Abstract.
Understanding patterns and trends of antimicrobial resistance (AMR) in Salmonella Typhi can guide empiric treatment recommendations and contribute to country decisions about typhoid conjugate vaccine (TCV) introduction. We systematically reviewed PubMed and Web of Science for articles reporting the proportion of Salmonella Typhi isolates resistant to individual antimicrobials worldwide from any time period. Isolates resistant to chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole were classified as multidrug resistant (MDR), and isolates that were MDR plus resistant to a fluoroquinolone and a third-generation cephalosporin were extensively drug resistant (XDR). Among the 198 articles eligible for analysis, a total of 55,459 Salmonella Typhi isolates were tested for AMR (median 80; range 2–5,191 per study). Of isolates from 2015 through 2018 in Asia, 1,638 (32.6%) of 5,032 were MDR, 167 (5.7%) of 2,914 were resistant to third-generation cephalosporins, and 148 (8.3%) of 1,777 were resistant to azithromycin. Two studies from Pakistan reported 14 (2.6%) of 546 isolates were XDR. In Africa, the median proportion of Salmonella Typhi isolates that were MDR increased each consecutive decade from 1990 to 1999 through 2010 to 2018. Salmonella Typhi has developed resistance to an increasing number of antimicrobial classes in Asia, where XDR Salmonella Typhi is now a major threat, whereas MDR has expanded in Africa. We suggest continued and increased surveillance is warranted to inform empiric treatment decisions and that AMR data be incorporated into country decisions on TCV introduction.
INTRODUCTION
Typhoid fever is a systemic infection caused by the bacterium Salmonella enterica subspecies enterica serovar Typhi (Salmonella Typhi) and is an important cause of illness and death worldwide, with an estimated 10.9 million new infections and 116,800 typhoid fever–related deaths occurring annually.1–3 Typhoid fever is difficult to distinguish from other causes of febrile illnesses, compounded by limited laboratory services in some low- and middle-income countries, making specific diagnosis and appropriate antimicrobial treatment challenging in routine practice.4–6
Historically, chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole were the first-line antimicrobial treatments for typhoid fever.7 However, multidrug resistant (MDR) Salmonella Typhi, defined as resistance to these three first-line drugs,8 was widespread by the late 1980s and 1990s, with reports from Pakistan, India, and other southern and Southeast Asian countries.9 Subsequently, ciprofloxacin became the drug of choice, but resistance appeared quickly, first in 199110 and in an outbreak in 1997.11 With the emergence of MDR Salmonella Typhi and MDR with fluoroquinolone resistance, third-generation cephalosporins, macrolides, and carbapenems have been used increasingly for the treatment of typhoid fever.7 Extensively drug resistant (XDR) Salmonella Typhi, defined as resistance to first-line antimicrobials, a fluoroquinolone, and a third-generation cephalosporin,12 was reported in Hyderabad, Pakistan, in 2016.13 Since then, the WHO has been notified of more than 10,365 infections with XDR Salmonella Typhi in Pakistan,14 and travel-associated infections have been reported in Canada,15 Denmark,16 Australia,17 and the United States.18 Hence, antimicrobial resistance (AMR) in Salmonella Typhi is a global threat.
Increasing access to safe drinking water, food, and improved sanitation are important measures for controlling the impact and spread of typhoid fever,19–21 but have elsewhere been associated with socioeconomic progress that has taken place over long time periods.22–24 Typhoid vaccines represent an important and accessible tool to avert illness and death in the short- to medium-term while water, food, and sanitation improvements take place. To assist countries with decision-making about typhoid conjugate vaccine (TCV) introduction and other control efforts, and to guide empiric management decisions for typhoid fever, we performed a comprehensive, systematic review of the literature to describe the prevalence and trends of AMR among Salmonella Typhi.
METHODS
Search strategy and study selection.
The systematic review protocol was registered with PROPSERO: International Prospective Register of Systematic Reviews (CRD42019131038) on May 10, 2019. Following the preferred reporting items for systematic reviews and meta-analyses,25 we searched two databases, PubMed from inception through April 16, 2019, and Web of Science from inception through April 17, 2019. Each search included key words of Typhi, typhoid, “enteric fever,” antimicrobial, susceptibility, and resistance (Supplemental Table S1). No restrictions were placed on location, date of specimen collection, or language of publication.
Epidemiologic studies of any design reporting antimicrobial susceptibility testing (AST) results of Salmonella Typhi isolated from human source, normally sterile site (e.g., blood, bone marrow) specimens, were included. Policy reports, commentaries, editorials, and conference abstracts were excluded, as were studies where we could not distinguish between isolates from normally sterile site specimens and isolates not from normally sterile site specimens (e.g., stool, urine). Studies that did not present sufficient detail to calculate the prevalence of AMR among all reported Salmonella Typhi isolates were also excluded. Studies reporting travel-associated infections were included only if we were able to identify the country where the infection was likely acquired.
Search results from each database were imported into Endnote X8 (Clarivate Analytics, Boston, MA). We also included studies identified from the reference lists of three previous reviews on bloodstream infections that also reported AST of Salmonella Typhi.26–28 Endnote was used to remove duplicates before a de-duplicated list of articles was uploaded to the online systematic review tool Rayyan (Qatar Computing Research Institute, Doha, Qatar).29 All subsequent processes were performed in parallel by two authors (C. S. M. and S. D. C.). We screened titles and abstracts for inclusion, and any article selected by at least one author was included for full-text review. We then screened each full-text article for inclusion, with discrepancies resolved through discussion or the involvement of a third author (J. A. C.). After establishing a final list of included full-text articles, two authors abstracted study characteristics and AST data in parallel using a shared Google Sheets spreadsheet (Google LLC, Mountain View, CA).
Data abstraction.
Study characteristics that were abstracted included first author, publication year, normally sterile specimen type, and country of specimen collection. When a study collected isolates from multiple locations within a country or in separate countries, we documented the additional locations as study sites. We abstracted data for the year of susceptibility testing; antimicrobial susceptibility testing (AST) interpretive criteria used, such as the Clinical and Laboratory Standards Institute (CLSI) or European Committee on AST, and year of criteria; AST method (e.g., agar dilution, disk diffusion); number of total isolates tested; and number of isolates that tested susceptible, intermediate, or resistant to a predefined list of antimicrobials. The predefined list was compiled from the CLSI M100 table of suggested antimicrobial agents for Enterobacteriaceae groups A and B.30 Because zone sizes or minimum inhibitory concentration (MIC) values were often not reported, we were unable to attempt to recategorize AST results for fluoroquinolones, third-generation cephalosporins, and azalides for the current CLSI interpretive criteria.31
In addition to recording susceptibility results to individual antimicrobials, data were also abstracted on the MDR phenotype, defined as resistance to chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole9; and the XDR phenotype, defined as resistance to ampicillin, chloramphenicol, trimethoprim–sulfamethoxazole, a fluoroquinolone, and a third-generation cephalosporin.12 Fluoroquinolones abstracted were ciprofloxacin or ofloxacin, and third-generation cephalosporins were ceftriaxone or cefotaxime. We subsequently classified study sites by UN geographic regions and subregions.32
Analyses.
