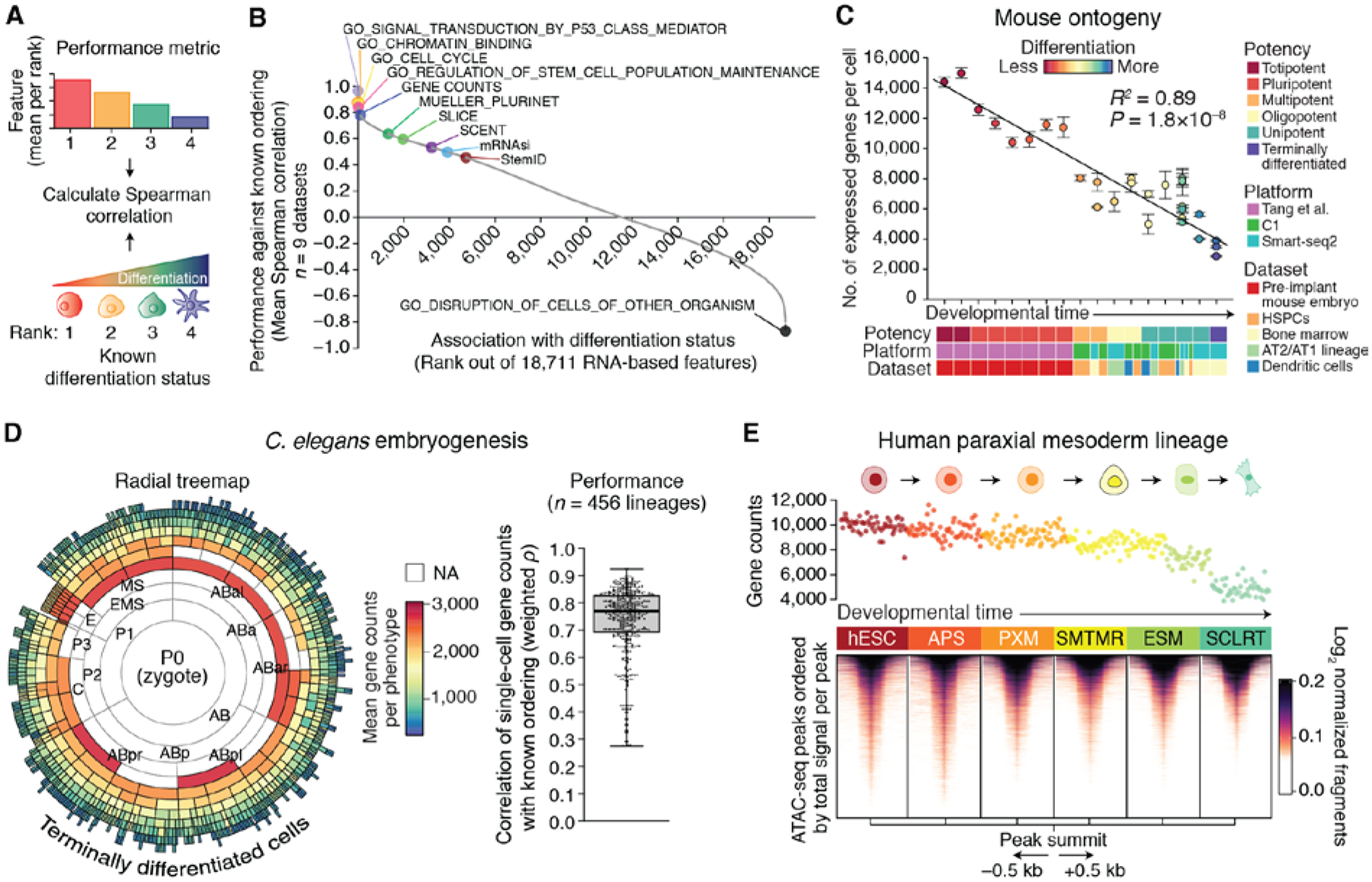
(A and B) In silico screen for correlates of cellular differentiation status in scRNA-seq data. (A) Depiction of the scoring scheme. Each phenotype was assigned a rank on the basis of its known differentiation status (less differentiated = lower rank), and the values of each RNA-based feature (fig. S1A) were mean-aggregated by rank for each dataset (higher value = lower rank). Performance was calculated as the mean Spearman correlation between known and predicted ranks across all nine training datasets (table S1). (B) Performance of all evaluated RNA-based features for predicting differentiation states in the training cohort, ordered by mean Spearman correlations (fig. S1 and table S2). (C) The developmental ordering of 30 mouse cell phenotypes across 17 developmental stages shown as a function of single-cell gene counts (table S3). Data are expressed as means ± 95% confidence intervals. The linear regression line and coefficient of determination (R2) are shown. (D) Performance of gene counts for ordering C. elegans embryogenesis. (Left) Radial tree map showing gene counts for each cell type with available scRNA-seq data from a recent study (48). NA, not available. Embryogenesis originates at the center of the plot [P0 (zygote)] and moves outwards towards terminally differentiated cells, with concentric rings representing sequential cell divisions. (Right) Boxplot showing weighted Spearman correlations between single-cell gene counts and developmental lineages with available transcriptomic data (n = 456). (E) Association between single-cell gene counts and chromatin accessibility in cells from an in vitro differentiation series of purified phenotypes from the human paraxial mesoderm lineage [Mesoderm (C1) dataset; table S1]. (Top) Association of single-cell gene counts with differentiation. Each point represents a cell colored by known phenotype (below). (Bottom) Heat map showing chromatin accessibility profiles for the same phenotypes as above. Peaks are centered by their summit, defined as the base with maximum coverage, shown within a window of 1 kb (±0.5 kb), and ordered top to bottom within each phenotype by decreasing total signal per peak. Cell type abbreviations are defined in materials and methods.
