Abstract
Simple Summary
Campylobacter species are among the major causes of bacterial foodborne infections. Here, we investigate, for the first time, class 1 integrons and associated gene cassettes among pan drug-resistant (PDR), extensively drug-resistant (XDR), and multidrug-resistant (MDR) Campylobacter species isolated from livestock animals and humans in Egypt. Our results revealed alarming PDR (2.55%) and inordinate XDR (68.94%) and MDR (28.5%) Campylobacter isolates. None of the examined isolates were pan-susceptible. The existence of a novel gene cassette array, namely aacC5-aadA7Δ4 and a putative phage tail tape measure protein on class 1 integrons of Campylobacter species is the most highlighted novelty of the current study. Evidence from this study showed the possibility of Campylobacter–bacteriophage interactions as well as treatment failure in animals and humans due to horizontal gene transfer mediated by class 1 integrons.
Abstract
Campylobacter species are common commensals in the gastrointestinal tract of livestock animals; thus, animal-to-human transmission occurs frequently. We investigated for the first time, class 1 integrons and associated gene cassettes among pan drug-resistant (PDR), extensively drug-resistant (XDR), and multidrug-resistant (MDR) Campylobacter species isolated from livestock animals and humans in Egypt. Campylobacter species were detected in 58.11% of the analyzed chicken samples represented as 67.53% Campylobacter jejuni (C. jejuni) and 32.47% Campylobacter coli (C. coli). C. jejuni isolates were reported in 51.42%, 74.28%, and 66.67% of examined minced meat, raw milk, and human stool samples, respectively. Variable antimicrobial resistance phenotypes; PDR (2.55%), XDR (68.94%), and MDR (28.5%) campylobacters were reported. Molecular analysis revealed that 97.36% of examined campylobacters were integrase gene-positive; all harbored the class 1 integrons, except one possessed an empty integron structure. DNA sequence analysis revealed the predominance of aadA (81.08%) and dfrA (67.56%) alleles accounting for resistance to aminoglycosides and trimethoprim, respectively. This is the first report of aacC5-aadA7Δ4 gene cassette array and a putative phage tail tape measure protein on class 1 integrons of Campylobacter isolates. Evidence from this study showed the possibility of Campylobacter–bacteriophage interactions and treatment failure in animals and humans due to horizontal gene transfer mediated by class 1 integrons.
Keywords: Campylobacter species, class 1 integrons, extensively drug-resistance, pan drug-resistance, gene cassette arrays
1. Introduction
Thermophilic Campylobacter species, particularly Campylobacter jejuni (C. jejuni) and Campylobacter coli (C. coli) pose veterinary and public health concerns due to their zoonotic potential, the enormous range of reservoir hosts, and persistence in the environment [1]. Consumption of contaminated food, especially poultry products, unpasteurized milk, and undercooked meat, as well as water, is a risk factor for C. jejuni and C. coli infections [2,3].
Most Campylobacter infections are self-limiting and require no therapeutic intervention other than supportive and rehydration therapy. However, prompt antimicrobial treatment is employed in immunocompromised individuals, patients whose symptoms are severe or persistent, intense or prolonged enteritis, cases of bacteremia, and those with extraintestinal infections [4]. Attention to the resistance of campylobacters has been launched due to the indiscriminate abuse of antibiotics [5]. Clinical, veterinary, and environmental surveys have shown that bacteria harboring integrons are frequently associated with the multidrug-resistant (MDR) phenotype [6]. However, the extensively drug-resistant (XDR) and pan drug-resistant (PDR) bacteria are epidemiologically significant not only due to their resistance to multiple antimicrobial agents but also to their ominous prospect of being resistant to almost all or all approved antimicrobial agents [7,8].
The integron is a site-specific recombination system capable of integrating mobile gene cassettes, which can be expressed and disseminated via horizontal gene transfer [9,10]. Class 1 integron includes two conserved segments (CSs), denoted as 5′- and 3′-CSs, flanking a gene cassette. An int1 gene encoding an integrase enzyme is located within the 5′-CS and is responsible for the recombination of a gene cassette [11]. The 3′-CS possesses qacE∆1 and sul1 genes encoding resistance to quaternary ammonium compounds and sulfonamide, respectively. Integrons can incorporate and express more than one gene cassette conferring resistance to multiple antimicrobial classes such as beta-lactams, aminoglycosides, trimethoprim, chloramphenicol, fosfomycin, macrolides, lincosamides, rifampicin, and quinolones [11].
