Abstract
Simple Summary
As DNA repair enzymes affect dynamics of liver damage and are involved in HBV viral replication, this study focused on the role of genetic variations within genes representing key DNA-repair pathways in HBV-induced liver cirrhosis. The obtained results have demonstrated that SNPs within XRCC1, ERCC2 genes may confer susceptibility to liver cirrhosis in chronic hepatitis B patients.
Abstract
Liver cirrhosis (LC), contributing to more than 1 million of deaths annually, is a major healthcare concern worldwide. Hepatitis B virus (HBV) is a major LC etiological factor, and 15% of patients with chronic HBV infection (CHB) develop LC within 5 years. Recently, novel host genetic determinants were shown to influence HBV lifecycle and CHB course. DNA repair enzymes can affect dynamics of liver damage and are involved in HBV covalently closed circular DNA (cccDNA) formation, an essential step for viral replication. This study aimed to evaluate the possible role of genes representing key DNA-repair pathways in HBV-induced liver damage. MALDI-TOF MS genotyping platform was applied to evaluate variations within XRCC1, XRCC4, ERCC2, ERCC5, RAD52, Mre11, and NBN genes. Apart from older age (p < 0.001), female sex (p = 0.021), portal hypertension (p < 0.001), thrombocytopenia (p < 0.001), high HBV DNA (p = 0.001), and high aspartate aminotransferase (AST) (p < 0.001), we found that G allele at rs238406 (ERCC2, p = 0.025), T allele at rs25487 (XRCC1, p = 0.012), rs13181 GG genotype (ERCC2, p = 0.034), and C allele at rs2735383 (NBN, p = 0.042) were also LC risk factors. The multivariate logistic regression model showed that rs25487 CC (p = 0.005) and rs238406 TT (p = 0.027) were independently associated with lower risk of LC. This study provides evidence for the impact of functional and potentially functional variations in key DNA-repair genes XRCC1 and ERCC2 in HBV-induced liver damage in a Caucasian population.
Keywords: HBV, liver cirrhosis, genetic polymorphism, DNA repair, XRCC1, ERCC2
1. Introduction
Liver cirrhosis (LC), the end-stage of different chronic liver diseases, has become one of the major causes of morbidity and mortality with more than 1 million deaths in 2010 according to the Global Burden of Disease (GBD) [1]. As the leading risk factor for the development of hepatocellular carcinoma (HCC), cirrhosis occurs in almost all patients with HCC, being additionally a limiting factor for anticancer therapy of hepatic, as well as non-hepatic malignancies [2]. Although LC is often asymptomatic, there is evidence demonstrating an increase in its prevalence worldwide [3,4]. The growing burden of LC is linked to increasing spread of its risk factors, including hepatitis B virus (HBV) and hepatitis C virus (HCV) infection, as well as non-alcoholic steatohepatitis (NASH) and and alcoholic steatohepatitis (ASH). The Global Hepatitis Report 2017 shows that HBV and HCV infections are responsible for 96% of the global mortality from hepatitis-related end-stage liver disease. In HBV-related cirrhosis, the 5-year cumulative HCC risk is around 15%.
The natural history of chronic hepatitis B is a complex interplay of viral, host, and environmental factors, and infected individuals can pass through several phases. According to the European Association for the Study of the Liver (EASL), HBeAg-positive chronic HBV infection is the first phase, which is characterized by high HBV DNA, persistent alanine aminotransferase (ALT) levels in upper limits of normal, the presence of serum hepatitis B e antigen (HBeAg), and no or minimal liver inflammation and/or fibrosis [5]. Low rates of spontaneous HBeAg loss are observed in this phase. In HBeAg-positive chronic hepatitis B (phase 2), apart from HBeAg positivity, high viral load and elevated ALT levels, moderate or severe liver inflammation, and/or clinical hepatitis is present [5,6]. Although the outcome of this phase varies, most of patients achieve HBeAg seroconversion and then enter HBeAg-negative chronic HBV infection (phase 3), or HBeAg-negative chronic hepatitis B (phase 4). Following seroconversion from serum antibodies to HBeAg (anti-HBe), HBeAg-negative carriers with undetectable or low levels of viral replication, normal ALT levels, and minimal histological lesions enter phase 3. These patients are at low risk of developing cirrhosis or HCC, but progression to CHB may still occur. Furthermore, a group of patients with moderate to high levels of serum HBV DNA, elevated or fluctuating ALT levels, and active liver necroinflammation and/or fibrosis undergo phase 4. Finally, some patients develop HBsAg-negative phase (phase 5, also known as “occult HBV infection”) with negative serum HBV surface antigen (HBsAg) and positive antibodies to HBV core antigen (anti-HBc), with or without detectable antibodies to HBsAg (anti-HBs). Those who developed cirrhosis before the HBsAg loss are at high risk of HCC as well as HBV reactivation due to immunosuppressive drug treatments [5,6,7].
Chronic liver inflammation affects many cellular pathways including dysregulation of DNA damage response (DDR). DDR plays a critical role in the maintenance of genomic stability and it protects from carcinogenesis by repairing DNA damage caused by multiple factors. Major DNA repair pathways are mismatch repair (MMR), nucleotide excision repair (NER), base excision repair (BER), homologous recombinational repair (HR), and non-homologous end joining (NHEJ). Most of these mechanisms have been previously implicated in HCC, and upregulation of DNA repair genes have been also shown to be associated with cirrhosis [8,9]. Recent experimental data have also linked the cellular DNA repair system with HBV replication cycle. In 2014, Königer et al. identified tyrosyl DNA phosphodiesterase-2 (TDP2) as the first host DNA-repair factor involved in HBV covalently closed circular DNA (cccDNA) biogenesis [10]. However, the additional steps towards cccDNA and the redundancy in DNA repair imply the involvement of numerous others. Moreover, several studies reporting impact of HBV expression on DNA repair support the notion that virus hijacks DDR proteins to successfully complete several steps of its life cycle [11,12,13,14,15]. Furthermore, different DNA tumor viruses have to cope with the DDR through multiple independent mechanisms, and HBV cannot remain an exception [16,17]. Moreover, the HBV interaction with DNA repair was previously suspected because of the association of HCC with chronic hepatitis B (CHB) and the known correlation of cancer with inappropriate DNA damage repair [15]. The interplay between the virus and DDR pathways can modulate to the course of HBV infection. This interaction is mainly determined by the efficiency of DDR machinery, which may be affected by single nucleotide polymorphisms (SNPs) in DNA repair genes [15,18]. These genetic alterations can influence individual DNA repair capacity, increasing the risk of developing liver cirrhosis or cancer, as well as directly affecting the course of HBV infection and HBV-induced pathogenesis.
Although numerous SNPs in genes encoding DNA repair have been identified, little is known about HBV infection-related DNA repair gene polymorphisms. Therefore, an effective analysis of DNA repair gene polymorphism during viral infection may represent a true advance in the study of HBV-induced liver cirrhosis. In this cross-sectional study, we aimed to determine whether polymorphisms in the key repair enzyme pathways BER (XRCC1), NER (ERCC2, ERCC5), HR (RAD52, Mre11, NBN), and NHEJ (XRCC4), which have been previously associated with HBV infection and/or susceptibility to cancer, interfere with HBV-induced liver cirrhosis.