This systematic screen revealed many known and unexpected correlates of differentiation status (Fig. 1B; fig. S1B; table S2). However, one feature in particular showed notable performance – the number of detectably expressed genes per cell (‘gene counts’). Appearing in the top 1% of the ranked list (104 out of 18,711), this data-driven feature compared favorably to well-established stem cell signatures, including cell cycle and pluripotency genes (14, 15), yet also showed evidence of unique biology and broader applicability. For example, regardless of whether we examined cycling cells, non-cycling cells, or published data from the earliest stages of human embryogenesis prior to the upregulation of pluripotency factors (21), gene counts generally decreased with successive stages of differentiation (fig. S2, A and B, left). Pluripotency genes, by contrast, showed an arc-like pattern early in human embryogenesis, characterized by progressively increasing expression until the emergence of embryonic stem cells, followed by decreasing expression (fig. S2B, right).
These findings suggested that gene counts might extend beyond isolated experimental systems to recapitulate the full spectrum of developmental potential. To test this possibility, we compiled, remapped, and normalized a set of in vivo mouse lineage trajectories based on five plate-based scRNA-seq experiments encompassing 5059 cells and 30 phenotypes that together spanned all major potency levels (22) (table S3 and materials and methods). Indeed, when averaged according to known phenotypes and assessed across independent studies, the association between gene counts and differentiation was maintained (R2 = 0.89, P = 1.8 × 10−8) (Fig. 1C and materials and methods). Notably, this relationship was also observed in other organisms, including Caenorhabditis elegans (Fig. 1D) and zebrafish (table S4), suggesting that it is a general feature of cellular ontogeny.
Because the transcriptional output of a cell is associated with its genome-wide chromatin profile, we next tested whether single-cell gene counts is ultimately a surrogate for global chromatin accessibility, which has been shown to decrease with differentiation in certain contexts (23–25). To do this, we compared single-cell gene counts derived from scRNA-seq data with paired bulk ATAC-seq (assay for transposase-accessible chromatin sequencing) profiles obtained from a recent study of in vitro mesodermal differentiation from human embryonic stem cells (hESCs) (26). Indeed, genome-wide chromatin accessibility was observed to progressively decrease with differentiation of hESCs into paraxial mesoderm and lateral mesoderm lineages (Fig. 1E; fig. S3, A and B). We observed strong concordance between the number of accessible peaks and the mean number of detectably expressed genes per phenotype (fig. S3A). Similar patterns were observed for lung adenocarcinoma cells jointly profiled by ATAC-seq and RNA-seq (sci-CAR) and for human hematopoiesis, supporting the generality of this result (fig. S3, C to E).
Development of CytoTRACE
The number of expressed genes per cell generally showed consistent performance with respect to key technical parameters and was generally correlated with mRNA content (figs. S4 to S7 and supplementary text). However, in some datasets, such as that for in vitro differentiation of hESCs into the gastrulation layers (27), the number of expressed genes per cell exhibited considerable intra-phenotypic variation (Fig. 2A, left). Indeed, when evaluated at a single-cell level, 412 predefined gene sets from our in silico screen outperformed gene counts (fig. S8A and table S2). Because scRNA-seq was designed to capture single-cell gene expression, we reasoned that genes whose expression patterns correlate with gene counts might better capture differentiation states. Indeed, by simply averaging the expression levels of genes that were most highly correlated with gene counts in each dataset (materials and methods), the resulting dataset-specific gene counts signature (GCS) became the top-performing measure in the screen, outranking every predefined gene set and computational tool that we assessed (fig. S8, A to D).
Fig. 2. Development and validation of CytoTRACE.