For each antimicrobial tested, we divided the number of resistant isolates by the total number of isolates tested and multiplied the resulting fraction by 100 to produce the proportion (%) of resistant isolates. When only the number of susceptible isolates was supplied and resistant isolates were not explicitly provided, we assumed that non-susceptible isolates were resistant. For example, an article described 90.0% of 100 isolates were susceptible to ampicillin, and we imputed data that 10 isolates in that study were resistant. No other data were imputed. Median proportions of isolates resistant by UN region and the chi-squared test for trend in proportions over four time periods (1970–1989, 1990–1999, 2000–2009, and 2010–2018) were calculated in R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria). Time periods were selected by decade, combining the 1970s and 1980s because of limited data available during those years. Time series histograms using 5-year interval periods were produced in Microsoft Excel 2016 (Microsoft Corporation, Redmond, WA) to illustrate the emergence of resistance patterns over time globally, in Asia, and in Africa. Intervals of 5 years were chosen for histograms to produce a more detailed time series than every 10 years. Maps were created using an online open-source map tool.33,34 In keeping with the aim to aid country-based decisions on TCV introduction and guide empiric management of typhoid fever, the decision to not perform a meta-analysis was made to avoid pooling data of multiple drugs from multiple countries. As a secondary analysis of published data, this study was exempt from institutional review board approval.
RESULTS
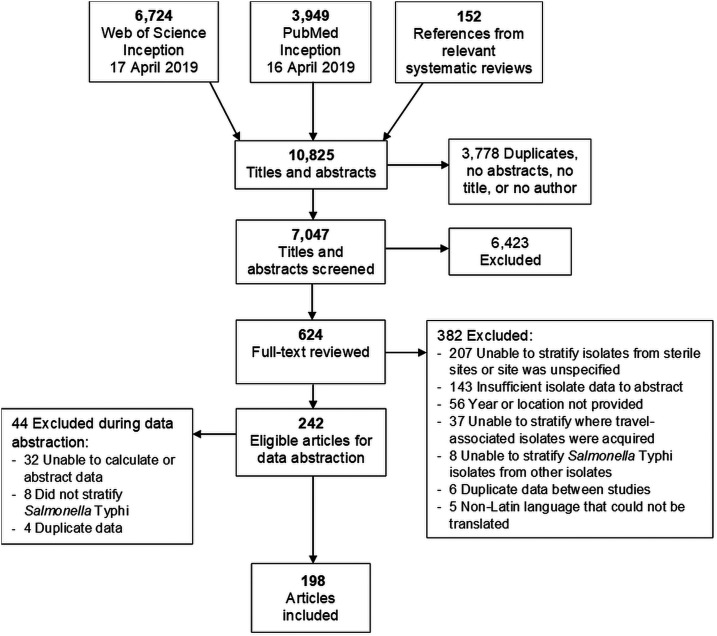
Our search of PubMed and Web of Science returned 6,724 and 3,949 articles, respectively (Figure 1). When including the references of relevant systematic reviews and then removing duplicates, we screened 7,047 titles and abstracts. Of these, 624 (8.9%) full-text articles were eligible for further review. We excluded 426 articles. The most common reason for article exclusion was failure to distinguish isolates that were collected from normally sterile sites from other sites, or the site was unspecified, resulting in 198 articles eligible for analysis (Supplemental Table S2).
Figure 1.
Preferred reporting items for systematic reviews and meta-analyses flow diagram of search strategy and selection of articles for antimicrobial resistance in Salmonella Typhi, 1972–2018. Some articles met the multiple exclusion criteria, and thus the sum of exclusion reasons is greater than the number of articles excluded.
Study characteristics.
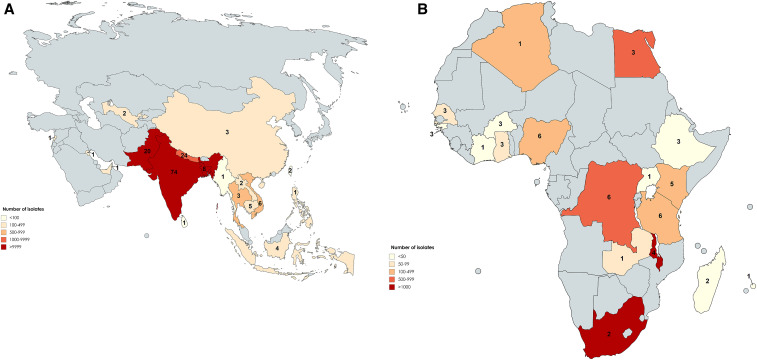
Studies reported on isolates collected from 1972 through 2018 in 38 countries and tested a total of 55,459 Salmonella Typhi isolates for AMR. By study, a median (range) of 80 (2–5,191) Salmonella Typhi isolates were tested. Among the 198 studies, four (2.0%) reported data collection in multiple locations, resulting in 216 study sites; 159 (73.6%) of 216 were located in 18 countries in the Asia region (Figure 2A), 54 (25.0%) in 18 countries in Africa (Figure 2B), two (0.9%) in the Americas, and one (0.5%) in Europe. There were 127 (58.8%) sites located in Southern Asia: 74 (34.3%) in India, 24 (11.1%) in Nepal, and 20 (9.3%) in Pakistan. Among isolates tested, 47,145 (85.0%) of 55,459 were from study sites located in Asia, 8,249 (14.9%) were from Africa, 43 (0.1%) were from the Americas, and 22 (< 0.1%) from Europe. Among isolates from Asia, 43,870 (93.1%) of 47,145 were from the Southern Asia subregion.
Figure 2.
(A) Number of study sites and isolates tested by country in the Asia region, 1972–2018 (created with MapChart). Number for each country denotes the number of study sites that reported data for Salmonella Typhi resistance. (B) Number of study sites and isolates tested by country in the Africa region, 1972–2018 (created with MapChart). Number for each country denotes the number of study sites that reported data for Salmonella Typhi resistance.