2. Results
2.1. Study Group Characteristics
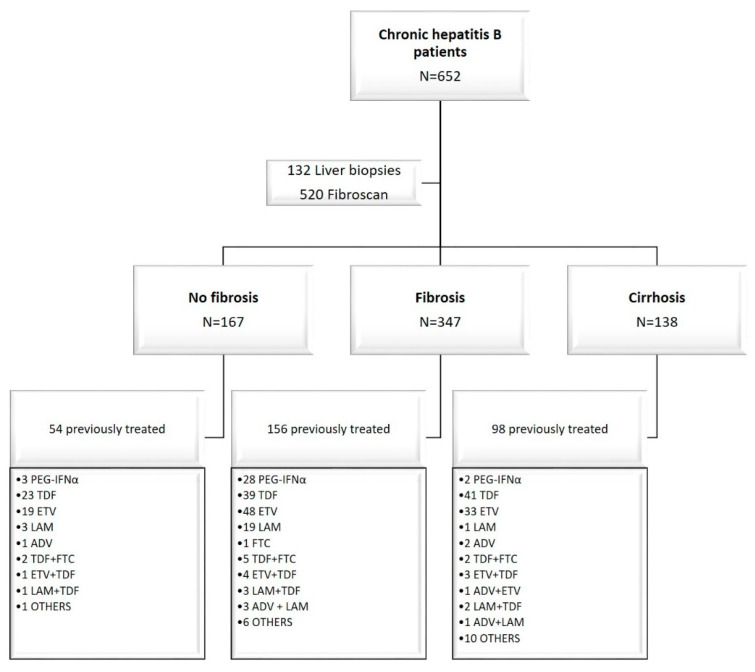
The study group consisted of 752 participants, including 652 compensated liver disease CHB patients and 100 healthy blood donors. The patients were enrolled in the study at the time of liver assessment. Among CHB patients, 308/652 (47.2%) patients were on antiviral therapy (<6 months prior to study enrollment). These patients received treatment according to Polish or French National Health Service recommendations: 33 (11%) received pegylated interferon alfa (PEG-IFNα), 230 (74.5%) received a nucleoside/nucleotide analogue (NA) monotherapy, and 45 (14.5%) received combined therapies (Figure 1). Table 1 summarizes the distribution of variables evaluated at study inclusion. A total of 138 (21.2%) CHB patients were affected with liver cirrhosis. Patients with liver cirrhosis were significantly older (p < 0.001) than patients with and without fibrosis and a control group. Men were predominant in the liver cirrhosis (72%) and fibrosis group (67%). The HBeAg and anti-HBeAg positivity rate in patients with cirrhosis was significantly higher than in participants without cirrhosis (Table 1).
Figure 1.
Disposition of enrolled patients. The liver assessment was performed at the time of enrollment in the study, and the treatment history was provided simultaneously. The median of treatment before the liver fibrosis assessment was 3 months. PEG-IFNα, pegylated interferon alfa; TDF, tenofovir; ETV, entecavir; LAM, lamivudine; ADV, adefovir; FTC, emtricitabine; OTHERS: ADV + ETV + LAM + TDF (2), PEG-IFNα + TDF (2), PEG-IFNα + ETV (2), ADV + LAM + TDF (2), vidarabine (1), famciclovir (1), PEG-IFNα + LAM + TDF (2), anti HBsAg monoclonal antibodies (2), anti HBsAg monoclonal antibodies + TDF (2), anti HBsAg monoclonal antibodies + ETV (1).
Table 1.
Characteristics of chronic hepatitis B patients with liver cirrhosis, hepatic fibrosis, and healthy controls.
| Variable | Chronic Hepatitis B | p-Value d | Healthy Controls (n = 100) |
p-Value e | ||
|---|---|---|---|---|---|---|
| No Fibrosis a (n = 167) |
Liver Fibrosis b (n = 347) |
Liver Cirrhosis c (n = 138) |
||||
| Age, years | 46 ± 1 | 50 ± 1 | 60 ± 0.9 | <0.0001 | 27.3 ± 0.9 | <0.0001 |
| Sex, % females | 49% | 33% | 28% | 0.00148 | 22% | 0.0031 |
| Ethnicity, % Caucasian | 100% | 100% | 100% | 1 | 100% | 1 |
| Alcohol consumption, % | 54% | 55% | 34% | 0.00015 | - | - |
| Previous treatment, % | 32% | 50% | 71% | <0.0001 | - | - |
| BMI, kg/m2 | 25.37 ± 0.39 | 25.67 ± 0.29 | 26.20 ± 0.41 | 0.1752 | - | - |
| Diabetes, % | 5% | 11% | 12% | 0.0175 | - | - |
| HT, % | 15% | 24% | 99% | <0.0001 | - | - |
| ALT, IU/L | 32.73 ± 1.64 | 37.43 ± 1.58 | 40.43 ± 2.71 | 0.0070 | - | - |
| AST, IU/L | 26.37 ± 1.17 | 29.68 ± 1.34 | 38.4 ± 2.2 | <0.0001 | - | - |
| ALB, IU/L | 41 ± 1.06 | 43.6 ± 0.25 | 40.8 ± 0.48 | 0.4838 | - | - |
| PLT, 109/L | 229 ± 4.58 | 208 ± 3.85 | 158 ± 6.06 | 0.0002 | - | - |
| HBV DNA, IU/mL | 8447 ± 3 828 | 15,015 ± 3 787 | 20,932 ± 269 | <0.0001 | - | - |
| HBeAg, % positive | 6% | 13% | 18% | 0.0014 | - | - |
| Anti-HBe, % positive | 92% | 82% | 66% | <0.0001 | - | - |
| Liver inflammation grade †,* | 1.5 (0–2) | 1.5 (1–3) | 1.5 (1.25–1.75) | 0.3017 | - | - |
| Liver fibrosis stage † | 0 (0–0) | 1 (0.5–3.5) | 4 (4–4) | <0.0001 | - | - |
Unless stated otherwise, data are shown as the mean ± standard error of the mean; † median value (interquartile range); * liver biopsy samples only (n = 132); a no scarring (stage 0); b fibrosis stages (I–III); c fibrosis stage IV; d liver cirrhosis vs. no fibrosis, e all CHB vs. healthy control group; p-values less than 0.05 are shown in bold. BMI, body mass index; HT, portal hypertension; ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALB, albumin; PLT, platelet count; CHB, chronic hepatitis B.
A total of nine variations within seven genes associated with the DNA repair pathways were successfully genotyped for all study participants (Table S1). The distribution of genotypes followed Hardy–Weinberg equilibrium (HWE) for the healthy volunteers (p > 0.05). For CHB patients, the distributions were not consistent with HWE (p < 0.05). Genotype distributions for all analyzed SNPs in patients with liver cirrhosis, liver fibrosis, no fibrosis, and healthy group are presented in Tables S2 and S3. Genotypic distribution of 6 analyzed SNPs (ERCC2: rs13181, rs238406; RAD52: rs7963551; XRCC1: rs25487; ERCC5: rs2018836; NBN: rs2735383) differed significantly between the CHB patients and the healthy control group (Table S2). Moreover, for rs238406 (ERCC2), rs25487 (XRCC1), and rs2735383 (NBN), we also found significant differences in genotypic distribution between the liver cirrhosis and no fibrosis group (Table S3). Evaluation of linkage disequilibrium (LD) pattern with the use of the r2 coefficient between pairs of analyzed SNPs showed that all of them were independent (r2 < 0.5).
In our study, rs13181 TT (ERCC2) and rs2018836 GG (ERCC5) genotypes were more common in patients with lower baseline HBV DNA levels (sex-adjusted GG, GT vs. TT: odds ratio (OR) = 1.97, 95% CI 1.35–2.88, p = 0.0004; sex-adjusted AA, AG vs. GG: OR = 1.94, 95% CI 1.34–2.81, p = 0.00039). XRCC1 rs25487 TT genotype was associated with lower liver enzyme levels in the study population (ALT: sex-adjusted CC vs. TT: OR = 1.91, 95% CI 1.22–2.99, p = 0.0042; AST: sex-adjusted CC vs. TT: OR = 2.26, 95% CI 1.67–4.36, p = 0.015). Additionally, carriers of NBN rs2735383 CC genotype had significantly lower ALT levels (sex-adjusted GG, GC vs. CC: OR = 1.45, 95% CI 1.05–1.99, p = 0.022).
2.2. Genetic Polymorphisms and the Susceptibility to CHB-Related Liver Diseases
Three genetic models, additive, dominant, and recessive models, were used to analyze the association between each SNP and liver cirrhosis. Univariate analyses of variables associated with liver cirrhosis are summarized in Table 2, with significant association observed for age, sex, alcohol consumption, treatment experience, hypertension, AST and PLT levels, baseline HBV viral load, rs13181, rs238406, rs25487, and rs2735383. After adjusting for sex and age, logistic regression analysis showed that CHB individuals carrying the rs238406 G allele (ERCC2) and rs25487 T allele (XRCC1) are at increased risk of developing cirrhosis and liver failure (Table S4). Next, a multiple logistic regression model was used to study the predictors of cirrhosis (Table 3, Table S5). Portal hypertension, thrombocytopenia (platelet count below 150,000), rs25487, and rs238406 were independently associated with liver cirrhosis.