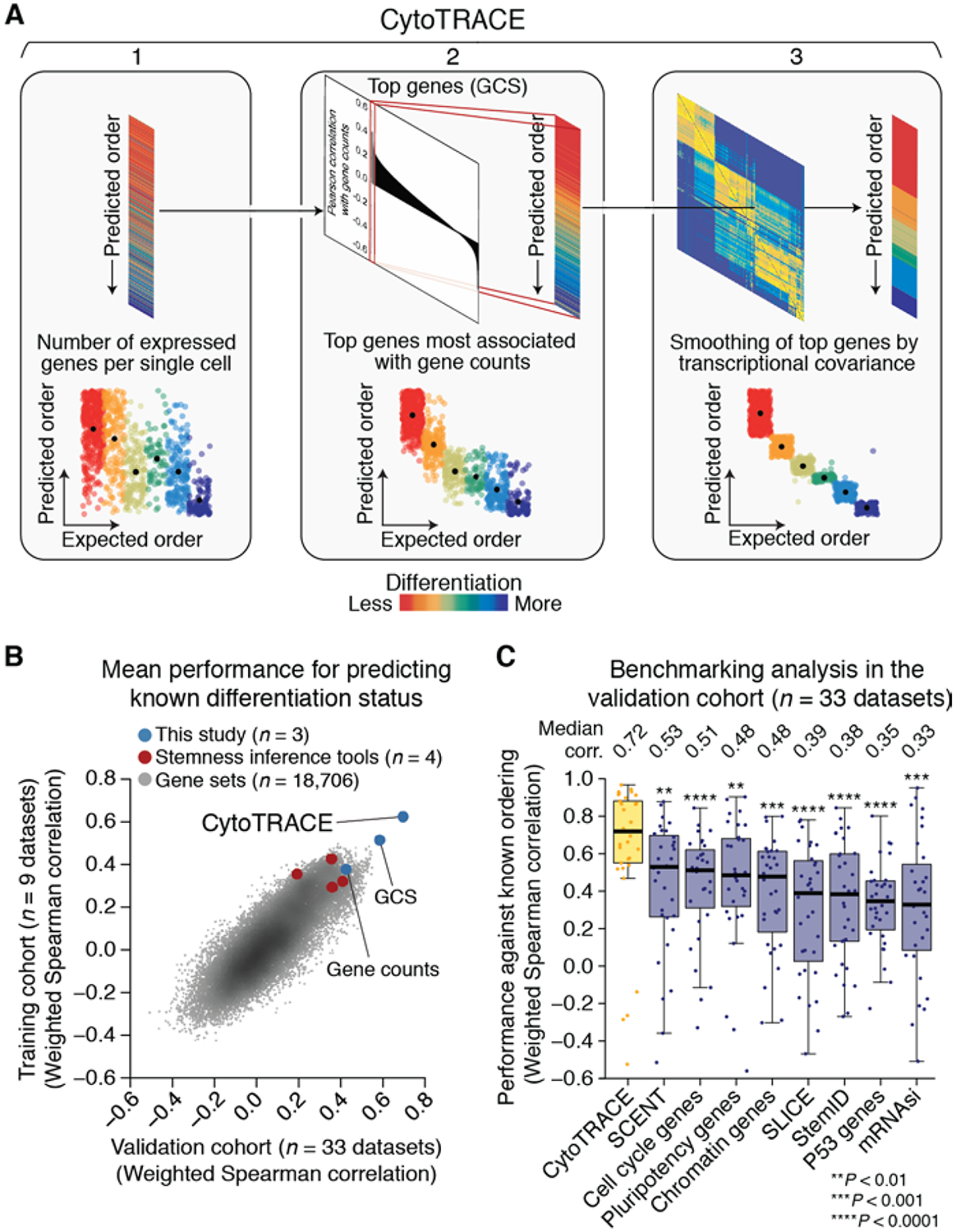
(A) Schematic overview of the CytoTRACE framework applied to the hESC in vitro differentiation (C1) dataset (materials and methods and table S1). (B) Scatterplot comparing the average performance of 18,706 annotated gene sets, four stemness inference methods, gene counts, GCS, and CytoTRACE in the training and validation cohorts (table S2). (C) Boxplots showing the single-cell performance of CytoTRACE against RNA-based features and methods in the validation cohort (n = 33 datasets; table S2). Each point represents the Spearman correlation, weighted by number of cells per phenotype, between predicted and known differentiation states for a given dataset, calculated as described in materials and methods. Statistical significance was assessed by a one-sided paired Wilcoxon signed-rank test against CytoTRACE (table S4).
GCS, like gene counts, is inherently insensitive to dropout events, is agnostic to prior knowledge of developmentally regulated genes, and is not solely attributable to multi-lineage priming (28) (fig. S9 and supplementary text) or a known molecular signature (e.g., pluripotency) (fig. S2B and table S5). Despite these characteristics, GCS was still moderately noisy in some datasets (e.g., Fig. 2A, center and fig. S8C). We therefore implemented a two-step procedure to directly smooth GCS on the basis of transcriptional covariance among single cells (Fig. 2A, right, and materials and methods). The resulting method, which we call CytoTRACE [for cellular (Cyto) Trajectory Reconstruction Analysis using gene Counts and Expression; https://cytotrace.stanford.edu], outperformed GCS and the other RNA-based features that we evaluated (fig. S8 and table S2).
Performance evaluation across tissues, species, and platforms
To validate our findings, we assembled an expanded compendium of 33 additional scRNA-seq datasets from 26 studies (fig. S10A, table S1, and materials and methods). These datasets represent diverse developmental and differentiation processes and consist of 141,267 single cells spanning 266 phenotypes, nine biological systems, five species [including two whole organisms (29, 30)], and nine scRNA-seq platforms (three droplet-based and six plate-based protocols, ranging from an average of ~10,000 unique molecular identifiers to ~1 million reads per cell, respectively; fig. S5A).
When assessed at the single-cell level, CytoTRACE outperformed all evaluated RNA-based features in the validation cohort (Fig. 2B), achieving a substantial gain in performance over the second highest-ranking approach (median rho = 0.72 versus 0.53 for the second-highest-ranking approach, P = 0.001) (Fig. 2C; fig. S10B; and table S2 and S4). Similar improvements were observed across many complex systems, including bone marrow differentiation (fig. S10C). In addition, CytoTRACE results were positively correlated with the direction of differentiation in 88% of datasets (P = 7 × 10–7, binomial test). These results were consistent with our findings for the training cohort (Fig. 2B and fig. S10D) and were robust to the use of smoothing (fig. S11). Moreover, no significant biases in performance were observed in relation to tissue type, species, the number of cells analyzed, time-series experiments versus snapshots of developmental states, or plate-based versus droplet-based technologies (fig. S12).