Previous studies reported class 1 integrons carried aminoglycoside (aadA and aacA4) and trimethoprim (dfr1 and dfr9) resistance gene cassettes in both C. jejuni and C. coli isolated from chicken house environment [12], poultry, and human sources [13,14,15,16] without showing the antimicrobial resistance profiles of the isolates. However, these genetic elements were not detected in an XDR C. jejuni CCARM 13,322 isolate recovered from a human case of diarrhea associated with international travel [17]. Hence, this study reports, for the first time, class 1 integrons and associated gene cassettes in thermophilic Campylobacter species isolated from livestock animals and humans showing variable antimicrobial resistance phenotypes.
2. Materials and Methods
2.1. Samples
A total of 550 samples comprising chickens and chicken products (n = 265), meat and meat products (n = 160), milk and milk products (n = 95) as well as human stools (n = 30) were collected during the period from January 2018 to December 2019. Samples of animal origins were obtained from various retail outlets, Zagazig city, Sharkia Governorate, Egypt. Human samples were collected from patients being affected by diarrhea and gastroenteritis, attending various private laboratories located in Zagazig city. The samples were transported immediately in an icebox to the bacteriology laboratory for further analysis. The animal study was approved by the Animal Welfare and Research Ethics Committee, Faculty of Veterinary Medicine, Zagazig University. The human study was conducted following the Ethics of the World Medical Association (Declaration of Helsinki) and was approved by the research ethics committee of the Faculty of Medicine, Zagazig University (ApprovalNoZU-IRB#2056-18-05-2019). The patients participating in the research study provided written informed consent.
2.2. Bacteriological Analysis and Molecular Identification
Isolation of Campylobacter species was performed under microaerobic conditions according to the protocol established by Vandepitte et al. [18]. Samples were enriched in Preston Campylobacter selective enrichment broth (Oxoid, Cambridge, UK) at 42 °C for 48 h. The enrichment broth was plated onto modified charcoal cefoperazone deoxycholate agar (mCCDA; Oxoid, Cambridge, UK) then transferred onto Columbia agar (Oxoid, Cambridge, UK) plates supplemented with 5% sterile defibrinated horse blood. Presumptive Campylobacter colonies were confirmed by oxidase, catalase, hippurate, and indoxyl acetate hydrolyses biochemical tests, in addition to testing their susceptibilities to nalidixic acid and cephalothin antimicrobials (30 mg/disc, each) [19]. The bacterial DNA was extracted using a QIAamp DNA Mini kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions. Polymerase chain reaction (PCR) amplifications of the 23S rRNA gene of Campylobacter species [20] in addition to mapA and ceuE genes of C. jejuni and C. coli, respectively, [21] were applied using oligonucleotide primers listed in Table S1.
2.3. Antimicrobial Susceptibility Testing
The antimicrobial susceptibilities of Campylobacter isolates were tested on Mueller–Hinton agar media (Oxoid-CM0337B, Cambridge, UK) supplemented with 5% sterile defibrinated horse blood under microaerobic conditions using the disc diffusion method [22] following the guidelines of the Clinical and Laboratory Standards Institute (CLSI) [23]. A panel of 25 standard antimicrobial discs (Oxoid, Cambridge, UK) within different 14 antimicrobial categories were examined including penicillins [ampicillin (AM; 10 µg) and amoxicillin (AX; 25 µg)], penicillin combinations [ampicillin-sulbactam (SAM; 20/10 µg) and amoxycillin-clavulanic acid (AMC; 20/10 µg)], cephalosporines [cephalothin (KF; 30 µg), cefoxitin (FOX; 30 µg), cefoperazone (CEP; 75 µg) and cefepime (FEP; 30 µg)], carbapenemes [meropenem (MEM; 10 µg)], monobactams [azetronam (ATM; 30 µg)], aminoglycosides [streptomycin (S; 10 µg), tobramycin (TOB; 10 µg), gentamycin (CN; 10 µg) and amikacin (AK; 30 µg)], macrolides [erythromycin (E; 15 µg), azithromycin (AZM; 15 µg) and clarithromycin (CLR; 15 µg)], quinolones [nalidixic acid (NA; 30 µg) and ciprofloxacin (CIP; 5 µg)], sulfonamides [sulfamethoxazole-trimethoprim (SXT; 23.75/1.25 μg)], amphenicols [chloramphenicol (C; 30 µg)], polypeptides [colistin (CT; 10 µg)], oxazolidones [lenzolid (LNZ; 30 µg)], lincosamides [clindamycin (DA; 2 µg)] and tetracyclines [doxycycline (DO; 30 µg)]. The interpretive criteria of CLSI (for most antimicrobials) [23] or the European Committee for Antimicrobial Susceptibility Testing (EUCAST) (for macrolides) were followed to classify Campylobacter isolates as susceptible, intermediate, or resistant [24].
