Abstract
Mitochondrial dysfunction, and consequently altered aerobic energy metabolism, is associated with numerous retinal diseases, including photoreceptor degeneration and diabetic retinopathy. Here, we describe a detailed protocol to directly measure oxygen consumption in the intact retina ex vivo using microplate-based fluorescence technology. We have used this method to assess preferred energy substrate for retinal tissue and suggested its application for investigating mechanisms of retinal disease.
Keywords: Aerobic respiration, Energy metabolism, Photoreceptor, Retinal degeneration, Mouse models
1. Introduction
Mitochondria are vital organelles that produce energy in the form of adenosine triphosphate (ATP) by oxidative phosphorylation (OXPHOS) in addition to regulating cellular metabolism, homeostasis, and apoptosis by maintaining a delicate balance between oxygen supply and consumption [1]. The density of mitochondria varies among cell types depending upon the metabolic need, and highly metabolically active cells (such as retinal photoreceptors) harbor a large number of mitochondria [1]. During aerobic respiration, molecular oxygen acts as the final electron acceptor at the end of the electron transport chain (ETC); thus, 90% of the cellular oxygen is metabolized in the mitochondria. However, this process also makes mitochondria a primary site for the generation of reactive oxygen species (ROS) [2]. Partial reduction of molecular oxygen (O2) produces two main species of ROS: the superoxide radical and hydrogen peroxide. The imbalance between generation and removal of the ROS may cause oxidative stress [3, 4].
The retina, with its high metabolic demand, has undergone spectacular adaptations during evolution to make its energy metabolism highly efficient and exclusive [5]. At the same time, unique metabolic demands make the retina highly vulnerable to alterations in energy metabolic pathways, including mitochondrial OXPHOS. Alterations in mitochondrial respiration and OXPHOS have been associated with neurodegeneration [6–9]. Mitochondrial dysfunction appears to be a convergent node in a number of retinal and macular degenerative diseases including retinitis pigmentosa [3, 10], age-related macular degeneration (AMD) [11] and diabetic retinopathy [12]. The retina is highly susceptible to oxidative stress [13] and becomes more prone to oxidation of nuclear and mitochondrial DNA, protein and lipid peroxidation [11]. Genetic association has been implicated between AMD and oxidative stress [14, 15]. Therefore, deeper insights into photoreceptor bioenergetics would provide a better understanding of mitochondrial defects in retinal degenerative diseases and reveal mechanisms by which genetic and environmental factors synergistically cause death of photoreceptors. Since the retina harbors multiple cell types, it is imperative to analyze mitochondrial function in context of whole retinal tissue with intact circuitry of different cells to understand how mitochondrial dysfunction correlates to retinal degeneration.
Mitochondrial function or energy metabolism can be assessed by quantification of mitochondrial respiration through measurement of oxygen consumption rate (OCR) [16]. For decades, the Clark electrode and other oxygen microelectrodes were adopted to measure mitochondrial oxygen consumption [17–20]. Experiments using double-barreled oxygen microelectrodes to measure oxygen tension and local voltages at differential tissue depths in cat retina indicated that light reduces oxygen consumption by almost 50% in the outer retina compared to the dark state [19]. This finding is corroborated by measurements in the intact rat retina using Clarktype O2 electrodes [20]. Furthermore, photoreceptors reportedly consume the highest amount of O2 in rat retina [21]. Another study using the same electrodes revealed that about half of all energy is used by NaK-A TPase pump for transport of Na+ in rabbit retina during the dark periods [18]. However, O2 electrodes have serious limitations because of the consumption of O2 during the experiment and the requirement of large sample volumes (usually in mL range) with continuous stirring that may lead to the loss of cellular/tissue context. In addition, only one retina can be used at a time for recording, thereby not permitting a concurrent high - throughput assessment of various experimental conditions [22].