To further evaluate CytoTRACE, we compared it with RNA velocity, a kinetic model that can predict future cell states but is limited to scRNA-seq data with continuous fate transitions (8). To analyze RNA velocity’s output, which consists of an individualized prediction for every cell (fig. S13), we identified all pairs of current and future cell states spanning a known shift in developmental potential (in the order of less to more, or vice versa). We then scored each predicted trajectory against known differentiation states on five datasets with continuous developmental processes (fig. S13B and materials and methods). To permit a fair comparison, CytoTRACE was evaluated on the same cells. Although both methods performed similarly on an embryonic chromaffin dataset from the RNA velocity study (8), CytoTRACE achieved higher accuracy overall (median of 74% versus 54%, respectively; fig. S13C). This was likely due to the short mRNA half-lives and developmental time scales assumed for the RNA velocity model (8) (supplementary text).
Having assessed performance on individual datasets, we next asked whether CytoTRACE could be applied across independent scRNA-seq datasets unified by batch correction. To address this, we leveraged mutual nearest neighbor and Gaussian kernel normalization techniques from Scanorama (31) (materials and methods). We then merged several datasets with this approach. Regardless of whether we integrated datasets profiled on different scRNA-seq platforms (Fig. 3A) or datasets containing developmentally distinct cell types (fig. S14), single-cell orderings predicted by CytoTRACE were accurate.
Fig. 3. Characterization of developmental hierarchies and quiescent stem cells using CytoTRACE.
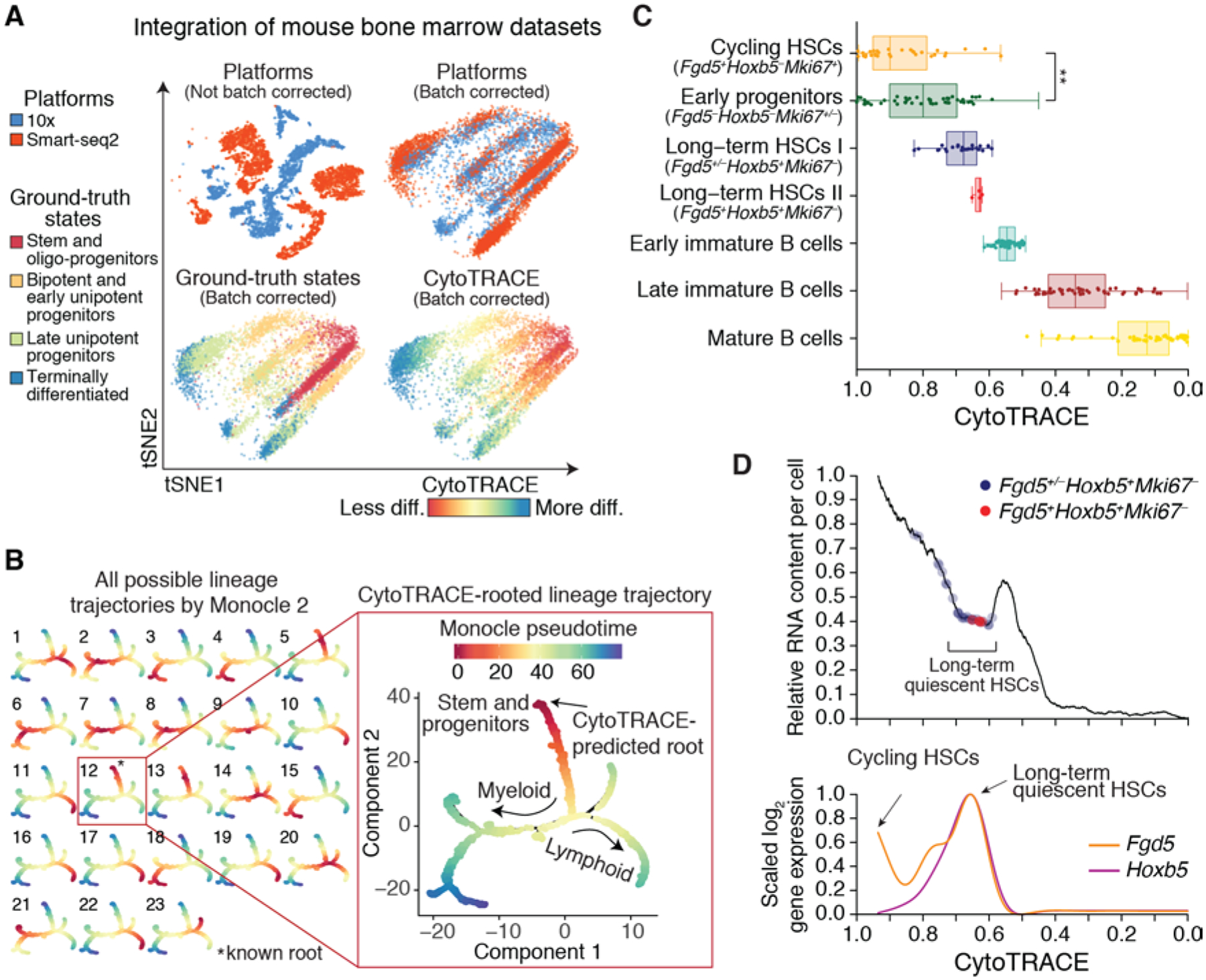
(A) Impact of batch correction (materials and methods) on two datasets of mouse bone marrow differentiation: Bone Marrow (10x) and Bone Marrow (Smart-seq2) (table S1). diff, differentiated. (B) Combined application of CytoTRACE and Monocle 2 to mouse bone marrow differentiation [Bone marrow (Smart-seq2) dataset] (table S1). (Left) Multi-lineage tree inferred by Monocle 2 showing all 23 possible pseudotimes when the root is unknown. (Right) Automatic selection of the correct root by CytoTRACE. (C and D) Prioritization of quiescent and cycling HSCs in index-sorted scRNA-seq data of mouse hematopoiesis [Bone Marrow (Smart-seq2) dataset] (table S1). All plots are identically ordered by CytoTRACE. (C) Boxplots showing CytoTRACE values for candidate cycling HSCs (n = 31), long-term or quiescent HSCs (n = 30), early immature B cells (n = 285), late immature B cells (n = 863), and mature B cells (n = 700). HSCs, long-term or quiescent HSCs, and proliferating cells were defined on the basis of expression of Fgd5 (49), Hoxb5 (35), and Mki67, respectively. Although boxplots represent all analyzed cells, a maximum of 50 cells per phenotype are displayed as points for clarity. Statistical significance was assessed by a two-sided Wilcoxon signed-rank test. **P = 0.003. (D) Top: RNA content per cell, shown as a function of CytoTRACE and displayed as the moving average of 200 cells. Bottom: Expression of Fgd5 and Hoxb5 displayed as a smoothing spline over the moving average of 200 cells. Data from monocytic and granulocytic lineages are consistent with the above results.