The multiple antimicrobial resistance (MAR) indices were calculated as previously reported [25]. Pan drug-resistance (resistance to all antimicrobial agents), extensive drug-resistance (resistance to all classes of antimicrobial agents except 2 or fewer), and multidrug-resistance (resistance to three or more classes of antimicrobial agents) were determined as reported elsewhere [26].
2.4. PCR Amplification of Class 1 Integrons and Associated Gene Cassettes
Campylobacter isolates exhibited variable antimicrobial resistance profiles (PDR, XDR, and MDR) were subjected to DNA extraction, using the QIAamp DNA Mini kit (Qiagen, Gmbh, Hilden, Germany) following the manufacturer’s recommendations. The isolates were screened for possession of the integrase gene as well as class 1 integrons using intI1 and hep primer sets, respectively [27,28] (Table S1). Isolates containing class 1 integrons were screened for the existence of contiguous resistance gene cassettes inserted in 5′ and 3′ conserved regions using 3′CS and 5′CS-targeted primers [29] (Table S1). The DNA of C. jejuni ATCC 33560 and sterile saline were included in all PCR assays as positive and negative controls, respectively.
2.5. Characterization of Gene Cassettes Arrays by DNA Sequencing
One of each amplified PCR product of repetitive distinct close size was selected, purified by PureLink PCR purification kit (Qiagen, Valencia, Spain) and sequenced using Big Dye Terminator V3.1 cycle sequencing kit (Perkin-Elmer Gmbh, Rodgau, Germany) in an Applied Biosystems 3130 genetic analyzer (California, USA). The resulting sequences were assembled using the SeqMan program within the Laser gene suite version 7 (DNAstar, Inc., Madison, WI, USA), then compared with the sequences in the GenBank database using the Basic Local Alignment Search Tool (http://www.ncbi.nlm.nih.gov/BLAST). The best BLAST hits on our query nucleotide sequences were selected based on the highest identity in the GenBank database. Alignment of the nucleotide sequences was performed using ClustalW sequence alignments (http://www.ebi.ac.uk/clustalw), then translation into amino acid sequences was performed using the ExPASy Translate Tool (http://us.expasy.org/, Swiss Institute of Bioinformatics SIB, Geneva, Switzerland). The novel complete gene cassette array (aacC5-aadA7Δ4) generated here assigned a new in number (in 1983) using the Integron Database INTEGRALL (http://integrall.bio.ua.pt/).
2.6. Bioinformatics and Statistical Analysis
Statistical Package for Social Sciences software (SPSS; v. 25, IBM, Armonk, NY, United States) was used for statistical analysis of data. Chi-squared test was used to determine if there were significant differences in the occurrence of antimicrobial resistance among different hosts (i.e., cattle, chicken, human) and between the two Campylobacter species being studied (C. jejuni and C. coli). P value was considered significant if <0.05. The overall distribution of the resistance phenotypes in Campylobacter isolates was visualized using a heat map. The clustering pattern of the isolates and the antimicrobial resistance phenotypes were determined by the hierarchical clustering dendrogram [30]. To predict the correlation among integron patterns and antimicrobial resistance phenotypes, correlation analyses were done on the raw data after conversion to a binary outcome (1 = variable presence, 0 = variable absence). The significance of the correlation was estimated at a significance level of 0.05. The variables ampicillin, amoxicillin, cephalothin, erythromycin, and sulfamethoxazole-trimethoprim were excluded from the analyses as they were identical among all isolates under study. The correlation analyses and visualization were done using R packages corrplot, heatmaply, hmisc, and ggpubr [31,32,33]. To estimate the similarities among Campylobacter isolates concerning various analyzed hosts (n = 3), the binary distances were calculated based on the presence or absence of certain integron patterns. This analysis was done using the functions dist and hlcust in the R environment.
2.7. Nucleotide Sequence Accession Numbers
DNA sequences generated in this study were submitted to GenBank and assigned the accession numbers of MT612446-MT612453.
3. Results
3.1. Prevalence of Campylobacter Species in Livestock Animals and Humans
As shown in Table 1, the overall occurrence rate of Campylobacter species was 42.72% (235/550), which significantly (p < 0.05) differed between species being 71.48% (168/235) for C. jejuni and 28.51% (67/235) for C. coli. Out of 265 samples of chicken origin, 154 (58.11%) Campylobacter isolates were detected, represented as 67.53% C. jejuni and 32.47% C. coli. The higher prevalence of C. jejuni was detected in chicken organs (61.54%), followed by cloacal swabs (57.14%) and chicken muscles (48.00%), while the isolation rate of C. coli from these sources was close to 30%, each. Moreover, C. jejuni were isolated from 18 of 35 (51.42%) minced meat samples, 26 of 35 (74.28%) raw milk, and 20 of 30 (66.67%) human stool samples, while C. coli were recorded by lower percentages. On the other hand, processed food products including chicken and meat luncheon, chicken and meat beef, smoked meat, canned milk, and canned and raw cheese were free from Campylobacter contamination. Campylobacter isolates yielded characteristic small, shiny, round, and gray colonies on mCCDA agar and no hemolysis on Columbia blood agar. All isolates were positive for oxidase, catalase, and nitrate reduction testing and exhibited sensitivity to nalidixic acid and resistance to cephalothin. C. jejuni isolates could hydrolyze indoxyl acetate and hippurate, while C. coli were indoxyl acetate-positive and hippurate-negative. Campylobacter isolates were further confirmed by PCR-based detection of the genus (23S rRNA) and species-specific (mapA for C. jejuni and ceuE for C. coli) genes.