A reliable technique to measure mitochondrial energy metabolism of acutely - derived mouse retina ex-vivo was recently developed in our laboratory [22]. This method is established on microplate-based, fluorescence technique that requires an extracellular flux (XF) analyzer (Seahorse XF Analyzer; Seahorse Bioscience, Billerica, MA, USA), and permits real - time comparative measurements of respiration of different samples in small volumes while eliminating the need for sample in suspension and for continuous stirring. This technology allows OCR (indicating respiration) and extracellular acidification rate (ECAR, reflecting glycolysis) measurements. Since tissues with high metabolic rates (e.g., heart and skeletal muscle) can use other substrates besides glucose [23, 24], we wanted to establish the preferred metabolic fuels in the retina [25]. The presence of normal vision in individuals with GLUT1 (the major glucose transporter) deficiency suggests that the retina might be able to utilize alternative energy substrates [26]. Furthermore, a mixture of several substrates including pyruvate, malate, glutamine, and leucine maintain OCR even in absence of glucose in mouse retina [27].
The detailed protocol being presented here is essentially based on Kooragayala et al. [22] with minor modifications and should allow direct measurements of changes in oxygen consumption in the intact retina ex- vivo. Here, we show the effect of different substrates on the retinal OCR. This protocol can also be used to evaluate other parameters of mitochondrial function, including ATP production and proton leak using ATP Synthase inhibitor, oligomycin.
2. Materials
2.1. Reagents
Ames’ medium (Sigma-Aldrich) supplemented with 12 mM NaCl buffered with 10 mM HEPES. Adjust to pH 7.35 ± 0.05 with NaOH and osmolarity to 285 ± 5 milliosmoles (mOsm) by adding water.
Basic salt solution (140 mM NaCl, 3.1 mM KCl, 1.24 mM MgSO4, 2 mM CaCl2, 10 mM HEPES, pH 7.35).
6 mM D-glucose, 0.15 and 1 mM pyruvate, 2 mM β-hydroxybutyrate (Sigma-Aldrich). After adding substrates to basic salt solution, adjust the osmolarity to 285 ± 5 mOsm (the final concentration of salts may therefore vary slightly).
Homogenization buffer (280 mM sucrose, 10 mM HEPES, 1 mM EDTA, and 1 mM EGTA). Adjust to pH 7.1 with KOH.
Cell-Tak (Corning Life Sciences).
0.1 M sodium bicarbonate solution (Sigma-Aldrich).
1 M NaOH solution (Sigma-Aldrich).
1% dodecyl Maltoside (Sigma-Aldrich).
0.1 mM potassium ferricyanide (Sigma-Aldrich).
0.5 mM potassium cyanide (Sigma-Aldrich).
10 mM sodium ascorbate (pH 7.4) (Sigma-Aldrich).
5 μM BAM15 (TimTec).
1 μM rotenone (Sigma-Aldrich).
2.2. Equipment
Seahorse XF24 Bioanalyzer (Agilent).
Seahorse XF24 Islet Capture FluxPak (Agilent).
Dissection microscope.
Dissecting forceps (Blunt and pointed) (Fine Science Tools).
Microscissors (Fine Science Tools).
Fine paintbrush.
1 mm biopsy puncher (Miltex).
100 mm glass petri dish.
CO2 asphyxiation chamber.
Advanced Model 3320 microosmometer (Advanced Instruments).
Spectrophotometer (Shimadzu 2700 UV-Vis; Shimadzu Corp., Kyoto, Japan).
2.3. Animals
1–3-month-old mice. We used Nrlp-GFP mice on C57BL/6 background.
3. Methods
To avoid cell death of retinal tissue, perform the entire procedure within 1.5–2 h.
3.1. Activation of Fluorophores
Incubate the cartridge plate with 1 mL Seahorse calibration medium at 37 °C overnight in a CO2-free incubator to activate the fluorophores.
3.2. Assay Conditions/Protocol
Set up the assay in the machine, as follows:
Measure respiration at baseline for five time points. A cycle of mix (3 min), wait (2 min) and measure (3 min) is optimal for obtaining a robust OCR readout.
After the baseline respiration, inject port A (BAM15).
Measure respiration for next four time points using the same cycle of mix, wait and measure after the addition of a mitochondrial uncoupler BAM15 from port A.
Inject port B (Rotenone).
After the addition of Complex I inhibitor Rotenone from port B, measure respiration for next four time points using the same cycle of mix, wait and measure.
Save this protocol.
3.3. Preparation of XF24 Islet Fluxpak
Neutralize Cell-Tak (20 μL) with 0.1 M sodium bicarbonate (171 μL) and with 1 M NaOH (9 μL).
Pipette 8 μL of this reaction mixture to the mesh inserts and rotate the pipette to distribute the Cell-Tak equally throughout the mesh insert.