Stem-cell-related genes and hierarchies
Given the ability of CytoTRACE to recover the direction of differentiation in nearly every evaluated dataset (supplementary text), we next explored its potential to identify markers of immature phenotypes without prior knowledge. By rank-ordering genes on the basis of their correlation with CytoTRACE, markers of immature cells were readily prioritized in 86% of benchmarking datasets (fig. S15A). These included well-established stem and progenitor markers, such as Kit and Stmn1 in mouse bone marrow (32) and Axin2 and Lgr5 in mouse intestinal crypts (33), underscoring the utility of CytoTRACE for the de novo discovery of developmentally regulated genes (fig. S15B and table S6).
Lineage relationships and their associated genes can also be determined by dedicated branch detection tools, such as Monocle 2; however, these approaches do not predict the starting point of the biological process. For example, when applied to 4,442 bone marrow cells, Monocle 2 identified 23 possible “roots” from which to calculate pseudotime values (Fig. 3B, left). By contrast, CytoTRACE readily identified the correct root without user input (Fig. 3B, right, and fig. S16, A and B). Integration of these methods facilitated automatic identification of lineage-specific regulatory factors and marker genes during granulocyte, monocyte, and B cell differentiation (fig. S16C). Similar results were obtained on mouse intestinal cells (fig. S16, D to F). Notably, other methods also showed strong performance when oriented by CytoTRACE (fig. S16G; table S4).
We next asked whether CytoTRACE could distinguish cycling and long-term or quiescent stem cells from their downstream progenitors (34). As these populations have been well-characterized in the bone marrow (3), we investigated this question in the mouse hematopoietic system. Although both cycling and quiescent hematopoietic stem cell (HSC) subpopulations (34) were correctly predicted to be less differentiated, only proliferative HSCs were significantly ranked above early progenitors (Fig. 3C). This result was not unexpected, however, because quiescent cells have reduced metabolic activity and low RNA content (1). By devising a simple approach to visualize inferred RNA content as a function of CytoTRACE (Fig. 3D, top), we observed a distinct valley in RNA abundance that coincided with elevated expression of Hoxb5, a marker of long-term or quiescent HSCs (35) (Fig. 3D, bottom). Since these cells could not be identified by gene counts or RNA content alone, this analysis confirmed the utility of CytoTRACE and demonstrates an approach for elucidating tissue-specific stem cells from scRNA-seq data.
Application to neoplastic disease
Increasing evidence suggests that human breast tumors contain less differentiated cells that are resistant to therapy and associated with the development of relapse and metastasis (10, 36). Subpopulations of tumor cells within the luminal progenitor (LP) epithelium are thought to give rise to aggressive basal-like breast cancers, such as triple-negative breast cancer (TNBC) (37), and possibly also to estrogen receptor positive (ER+) breast cancers (38). However, the differentiation states and tumor-initiating properties of LP subsets remain incompletely understood.
To determine whether CytoTRACE can provide insights into immature LP cells and their associated genes in breast cancer, we performed scRNA-seq profiling of breast tumor epithelial cells and adjacent normal epithelial cells from eight patients with triple-negative (n = 2) or ER+ (n = 6) breast cancer. Using a Smart-seq2 protocol combined with fluorescence-activated cell sorting (FACS), we index-sorted and sequenced cells from three major human epithelial subpopulations: basal (CD49fhighEPCAMmed-low), luminal progenitor (CD49fhighEPCAMhigh), and mature luminal (ML) subpopulations (CD49flowEPCAMhigh) (fig. S17A; table S7). After removing low quality cells and applying principal component analysis to visualize the data, we confirmed three well-separated clusters of basal, LP, and ML cells, each with characteristic expression patterns of previously described lineage markers (Fig. 4A and fig. S17B). No obvious clustering was observed for tumor versus normal cell differences or by patient (Fig. 4A; fig. S18A).
Fig. 4. Identification of immature cell markers in normal and malignant human breast LPs using CytoTRACE.