Table 2.
Results of univariate analyses for liver cirrhosis risk in chronic hepatitis B patients.
| Variable | n (%) | OR | 95% CI | p-Value |
|---|---|---|---|---|
|
Age >50 years ≤50 years |
346 (53%) 306 (47%) |
4.19 | 2.71–6.48 | <0.001 |
|
Sex Female Male |
242 (37%) 410 (63%) |
1.59 | 1.07–2.36 | 0.021 |
|
Alcohol consumption YES NO |
332 (51%) 320 (49%) |
2.37 | 1.58–3.55 | <0.001 |
|
Previous treatment YES NO |
308 (47%) 344 (53%) |
2.64 | 1.73–4.02 | <0.001 |
|
Portal hypertension YES NO |
254 (39%) 398 (61%) |
515 | 71–3756 | <0.001 |
|
AST >50 IU/L ≤50 IU/L |
118(18%) 534 (82%) |
3.10 | 1.76–58.44 | <0.001 |
|
ALT >42 IU/L ≤42 IU/L |
98 (15%) 554 (85%) |
1.22 | 0.80–1.84 | 0.34 |
|
PLT >150 IU/L ≤150 IU/L |
502 (77%) 150 (23%) |
4.96 | 3.18–7.73 | <0.001 |
|
HBV DNA >6000 kIU/mL ≤6000 kIU/ml |
529 (81%) 123 (19%) |
3.75 | 1.70–8.26 | 0.001 |
|
AST/ALT >1 ≤1 |
254 (39%) 398 (61%) |
1.73 | 1.18–2.53 | 0.004 |
| XRCC1 rs25487 | ||||
| CC vs. TT | 294 (74%) vs. 104 (26%) | 1.58 | 0.95–2.60 | 0.069 |
| CC vs. CT, TT | 294 (45%) vs. 358 (55%) | 0.61 | 0.42–0.89 | 0.012 |
| TT vs. CT, CC | 104 (16%) vs. 548 (84%) | 1.03 | 0.63–1.70 | 0.89 |
| ERCC2 rs238406 | ||||
| GG vs. TT | 235 (60.5%) vs. 154 (39.5%) | 1.58 | 0.91–2.72 | 0.099 |
| GG vs. GT, TT | 235 (36%) vs. 417 (64%) | 1.14 | 0.75–1.73 | 0.54 |
| TT vs. GT, GG | 154 (23.5%) vs. 498 (76.5%) | 0.53 | 0.30–0.92 | 0.025 |
| ERCC2 rs13181 | ||||
| TT vs. GG | 303 (72%) vs. 117 (28%) | 4.27 | 1.11–16.42 | 0.034 |
| GG vs. GT, TT | 117 (18%) vs. 535 (82%) | 1.35 | 0.86–2.14 | 0.19 |
| TT vs. GT, GG | 303 (46.5%) vs. 349 (53.5%) | 0.91 | 0.63–1.33 | 0.64 |
| ERCC5 rs2018836 | ||||
| GG vs. AA | 333 (79%) vs. 88 (21%) | 1.28 | 0.77–2.12 | 0.33 |
| GG vs. GA, AA | 333 (51%) vs. 319 (49%) | 1.41 | 0.86–2.30 | 0.17 |
| AA vs. GA, GG | 88 (13.5%) vs. 564 (86.5%) | 0.92 | 0.61–1.32 | 0.65 |
| RAD 52 rs7963551 | ||||
| TT vs. GG | 492 (87%) vs. 72 (13%) | 1.04 | 0.80–1.25 | 0.97 |
| TT vs. TG, GG | 492 (75.5%) vs. 160 (24.5%) | 0.97 | 0.61–1.54 | 0.91 |
| GG vs. TG, TT | 72 (11%) vs. 580 (89%) | 0.96 | 0.51–1.81 | 0.90 |
| MRE 11 rs496797 | ||||
| TT vs. CC | 163 (44%) vs. 209 (56%) | 0.67 | 0.42–1.09 | 0.11 |
| TT vs. TC, CC | 163 (25%) vs. 489 (75%) | 1.17 | 0.78–1.76 | 0.44 |
| CC vs. TC, TT | 209 (32%) vs. 443 (68%) | 0.68 | 0.45–1.003 | 0.067 |
| MRE 11 rs535801 | ||||
| CC vs. TT | 316 (77%) vs. 95 (23%) | 1.10 | 0.67–1.82 | 0.70 |
| CC vs. CT, TT | 316 (48.5%) vs. 336 (51.5%) | 1.04 | 0.72–1.50 | 0.84 |
| TT vs. CT, CC | 95 (14.5%) vs. 557 (85.5%) | 1.30 | 0.80–2.10 | 0.29 |
| NBN rs2735383 | ||||
| CC vs. GG | 336 (77%) vs. 101 (23%) | 0.59 | 0.35–1.01 | 0.056 |
| CC vs. CG, GG | 336 (51.5%) vs. 316 (48.5%) | 1.29 | 0.88–1.89 | 0.18 |
| GG vs. CG, CC | 101 (15.5%) vs. 551 (84.5%) | 0.53 | 0.29–0.98 | 0.042 |
| XRCC4 rs1805377 | ||||
| GG vs. AA | 522 (97.5%) vs. 13 (2.5%) | 1.51 | 0.71–3.21 | 0.28 |
| GG vs. GA, AA | 522 (80%) vs. 130 (20%) | 0.85 | 0.54–1.32 | 0.46 |
| AA vs. GA, GG | 13 (2%) vs. 639 (98%) | 2.15 | 7.44–6.22 | 0.16 |
p-values less than 0.05 are marked in bold. CI, confidence interval; OR, odds ratio.
Table 3.
Final multiple logistic regression model for liver cirrhosis risk in chronic hepatitis B patients.
| Variable | n (%) | OR | 95% CI | p-Value |
|---|---|---|---|---|
|
Portal hypertension YES NO |
254 (39%) 398 (61%) |
446 | 58–3425 | <0.001 |
|
Previous treatment YES NO |
308 (47%) 344 (53%) |
3.88 | 1.56–9.67 | 0.003 |
|
PLT >150 IU/L ≤150 IU/L |
502 (77%) 150 (23%) |
4.76 | 1.97–11.67 | <0.001 |
| XRCC1 rs25487 | ||||
| CC vs. CT, TT | 294 (45%) vs. 358 (55%) | 0.27 | 0.13–0.57 | 0.005 |
| ERCC2 rs238406 | ||||
| TT vs. GT, GG | 154 (23.5) vs. 498 (76.5) | 0.32 | 0.12–0.88 | 0.027 |
p-values less than 0.05 are marked in bold.
Corresponding analysis was made to investigate associations between evaluated variabilities and the liver fibrosis grade. rs238406 TT (ERCC2), rs7963551 TT (RAD52), and rs25487 CC (XRCC1) were identified as predictors of lower fibrosis stage (S ≤ 2) (Table 4, Table S6). However, rs238406TT and rs25487 CC remained significant predictors in a multiple logistic regression model (Table 5, Table S7).
Table 4.
Results of univariate analyses for the risk of advanced liver fibrosis † in chronic hepatitis B patients.