Allow a minimum of 25 min for adsorption.
Wash each mesh insert with Ames’ medium to remove excess sodium bicarbonate.
Keep the mesh insert aside for later use.
3.4. Preparation of Compounds
3.5. Retinal Discs Preparation
Euthanize light-adapted mice at a specific time (e.g., between 12 and 2 PM) by CO2 asphyxiation.
Enucleate eyes rapidly and place into ice-cold Ames’ medium in a petri - dish.
Keep this petri - dish on ice under a dissecting microscope until the end of the experiment.
Dissect the cornea and lens from the posterior segment of the eye. Remove vitreous humor from the retina using forceps. Pull the scleral muscle layer away from the neural retina to get an intact retinal eyecup free of retinal pigment epithelium (see Note 3).
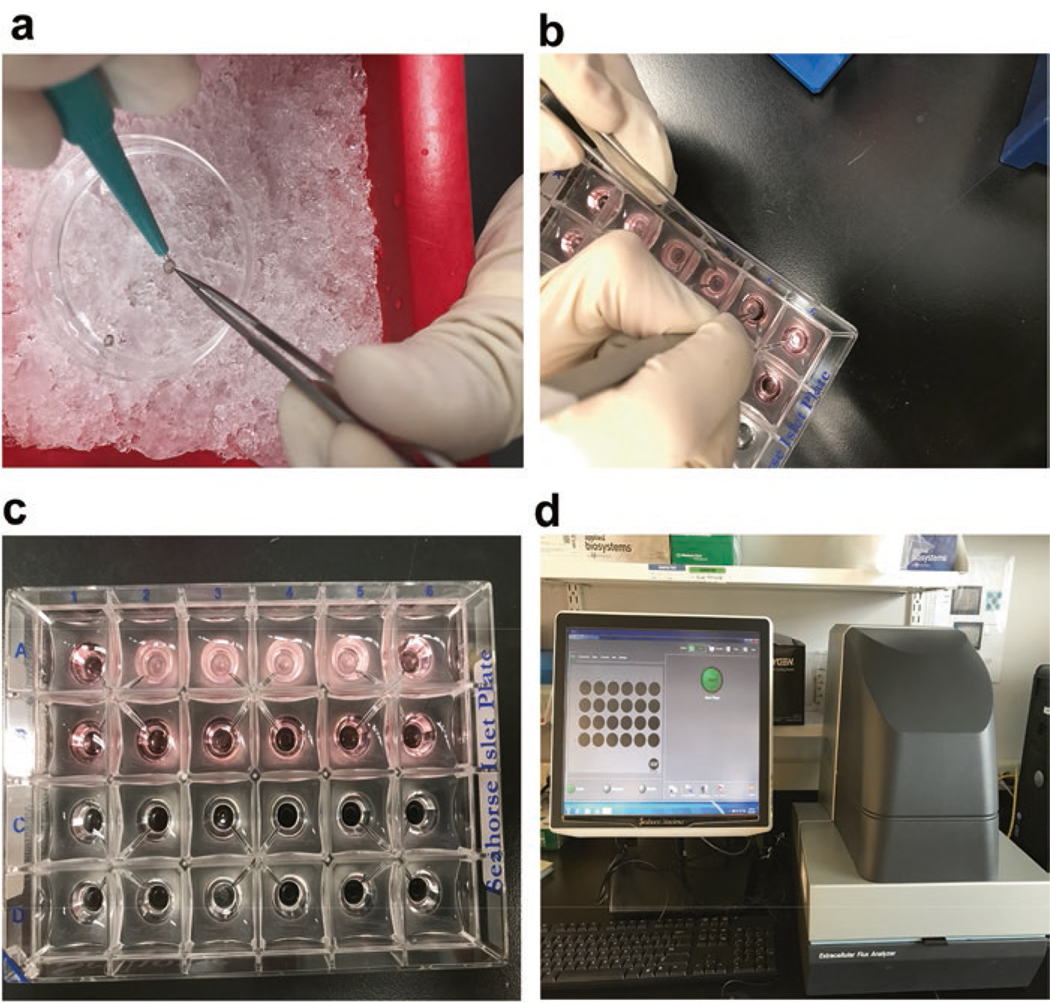
Use 1 mm diameter biopsy puncher to cut disks of neural retina on the glass petri - dish. Prepare three sections from each retina, equidistant from the optic nerve head (Fig. 1a; see Note 4).