Table 1.
Prevalence of Campylobacter species isolated from animal and human sources.
| Source (No.) | Sample Type (No.) | Overall Prevalence of Campylobacter Isolates No. (%) |
Campylobacter Species No. (%) |
p Value | |
|---|---|---|---|---|---|
| C. jejuni | C. coli | ||||
| Chickens and chicken products (265) | Cloacal swab (70) | 60 (85.71) | 40 (57.14) | 20 (28.57) | 0.001 |
| Breast muscle (25) | 18 (72.00) | 14 (56.00) | 4 (16.00) | 0.003 | |
| Thigh chicken muscle (25) | 16 (64.00) | 10 (40.00) | 6 (24.00) | 0.225 | |
| Liver (25) | 25 (100.00) | 16 (64.00) | 9 (36.00) | 0.048 | |
| Spleen (20) | 18 (90.00) | 14 (70.00) | 4 (20.00) | 0.001 | |
| Intestine (20) | 17 (85.00) | 10 (50.00) | 7 (35.00) | 0.337 | |
| Chicken beef (40) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Chicken luncheon (40) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Meat and meat products (160) | Minced meat (35) | 28 (80.00) | 18 (51.43) | 10 (28.57) | 0.05 |
| Smoked meat (45) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Meat beef (40) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Meat luncheon (40) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Dairy products (95) | Raw milk (35) | 29 (82.86) | 26 (74.29) | 3 (8.57) | <0.0001 |
| Canned milk (20) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Canned cheese (20) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Raw cheese (20) | 0 (0.00) | 0 (0.00) | 0 (0.00) | NE | |
| Human (30) | Stool (30) | 24 (80.00) | 20 (66.67) | 4 (13.33) | <0.0001 |
| Total | 550 | 235 (42.73) | 168 (71.49) | 67 (28.51) | <0.0001 |
NE, not estimated, p values < 0.05 are statistically significant.
3.2. Antimicrobial Resistance Profiles
The in vitro antimicrobial susceptibilities of 235 Campylobacter isolates comprising 168 C. jejuni and 67 C. coli against 25 antimicrobial agents are summarized in Table 2. The results revealed that all Campylobacter isolates originating from animal and human sources were resistant to amoxicillin, ampicillin, erythromycin, cephalothin, and sulfamethoxazole-trimethoprim (100%, each). Moreover, high levels of resistance were recorded for clarithromycin (100% and 97%), clindamycin (96.4% and 95.5%), nalidixic acid (90.5% and 86.6%), amoxycillin-clavulanic acid (89.3% and 80.6%), cefepime (88.1% and 83.6%), doxycycline (86.3% and 86.5%), colistin (83.9% and 88%) and chloramphenicol (83.3% and 80.6%) for C. jejuni and C. coli isolates, respectively. On the other hand, lower resistance rates were reported for amikacin (21.4% and 20.9%) and cefoxitin (26.8% and 43.2%) against C. jejuni and C. coli isolates, respectively. Of note, C. jejuni and C. coli were resistant to meropenem with alarming percentages (19.6% and 32.8%, respectively). Statistical analysis revealed significant differences in the resistance of Campylobacter species isolated from different sources to the most tested antimicrobials (p < 0.05) except for ampicillin-sulbactam that showed non-significant variation (p > 0.05). However, non-significant differences (p > 0.05) were reported between resistance of C. jejuni and C. coli to almost half of the examined antimicrobial agents.
Table 2.