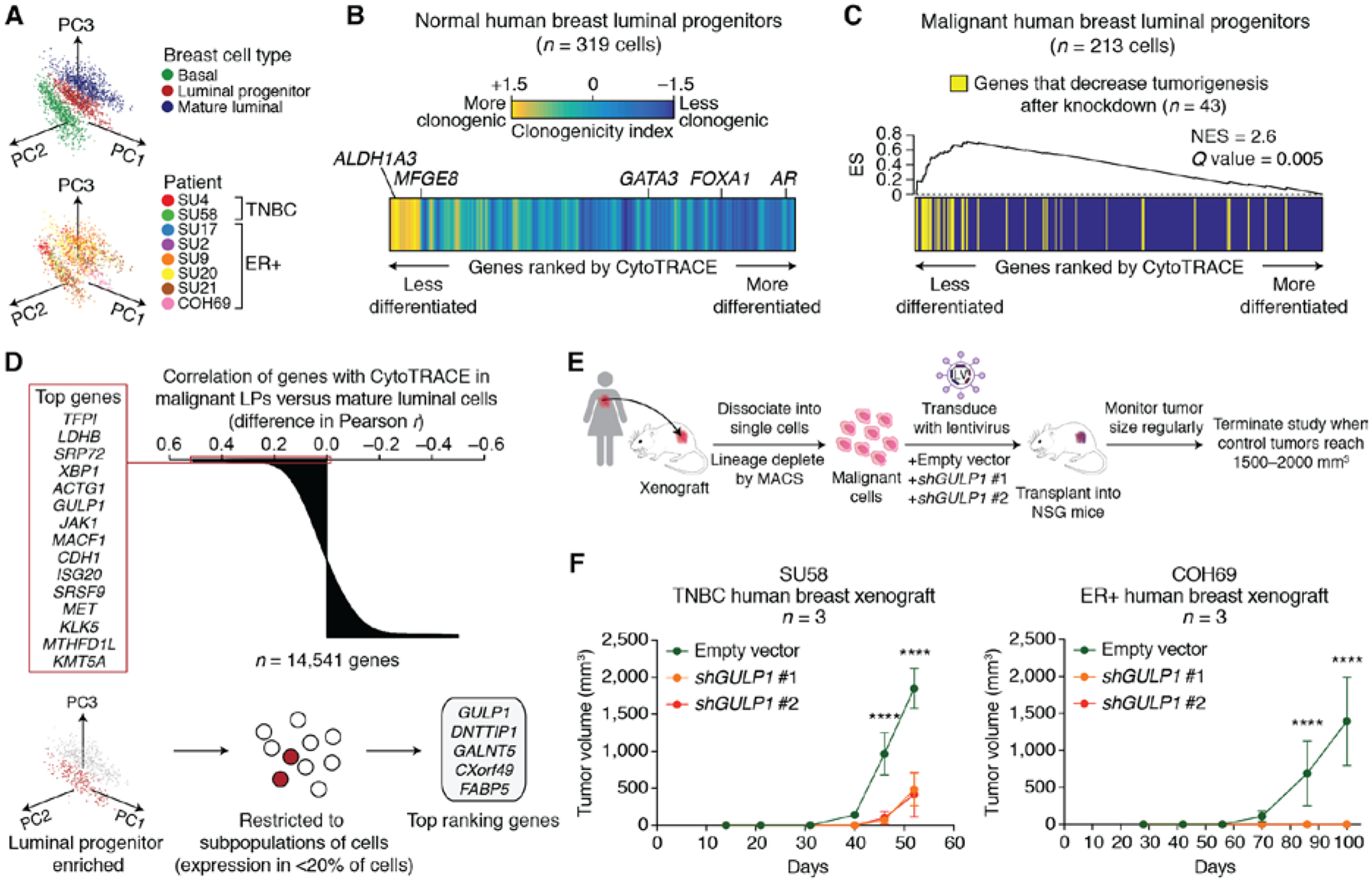
(A) Principal component analysis of scRNA-seq profiles from 1902 human breast epithelial cells, colored according to subpopulations (top) and patient (bottom). (B) Heat map showing genes from adjacent normal LPs rank-ordered by their Pearson correlation with CytoTRACE and colored according to a clonogenicity index, defined as the log2 fold change in expression between highly and lowly clonogenic LPs from normal human breast (39) (materials and methods). The clonogenicity index is displayed as a moving average of 200 genes. Key genes associated with less (ALDH1A3, MFGE8) and more (GATA3, FOXA1, AR) differentiated normal LPs are indicated. (C) Enrichment of genes associated with human breast tumorigenesis [RNAi dropout viability screen (41)] within a ranked list of genes expressed by malignant LPs, rank-ordered by their Pearson correlation with CytoTRACE. Enrichment was calculated with preranked gene set enrichment analysis. NES, normalized enrichment score; ES, enrichment score. (D) Identification of candidate tumorigenic genes associated with immature malignant human LPs. (Top) Genes rank-ordered by the difference in their Pearson correlations with CytoTRACE in malignant LPs versus malignant mature luminal cells. The top 15 genes that are predicted to be specifically associated with less differentiated LPs are indicated on the left. (Bottom) Schema for the identification of genes that are ranked as above, but that are also more highly expressed in malignant LPs than MLs (log2 fold change > 0; Benjamini-Hochberg adjusted P < 0.05, unpaired two-sided t-test) and that are expressed by a subpopulation of LPs (<20% of cells). The top 5 filtered genes are shown (right). (E) Schema for shRNA knockdown of GULP1 in a human breast cancer xenograft model. (F) Growth of human breast cancer xenografts from two patients, one with TNBC (left) and one with ER+ luminal-type cancer (right), after lentiviral transduction with empty vector or shRNA targeting GULP1. Tumor volumes after knockdown with shGULP1 #1 (orange) and shGULP1 #2 (red) were indistinguishable in COH69 xenografts (right). Data are expressed as means ± SD (n = 3 mice). Statistical significance was assessed by a two-way ANOVA. **** P < 0.0001.