| Variable | n (%) | OR | 95% Cl | p-Value |
|---|---|---|---|---|
|
Age >50 years ≤50 years |
346 (53%) 306 (47%) |
2.98 | 2.15–4.12 | <0.001 |
|
Sex Female Male |
242 (37%) 410 (63%) |
0.39 | 0.28–0.55 | <0.001 |
|
Alcohol consumption YES NO |
332 (51%) 320 (49%) |
1.70 | 1.21–2.40 | 0.002 |
|
Previous treatment YES NO |
308 (47%) 344 (53%) |
3.19 | 2.23–4.57 | <0.001 |
|
Portal hypertension YES NO |
254 (39%) 398 (61%) |
7.32 | 4.98–10.76 | <0.001 |
|
AST >50 IU/L ≤50 IU/L |
118 (18%) 534 (82%) |
3.76 | 2.07–6.84 | <0.001 |
|
ALT >42 IU/L ≤42 IU/L |
98 (15%) 554 (85%) |
0.62 | 0.44–0.87 | 0.006 |
|
PLT >150 IU/L ≤150 IU/L |
502 (77%) 150 (23%) |
6.64 | 4.11–10.74 | <0.001 |
|
HBV DNA >6000 kIU/mL ≤6000 kIU/mL |
529 (81%) 123 (19%) |
1.76 | 1.13–2.73 | 0.011 |
|
AST/ALT >1 ≤1 |
254 (39%) 398 (61%) |
1.44 | 1.02–2.04 | 0.036 |
| XRCC1 rs25487 | ||||
| CC vs. TT | 294 (45) vs. 104 (16) | 1.26 | 0.82–1.94 | 0.29 |
| CC vs. CT, TT | 294 (45%) vs. 358 (55%) | 0.69 | 0.51–0.95 | 0.024 |
| TT vs. CT, CC | 104 (16%) vs. 548 (84%) | 0.78 | 0.49–1.22 | 0.27 |
| ERCC2 rs238406 | ||||
| GG vs. TT | 235 (36) vs. 154 (23.5) | 0.92 | 0.59–1.42 | 0.70 |
| GG vs. GT, TT | 235 (36) vs. 417 (64) | 1.04 | 0.76–1.42 | 0.79 |
| TT vs. GT, GG | 154 (23.5) vs. 498 (76.5) | 0.91 | 0.97–1.44 | 0.68 |
| ERCC2 rs13181 | ||||
| TT vs. GG | 303 (46.5) vs. 117 (17) | 2.85 | 0.66–12.39 | 0.16 |
| GG vs. GT, TT | 117 (18) vs. 535 (82) | 1.17 | 0.78–1.76 | 0.43 |
| TT vs. GT, GG | 303 (46.5) vs. 349 (53.5) | 0.90 | 0.66–1.23 | 0.53 |
| ERCC5 rs2018836 | ||||
| GG vs. AA | 333 (51) vs. 88 (13.5) | 0.73 | 0.47–1.14 | 0.17 |
| GG vs. GA, AA | 333 (51) vs. 319 (49) | 1.04 | 0.73–1.49 | 0.80 |
| AA vs. GA, GG | 88 (13.5) vs. 564 (86.5) | 0.64 | 0.42–0.96 | 0.031 |
| RAD 52 rs7963551 | ||||
| TT vs. GG | 492 (75.5) vs. 72 (11) | 1.67 | 1.04–2.68 | 0.032 |
| TT vs. TG, GG | 492 (75.5) vs. 160 (24.5) | 0.66 | 0.46–0.97 | 0.034 |
| GG vs. TG, TT | 72 (11) vs. 580 (89) | 1.54 | 0.94–2.54 | 0.08 |
| MRE 11 rs496797 | ||||
| TT vs. CC | 163 (25) vs. 209 (32) | 0.94 | 0.63–1.42 | 0.79 |
| TT vs. TC, CC | 163 (25) vs. 489 (75) | 0.88 | 0.62–1.26 | 0.49 |
| CC vs. TC, TT | 209 (32) vs. 443 (68) | 0.83 | 0.59–1.16 | 0.28 |
| MRE 11 rs535801 | ||||
| CC vs. TT | 316 (48.5) vs. 95 (14.5) | 0.86 | 0.56–1.30 | 0.47 |
| CC vs. CT, TT | 316 (48.5) vs. 336 (51.5) | 1.15 | 0.84–1.57 | 0.37 |
| TT vs. CT, CC | 95 (14.5) vs. 557 (85.5) | 0.95 | 0.62–1.46 | 0.83 |
| NBN rs2735383 | ||||
| CC vs. GG | 336 (51.5) vs. 101 (15.5) | 1.03 | 0.68–1.57 | 0.88 |
| CC vs. CG, GG | 336 (51.5) vs. 316 (48.5) | 0.88 | 0.64–1.21 | 0.45 |
| GG vs. CG, CC | 101 (15.5) vs. 551 (84.5) | 0.55 | 0.55–1.31 | 0.46 |
| XRCC4 rs1805377 | ||||
| GG vs. AA | 522 (80) vs. 13 (2) | 1.23 | 0.63–2.39 | 0.54 |
| GG vs. GA, AA | 522 (80) vs. 130 (20) | 0.84 | 0.57–1.24 | 0.39 |
| AA vs. GA, GG | 13 (2) vs. 639 (98) | 0.81 | 0.27–2.41 | 0.71 |
CI, confidence interval; MAF, minor allele frequency. p-values less than 0.05 are marked in bold. † S > 2.
Table 5.
Results of multivariate analyses for advanced † liver fibrosis.
| Variable | n (%) | OR | 95% Cl | p-Value |
|---|---|---|---|---|
|
Sex Female Male |
242 (37%) 410 (63%) |
0.48 | 0.28–0.84 | 0.009 |
|
Previous treatment YES NO |
308 (47%) 344 (53%) |
2.30 | 1.40–3.78 | <0.001 |
|
Portal hypertension YES NO |
254 (39%) 398 (61%) |
6.60 | 4.00–10.89 | <0.001 |
|
AST >50 IU/L ≤50 IU/L |
118 (18%) 534 (82%) |
2.44 | 1.06–5.62 | 0.034 |
|
PLT >150 IU/L ≤150 IU/L |
502 (77%) 150 (23%) |
3.84 | 2.02–7.26 | <0.001 |
| XRCC1 rs25487 | ||||
| CC vs. CT, TT | 294 (45%) vs. 358 (55%) | 0.51 | 0.31–0.85 | 0.009 |
| ERCC2 rs238406 | ||||
| TT vs. GT, GG | 154 (23.5) vs. 498 (76.5) | 0.47 | 0.27–0.81 | 0.006 |
CI, confidence interval; OR, odds ratio; p-values less than 0.05 are marked in bold. † S > 2. p-values less than 0.05 are marked in bold
3. Discussion
In the present study, we investigated the association between the SNPs within DNA repair enzymes associated with HBV infection and/or susceptibility to cancer and HBV-related liver cirrhosis. We showed that SNPs within XRCC1 and ERCC2 genes are independently associated with increased risk of cirrhosis. Liver cirrhosis represents the irreversible final fibrosis stage of the liver, being the consequence of wound healing response to chronic liver injury. One of the leading cause of hepatic disease is HBV infection, which was responsible for 66% of viral hepatitis-related deaths in 2015. The unsteady rates of development of liver damage under similar risk factors as well as the variability in the pathogenesis of HBV infection demonstrate that the molecular mechanism of HBV-induced cirrhosis is not fully understood. Recently, a number of functional genetic polymorphisms that likely increase the risk of liver fibrosis progression were described, which, together with other external factors such as alcohol intake, body mass index (BMI), diabetes, or hepatitis could be helpful in the determination of risk profile for the individual patient [19,20,21,22,23,24]. Moreover, host genetic background is undoubtedly involved in the development of HBV-related liver diseases [25]. As HBV exploits the cellular DDR system for RC-DNA to cccDNA conversion, the genes encoding host DNA repair enzymes may be potential candidates to predict the progression of HBV-mediated disease severity [15,18].
In this study, we investigated nine genetic polymorphisms in a cohort of CHB patients and in healthy blood donors. We observed significant differences in genotype distribution for six SNPs between CHB patients and healthy individuals. Three genetic variations (ERCC2: rs238406, XRCC1: rs25487, NBN: rs2735383) had diverse frequency in CHB patients with liver cirrhosis and with no fibrosis, highlighting their possible role in HBV-induced liver disease progression. The homozygous TT genotype at ERCC2 rs238406, CC at XRCC1 rs25487, and GG at NBN rs2735383 were significantly associated with a lower risk of cirrhosis. As compared to the no fibrosis group, the cirrhotic group harbored a higher frequency of ERCC2 rs238406 G allele, which appeared to be a risk factor for cirrhosis, and lower numbers of the variant XRCC1 rs25487 C allele, which had a protective effect. Additionally, both alleles together with RAD52 rs7963551 TT were more common in patients with lower liver fibrosis stage, confirming their role against liver damage progression.