Antimicrobial resistance pattern of Campylobacter species isolated from different sources.
| AMA | Number of C. jejuni Isolates | Number of C. coli Isolates | p Value | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chickens (n = 104) | Raw Milk (26) | Minced Meat (18) | Human Stool (20) | Total (168) | Chickens (n = 50) | Raw Milk (3) | Minced Meat (10) | Human Stool (4) | Total (67) | |||||||
| Muscle (24) | Internal Organs (40) | Cloacal Swabs (40) | Muscle (10) | Internal Organs (20) | Cloacal Swabs (20) | Various Hosts | Campylobacter Species | |||||||||
| AX | 24 | 40 | 40 | 26 | 18 | 20 | 168 | 10 | 20 | 20 | 3 | 10 | 4 | 67 | NA | NA |
| (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | |||
| AM | 24 | 40 | 40 | 26 | 18 | 20 | 168 | 10 | 20 | 20 | 3 | 10 | 4 | 67 | NA | NA |
| (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | |||
| SAM | 15 | 27 | 20 | 11 | 10 | 10 | 93 | 0.00 | 11 | 16 | 3 | 1 | 0.00 | 31 | 0.1022 | 0.2 |
| (62.50) | (67.50) | (50.00) | (42.31) | (55.55) | (50.00) | (55.36) | (0.00) | (55.00) | (80.00) | (100.00) | (10.00) | (0.00) | (46.27) | |||
| AMC | 24 | 34 | 34 | 26 | 17 | 15 | 150 | 7 | 20 | 18 | 3 | 10 | 0.00 | 58 | <0.0001 | 0.55 |
| (100.00) | (85.00) | (85.00) | (100.00) | (94.44) | (75.00) | (89.29) | (70.00) | (100.00) | (90.00) | (100.00) | (100.00) | (0.00) | (86.57) | |||
| KF | 24 | 40 | 40 | 26 | 18 | 20 | 168 | 10 | 20 | 20 | 3 | 10 | 4 | 67 | NA | NA |
| (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | |||
| FOX | 8 | 4 | 18 | 17 | 12 | 9 | 68 | 0.00 | 4 | 10 | 0.00 | 0.00 | 4 | 18 | 0.002 | 0.05 |
| (33.33) | (10.00) | (45.00) | (65.38) | (66.67) | (45.00) | (40.48) | (0.00) | (20.00) | (50.00) | (0.00) | (0.00) | (100.00) | (26.87) | |||
| CEP | 14 | 34 | 19 | 26 | 12 | 18 | 123 | 10 | 20 | 12 | 3 | 9 | 4 | 58 | 0.0068 | 0.02 |
| (58.33) | (85.00) | (47.50) | (100.00) | (66.67) | (90.00) | (73.21) | (100.00) | (100.00) | (60.00) | (100.00) | (90.00) | (100.00) | (86.57) | |||
| FEP | 16 | 38 | 39 | 17 | 10 | 20 | 140 | 8 | 19 | 20 | 3 | 6 | 4 | 60 | <0.0001 | 0.2 |
| (66.67) | (95.00) | (97.50) | (65.38) | (55.56) | (100.00) | (83.33) | (80.00) | (95.00) | (100.00) | (100.00) | (60.00) | (100.00) | (89.55) | |||
| MEM | 1 | 2 | 20 | 0.00 | 6 | 4 | 33 | 0.00 | 14 | 8 | 0.00 | 0.00 | 0.00 | 22 | 0.0124 | 0.03 |
| (4.17) | (5.00) | (50.00) | (0.00) | (33.33) | (20.00) | (19.64) | (0.00) | (70.00) | (40.00) | (0.00) | (0.00) | (0.00) | (32.84) | |||
| ATM | 14 | 39 | 36 | 26 | 17 | 10 | 142 | 0.00 | 14 | 12 | 3 | 10 | 4 | 43 | <0.0001 | 0.0006 |
| (58.33) | (97.50) | (90.00) | (100.00) | (94.44) | (50.00) | (84.52) | (0.00) | (70.00) | (60.00) | (100.00) | (100.00) | (100.00) | (64.18) | |||
| S | 22 | 16 | 34 | 12 | 7 | 14 | 105 | 4 | 17 | 18 | 0.00 | 2 | 3 | 44 | <0.0001 | 0.6 |
| (91.67) | (40.00) | (85.00) | (46.15) | (38.89) | (70.00) | (62.50) | (40.00) | (85.00) | (90.00) | (0.00) | (20.00) | (75.00) | (65.67) | |||
| TOB | 22 | 16 | 30 | 8 | 6 | 13 | 95 | 4 | 17 | 17 | 0.00 | 0.00 | 4 | 42 | <0.0001 | 0.38 |
| (91.67) | (40.00) | (75.00) | (30.77) | (33.33) | (65.00) | (56.55) | (40.00) | (85.00) | (85.00) | (0.00) | (0.00) | (100.00) | (62.69) | |||
| CN | 24 | 24 | 36 | 14 | 2 | 9 | 109 | 6 | 17 | 14 | 0.00 | 5 | 4 | 46 | <0.0001 | 0.58 |
| (100.00) | (60.00) | (90.00) | (53.85) | (11.11) | (45.00) | (64.88) | (60.00) | (85.00) | (70.00) | (0.00) | (50.00) | (100.00) | (68.66) | |||