To validate the ability of CytoTRACE to define LP differentiation states, we started by rank-ordering genes expressed in adjacent normal LPs by their Pearson correlation with CytoTRACE. We found that previously described marker genes of less-differentiated normal LPs [ALDH1A3 and MFGE8) (39)] and more-differentiated normal LPs [GATA3, FOXA1, and AR (39, 40)] were successfully enriched by this approach (Fig. 4B). Moreover, genes that were up-regulated in highly clonogenic normal LPs (39) were skewed toward genes predicted to mark less-differentiated cells (Fig. 4B).
We next sought to identify LP genes associated with tumorigenesis. We first ordered genes expressed in malignant LPs by their Pearson correlation with CytoTRACE. In this rank-ordered list, we observed a significant enrichment of genes whose knockdown by RNA interference (RNAi) led to decreased viability of tumor cells in patient-derived xenograft (PDX) models of TNBC (41) (Q = 0.002, gene set enrichment analysis) (Fig. 4C; fig. S18, B and C, and table S8). Moreover, when we applied CytoTRACE to prioritize genes in tumor LPs compared to tumor MLs, the latter of which are developmentally downstream of LPs in normal breast (39), the top 15 genes included known members of tumorigenic pathways in breast cancer [e.g., MET and JAK1 (42, 43)], as well as unknown candidates (e.g., GULP1) (Fig. 4D, top). We focused on genes that were (i) more highly expressed in tumor LPs than MLs and (ii) expressed in a subpopulation of tumor LPs (<20% of cells) (Fig. 4D, bottom). After applying this filter, GULP1 emerged as the top candidate gene (Fig. 4D, bottom right, and fig. S18C).
GULP1 is an engulfment adaptor protein (44) and its murine homolog is a specific marker of mouse HSCs, suggesting a conserved role of this gene in other immature cell states (fig. S19A). We measured the effect of GULP1 knockdown on the proliferation of metastatic TNBC cell lines, MDA-MB-231 and MDA-MB-468 (fig. S19, B to E). We found that GULP1 knockdown reduced proliferation of both cell lines as measured by a colorimetric assay for metabolic activity (fig. S19E). In addition, GULP1 knockdown in PDXs (n=2) either inhibited tumor growth (TNBC sample) or fully abrogated tumor growth (ER+ sample) (Fig. 4, E and F). These data suggest a possible role for GULP1 in human breast cancer tumorigenesis.
Discussion
Efforts to characterize single-cell transcriptomes in diverse tissues, organs, and whole organisms have underscored the need for RNA-based determinants of developmental potential. In our analysis of RNA-based features across numerous developmental processes, we observed that the number of detectably expressed genes per cell powerfully associates with cellular differentiation status.
Although previous studies have demonstrated a global reduction in chromatin accessibility and/or plasticity during lineage commitment in specific developmental settings [e.g., embryonic stem cells, intestinal stem cells, and neural stem cells (23–25)], this work extends the scope of this result and quantitatively links it to single-cell gene counts. Moreover, as has been previously shown (45), our results indicate that variability in gene counts between phenotypically identical single cells is not exclusively due to drop-out events, but also due to differential sampling of the transcriptome (fig. S4). Our results are therefore consistent with a model in which less mature cells maintain looser chromatin to permit wider sampling of the transcriptome, while more differentiated cells generally restrict chromatin accessibility and transcriptional diversity as they specialize (Fig. 1E and fig. S3) (46). Theoretically, this model can be represented by “attractor states” within a genome-wide gene regulatory network (47). In this context, differentiating cells descend toward stable regions of the network (attractor states), characterized by restricted gene expression, whereas metastable cells broadly sample the network, maintaining higher differentiation potential (47). Future investigations of this phenomenon, and its relationship to single-cell gene counts, may reveal new mechanisms of stem cell regulation and lineage commitment. However, further studies will be needed to confirm the validity of this model across additional tissue compartments, developmental time points, and phenotypic states.
In summary, we have shown that the number of expressed genes per cell is a hallmark of developmental potential. By exploiting this property of scRNA-seq data, we developed a general framework for resolving single-cell differentiation hierarchies. We envision that our approach will complement existing scRNA-seq analysis strategies, with implications for the identification of immature cells and their developmental trajectories in complex tissues throughout multicellular life.
Supplementary Material
Acknowledgments:
We thank A. Chaudhuri, M. Vahid, T. Raveh, A. Gentles, and A. Alizadeh for critical feedback on the manuscript. We are grateful to S. Bobo for assistance with patient specimen acquisition, R. Sinha and C.K.F. Chan for provision of data, C.L. Liu for assistance with the website, P. Lovelace and S. Weber for assistance with FACS, and S. Sim for assistance with scRNA-seq libraries.
Funding: This work was supported by grants from the National Cancer Institute (A.M.N., R00CA187192-03; M.F.C., 5R01CA100225-09; G.S.G., PHS Grant Number CA09302), the Stinehart-Reed foundation (A.M.N.), the Stanford Bio-X Interdisciplinary Initiatives Seed Grants Program (IIP) (A.M.N., M.F.C.), the Virginia and D.K. Ludwig Fund for Cancer Research (A.M.N., M.F.C), the U.S. Department of Defense (M.F.C., W81XWH-11-1-0287 and W81XWH-13-1-0281; S.S.S., W81XWH-12-1-0020), a National Science Foundation Graduate Research Fellowship (DGE-1656518 to M.J.B.), Stanford Bio-X Bowes Graduate Student Fellowship (G.S.G.), and the Stanford Medical Science Training Program (G.S.G.).