HBV is a non-cytopathic virus; therefore, liver damage in CHB is associated with host immune response attempting to eliminate the virus [26]. The pathophysiology of HBV-induced fibrosis is related to chronic activation of wound healing mainly as a result of necroinflammation driven by the secondary recruitment of mononuclear cells to the site of infection [27]. Evidence specifically on the role of DNA repair genes in fibrosis progression is scarce. However, the host DNA enzymes and repair machinery is crucial for HBV to replicate. The tyrosyl-DNA phosphodiesterase 2 (TDP2) [10], and more recently reported DNA topoisomerases [28] as well as ataxia telangiectasia mutated pathway and Rad3-related pathway and signaling factor CHK1 (ATR-CHK1) [29], which were shown to take part in the synthesis of HBV cccDNA, may serve as examples. Nonetheless, the detailed mechanisms of HBV DNA turnover, and specifically cccDNA formation, inside the hepatocyte are still to be elucidated, and the possible role of another host factor is postulated [15,18]. Thus, one may speculate that altering the pathways responsible for DNA maintenance might affect viral lifecycle, and, in consequence, relate to the dynamics of generated liver damage.
The X-ray repair cross complementing 1 protein (XRCC1) plays a crucial role in the coordination of two overlapping repair pathways, SSBR and BER [30]. The XRCC1 gene, located on chromosome 19q13.2, along with its genetic diversity were widely studied in the past decade. A number of studies reported that XRCC1 polymorphism is associated with different types of cancer including lung, esophageal, breast, bladder, gastrointestinal, as well as hepatocellular carcinoma [31,32,33,34]. However, results regarding association between rs25487 [C/T] (Arg399Gln) and HCC remain inconclusive. Some studies demonstrated rs25487 as a risk factor of HCC [35,36], whereas others found no such associations [37,38]. In our study, XRCC1 rs25487 was demonstrated to be a strong prognostic factor for liver cirrhosis and advanced liver fibrosis. To the best of our knowledge, this is the third study where the association between XRCC1 rs25487 and the risk of HBV-related liver cirrhosis were analyzed. Bose et al. showed that the XRCC1 genotype alters the risk of HBV-related liver disease [39]. However, this study was conducted on a relatively small group of patients (25 patients with cirrhosis). Another study, performed on Brazilian patients with viral hepatitis, confirmed these findings, but again the study group was relatively small (27 HBV-induced cirrhotic patients) [40]. Even so, both studies present the same hypothesis that XRCC1 399 codon has an impact on development of HBV-induced liver cirrhosis. The role of this polymorphism can be explained by the fact that genetic variation at this site contributes to the amino acid change at evolutionarily conserved region, and therefore has an influence on XRCC1 function [41]. In fact, XRCC1 rs25487 polymorphism was associated with less efficient DNA repair, which, in consequence, may result in hepatocyte apoptosis [42]. Slyskova et al. demonstrated that individuals with rs25487 TT display a three- to fourfold reduced DNA repair capacity in comparison to CC carriers, which were at lower risk of cirrhosis development in our study [43]. Additionally, the T allele was associated with an increase of chromosomal deletions indicative of defects in BER pathway and with a lower survival time of patients with different types of cancer [44,45,46]. Moreover, in vitro and in vivo studies demonstrated that HBV can induce reactive oxygen species (ROS) production that causes oxidative stress resulting in hepatocyte injury [47,48,49]. As XRCC1 is one of the main players in the repair of ROS-induced lesions [50], it brings us to the conclusion that affecting protein function rs25487 may have an impact on HBV-induced liver fibrosis development.
The DNA helicase encoded by the excision repair cross-complementing group 2 gene (ERCC2), also called xeroderma pigmentosum group D (XPD), is involved in basal cellular transcription and NER of DNA lesions [51]. To date, a number of studies have implicated the role of genetic polymorphism within ERCC2 gene in various cancers, such as gastric, pancreatic, glioma, bladder, and hepatocellular carcinoma [52,53,54,55,56,57]. The most studied polymorphisms rs13181 (Lys751Gln) and rs238406 (Arg156Arg) were demonstrated to alter the function of encoded protein, and, in consequence, influence on DNA repair capacity [58,59]. A recent meta-analysis demonstrated that ERCC2 rs13181 GG genotype has a positive prognostic value in predicting the overall survival of HCC patients [56]. On the other hand, rs238406 TT genotype was associated with an increased overall cancer risk [60]. In our study, both SNPs were associated with liver cirrhosis development. This effect was stronger for rs238406 G allele, which was identified as a risk factor for both liver cirrhosis and advanced fibrosis. To our knowledge, this is the first study that examines the influence of rs238406 on hepatic cirrhosis. Because rs238406 does not change the amino acid residue (Arg156Arg), it should likely have no effect on the function of ERCC2 enzyme. However, latest studies showed that such a silent mutation may affect gene expression, protein levels, and can also modify the structure and functionality of proteins [61]. Moreover, ERCC2 expression levels may be used to distinguish between HBV-related HCC tumor and paracancerous tissues [62], which confirms its role in HBV-related liver damage. Moreover, HBV can modulate DNA damage by inhibiting several DNA damage response proteins, including ERCC2, through viral HBx [63]. HBx has shown to directly bind the transcription factor IIH (TFIIH) component xeroderma pigmentosum complementation group D (XPD/ERCC2) [64]. Therefore, any genetic variations within ERCCA2 may cause structural changes in the interaction site or affect the HBx binding affinity to ERCC2, depressing the DNA repair capability. Very recently, a first connection between genetic variations within ERCC-related genes and cirrhosis was reported [65]. Significant differences in genotype distribiution between control, cirrhosis, and liver cancer groups were demonstrated for ERCC5 variants. Moreover, it was proposed that two ERCC5 gene polymorphisms (rs229614 and rs2296148) may be important targets for cirrhosis.
Despite the relatively large sample size, this study has some limitations. First, we did not examine the influence of analyzed SNPs on mRNA levels because of the tissue access constraints. Therefore, the exact mechanisms by which the SNPs regulate the gene transcription activity and/or impact protein activity should be verified. Second, we analyzed only nine SNPs within seven DDR genes, and this may be insufficient to fully predict the DDR contribution to overall risk of HBV-induced liver cirrhosis. Additionally, we lacked some potentially useful characteristics of the patients, such as duration of CHB or phase of infection, which may undoubtedly affect the severity of liver damage. Moreover, as currently noninvasive, elastography-based assessment of liver fibrosis is commonly used in clinical practice, our cohort constituted both patients who underwent liver biopsy and individuals with fibroscan analysis conducted as a noninvasive alternative to study liver fibrosis.
4. Materials and Methods
4.1. Patients
Adult patients with confirmed CHB infection (HBsAg positive for more than 6 months) were enrolled from the ANRS CO22 HEPATHER cohort (ClinicalTrials.gov registry number: NCT01953458) and Department of Infectious Diseases, Medical University of Gdansk, and the Hepatology Outpatients Clinic Pomeranian Centre for Infectious Diseases and Tuberculosis in Gdansk. Patients were enrolled in the study at the time of liver fibrosis assessment. Only individuals who were treatment-naïve or received only short-term ant-HBV treatment (<6 months) were included in the study. Additionally, a control group containing uninfected subjects seronegative to human immunodeficiency virus (HIV), HBV, and HCV from the Gdansk Regional Centre of Blood Donations and Hemotherapy.
Patients were diagnosed on the basis of the diagnostic criteria of guidance [66]. All serum samples underwent standard procedure in local clinical center laboratory, including tests for serum HBV surface antigen (HBsAg), HBV surface antibody (HBsAb), HBeAg, HBV e antibody (HBeAb), HBV core antibody (HBcAb), and HBV DNA levels. The patients with co-infection with HIV, HCV, or HDV were excluded from this study.
Fibrosis scores were assessed when patients were included in the cohort. For 132 patients who had liver biopsy conducted, we assessed degree of fibrosis and inflammation activity according to the Scheuer scoring system (F0–F4) [67]. Non-invasive transient elastography by using FibroScan (Echosens, Paris, France) was used to diagnose liver fibrosis for 520 patients in whom liver biopsy was not performed. Fibrosis and cirrhosis were staged according to the METAVIR scoring system [68]. The liver fibrosis group consisted of the patients who had stage I to stage III fibrosis according to the METAVIR or Scheuer scores. All individuals that had fibrosis stage IV were classified as being in the liver cirrhosis group.
All procedures followed were in compliance with the Declaration of Helsinki and were approved by the Local Independent Bioethics Committee at the Medical University of Gdansk. All enrolled subjects gave written informed consent for their participation.