| AK | 18 | 0.00 | 14 | 0.00 | 6 | 6 | 44 | 6 | 3 | 6 | 0.00 | 00.00 | 4 | 19 | 0.0032 | 0.73 |
| (75.00) | (0.00) | (35.00) | (0.00) | (33.33) | (30.00) | (26.19) | (60.00) | (15.00) | (30.00) | (0.00) | (0.00) | (100.00) | (28.36) | |||
| E | 24 | 40 | 40 | 26 | 18 | 20 | 168 | 10 | 20 | 20 | 3 | 10 | 4 | 67 | NA | NA |
| (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | |||
| AZM | 15 | 33 | 32 | 26 | 18 | 20 | 144 | 4 | 20 | 14 | 3 | 10 | 4 | 55 | <0.0001 | 0.48 |
| (62.50) | (82.50) | (80.00) | (100.00) | (100.00) | (100.00) | (85.71) | (40.00) | (100.00) | (70.00) | (100.00) | (100.00) | (100.00) | (82.09) | |||
| CLR | 24 | 40 | 6 | 26 | 18 | 20 | 134 | 10 | 20 | 2 | 3 | 10 | 0.00 | 45 | <0.0001 | 0.04 |
| (100.00) | (100.00) | (15.00) | (100.00) | (100.00) | (100.00) | (79.76) | (100.00) | (100.00) | (10.00) | (100.00) | (100.00) | (0.00) | (67.16) | |||
| CIP | 24 | 16 | 32 | 26 | 1 | 11 | 110 | 6 | 9 | 16 | 0.00 | 9 | 0.00 | 40 | <0.0001 | 0.4 |
| (100.00) | (40.00) | (80.00) | (100.00) | (5.56) | (55.00) | (65.48) | (60.00) | (45.00) | (80.00) | (0.00) | (90.00) | (0.00) | (59.70) | |||
| NA | 24 | 33 | 40 | 26 | 17 | 10 | 150 | 6 | 20 | 18 | 0.00 | 10 | 4 | 58 | <0.0001 | 0.55 |
| (100.00) | (82.50) | (100.00) | (100.00) | (94.44) | (50.00) | (89.29) | (60.00) | (100.00) | (90.00) | (0.00) | (100.00) | (100.00) | (86.57) | |||
| SXT | 24 | 40 | 40 | 26 | 18 | 20 | 168 | 10 | 20 | 20 | 3 | 10 | 4 | 67 | <0.0001 | 0.005 |
| (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | |||
| C | 24 | 26 | 32 | 26 | 17 | 16 | 141 | 6 | 16 | 16 | 3 | 5 | 0.00 | 46 | 0.0461 | 0.0088 |
| (100.00) | (65.00) | (80.00) | (100.00) | (94.44) | (80.00) | (83.93) | (60.00) | (80.00) | (80.00) | (100.00) | (50.00) | (0.00) | (68.66) | |||
| CT | 14 | 28 | 34 | 26 | 18 | 11 | 131 | 6 | 16 | 18 | 3 | 10 | 4 | 57 | <0.0001 | 0.21 |
| (58.33) | (70.00) | (85.00) | (100.00) | (100.00) | (55.00) | (77.98) | (60.00) | (80.00) | (90.00) | (100.00) | (100.00) | (100.00) | (85.07) | |||
| LNZ | 24 | 39 | 28 | 26 | 14 | 13 | 144 | 10 | 9 | 18 | 0.00 | 10 | 0.00 | 47 | 0.0012 | 0.005 |
| (100.00) | (97.50) | (70.00) | (100.00) | (77.78) | (65.00) | (85.71) | (100.00) | (45.00) | (90.00) | (0.00) | (100.00) | (0.00) | (70.15) | |||
| DA | 23 | 40 | 39 | 26 | 18 | 20 | 166 | 10 | 20 | 20 | 3 | 10 | 0.00 | 63 | <0.0001 | 0.03 |
| (95.83) | (100.00) | (97.50) | (100.00) | (100.00) | (100.00) | (98.81) | (100.00) | (100.00) | (100.00) | (100.00) | (100.00) | (0.00) | (94.03) | |||
| DO | 24 | 37 | 24 | 26 | 18 | 20 | 149 | 10 | 20 | 18 | 3 | 10 | 0.00 | 61 | 0.0103 | 0.59 |
| (100.00) | (92.50) | (60.00) | (100.00) | (100.00) | (100.00) | (88.69) | (100.00) | (100.00) | (90.00) | (100.00) | (100.00) | (0.00) | (91.04) | |||
Values represent number of Campylobacter isolates (%), p values were calculated based on Chi-squared test; p values < 0.05 are statistically significant; p values < 0.01 are highly significant; AMA, antimicrobial agent; AX, amoxicillin; AM, ampicillin; SAM, ampicillin-sulbactam; AMC, amoxycillin-clavulanic acid; KF, cephalothin; FOX, cefoxitin; CEP, cefoperazone; FEP, cefepime; MEM, meropenem; ATM, aztreonam; S, streptomycin; TOB, tobramycin; CN, gentamicin; AK, amikacin; E, erythromycin; AZM, azithromycin; CLR, clarithromycin; CIP, ciprofloxacin; NA, nalidixic acid; SXT, sulfamethoxazole-trimethoprim; C, chloramphenicol; CT, colistin; LNZ, lenzolid; DA, clindamycin; DO, doxycycline; NA, non-applicable.