Of the 198 studies, 130 (65.7%) reported AST using CLSI interpretive criteria (Table 1). Among studies using CLSI criteria, 82 (63.1%) were before the 2012 breakpoint changes for fluoroquinolones.35 Ten (5.1%) studies used a combination of two AST interpretive criteria, and 59 (29.8%) studies did not report which AST interpretive criteria were used. Regarding laboratory antimicrobial susceptibility methods and findings, 173 (87.4%) of 198 studies used a disc diffusion method (e.g., the Kirby–Bauer test, Stokes’ method), whereas 82 (41.1%) used outputs from MIC techniques (e.g., broth micro-dilution, E-test, agar dilution) to determine resistant, intermediate, and susceptible isolates. Twelve (6.1%) used automated methods; 78 (39.4%) reported using a combination of a disc diffusion, MIC, or automated method; and nine (4.5%) did not report how susceptibility was determined.
Table 1.
Interpretive criteria and laboratory testing method used among 198 included studies of antimicrobial resistance of Salmonella Typhi, global, 1972 through 2018
| Interpretive criteria | Number of studies (% of 198 included*) |
|---|---|
| CLSI | 83 (41.9) |
| Pre-2012 | 35 (17.7) |
| 2012 or later | 38 (19.2) |
| Specific year unreferenced† | 10 (5.1) |
| Not reported | 59 (29.8) |
| The National Committee for Clinical Laboratory Standards (now CLSI as of 2005) | 47 (23.7) |
| The European Committee on Antimicrobial Susceptibility Testing | 10 (5.1) |
| The British Society for Antimicrobial Chemotherapy | 7 (3.5) |
| The French Microbiology Society | 2 (1.0) |
| Laboratory antimicrobial susceptibility testing method | |
| Disc diffusion‡ | 173 (87.4) |
| Minimum inhibitory concentration§ | 82 (41.4) |
| Combination of at least two methods | 78 (39.4) |
| Automated | 12 (6.1) |
| Not reported | 9 (4.5) |
CLSI = Clinical and Laboratory Standards Institute.
Some studies used multiple interpretive criteria or testing strategies for different antimicrobials; thus, numbers will exceed the total number of studies included.
Two studies specified multiple CLSI years covering both pre-2012 and 2012 or later.
The Kirby–Bauer test and the Stokes method.
E-test, agar dilution, and broth dilution.
Overall AMR.
Among all Salmonella Typhi isolates, 9,056 (25.9%) of 34,996 were resistant to chloramphenicol, 13,481 (38.8%) of 34,783 to ampicillin, and 13,366 (37.9%) of 35,270 to trimethoprim–sulfamethoxazole (Table 2). Of isolates, 12,666 (35.5%) of 35,659 were MDR, 9,495 (64.7%) of 14,671 were nalidixic acid resistant, and 5,406 (15.0%) and 6,979 (19.4%) of 35,975 were ciprofloxacin resistant and intermediate, respectively. Of isolates, 450 (1.3%) of 35,302 were resistant to ceftriaxone and 270 (4.5%) of 6,043 to azithromycin.
Table 2.
Salmonella Typhi antimicrobial susceptibility testing profiles, global, 1972–2018
| Antimicrobial class and agent* | Susceptible | Intermediate | Resistant | Total tested | Percent of isolates resistant |
|---|---|---|---|---|---|
| Traditional first-line | |||||
| Chloramphenicol | 25,907 | 33 | 9,056 | 34,996 | 25.9 |
| Ampicillin | 21,197 | 105 | 13,481 | 34,783 | 38.8 |
| Amoxicillin | 1,569 | 34 | 2,525 | 4,128 | 61.2 |
| Amoxicillin–clavulanic acid | 1,184 | 1 | 103 | 1,288 | 8.0 |
| Trimethoprim–sulfamethoxazole | 21,896 | 8 | 13,366 | 35,270 | 37.9 |
| Quinolone | |||||
| Nalidixic acid | 5,084 | 92 | 9,495 | 14,671 | 64.7 |
| Fluoroquinolone | |||||
| Ciprofloxacin | 23,590 | 6,979 | 5,406 | 35,975 | 15.0 |
| Ofloxacin | 8,095 | 389 | 4,106 | 12,590 | 32.6 |
| Third-generation cephalosporin | |||||
| Ceftriaxone | 34,771 | 81 | 450 | 35,302 | 1.3 |
| Cefotaxime | 5,072 | 45 | 468 | 5,585 | 8.4 |
| Macrolide | |||||
| Azithromycin | 5,759 | 14 | 270 | 6,043 | 4.5 |
| Carbapenem | |||||
| Meropenem | 813 | 0 | 21 | 834 | 2.5 |
| Aminoglycoside | |||||
| Gentamicin† | 5,477 | 16 | 676 | 6,169 | 11.0 |
| Tetracycline | 2,068 | 24 | 1,435 | 3,527 | 40.7 |
| Multidrug resistant (MDR) | – | – | 12,666 | 35,659 | 35.5 |
| Extensively drug resistant (XDR) | – | – | 14 | 546 | 2.6 |
MDR = phenotype defined as resistance to chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole. XDR = phenotype defined as resistance to ampicillin, chloramphenicol, trimethoprim–sulfamethoxazole, a fluoroquinolone, and a third-generation cephalosporin.
Antimicrobial susceptibility profiles not recategorized to current Clinical and Laboratory Standards Institute break points.
Susceptibility of gentamicin not recommended to be reported or used therapeutically for Salmonella Typhi by the Clinical Laboratory Standards Institute.