4.2. SNP Genotyping
The MagNa Pure LC DNA Isolation System was used for genomic DNA extraction. A sample preparation followed the standard manufacturer protocol for MagNA Pure Compact Nucleic Acid Isolation Kit I (Roche, Mannheim, Germany). Gene analysis of XRCC1 (rs25487), ERCC2 (rs238406, rs13181), ERCC5 (rs2018836), RAD52 (rs7963551), Mre11 (rs496797, rs535801), NBN (rs2735383), and XRCC4 (rs1805377) was carried out by mass spectrometry analysis. The Mass Assay Designer software package (v.4.0) was used to design 9 specific PCR primer pairs and 9 extension primer sequences (Table S1). The manufacturer’s standard protocol was followed for iPLEX Pro chemistry (Agena Bioscience, San Diego, CA, USA). In brief, genomic DNA was amplified resulting in 10 different products containing SNPs of interest, followed by a shrimp alkaline phosphatase treatment to remove the excess of nucleotides. Next, the single-base extension reaction was performed with mass-modified terminator nucleotides, and after desalting, the obtained products were dispensed onto a 96 SpectroCHIP array using an RS1000 Nanodispenser. SNPs variations were distinguished with MALDI-TOF mass spectrometry on the basis of different molecular weights and analyzed by MassARRAY Typer Analyzer 4.0 software.
4.3. Statistical Analysis
Statistical analyses were performed using STATISTICA software version 13.3 (StatSoft, Tulsa, OK, USA). The linkage disequilibrium analysis and deviations from Hardy–Weinberg equilibrium of analyzed SNPs were conducted by the MIDAS software. The relation between categorical vs. categorical variables were evaluated by chi-squared or Fisher’s exact test, as appropriate. Logistic regression analysis was used to evaluate the contribution of genetic and nongenetic factors under the dominant, recessive, and additive models. A backward stepwise regression approach was applied when building multivariate models. All of the p-values presented were two-sided and only p < 0.05 was considered significant. A Benjamini–Hochberg procedure was applied to account for multiple testing. Statistical power was calculated post hoc using the G*Power 3.1.9.4.
5. Conclusions
In conclusion, this study shows that functional and potentially functional SNPs within XRCC1 and ERCC2 genes may confer susceptibility to HBV-associated liver cirrhosis. Moreover, ERCC2 rs238406, acting as a biomarker of high liver fibrosis stage, suggests an important role of the functional ERCC2 SNPs in the etiology of HBV-induced liver damage in a Caucasian population. Considering limitations of this study, future studies should explore more genes, more SNPs, and interaction with environmental/behavioral factors such as the alcohol use.
Acknowledgments
We thank participants and participating clinicians of the ANRS CO22 HEPATHER cohort at each study site. We thank F. Carrat, S. Pol, and H. Fontaine, the coordinators of the cohort; C. Dorival, the project manager; and the ANRS CO22 HEPATHER scientific committee. The ANRS CO22 HEPATHER cohort is sponsored and funded by Inserm-ANRS (France REcherche Nord&sud Sida-vih Hepatites); ANR (Agence Nationale de la Recherche); DGS (Direction Générale de la Santé); and MSD, Janssen, Gilead, AbbVie, BMS and Roche.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/11/3295/s1. Table S1: Primers used for MALDI-TOF MassARRAY single-nucleotide polymorphism genotyping. Table S2: Genotypic distribution of analyzed SNPs among chronic hepatitis B patients and a healthy control group. Table S3: Genotypic distribution of analyzed SNPs among chronic hepatitis B patients with liver cirrhosis, hepatic fibrosis, and a no fibrosis group. Table S4: Multiple logistic regression model for liver cirrhosis risk in chronic hepatitis B patients. Table S5: Mann-Whitney U-test analysis levels of selected variabilities in the plasma of patients with and without liver cirrhosis. Table S6: Mann-Whitney U-test analysis levels of selected variabilities in the plasma of patients with and without advanced liver fibrosis. Table S7: Multiple logistic regression model for liver cirrhosis risk in chronic hepatitis B patients.
Author Contributions
Conceptualization, M.R.; data curation, M.R., A.W., A.S., T.R., P.S., M.D. and E.R.V.; formal analysis, K.P.B.; funding acquisition, K.P.B.; investigation, M.R., A.W., A.S. and T.R.; methodology, M.R. and A.W.; project administration, K.P.B.; resources, P.S. and T.F.B.; software, M.R. and M.D.; supervision, M.R. and K.P.B.; validation, M.R. and A.W.; visualization, M.R.; writing—original draft, M.R.; writing—review and editing, M.R. and A.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Centre for Research and Development (NCBR, grant no. INFECT-ERA/01/2014); the French National Research Agency (ANR, grant no. ANR-13-IFEC-0006-02) in the frame of the European consortium Infect-ERA hepBccc; and the MOBI4Health project, which is funded from the European Union’s Seventh Framework Programme under grant agreement no. 316094. The APC was funded by the University of Gdansk.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Mokdad A.A., Lopez A.D., Shahraz S., Lozano R., Mokdad A.H., Stanaway J., Murray C.J., Naghavi M. Liver cirrhosis mortality in 187 countries between 1980 and 2010: A systematic analysis. BMC Med. 2014;12:145. doi: 10.1186/s12916-014-0145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanda T., Takahashi K., Nakamura M., Nakamoto S., Wu S., Haga Y., Sasaki R., Jiang X., Yokosuka O. Androgen Receptor Could Be a Potential Therapeutic Target in Patients with Advanced Hepatocellular Carcinoma. Cancers. 2017;9:43. doi: 10.3390/cancers9050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2015 Risk Factors Collaborators Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet. 2016;388:1659–1724. doi: 10.1016/S0140-6736(16)31679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.GBD 2016 Risk Factors Collaborators Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks, 1990–016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1345–1422. doi: 10.1016/S0140-6736(17)32366-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lampertico P., Agarwal K., Berg T., Buti M., Janssen H.L.A., Papatheodoridis G., Zoulim F., Tacke F. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 2017;67:370–398. doi: 10.1016/j.jhep.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Terrault N.A., Lok A.S.F., McMahon B.J., Chang K.-M., Hwang J.P., Jonas M.M., Brown R.S., Bzowej N.H., Wong J.B. Update on Prevention, Diagnosis, and Treatment of Chronic Hepatitis B: AASLD 2018 Hepatitis B Guidance. Clin. Liver Dis. 2018;12:33–34. doi: 10.1002/cld.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlachogiannakos J., Papatheodoridis G.V. Hepatitis B: Who and when to treat? Liver Int. 2018;38:71–78. doi: 10.1111/liv.13631. [DOI] [PubMed] [Google Scholar]
- 8.Yang S.-F., Chang C.-W., Wei R.-J., Shiue Y.-L., Wang S.-N., Yeh Y.-T. Involvement of DNA damage response pathways in hepatocellular carcinoma. Biomed. Res. Int. 2014;2014:153867. doi: 10.1155/2014/153867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee N., Walker G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017;58:235–263. doi: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Königer C., Wingert I., Marsmann M., Rösler C., Beck J., Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc. Natl. Acad. Sci. USA. 2014;111:E4244–E4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko H.-L., Ren E.-C. Novel poly (ADP-ribose) polymerase 1 binding motif in hepatitis B virus core promoter impairs DNA damage repair. Hepatology. 2011;54:1190–1198. doi: 10.1002/hep.24502. [DOI] [PubMed] [Google Scholar]
- 12.Chung Y.-L. Defective DNA damage response and repair in liver cells expressing hepatitis B virus surface antigen. FASEB J. 2013;27:2316–2327. doi: 10.1096/fj.12-226639. [DOI] [PubMed] [Google Scholar]