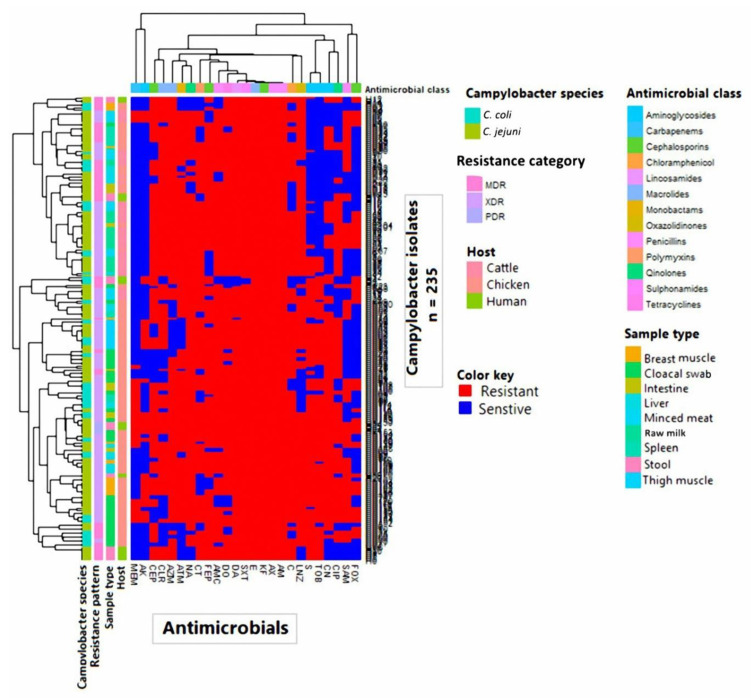
As shown in Figure 1 and Table S2, the antibiogram analysis revealed that Campylobacter isolates showed resistance to 11–25 antimicrobial agents with MAR indices ranged from 0.44 to 1.00 and demonstrated 93 distinct resistance patterns. The antibiotype 55 was the most prevalent among the analyzed isolates (n = 8; 3.40%) (Table S2).
Figure 1.
Overall distribution and clustering of Campylobacter isolates (n = 235) under study and the patterns of their antimicrobial resistance. Different Campylobacter species, hosts, sample types, antimicrobial classes, and resistance categories are shown for each isolate as color codes. The heat map represents the hierarchical clustering of the isolates and the antimicrobial classes.
The PDR, XDR, and MDR patterns were reported among the analyzed isolates (Table 3 and Figures S1 and S2). In total, 2.55% (6/235) of Campylobacter isolates exhibited PDR patterns being resistant to all tested antimicrobial agents. The XDR profiles were extremely increased among analyzed isolates with a percentage of 68.94% (162/235). However, 28.5% (67/235) of the isolates showed MDR patterns. None of the examined Campylobacter isolates was pan-susceptible. Regarding the isolation source, most C. jejuni (68.26%) and C. coli (74%) isolates originated from chicken samples were XDR. All C. jejuni and C. coli isolates recovered from raw milk showed XDR and MDR patterns, respectively. Moreover, all C. coli isolated from human stool and minced meat exhibited MDR and XDR profiles, respectively, while 88.9% of C. jejuni originated from the minced meat were XDR. The PDR Campylobacter isolates originated from chicken cloacal swabs (n = 2) and human stool (n = 4).
Table 3.