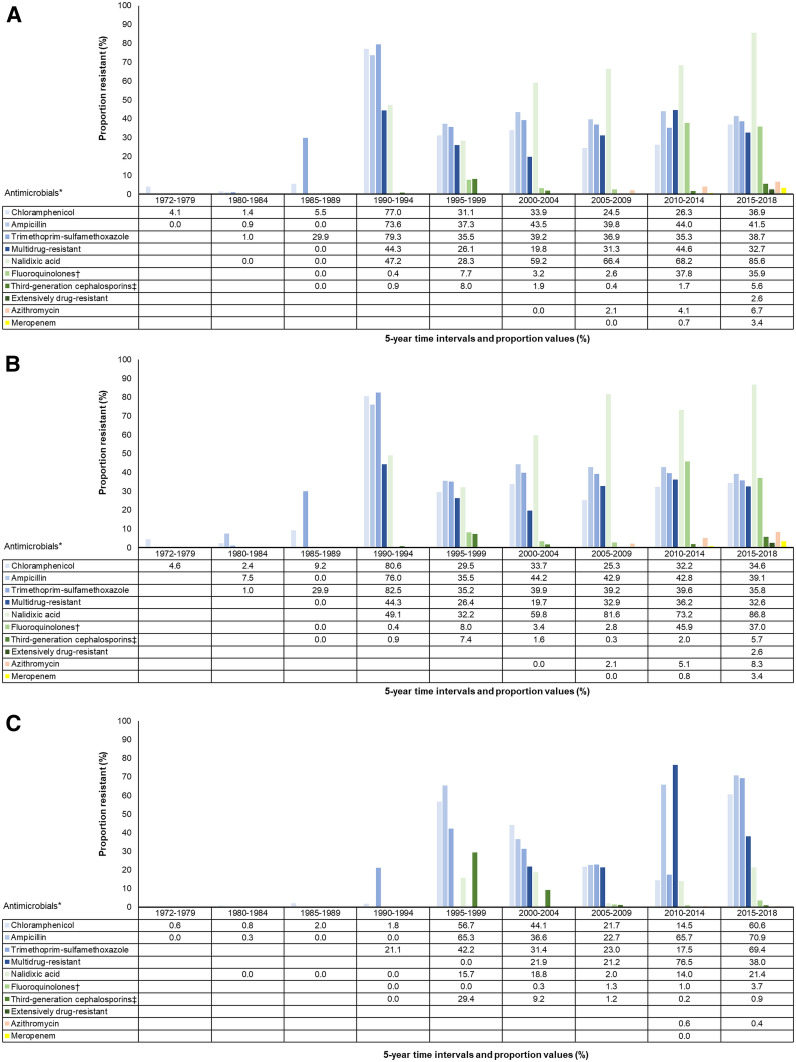
From 1990 through 1994, 955 (77.0%) of 1,241, 793 (73.6%) of 1,077, and 871 (79.3%) of 1,098 Salmonella Typhi isolates were resistant to chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole, respectively (Figure 3A). During the same period from 1990 through 1994, 1,205 (44.3%) of 2,719 were MDR. Of isolates from 2010 through 2014, 5,981 (44.6%) of 13,416 were MDR, and from 2015 through 2018 1,679 (32.7%) of 5,140 were MDR. Of isolates in the time periods 2005–2009, 2010–2014, and 2015–2018, 27 (2.1%) of 1,279, 93 (4.1%) of 2,263, and 150 (6.7%) of 2,247 were resistant to azithromycin, respectively.
Figure 3.
(A) Antimicrobial resistant Salmonella Typhi isolates worldwide, 1972–2018. (B) Antimicrobial resistant Salmonella Typhi isolates in Asia, 1972–2018. (C) Antimicrobial resistant Salmonella Typhi isolates in Africa, 1972–2018. Full data provided in Supplement Table S4. *Ordered chronologically by antimicrobial agent introduction and grouped by color by multidrug resistant and extensively drug resistant phenotypes. †Fluoroquinolone includes ciprofloxacin and ofloxacin. ‡Third-generation cephalosporin includes ceftriaxone and cefotaxime.
Antimicrobial resistance in Asia.
When stratified by time periods 1972–1989, 1990–1999, 2000–2009, and 2010–2018 in the Asia region, the median (interquartile range) proportion of Salmonella Typhi isolates that were MDR was 0.0% (0.0%), 31.2% (22.9–47.0%), 16.2% (6.1–35.6%), and 5.5% (2.0–24.3%), respectively (Table 3). By time period, ciprofloxacin resistance was found in zero (0.0%) of 61, 219 (3.7%) of 5,912, 439 (3.1%) of 14,040, and 4,670 (41.1%) of 11,349 (χ2 5,376, P < 0.001) isolates. By time period, ceftriaxone resistance was found in zero (0.0%) of 203, 124 (2.5%) of 4,898, 47 (0.4%) of 12,761, and 266 (1.9%) of 13,970 (χ2 2.3, P = 0.133) isolates.
Table 3.
Proportion of Salmonella Typhi isolates resistant to antimicrobials in Asia over four time periods from 1972 through 2018
| Antimicrobial* | 1972–1989 | 1990–1999 | 2000–2009 | 2010–2018 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n/N | % | Median proportion, % (IQR) | n/N | % | Median proportion, % (IQR) | n/N | % | Median proportion, % (IQR) | n/N | % | Median proportion, % (IQR) | χ2 Trend of proportion (P-value) | |
| Chloramphenicol | 210/4,525 | 4.6 | 4.0 (0.8–31.4) | 1,530/3,134 | 48.8 | 57.6 (37.7–78.0) | 2,418/8,149 | 29.7 | 20.8 (6.2–40.9) | 3,770/11,261 | 33.5 | 5.7 (2.1–31.3) | 649.9 (< 0.001) |
| Ampicillin | 16/225 | 7.1 | 0.0 (0.0–5.1) | 1,476/2,969 | 49.7 | 57.8 (36.4–68.8) | 4,044/9,319 | 43.4 | 38.5 (17.0–53.6) | 6,755/16,304 | 41.4 | 15.6 (5.0–47.7) | 17.5 (< 0.001) |
| Trimethoprim–sulfamethoxazole | 48/350 | 13.7 | 0.6 (0.0–9.0) | 1,533/2,956 | 51.9 | 54.3 (33.5–80.9) | 4,142/10,505 | 39.4 | 30.6 (9.9–50.0) | 6,428/16,799 | 38.3 | 6.8 (3.0–34.9) | 47.1 (< 0.001) |
| Multidrug resistant (MDR) | 0/61 | 0.0 | 0.0 (0.0) | 2,000/5,731 | 34.9 | 31.2 (22.9–47.0) | 2,780/10,214 | 27.2 | 16.2 (6.1–35.6) | 5,487/15,661 | 35.0 | 5.5 (2.0–24.3) | 23.1 (< 0.001) |
| Nalidixic acid | NR | – | NR | 390/874 | 44.6 | 24.5 (5.7–48.6) | 5,338/7,360 | 72.5 | 75.0 (60.4–86.9) | 3,667/4,868 | 75.3 | 93.1 (78.1–96.9) | 185.9 (< 0.001) |
| Ciprofloxacin | 0/61 | 0.0 | 0.0 (0.0) | 219/5,912 | 3.7 | 0.0 (0.0–5.0) | 439/14,040 | 3.1 | 0.0 (0.0–4.2) | 4,670/11,349 | 41.1 | 19.5 (3.5–75.4) | 5,376 (< 0.001) |
| Ceftriaxone | 0/203 | 0.0 | 0.0 (0.0) | 124/4,898 | 2.5 | 0.0 (0.0) | 47/12,761 | 0.4 | 0.0 (0.0) | 266/13,970 | 1.9 | 0.0 (0.0–0.1) | 2.3 (0.133) |
| Azithromycin | NR | – | NR | NR | – | NR | 27/1,533 | 1.8 | 0.0 (0.0) | 238/3,556 | 6.7 | 0.0 (0.0–5.0) | 51.8 (< 0.001) |
IQR = interquartile range; MDR = phenotype defined as resistance to chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole; NR = no eligible studies reporting data for year period and drug.