- 13.Kitamura K., Wang Z., Chowdhury S., Simadu M., Koura M., Muramatsu M. Uracil DNA Glycosylase Counteracts APOBEC3G-Induced Hypermutation of Hepatitis B Viral Genomes: Excision Repair of Covalently Closed Circular DNA. PLoS Pathog. 2013;9:e1003361. doi: 10.1371/journal.ppat.1003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricardo-Lax I., Ramanan V., Michailidis E., Shamia T., Reuven N., Rice C.M., Shlomai A., Shaul Y. Hepatitis B virus induces RNR-R2 expression via DNA damage response activation. J. Hepatol. 2015;63:789–796. doi: 10.1016/j.jhep.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Schreiner S., Nassal M. A Role for the Host DNA Damage Response in Hepatitis B Virus cccDNA Formation-and Beyond? Viruses. 2017;9:125. doi: 10.3390/v9050125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgs M.R., Chouteau P., Lerat H. “Liver let die”: Oxidative DNA damage and hepatotropic viruses. J. Gen. Virol. 2014;95:991–1004. doi: 10.1099/vir.0.059485-0. [DOI] [PubMed] [Google Scholar]
- 17.Hollingworth R., Skalka G., Stewart G., Hislop A., Blackbourn D., Grand R. Activation of DNA Damage Response Pathways during Lytic Replication of KSHV. Viruses. 2015;7:2908–2927. doi: 10.3390/v7062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez-Moreno A., Garaigorta U. Hepatitis B Virus and DNA Damage Response: Interactions and Consequences for the Infection. Viruses. 2017;9:304. doi: 10.3390/v9100304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrier P., Debette-Gratien M., Jacques J., Loustaud-Ratti V. Cirrhotic patients and older people. World J. Hepatol. 2019;11:663–677. doi: 10.4254/wjh.v11.i9.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith A., Baumgartner K., Bositis C. Cirrhosis: Diagnosis and Management. Am. Fam. Phys. 2019;100:759–770. [PubMed] [Google Scholar]
- 21.Chen V.L., Chen Y., Du X., Handelman S.K., Speliotes E.K. Genetic variants that associate with liver cirrhosis have pleiotropic effects on human traits. Liver Int. 2019 doi: 10.1111/liv.14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Youssef S.S., Abbas E.A.E.R., Youness R.A., Elemeery M.N., Nasr A.S., Seif S. PNPLA3 and IL 28B signature for predicting susceptibility to chronic hepatitis C infection and fibrosis progression. Arch. Physiol. Biochem. 2019 doi: 10.1080/13813455.2019.1694039. [DOI] [PubMed] [Google Scholar]
- 23.Thanapirom K., Suksawatamnuay S., Sukeepaisarnjaroen W., Tangkijvanich P., Thaimai P., Wasitthankasem R., Poovorawan Y., Komolmit P. Genetic associations of Vitamin D receptor polymorphisms with advanced liver fibrosis and response to pegylated interferon-based therapy in chronic hepatitis C. PeerJ. 2019;2019 doi: 10.7717/peerj.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abshagen K., Berger C., Dietrich A., Schütz T., Wittekind C., Stumvoll M., Blüher M., Klöting N. A Human REPIN1 Gene Variant: Genetic Risk Factor for the Development of Nonalcoholic Fatty Liver Disease. Clin. Transl. Gastroenterol. 2020;11 doi: 10.14309/ctg.0000000000000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akcay I.M., Katrinli S., Ozdil K., Doganay G.D., Doganay L. Host genetic factors affecting hepatitis B infection outcomes: Insights from genome-wide association studies. World J. Gastroenterol. 2018;24:3347–3360. doi: 10.3748/wjg.v24.i30.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guidotti L.G., Chisari F.V. Immunobiology and pathogenesis of viral hepatitis. Annu. Rev. Pathol. 2006;1:23–61. doi: 10.1146/annurev.pathol.1.110304.100230. [DOI] [PubMed] [Google Scholar]
- 27.Rehermann B. Pathogenesis of chronic viral hepatitis: Differential roles of T cells and NK cells. Nat. Med. 2013;19:859–868. doi: 10.1038/nm.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheraz M., Cheng J., Tang L., Chang J., Guo J.-T. Cellular DNA Topoisomerases Are Required for the Synthesis of Hepatitis B Virus Covalently Closed Circular DNA. J. Virol. 2019;93 doi: 10.1128/JVI.02230-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J., Luckenbaugh L., Hu H., Yan Z., Gao L., Hu J. Involvement of host atr-chk1 pathway in hepatitis b virus covalently closed circular dna formation. Am. Soc. Microbiol. 2020;11 doi: 10.1128/mBio.03423-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nazarkina Z.K., Khodyreva S.N., Marsin S., Lavrik O.I., Radicella J.P. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair (Amst) 2007;6:254–264. doi: 10.1016/j.dnarep.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Guan Q., Chen Z., Chen Q., Zhi X. XRCC1 and XPD polymorphisms and their relation to the clinical course in hepatocarcinoma patients. Oncol. Lett. 2017;14:2783–2788. doi: 10.3892/ol.2017.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feki-Tounsi M., Khlifi R., Louati I., Fourati M., Mhiri M.N., Hamza-Chaffai A., Rebai A. Polymorphisms in XRCC1, ERCC2, and ERCC3 DNA repair genes, CYP1A1 xenobiotic metabolism gene, and tobacco are associated with bladder cancer susceptibility in Tunisian population. Environ. Sci. Pollut. Res. 2017;24:22476–22484. doi: 10.1007/s11356-017-9767-x. [DOI] [PubMed] [Google Scholar]
- 33.Isakova J., Talaibekova E., Aldasheva N., Vinnikov D., Aldashev A. The association of polymorphic markers Arg399Gln of XRCC1 gene, Arg72Pro of TP53 gene and T309G of MDM2 gene with breast cancer in Kyrgyz females. BMC Cancer. 2017;17 doi: 10.1186/s12885-017-3762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tasnim T., Al-Mamun M.M.A., Nahid N.A., Islam M.R., Apu M.N.H., Bushra M.U., Rabbi S.N.I., Nahar Z., Chowdhury J.A., Ahmed M.U., et al. Genetic variants of SULT1A1 and XRCC1 genes and risk of lung cancer in Bangladeshi population. Tumor Biol. 2017;39 doi: 10.1177/1010428317729270. [DOI] [PubMed] [Google Scholar]
- 35.Li J., Li Z., Feng L., Guo W., Zhang S. Polymorphisms of DNA repair gene XRCC1 and hepatocellular carcinoma risk among East Asians: A meta-analysis. Tumor Biol. 2013;34:261–269. doi: 10.1007/s13277-012-0546-5. [DOI] [PubMed] [Google Scholar]
- 36.Guo L.Y., Jin X.P., Niu W., Li X.F., Liu B.H., Wang Y.L. Association of XPD and XRCC1 genetic polymorphisms with hepatocellular carcinoma risk. Asian Pac. J. Cancer Prev. 2012;13:4423–4426. doi: 10.7314/APJCP.2012.13.9.4423. [DOI] [PubMed] [Google Scholar]
- 37.Zeng X.Y., Huang J.M., Xu J.W., Xu Y., Yu H.P., Ji L., Qiu X.Q. Meta-analysis demonstrates lack of a relationship between XRCC1-399 gene polymorphisms and susceptibility to hepatocellular carcinoma. Genet. Mol. Res. 2013;12:1916–1923. doi: 10.4238/2013.March.15.5. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y.H., Wang B., Xu B.P., Jiang D.N., Zhao D.M., Ji M.R., Zhou L., Li X., Lu C.Z. The association of six non-synonymous variants in three DNA repair genes with hepatocellular carcinoma risk: A meta-analysis. J. Cell. Mol. Med. 2016;20:2056–2063. doi: 10.1111/jcmm.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bose S., Tripathi D.M., Sukriti, Sakhuja P., Kazim S.N., Sarin S.K. Genetic polymorphisms of CYP2E1 and DNA repair genes HOGG1 and XRCC1: Association with hepatitis B related advanced liver disease and cancer. Gene. 2013;519:231–237. doi: 10.1016/j.gene.2013.02.025. [DOI] [PubMed] [Google Scholar]
- 40.Almeida Pereira Leite S.T., Marques-Guimarães N., Silva-Oliveira J.C., Dutra-Souto F.J., Alves-dos-Santos R., Bassi-Branco C.L. The X-ray repair cross complementing protein 1 (XRCC1) rs25487 polymorphism and susceptibility to cirrhosis in Brazilian patients with chronic viral hepatitis. Ann. Hepatol. 2013;12:733–739. doi: 10.1016/S1665-2681(19)31314-6. [DOI] [PubMed] [Google Scholar]
- 41.Wei B., Zhou Y., Xu Z., Ruan J., Zhu M., Jin K., Zhou D., Hu Q., Wang Q., Wang Z., et al. XRCC1 Arg399Gln and Arg194Trp polymorphisms in prostate cancer risk: A meta-analysis. Prostate Cancer Prostatic Dis. 2011;14:225–231. doi: 10.1038/pcan.2011.26. [DOI] [PubMed] [Google Scholar]