With the help of a fine paintbrush, place the retinal sections a little off-center with the ganglion cell layer side down onto pretreated XF24 Islet Fluxpak mesh inserts.
Add 675 μL Ames’ medium in the XF24 Islet plate and keep on ice.
Place the mesh inserts with retina punch in this XF24 Islet plate (Fig. 1b).
Rotate the mesh insert with the help of blunt and pointed forceps to position the retinal disc directly under the O2 sensor (Fig. 1c).
Collect retinal discs from 4 to 12 mouse retinas in one dish for a specific experimental condition (see Note 5).
For statistical analysis, OCR measurements from each disc are considered as independent values.
Fig. 1.
Preparation of retinal discs for XF24 Islet Fluxpak. (a) 1 mm retinal disks were prepared from acutely isolated retina on ice in Ames’ medium with the help of a biopsy puncher and a fine forcep. (b) The mesh insert with retinal disk was placed in the XF24 Islet plate and it was rotated with the help of blunt and pointed forceps to position the retinal disc directly under the O2 sensor. (c) The picture shows Seahorse XF24 Islet plate with mesh inserts in “A” row. (d) Seahorse XF24 analyzer and monitor. After setting up the protocol, the screen shows that the machine is ready to start the experiment
3.6. Calibration of Compounds and OCR Measurements
Add 75 μL BAM15 (final concentration 5 μM) to the injection port “A” of the cartridge plate.
Add 75 μL Rotenone (final concentration 1 μM) to the injection port “B” of the cartridge plate.
Load this cartridge plate into the Seahorse XF analyzer and run the experiment for automatic calibration (Fig. 1d).
At the end of the calibration, replace the cartridge plate with XF24 Islet plate with retina (see Note 6).
Start the experiment again for the measurement of OCR (see Note 5).
3.7. Substrates Used for Evaluation
Prepare a basic salt solution and supplement this solution with defined combinations and concentrations of substrates.
Add 675 μL of this basic buffer in the XF24 Islet plate to test the effect of substrates on OCR (Fig. 2a, b).
Use the same volume of Ames’ medium as a control without retinal punch.
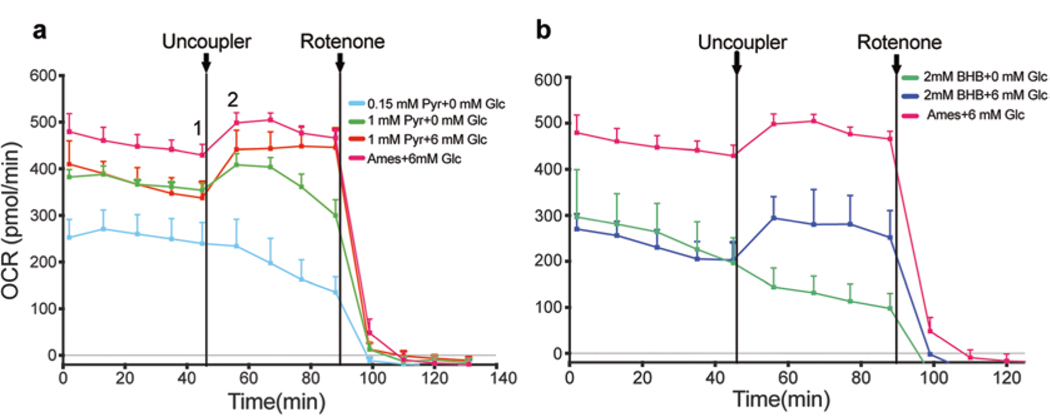
Fig. 2.
Comparison of oxygen consumption traces in basic salt solution with different concentrations of substrates. (a) Pyruvate was used as a substrate at 0.15 mM and 1 mM concentration with or without glucose. At 0.15 mM, pyruvate was insufficient to maintain OCR. Addition of glucose along with pyruvate (1 mM) yielded relatively stable OCR values compared to pyruvate alone; however, the baseline OCR is lower compared to Ames’ medium. (b) β-hydroxybutyrate (2 mM) was used as a substrate either alone or in presence of glucose (6 mM). β-hydroxybutyrate alone could not sustain OCR but the addition of glucose stabilized the OCR values. However, the baseline OCR was lower than Ames’ medium. These OCR readings were not normalized with cytochrome a
3.8. Cytochrome a Quantification for Normalization (As Described Previously [22])
Determine cytochrome a content in retinas spectrophotometrically in a spectrophotometer.