Footnotes
Competing interests: G.S.G., S.S.S, M.F.C., and A.M.N. are inventors on a provisional patent application filed by Stanford University (US 62/852,231) that covers methods described in this work.
Data and materials availability: Details of publicly available datasets are provided in methods and table S1. Single-cell RNA-seq expression data generated in this study are available at https://cytotrace.stanford.edu and have been deposited with the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession code GSE138536.
Supplementary Materials:
Materials and Methods
Supplementary Text
Figures S1–S19
Tables S1–S8
References (50–128)
REFERENCES AND NOTES
- 1.Visvader JE, Clevers H, Tissue-specific designs of stem cell hierarchies. Nat Cell Biol 18, 349–355 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Kretzschmar K, Watt FM, Lineage tracing. Cell 148, 33–45 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Seita J, Weissman IL, Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med 2, 640–653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sulston JE, Schierenberg E, White JG, Thomson JN, The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol 100, 64–119 (1983). [DOI] [PubMed] [Google Scholar]
- 5.Kester L, van Oudenaarden A, Single-Cell Transcriptomics Meets Lineage Tracing. Cell Stem Cell 23, 166–179 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Saelens W, Cannoodt R, Todorov H, Saeys Y, A comparison of single-cell trajectory inference methods. Nat Biotechnol 37, 547–554 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Qiu X et al. , Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.La Manno G et al. , RNA velocity of single cells. Nature 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin J et al. , Single-Cell RNA-Seq with Waterfall Reveals Molecular Cascades underlying Adult Neurogenesis. Cell Stem Cell 17, 360–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke MF, Clinical and Therapeutic Implications of Cancer Stem Cells. N Engl J Med 380, 2237–2245 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Grun D et al. , De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell 19, 266–277 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teschendorff AE, Enver T, Single-cell entropy for accurate estimation of differentiation potency from a cell’s transcriptome. Nat Commun 8, 15599 (2017). doi: 10.1038/ncomms15599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo M, Bao EL, Wagner M, Whitsett JA, Xu Y, SLICE: determining cell differentiation and lineage based on single cell entropy. Nucleic Acids Res 45, e54 (2017). doi: 10.1093/nar/gkw1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller FJ et al. , Regulatory networks define phenotypic classes of human stem cell lines. Nature 455, 401–405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malta TM et al. , Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 173, 338–354 e315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liberzon A et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerstein MB et al. , Architecture of the human regulatory network derived from ENCODE data. Nature 489, 91–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lachmann A et al. , ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics 26, 2438–2444 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Treutlein B et al. , Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macaulay IC et al. , Single-Cell RNA-Sequencing Reveals a Continuous Spectrum of Differentiation in Hematopoietic Cells. Cell Rep 14, 966–977 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan L et al. , Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20, 1131–1139 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Jaenisch R, Young R, Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aughey GN, Estacio Gomez A, Thomson J, Yin H, Southall TD, CATaDa reveals global remodelling of chromatin accessibility during stem cell differentiation in vivo. Elife 7, e32341 (2018). doi: 10.7554/eLife.32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez NC et al. , Widespread Chromatin Accessibility at Repetitive Elements Links Stem Cells with Human Cancer. Cell Rep 17, 1607–1620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pekowska A et al. , Gain of CTCF-Anchored Chromatin Loops Marks the Exit from Naive Pluripotency. Cell Syst 7, 482–495 e410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh KM et al. , Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 166, 451–467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu LF et al. , Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biol 17, 173 (2016). doi: 10.1186/s13059-016-1033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang S, Guo YP, May G, Enver T, Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol 305, 695–713 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Plass M et al. , Cell type atlas and lineage tree of a whole complex animal by single-cell transcriptomics. Science 360, eaaq1723 (2018). doi: 10.1126/science.aaq1723. [DOI] [PubMed] [Google Scholar]
- 30.Farrell JA et al. , Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018). doi: 10.1126/science.aar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hie B, Bryson B, Berger B, Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat Biotechnol 37, 685–691 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabula Muris C et al. , Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haber AL et al. , A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson A et al. , Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Chen JY et al. , Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature 530, 223–227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pattabiraman DR et al. , Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science 351, aad3680 (2016). doi: 10.1126/science.aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim E et al. , Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med 15, 907–913 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Polyak K, Breast cancer: origins and evolution. J Clin Invest 117, 3155–3163 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shehata M et al. , Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast Cancer Res 14, R134 (2012). doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z, GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell 127, 1041–1055 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsieh RW et al. , CDK19 is a Regulator of Triple-Negative Breast Cancer Growth. bioRxiv, 317776 (2018). doi: 10.1101/317776. [DOI] [Google Scholar]
- 42.Graveel CR, Tolbert D, Vande Woude GF, MET: a critical player in tumorigenesis and therapeutic target. Cold Spring Harb Perspect Biol 5, a009209 (2013). doi: 10.1101/cshperspect.a009209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barkauskas CE et al. , Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu QA, Hengartner MO, Human CED-6 encodes a functional homologue of the Caenorhabditis elegans engulfment protein CED-6. Curr Biol 9, 1347–1350 (1999). [DOI] [PubMed] [Google Scholar]
- 45.Marinov GK et al. , From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res 24, 496–510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melcer S, Meshorer E, Chromatin plasticity in pluripotent cells. Essays In Biochemistry 48, 245–262 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Huang S, Eichler G, Bar-Yam Y, Ingber DE, Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys Rev Lett 94, 128701 (2005). doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- 48.Packer JS et al. , A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science, eaax1971 (2019). doi: 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gazit R et al. , Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med 211, 1315–1331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