- 42.Bernstein C., Bernstein H., Payne C.M., Garewal H. DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: Fail-safe protection against carcinogenesis. Mutat. Res. Rev. Mutat. Res. 2002;511:145–178. doi: 10.1016/S1383-5742(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 43.Slyskova J., Dusinska M., Kuricova M., Soucek P., Vodickova L., Susova S., Naccarati A., Tulupova E., Vodicka P. Relationship between the capacity to repair 8-oxoguanine, biomarkers of genotoxicity and individual susceptibility in styrene-exposed workers. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2007;634:101–111. doi: 10.1016/j.mrgentox.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 44.Au W.W., Salama S.A., Sierra-Torres C.H. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ. Health Perspect. 2003;111:1843–1850. doi: 10.1289/ehp.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han X., Xing Q., Li Y., Sun J., Ji H., Pan H., Jun L. Study on the DNA repair gene XRCC1 and XRCC3 polymorphism in prediction and prognosis of hepatocellular carcinoma risk. Hepatogastroenterology. 2012;59:2285–2289. doi: 10.5754/hge12096. [DOI] [PubMed] [Google Scholar]
- 46.Wang S., Wang J., Bai Y., Wang Q., Liu L., Zhang K., Hong X., Deng Q., Zhang X., He M., et al. The genetic variations in DNA repair genes ERCC2 and XRCC1 were associated with the overall survival of advanced non-small-cell lung cancer patients. Cancer Med. 2016;5:2332–2342. doi: 10.1002/cam4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ha H.L., Shin H.J., Feitelson M.A., Yu D.Y. Oxidative stress and antioxidants in hepatic pathogenesis. World J. Gastroenterol. 2010;16:6035–6043. doi: 10.3748/wjg.v16.i48.6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xianyu J., Feng J., Yang Y., Tang J., Xie G., Fan L. Correlation of oxidative stress in patients with HBV-induced liver disease with HBV genotypes and drug resistance mutations. Clin. Biochem. 2018;55:21–27. doi: 10.1016/j.clinbiochem.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 49.Ma N., Liu W., Zhang X., Gao X., Yu F., Guo W., Meng Y., Gao P., Zhou J., Yuan M., et al. Oxidative Stress-Related Gene Polymorphisms Are Associated With Hepatitis B Virus-Induced Liver Disease in the Northern Chinese Han Population. Front. Genet. 2020;10 doi: 10.3389/fgene.2019.01290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krokan H.E., Bjørås M. Base excision repair. Cold Spring Harb. Perspect. Biol. 2013;5:1–22. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh A., Compe E., Le May N., Egly J.M. TFIIH subunit alterations causing xeroderma pigmentosum and trichothiodystrophy specifically disturb several steps during transcription. Am. J. Hum. Genet. 2015;96:194–207. doi: 10.1016/j.ajhg.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li M., Zhao Y., Zhao E., Wang K., Lu W., Yuan L. Predictive Value of Two Polymorphisms of ERCC2, rs13181 and rs1799793, in Clinical Outcomes of Chemotherapy in Gastric Cancer Patients: A meta-analysis. Dis. Markers. 2018;2018 doi: 10.1155/2018/3947626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai P., Li J., Li W., Qin X., Wu X., Di W., Zhang Y., Kumar S. Genetic polymorphisms and pancreatic cancer risk: A PRISMA-compliant systematic review and meta-analysis. Medicine. 2019;98 doi: 10.1097/MD.0000000000016541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Z., Kong J., Kong Y., Cai F., Xu X., Liu J., Wang S. Association of XPD Asp312Asn polymorphism and response to oxaliplatin-based first-line chemotherapy and survival in patients with metastatic colorectal cancer. Adv. Clin. Exp. Med. 2019;28:1459–1468. doi: 10.17219/acem/108552. [DOI] [PubMed] [Google Scholar]
- 55.Reis H., Szarvas T. Predictive biomarkers in bladder cancer. Pathologe. 2019 doi: 10.1007/s00292-019-00688-5. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y., Zhao E., Zhang J., Chen Y., Ma J., Li H. A comprehensive evaluation of the association between polymorphisms in XRCC1, ERCC2, and XRCC3 and Prognosis in Hepatocellular Carcinoma: A Meta-Analysis. J. Oncol. 2019;2019 doi: 10.1155/2019/2408946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tavares C.B., Gomes-Braga F.D., Sousa E.B., Brito J.N.P.d.O., Melo M.d.A., Campelo V., Net F.M., de Araújo R.M.L., Kessler I.M., SousaJúnior L.d.M., et al. Association between single nucleotide polymorphisms and glioma risk: A systematic literature review. Cancer Investig. 2020:1–35. doi: 10.1080/07357907.2020.1719502. [DOI] [PubMed] [Google Scholar]
- 58.Xiao S., Cui S., Lu X., Guan Y., Li D., Liu Q., Cai Y., Jin C., Yang J., Wu S., et al. The ERCC2/XPD Lys751Gln polymorphism affects DNA repair of benzo[a]pyrene induced damage, tested in an in vitro model. Toxicol. Vitr. 2016;34:300–308. doi: 10.1016/j.tiv.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 59.Zhou F., Zhu M., Wang M., Qiu L., Cheng L., Jia M., Xiang J., Wei Q. Genetic variants of DNA repair genes predict the survival of patients with esophageal squamous cell cancer receiving platinum-based adjuvant chemotherapy. J. Transl. Med. 2016;14 doi: 10.1186/s12967-016-0903-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu H., Li S., Hu X., Qin W., Wang Y., Sun T., Wu Z., Wang X., Lu S., Xu D., et al. Associations of mRNA expression of DNA repair genes and genetic polymorphisms with cancer risk: A bioinformatics analysis and meta-analysis. J. Cancer. 2019;10:3593–3607. doi: 10.7150/jca.30975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fernández-Calero T., Cabrera-Cabrera F., Ehrlich R., Marín M. Silent Polymorphisms: Can the tRNA Population Explain Changes in Protein Properties? Life. 2016;6:9. doi: 10.3390/life6010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L., Xu M., Cui C.B., Wei P.H., Wu S.Z., Cen Z.J., Meng X.X., Huang Q.G., Xie Z.C. Diagnostic and prognostic values of the mRNA expression of excision repair cross-complementation enzymes in hepatitis B virus-related hepatocellular carcinoma. Cancer Manag. Res. 2018;10:5313–5328. doi: 10.2147/CMAR.S179043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nagahashi M., Matsuda Y., Moro K., Tsuchida J., Soma D., Hirose Y., Kobayashi T., Kosugi S.I., Takabe K., Komatsu M., et al. DNA damage response and sphingolipid signaling in liver diseases. Surg. Today. 2016;46:995–1005. doi: 10.1007/s00595-015-1270-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qadri I., Conaway J.W., Conaway R.C., Schaack J., Siddiqui A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc. Natl. Acad. Sci. USA. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang G., Yang Y., Ma X., Huang L., Li W., Song X., Zhang H., Liu W., Lu J. Association of ERCC5 Genetic Polymorphisms With Cirrhosis and Liver Cancer. Technol. Cancer Res. Treat. 2020;19 doi: 10.1177/1533033820943244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terrault N.A., Lok A.S.F., McMahon B.J., Chang K.-M., Hwang J.P., Jonas M.M., Brown R.S., Bzowej N.H., Wong J.B., Wong J. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology. 2018;67:1560–1599. doi: 10.1002/hep.29800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Scheuer P.J. Classification of chronic viral hepatitis: A need for reassessment. J. Hepatol. 1991;13:372–374. doi: 10.1016/0168-8278(91)90084-O. [DOI] [PubMed] [Google Scholar]
- 68.Carrat F., Fontaine H., Dorival C., Simony M., Diallo A., Hezode C., De Ledinghen V., Larrey D., Haour G., Bronowicki J.P., et al. Clinical outcomes in patients with chronic hepatitis C after direct-acting antiviral treatment: A prospective cohort study. Lancet. 2019;393:1453–1464. doi: 10.1016/S0140-6736(18)32111-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.