Homogenize four to eight retinas by pipetting in 0.1 mL of ice-cold homogenization buffer.
Add 0.9 mL of the same buffer containing 1% dodecyl malto-side to the homogenate and vortex for 45 s.
Centrifuge for 2 min at 16,000 × g, and transfer the supernatant to a plastic cuvette.
Add 0.1 mM potassium ferricyanide for the full oxidation of the cytochromes and collect and average five spectra in the wavelength range of 500–650 nm.
Add 0.5 mM of potassium cyanide and 10 mM sodium ascorbate to reduce cytochrome a, and collect and average five reduced spectra.
Subtract the oxidized spectrum from the reduced spectrum and calculate the absorbance difference at 605 nm after manually tracing a baseline from 575 and 630 nm.
For obtaining cytochrome a concentration, use an extinction coefficient of 10.8 mM−1 cm−1.
Use minimum four retinas to estimate the total cytochrome a content.
Use the surface area of a retinal disc relative to whole retina to calculate the cytochrome a content/disc when normalizing OCR.
3.9. Data Analysis
Analysis of Seahorse XF analyzer data involves the following steps:
Open the .xls file in Seahorse XF analyzer and convert the raw OCR values using the Seahorse Akos algorithm and save this file [28].
Open this new .xls file with Akos values and label the conditions of the experiment.
Normalize the Akos values by dividing with total cytochrome a values.
Plot these raw values into GraphPad Prism for making line graph for showing the baseline and uncoupled respiration.
Subtract residual OCR obtained after addition of rotenone (<1% of maximal OCR) from all points.
-
Calculate the Basal/Max (%) and maximal reserve capacity (MRC) as shown below. MRC is defined as the differential between maximal (Fig. 2a, graph point 2) and basal O2 consumption (Fig. 2a, graph point 1).
Basal/Max (%) = (Value at last time point of baseline respiration/(Maximum value after the addition of uncoupler)) × 100.
MRC = 100 − (Basal/Max ratio).
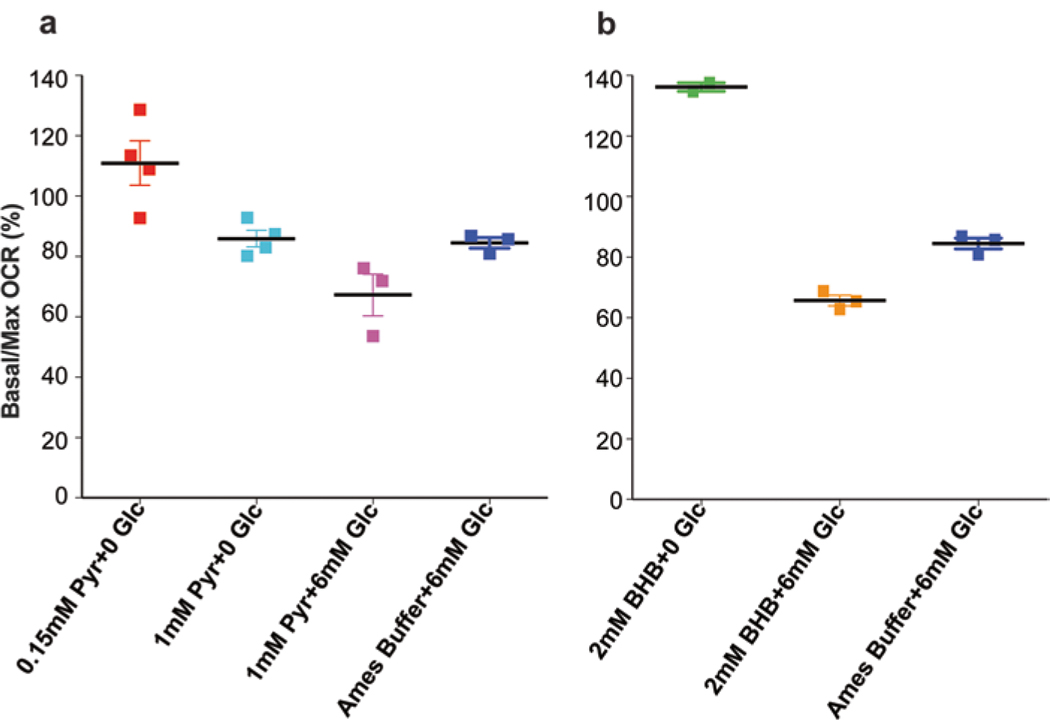
Plot Basal/Max (%) as scatter dot plot (Fig. 3a, b) and MRC as histograms.