Antimicrobial susceptibility profiles not recategorized to current Clinical and Laboratory Standards Institute break points.
Of isolates from 2015 through 2018, 1,638 (32.6%) of 5,032 were MDR, 167 (5.7%) of 2,914 were resistant to third-generation cephalosporins, 148 (8.3%) of 1,777 to azithromycin, and 20 (3.4%) of 594 to meropenem (Figure 3B). Two studies from Pakistan reported 14 (2.6%) of 546 isolates were XDR.36,37
Antimicrobial resistance in Africa.
Two studies tested 3,327 isolates during the time period 1972 through 1989, and neither study reported testing trimethoprim–sulfamethoxazole, MDR, ciprofloxacin, ceftriaxone, or azithromycin (Table 4).38,39 In the time periods 1990–1999, 2000–2009, and 2010–2018, the median (interquartile range) proportion of MDR Salmonella Typhi isolates was 0.0% (0.0%), 0.0% (0.0–25.5%), and 38.0% (30.4–85.7%), respectively. Of isolates during the same three time periods, ciprofloxacin resistance was found in zero (0.0%) of 133, 14 (1.2%) of 1,209, and 39 (1.2%) of 3,228 (χ2 0.7, P = 0.415), respectively. Of isolates, ceftriaxone resistance was found in zero (0.0%) of 55, eight (0.7%) of 1,160, and five (0.2%) of 2,212 (χ2 2.9, P = 0.09), respectively. No eligible studies in Africa reported azithromycin resistance until the 2010–2018 period. Of isolates in 2010–2018, five (0.5%) of 954 were azithromycin resistant. One study reported zero (0.0%) of seven isolates resistant to meropenem in 2013.40
Table 4.
Proportion of Salmonella Typhi isolates resistant to antimicrobials in Africa over four time periods from 1972 through 2018
| 1972–1989 | 1990–1999 | 2000–2009 | 2010–2018 | χ2 Trend of proportion (P-value) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antimicrobial* | n/N | % | Median proportion, % (IQR) | n/N | % | Median proportion, % (IQR) | n/N | % | Median proportion, % (IQR) | n/N | % | Median proportion, % (IQR) | |
| Chloramphenicol | 6/3,313 | 0.2 | 0.0 (0.0–0.2) | 81/157 | 51.6 | 0.0 (0.0–0.0) | 319/1,276 | 25.0 | 49.7 (4.9–97.6) | 784/1,155 | 67.9 | 66.7 (0.0–74.1) | 2,251.9 (< 0.001) |
| Ampicillin | 35/3,313 | 1.1 | 0.6 (0.4–1.7) | 69/177 | 39.0 | 0.0 (0.0–2.1) | 294/1,206 | 24.4 | 31.8 (5.8–72.5) | 730/3,166 | 23.1 | 33.3 (0.0–76.7) | 675.7 (< 0.001) |
| Trimethoprim–sulfamethoxazole | NR | – | NR | 55/159 | 34.6 | 0.0 (0.0) | 310/1,273 | 24.4 | 40.4 (7.7–80.0) | 850/3,163 | 26.9 | 49.4 (0.2–79.9) | 0.002 (0.996) |
| Multidrug resistant (MDR) | NR | – | NR | 0/37 | 0.0 | 0.0 (0.0) | 226/1,060 | 21.3 | 0.0 (0.0–25.5) | 2,173/2,895 | 75.1 | 38.0 (30.4–85.7) | 976.6 (< 0.001) |
| Nalidixic acid | 0/14 | 0.0 | 0.0 (0.0) | 11/95 | 11.6 | 0.0 (0.0–13.2) | 29/1,059 | 2.7 | 1.5 (0.0–1) | 51/358 | 14.2 | 15.4 (3.5–21.4) | 25.5 (< 0.001) |
| Ciprofloxacin | NR | – | NR | 0/133 | 0.0 | 0.0 (0.0) | 14/1,209 | 1.2 | 0.0 (0.0) | 39/3,228 | 1.2 | 0.0 (0.0–4.0) | 0.7 (0.415) |
| Ceftriaxone | NR | – | NR | 0/55 | 0.0 | 0.0 (0.0) | 8/1,160 | 0.7 | 0.0 (0.0) | 5/2,212 | 0.2 | 0.0 (0.0) | 2.9 (0.09) |
| Azithromycin | NR | – | NR | NR | NR | NR | NR | 5/954 | 0.5 | 0.0 (0.0–0.5) | † | ||
IQR = interquartile range; NR = no eligible study reporting data for year period and drug.
Antimicrobial susceptibility profiles not recategorized to current Clinical and Laboratory Standards Institute break points.
Unable to calculate; MDR = phenotype defined as resistance to chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole.
Among studies reporting MDR isolates, one study reported zero (0.0%) of 37 MDR isolates from 1995 through 1999.41 Of isolates during the periods 2000–2004, 2005–2009, and 2010–2014, 28 (21.9%) of 128, 198 (21.2%) of 932, and 2,132 (76.5%) of 2,787 isolates were MDR, respectively (Figure 3C). From 2015 through 2018, one study in Democratic Republic of the Congo reported 41 (38.0%) of 108 isolates were MDR.42 Among isolates from 2015 through 2018, 357 (60.6%) of 589 were resistant to chloramphenicol, 343 (70.9%) of 484 to ampicillin, and 396 (69.4%) of 571 to trimethoprim–sulfamethoxazole. Third-generation cephalosporin resistance was found in 35 (29.4%) of 119, 24 (9.2%) of 262, 13 (1.2%) of 1,111, four (0.2%) of 2,416, and one (0.9%) of 111 in 1995–1999, 2000–2004, 2005–2009, 2010–2014, and 2015–2018, respectively. No eligible studies from Africa reported XDR isolates.
Other typhoidal and non-typhoidal Salmonella.