Fig. 3.
Different substrates reveal differences in reserve capacity. (a) Pyruvate was used as a substrate at 0.15 mM and 1 mM concentration with or without glucose. (b) β-hydroxybutyrate was used at 2 mM concentration with or without glucose. Optimal reserve capacity was obtained after the addition of glucose to either substrate. Adult mouse retina was used for all experiments
Acknowledgment
The authors are supported by the Intramural Research Program (EY000450 and EY000546) of the National Eye Institute, National Institutes of Health, Bethesda, MD, USA. YKA is supported in part by Department of Science and Technology INSPIRE Faculty award (Govt. of India). We thank Drs. Tiziana Cogliati and Jacob Nellissery for comments on the manuscript and Drs. Shan Chen and Raul Covian for technical advice.
4 Notes
We used BAM15 instead of the commonly used uncoupler, trifluoromethoxyphenyl hydrazone (FCCP). In our previous study [22], BAM15 showed better efficiency in uncoupling mitochondrial proton flux and with persistent cell viability up to 50 μM [22].
During the experiment, keep working stocks of compounds at RT and bring Ames’ medium to 37 °C.
Keep petri - dish containing retina on ice throughout the experiment.
Collect only live, circular, intact retinal discs (by visual inspection).
The protocol, once started, should be completed within 1.5–2 h.
After placing the XF24 Islet Fluxpak plate inside the seahorse XF analyzer, the tissue is maintained at 37 °C until the end of the experiment.
References
- 1.Barot M, Gokulgandhi MR, Mitra AK (2011) Mitochondrial dysfunction in retinal diseases. Curr Eye Res 36(12):1069–1077. 10.3109/02713683.2011.607536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribas V, Garcia-Ruiz C, Fernandez-Checa JC (2014) Glutathione and mitochondria. Front Pharmacol 5:151 10.3389/fphar.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlachantoni D, Bramall AN, Murphy MP et al. (2011) Evidence of severe mitochondrial oxidative stress and a protective effect of low oxygen in mouse models of inherited photoreceptor degeneration. Hum Mol Genet 20(2):322–335. 10.1093/hmg/ddq467 [DOI] [PubMed] [Google Scholar]
- 4.Osorio-Paz I, Uribe-Carvajal S, Salceda R (2015) In the early stages of diabetes, rat retinal mitochondria undergo mild uncoupling due to UCP2 activity. PLoS One 10(5):e0122727. 10.1371/journal.pone.0122727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurley JB, Chertov AO, Lindsay K et al. (2014) Energy metabolism in the vertebrate retina In: Furukawa T, Hurley J, Kawamura S (eds) Vertebrate photoreceptors. Springer, Japan, pp 91–137 [Google Scholar]
- 6.Wright AF, Jacobson SG, Cideciyan AV et al. (2004) Lifespan and mitochondrial control of neurodegeneration. Nat Genet 36(11):1153–1158. 10.1038/ng1448 [DOI] [PubMed] [Google Scholar]
- 7.Bossy-Wetzel E, Schwarzenbacher R, Lipton SA (2004) Molecular pathways to neurodegeneration. Nat Med 10(Suppl):S2–S9. 10.1038/nm1067 [DOI] [PubMed] [Google Scholar]
- 8.Orrenius S, Gogvadze V, Zhivotovsky B (2007) Mitochondrial oxidative stress: implications for cell death. Annu Rev Pharmacol Toxicol 47:143–183. 10.1146/annurev.pharmtox.47.120505.105122 [DOI] [PubMed] [Google Scholar]
- 9.Scheibye-Knudsen M, Fang EF, Croteau DL, Wilson DM 3rd, Bohr VA (2015) Protecting the mitochondrial powerhouse. Trends Cell Biol 25 (3):158–170. doi: 10.1016/j.tcb.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright AF, Chakarova CF, Abd El-Aziz MM, Bhattacharya SS (2010) Photoreceptor degeneration: genetic and mechanistic dissection of a complex trait. Nat Rev Genet 11(4):273–284. 10.1038/nrg2717 [DOI] [PubMed] [Google Scholar]
- 11.Jarrett SG, Boulton ME (2012) Consequences of oxidative stress in age-related macular degeneration. Mol Asp Med 33(4):399–417. 10.1016/j.mam.2012.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camara AK, Lesnefsky EJ, Stowe DF (2010) Potential therapeutic benefits of strategies directed to mitochondria. Antioxid Redox Signal 13(3):279–347. 10.1089/ars.2009.2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujihara M, Nagai N, Sussan TE, Biswal S, Handa JT (2008) Chronic cigarette smoke causes oxidative damage and apoptosis to retinal pigmented epithelial cells in mice. PLoS One 3(9):e3119 10.1371/journal.pone.0003119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fritsche LG, Fariss RN, Stambolian D, Abecasis GR, Curcio CA, Swaroop A (2014) Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet 15:151–171. 10.1146/annurev-genom-090413-025610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fritsche LG, Igl W, Bailey JN et al. (2016) A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 48(2):134–143. 10.1038/ng.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brand MD, Nicholls DG (2011) Assessing mitochondrial dysfunction in cells. Biochem J 435(2):297–312. 10.1042/BJ20110162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Futterman S, Kinoshita JH (1959) Metabolism of the retina. I. Respiration of cattle retina. J Biol Chem 234(4):723–726 [PubMed] [Google Scholar]
- 18.Ames A 3rd, Li YY, Heher EC, Kimble CR (1992) Energy metabolism of rabbit retina as related to function: high cost of Na+ transport. J Neurosci 12(3):840–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linsenmeier RA (1986) Effects of light and darkness on oxygen distribution and consumption in the cat retina. J Gen Physiol 88(4):521–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medrano CJ, Fox DA (1995) Oxygen consumption in the rat outer and inner retina: light- and pharmacologically-induced inhibition. Exp Eye Res 61(3):273–284 [DOI] [PubMed] [Google Scholar]
- 21.DY Y, Cringle SJ (2005) Retinal degeneration and local oxygen metabolism. Exp Eye Res 80(6):745–751. 10.1016/j.exer.2005.01.018 [DOI] [PubMed] [Google Scholar]
- 22.Kooragayala K, Gotoh N, Cogliati T et al. (2015) Quantification of oxygen consumption in retina ex vivo demonstrates limited reserve capacity of photoreceptor mitochondria. Invest Ophthalmol Vis Sci 56(13):8428–8436. 10.1167/iovs.15-17901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC (2010) Myocardial fatty acid metabolism in health and disease. Physiol Rev 90(1):207–258. 10.1152/physrev.00015.2009 [DOI] [PubMed] [Google Scholar]
- 24.Malgoyre A, Chabert C, Tonini J, Koulmann N, Bigard X, Sanchez H (2017) Alterations to mitochondrial fatty-acid use in skeletal muscle after chronic exposure to hypoxia depend on metabolic phenotype. J Appl Physiol 122(3):666–674. 10.1152/japplphysiol.00090.2016 [DOI] [PubMed] [Google Scholar]
- 25.Joyal JS, Sun Y, Gantner ML et al. (2016) Retinal lipid and glucose metabolism dictates angiogenesis through the lipid sensor Ffar1. Nat Med 22(4):439–445. 10.1038/nm.4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klepper J, Willemsen M, Verrips A et al. (2001) Autosomal dominant transmission of GLUT1 deficiency. Hum Mol Genet 10(1):63–68 [DOI] [PubMed] [Google Scholar]
- 27.Chertov AO, Holzhausen L, Kuok IT et al. (2011) Roles of glucose in photoreceptor survival J Biol Chem 286(40):34700–34711. 10.1074/jbc.M111.279752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerencser AA, Neilson A, Choi SW et al. (2009) Quantitative microplate-based respirometry with correction for oxygen diffusion. Anal Chem 81(16):6868–6878. 10.1021/ac900881z [DOI] [PMC free article] [PubMed] [Google Scholar]