Among 12,850 other typhoidal Salmonella isolates tested, 10,464 (81.4%) were Salmonella serovar Paratyphi A, 1,305 (10.2%) Salmonella Paratyphi A or Paratyphi B, 1,012 (7.9%) unspecified Salmonella “Paratyphi,” 68 (0.5%) Salmonella Paratyphi B, and one (< 0.1%) Salmonella Paratyphi C. Of 1,462 non-typhoidal S. enterica isolates tested, 793 (54.2%) were Salmonella Typhimurium and 669 (45.8%) were Salmonella Enteritidis. Of Salmonella Paratyphi A isolates, 650 (8.9%) of 7,335 were MDR, 3,027 (32.4%) of 9,332 were resistant to fluoroquinolones, and 45 (2.1%) of 2,184 were resistant to azithromycin. Antimicrobial susceptibility testing data for typhoidal Salmonella other than serovar Typhi are presented in Supplemental Table S4.
DISCUSSION
We demonstrate that AMR among Salmonella Typhi isolates is a substantial problem in countries in Asia and Africa. Multidrug resistant Salmonella Typhi remains prevalent in Asia, with resistance developing to an increasing number of antimicrobial classes such that XDR Salmonella Typhi is now a major threat. MDR Salmonella Typhi is a growing problem in Africa.
The first AMR phenotype to appear in our review was resistance to chloramphenicol in a study from Vietnam in 1972.43 We show clear evidence of increasing prevalence of resistance to traditional first-line antimicrobials in the 1980s and a substantial increased prevalence of MDR in the 1990s. Because third-generation cephalosporins and azithromycin were not widely used or available for the treatment of typhoid fever from 1970 through 1989,7,44 data were lacking on susceptibility to these drugs during earlier time periods.
When stratified by region, resistance to the three traditional first-line antimicrobials and MDR appear later in Africa compared with Asia. This difference is consistent with phylogenetic analyses of whole-genome sequencing data, suggesting the introduction of the MDR H58 haplotype from Asia to Africa.45–47 As there were no eligible studies located in Oceania, we could not address AMR in this region. The H58 haplotype was identified in Fiji as early as 1992.45 However, the H58 haplotype has so far not expanded in Fiji where most Salmonella Typhi isolates remain susceptible to traditional first-line antimicrobials.48,49 We observed a decline in the median prevalence of resistance to each of the traditional first-line antimicrobials and in MDR across Asia, corroborating reports of increasing prevalence of chloramphenicol-susceptible strains in areas that previously documented a high prevalence of resistance to the drug.50–53
The trend of resistance to a growing number of antimicrobial classes is alarming, including recently to azithromycin37,54,55 and outbreaks of XDR Salmonella Typhi.12–14 Our review captured one study in Pakistan36 and one in Indonesia56 reporting meropenem resistance in Salmonella Typhi. To our knowledge, these would be the first reports of carbapenem resistance in Salmonella Typhi and cause for great concern. We recommend that this finding be confirmed with additional testing of the isolates. There were no eligible articles in our review reporting XDR Salmonella Typhi from Africa. However, if the history of the spread of MDR Salmonella Typhi applies, XDR Salmonella Typhi is likely to be introduced and spread in Africa in due course. Resistance to multiple antimicrobial classes is likely being driven by indiscriminate antimicrobial use, and weak stewardship practices in the community and in healthcare facilities.57
Because we sought to produce a comprehensive review of AMR to assist countries with decision-making about TCV introduction and to guide empiric management decisions, we decided a priori against conducting a meta-analysis that would produce pooled prevalence estimates. Furthermore, because the overwhelming majority of eligible studies were located in Southern Asia and reported a broad range of antimicrobials, pooled estimate would be of little value when evaluating AMR in Salmonella Typhi for a specific country that was not part of the pooled analysis.
An earlier systematic review and meta-analysis of enteric fever produced pooled estimates for MDR and fluoroquinolone resistance.58 The earlier review and ours both demonstrate that although data are limited for known typhoid-endemic areas, antimicrobial resistant Salmonella Typhi is becoming more prevalent. However, our review also had a number of additional strengths. First, Browne and others classified intermediate organisms as resistant. According to the CLSI, the intermediate category “implies clinical efficacy in anatomical sites where the drugs are physiologically concentrated” and the resistant category “implies clinical efficacy of the agent against the isolate has not been reliably shown in treatment studies.”59 In keeping with this guidance, we did not categorize intermediate as a category of resistant isolates, and thus avoided possible bias toward inflation in the number of resistant isolates. Second, we classified isolates as MDR if the study authors clearly defined MDR isolates as resistant to chloramphenicol, ampicillin, and trimethoprim–sulfamethoxazole, and we could ascertain that resistance to these three first-line antimicrobials was present. Third, we placed no restrictions on the number of isolates the study tested for resistance. Fourth, we required studies to report the dates for data collection. If a study did not report dates, it was excluded, and we did not impute dates based on study publication date. By not pooling the proportions, recategorizing intermediate isolates, or imputing data, we believe our review presents raw data that are readily accessible to decision-makers.
Our study had a number of limitations. First, a substantial proportion of included studies did not report the interpretive criteria used, and we were unable to make adjustments to the interpretive criteria based on the current CLSI or other breakpoint guidelines because we did not have access to zone sizes or MIC values for most studies. Notably, the CLSI interpretive criteria for Salmonella Typhi have changed over time, including to fluoroquinolones, third-generation cephalosporins, and the introduction of azalide break points.31 These changes may have generated artefactual variation in the prevalence of resistance over time. Second, without having a full classification of susceptible, intermediate, and resistant isolates in every study, we were also limited by having to impute data on resistant isolates in studies that only reported those that were susceptible or that only provided the proportion and total number of isolates tested. Third, our data comprised mostly hospital and healthcare facility–based studies that may overrepresent AMR. Finally, our global AMR data are driven by the predominance of studies located in Asia, particularly India, Nepal, and Pakistan. Several UN regions and subregions had no eligible studies or were underrepresented. Higher income countries in Northern America and Northern, Southern, and Western Europe usually rely on national surveillance systems to report AMR.60,61 No eligible studies were identified from Oceania, Central America, Caribbean, or Eastern Europe. Although studies were included from South America, Northern, Middle, and Southern Africa, and Eastern, Central, and Western Asia, these were limited in number and dominated by studies in Eastern Africa, and Southeastern and Southern Asia.
The prevalence of resistance among Salmonella Typhi isolates is growing, especially the XDR phenotype in Asia and the MDR phenotype in Africa. Isolates resistant to many classes of antimicrobials pose a substantial threat to global health. Typhoid control efforts should be expanded, including the introduction of TCV which has been demonstrated to reduce typhoid fever incidence in endemic areas62 and was recommended to combat and control XDR.63 Where XDR Salmonella Typhi is present, azithromycin and carbapenems remain effective for uncomplicated and complicated typhoid fever, respectively. We encourage the implementation of robust stewardship and surveillance programs to inform empiric treatment decisions and reduce AMR.
Supplemental tables
Acknowledgments:
We would like to thank Ariella P. Dale for her contribution to screening full text and extracting study characteristics, and Chuen Yen Hong for translating articles in Chinese text.
Note: Supplemental tables appear at www.ajtmh.org.
REFERENCES
- 1.GBD 2017 Typhoid and Paratyphoid Collaborators 2019. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis 19: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchello CS, Hong CY, Crump JA, 2019. Global typhoid fever incidence: a systematic review and meta-analysis. Clin Infect Dis 68 (Suppl 2): S105–S116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.GBD 2017 Causes of Death Collaborators , 2018. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392: 1736–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.World Health Organization , 2003. Background Document: The Diagnosis, Treatment and Prevention of Typhoid Fever. Geneva, Switzerland, WHO. [Google Scholar]
- 5.Bhutta ZA, 2006. Current concepts in the diagnosis and treatment of typhoid fever. BMJ 333: 78–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uneke CJ, 2008. Concurrent malaria and typhoid fever in the tropics: the diagnostic challenges and public health implications. J Vector Borne Dis 45: 133–142. [PubMed] [Google Scholar]
- 7.Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM, 2015. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev 28: 901–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowe B, Ward LR, Threlfall EJ, 1991. Treatment of multiresistant typhoid fever. Lancet 337: 1422. [DOI] [PubMed] [Google Scholar]
- 9.Rowe B, Ward LR, Threlfall EJ, 1997. Multidrug-resistant Salmonella Typhi: a worldwide epidemic. Clin Infect Dis 24 (Suppl 1): S106–S109. [DOI] [PubMed] [Google Scholar]
- 10.Umasankar S, Wall RA, Berger J, 1992. A case of ciprofloxacin-resistant typhoid fever. Commun Dis Rep CDR Rev 2: R139–R140. [PubMed] [Google Scholar]
- 11.Threlfall EJ, Murdoch DA, Banatvala NA, Bone A, Shoismatulloev BI, Ward LR, 1998. Epidemic ciprofloxacin-resistant Salmonella Typhi in Tajikistan. Lancet 351: 339. [DOI] [PubMed] [Google Scholar]
- 12.Klemm EJ, et al. 2018. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio 9: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qamar FN, et al. 2018. Outbreak investigation of ceftriaxone-resistant Salmonella enterica serotype Typhi and its risk factors among the general population in Hyderabad, Pakistan: a matched case-control study. Lancet Infect Dis 18: 1368–1376. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization , 2019. Drug resistant Salmonella infections in Pakistan: update. Wkly Epidemiol Monit 12. [Google Scholar]
- 15.Wong W, Al H, Patel S, Yau Y, Eshaghi A, Zittermann S, Tattum L, Morris SK, 2019. The first Canadian pediatric case of extensively drug-resistant Salmonella Typhi originating from an outbreak in Pakistan and its implication for empiric antimicrobial choices. IDCases 15: e00492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engsbro AL, Jespersen HSR, Goldschmidt MI, Mollerup S, Worning P, Schou M, Westh H, Schneider UV, 2019. Ceftriaxone-resistant Salmonella enterica serotype Typhi in a pregnant traveller returning from Karachi, Pakistan to Denmark. Euro Surveill 24: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howard-Jones A, Kesson AM, Outhred AC, Britton PN, 2019. First reported case of extensively drug-resistant typhoid in Australia. Med J Aust 211: 286–286.e1. [DOI] [PubMed] [Google Scholar]
- 18.Chatham-Stephens K, et al. 2019. Emergence of extensively drug-resistant Salmonella Typhi infections among travelers to or from Pakistan — United States, 2016–2018. Morb Mortal Wkly Rep 68: 11–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crump JA, 2019. Progress in typhoid fever epidemiology. Clin Infect Dis 68 (Suppl 1): S4–S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pitzer VE, Feasey NA, Msefula C, Mallewa J, Kennedy N, Dube Q, Denis B, Gordon MA, Heyderman RS, 2015. Mathematical modeling to assess the drivers of the recent emergence of typhoid fever in Blantyre, Malawi. Clin Infect Dis 61 (Suppl 4): S251–S258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luby SP, Faizan MK, Fisher-Hoch SP, Syed A, Mintz ED, Bhutta ZA, McCormick JB, 1998. Risk factors for typhoid fever in an endemic setting, Karachi, Pakistan. Epidemiol Infect 120: 129–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luby SP, 2018. Urban slums: a supportive ecosystem for typhoidal salmonellae. J Infect Dis 218 (Suppl 4): S250–S254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhutta ZA, Zaidi AKM, Pangestu T, 2018. Foreword: reducing typhoid burden within a generation. Am J Trop Med Hyg 99: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crump JA, Mintz ED, 2010. Global trends in typhoid and paratyphoid fever. Clin Infect Dis 50: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moher D, Liberati A, Tetzlaff J, Altman DG, 2009. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339: b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marchello CS, Dale AP, Pisharody S, Rubach MP, Crump JA, 2019. A systematic review and meta-analysis of the prevalence of community-onset bloodstream infections among hospitalized patients in Africa and Asia. Antimicrob Agents Chemother 64: e01974-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy EA, Shaw AV, Crump JA, 2010. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis 10: 417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deen J, von Seidlein L, Andersen F, Elle N, White NJ, Lubell Y, 2012. Community-acquired bacterial bloodstream infections in developing countries in south and southeast Asia: a systematic review. Lancet Infect Dis 12: 480–487. [DOI] [PubMed] [Google Scholar]
- 29.Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A, 2016. Rayyan–a web and mobile app for systematic reviews. Syst Rev 5: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstein MP, Limbago B, Patel JB, Mathers AJ, Campeau S, Mazzulli T, Eliopoulos GM, Patel R, Galas MF, Richter SS, 2018. M100 Performance Standards for Antimicrobial Susceptibility Testing, 29th edition Wayne, PA: CLSI. [Google Scholar]
- 31.Humphries RM, Abbott AN, Hindler JA, 2019. Understanding and addressing CLSI breakpoint revisions: a primer for clinical laboratories. J Clin Microbiol 57: e00203-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.United Nations Statistics Division , 2019. Standard Country or Area Codes for Statistical Use (M49). Available at: https://unstats.un.org/unsd/methodology/m49/. Accessed February 5, 2019. [Google Scholar]
- 33.Mapchart.net , 2020. MapChart: Africa–Countries. Available at: https://mapchart.net/africa.html. Accessed April 3, 2020. [Google Scholar]
- 34.Mapchart.net , 2020. MapChart: Asia-Countries. Available at: https://mapchart.net/asia.html. Accessed April 3, 2020. [Google Scholar]
- 35.CLSI , 2012. Performance Standards for Antimicrobial Susceptibility Testing, 22nd edition Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 36.Laghari GS, Hussain Z, Hussain SZM, Kumar H, Uddin SMM, Haq A, 2019. Antimicrobial susceptibility patterns of Salmonella species in southern Pakistan. Cureus 11: e4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed Shaikh A, Shaikh A, Tahir A, 2019. Antimicrobial resistance trends of typhoidal Salmonellae in southern Pakistan. RMJ 44: 7–10. [Google Scholar]
- 38.Ako-Nai AK, Taiwo O, Ebri A, Adeniran MO, 1990. Bacterial isolates involved in cases of septicaemia in a Nigerian hospital. East Afr Med J 67: 407–412. [PubMed] [Google Scholar]
- 39.Coovadia YM, Van den Ende J, 1987. Chloramphenicol-resistant Salmonella Typhi in Durban, South Africa. Trop Geogr Med 39: 64–66. [PubMed] [Google Scholar]
- 40.Onken A, Said AK, Jørstad M, Jenum PA, Blomberg B, 2015. Prevalence and antimicrobial resistance of microbes causing bloodstream infections in Unguja, Zanzibar. PLoS One 10: e0145632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feasey NA, et al. 2015. Rapid emergence of multidrug resistant, H58-lineage Salmonella Typhi in Blantyre, Malawi. PLoS Negl Trop Dis 9: e0003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tack B, et al. 2019. Salmonella Typhi from blood cultures in the Democratic Republic of the Congo: a 10-year surveillance. Clin Infect Dis 68 (Suppl 2): S130–S137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown JD, Duong Hong MO, Rhoades ER, 1975. Chloramphenicol-resistant Salmonella Typhi in Saigon. JAMA 231: 162–166. [PubMed] [Google Scholar]
- 44.Mirza SH, Beechmg NJ, Hart CA, 1996. Multi-drug resistant typhoid: a global problem. J Med Microbiol 44: 317–319. [DOI] [PubMed] [Google Scholar]
- 45.Wong VK, et al. 2015. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet 47: 632–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong VK, et al. 2016. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kariuki S, et al. 2010. Typhoid in Kenya is associated with a dominant multidrug-resistant Salmonella enterica Serovar Typhi haplotype that is also widespread in Southeast Asia. J Clin Microbiol 48: 2171–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Getahun A, Parry CM, Crump JA, Rosa V, Jenney A, Naidu R, Mulholland K, Strugnell RA, 2019. A retrospective study of patients with blood culture-confirmed typhoid fever in Fiji during 2014–2015: epidemiology, clinical features, treatment and outcome. Trans R Soc Trop Med Hyg 113: 764–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson CN, et al. 2014. Typhoid fever in Fiji: a reversible plague? Trop Med Int Health 19: 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lakshmi V, Ashok R, Susmita J, Shailaja VV, 2006. Changing trends in the antibiograms of Salmonella isolates at a tertiary care hospital in Hyderabad. Indian J Med Microbiol 24: 45–48. [DOI] [PubMed] [Google Scholar]
- 51.Capoor MR, Nair D, 2010. Quinolone and cephalosporin resistance in enteric fever. J Glob Infect Dis 2: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar Y, Sharma A, Mani KR, 2009. High level of resistance to nalidixic acid in Salmonella enterica serovar Typhi in central India. J Infect Dev Ctries 3: 467–469. [DOI] [PubMed] [Google Scholar]
- 53.Shetty AK, Shetty IN, Furtado ZV, Antony B, Boloor R, 2012. Antibiogram of Salmonella isolates from blood with an emphasis on nalidixic acid and chloramphenicol susceptibility in a tertiary care hospital in Coastal Karnataka: a prospective study. J Lab Physicians 4: 74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewis MD, Serichantalergs O, Pitarangsi C, Chuanak N, Mason CJ, Regmi LR, Pandey P, Laskar R, Shrestha CD, Malla S, 2005. Typhoid fever: a massive, single-point source, multidrug-resistant outbreak in Nepal. Clin Infect Dis 40: 554–561. [DOI] [PubMed] [Google Scholar]
- 55.Jain S, Das Chugh T, 2013. Antimicrobial resistance among blood culture isolates of Salmonella enterica in New Delhi. J Infect Dev Ctries 7: 788–795. [DOI] [PubMed] [Google Scholar]
- 56.Hardjo Lugito NP, Cucunawangsih, 2017. Antimicrobial resistance of Salmonella enterica serovars Typhi and Paratyphi isolates from a general hospital in Karawaci, Tangerang, Indonesia: a five-year review. Int J Microbiol 2017: 6215136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.World Health Organization , 2015. Global Action Plan on Antimicrobial Resistance. Geneva, Switzerland: WHO. [Google Scholar]
- 58.Browne AJ, et al. 2020. Drug-resistant enteric fever worldwide, 1990 to 2018: a systematic review and meta-analysis. BMC Med 18: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.CLSI , 2020. Performance Standards for Antimicrobial Susceptibility Testing, 30th edition Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 60.CDC , 2020. National Antimicrobial Resistance Monitoring System (NARMS) Now: Human Data. Atlanta, GA: U.S. Department of Health and Human Services, CDC; Available at: https://www.cdc.gov/narmsnow. Accessed January 6, 2020. [Google Scholar]
- 61.ECDC , 2020. European Antimicrobial Resistance Surveillnce Network (EARS-Net). Sweden: European Centre for Disease Prevention and Control (ECDC) Available at: http://www.ecdc.europa.eu/en/healthtopics/antimicrobial_resistance/database/Pages/database.aspx. Accessed January 6, 2020. [Google Scholar]
- 62.Shakya M, et al. 2019. Phase 3 efficacy analysis of a typhoid conjugate vaccine trial in Nepal. N Engl J Med 381: 2209–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrews JR, Qamar FN, Charles RC, Ryan ET, 2018. Extensively drug-resistant typhoid–are conjugate vaccines arriving just in time? N Engl J Med 379: 1493–1495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.