Occurrence of MDR, XDR and PDR categories in Campylobacter isolates from different sources.
| Resistance Category | Resistance to Antimicrobial Class (n = 14) | Resistance to Antimicrobial Agent (n = 25) | No. of Resistant Campylobacter Isolates (Source) | |
|---|---|---|---|---|
| C. jejuni (n = 168) | C. coli (n = 67) | |||
| MDR (n = 67) |
7 | 11 | 0 | 2 (chicken muscle) |
| 12 | 0 | 1 (chicken muscle) | ||
| 14 | 0 | 1 (human stool) | ||
| 15 | 0 | 3 (human stool) | ||
| 8 | 13 | 0 | 1 (chicken muscle) | |
| 10 | 13 | 2 (cloacal swab) | 0 | |
| 14 | 1 (minced meat) | 0 | ||
| 15 | 8 (chicken organ) | 0 | ||
| 16 | 2 (cloacal swab), 9 (human stool) | 3 (raw milk) | ||
| 17 | 5 (human stool), 1 (cloacal swab) | 2 (cloacal swab) | ||
| 11 | 15 | 1 (minced meat) | 2 (cloacal swab) | |
| 16 | 4 (cloacal swab), 2 (chicken organ) | 0 | ||
| 17 | 8 (chicken organ), 2 (cloacal swab) | 3 (chicken organ) | ||
| 18 | 1 (cloacal swab) | 0 | ||
| 19 | 1 (chicken muscle) | 2 (cloacal swab) | ||
| XDR (n = 162) |
12 | 16 | 2 (cloacal swab) | 0 |
| 17 | 3 (minced meat) | 4 (minced meat) | ||
| 18 | 2 (chicken muscle), 5 (chicken organ), 4 (cloacal swab) | 4 (minced meat), 2 (chicken muscle), 2 (cloacal swab) | ||
| 19 | 5 (chicken muscle), 5 (chicken organ), 2 (cloacal swab), 4 (minced meat) | 1 (minced meat), 1 (chicken organ), 2 (cloacal swab) | ||
| 20 | 5 (raw milk), 2 (chicken muscle), 2 (cloacal swab), 2 (chicken organ), 2 (human stool) | 6 (chicken organ), 4 (chicken muscle) | ||
| 21 | 2 (chicken muscle) | 4 (chicken organ) | ||
| 22 | 4 (chicken muscle) | 0 | ||
| 23 | 3 (chicken muscle) | 0 | ||
| 13 | 18 | 1 (minced meat) | 1 (minced meat), 2 (cloacal swab) | |
| 19 | 9 (raw milk), 1 (chicken muscle), 3 (chicken organ) | 1 (cloacal swab) | ||
| 20 | 2 (minced meat), 5 (raw milk) | 1 (cloacal swab) | ||
| 21 | 6 (raw milk), 3 (chicken muscle), 6 (cloacal swab) | 3 (chicken organ) | ||
| 22 | 1 (raw milk), 5 (chicken organ), 2 (cloacal swab) | 0 | ||
| 23 | 1 (chicken muscle) | 0 | ||
| 14 | 18 | 1 (cloacal swab) | 0 | |
| 19 | 1 (cloacal swab) | 0 | ||
| 20 | 6 (minced meat) | 0 | ||
| 21 | 1 (cloacal swab) | 0 | ||
| 22 | 3 (cloacal swab) | 0 | ||
| 23 | 2 (cloacal organ) | 3 (chicken organ), 4 (cloacal swab) | ||
| 24 | 2 (cloacal swab) | 2 (cloacal swab) | ||
| PDR (n = 6) |
14 | 25 | 4 (human stool), 2 (cloacal swab) | 0 |
MDR, multiple drug-resistance; XDR, extensively drug-resistance; PDR, pan drug-resistance.
3.3. Screening for Class 1 Integrons and Characterization of Associated Gene Cassettes in Campylobacter Isolates
Thirty-eight Campylobacter isolates (28 C. jejuni, and 10 C. coli) categorized as MDR (n = 5), XDR (n = 31), and PDR (n = 2) representing all sample origins and being resistant to at least 15 antimicrobial agents were screened for the possession of class 1 integrons using PCR assay. Overall, 37 of 38 (97.36%) examined isolates were positive for the integrase gene (intI1), all harbored class 1 integrons carrying gene cassettes of varying sizes ranging from 349 to 2600 bp. Only one C. jejuni isolate (code No. 16) possessed an empty integron structure with no gene cassettes inserted between its conserved segments (Table 4). Eight repetitive distinct gene cassettes were selected among integron positive isolates for DNA sequencing. Other gene cassette arrays were identified according to their PCR product sizes based on relevant previously published data.
Table 4.
Antimicrobial resistance patterns and gene cassette arrays carried by class 1 integrons in MDR, XDR and PDR Campylobacter isolates recovered from different sources (n = 38).
| Isolate No. | Code No. | Campylobacter Species | Source | Antimicrobial Resistance Pattern | Resistance to Antimicrobial Agents | Resistance to Antimicrobial Classes | Antimicrobial Resistance Type | intI1/Class 1 Integron | Resistance Gene(s) in Class 1 Integron (Size in bp) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CK105 | C. coli | Chicken intestine | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, MEM, ATM, S, TOB, CN, E, AZM, CLR, NA, SXT, CT, DA, DO | 21 | 12 | XDR | +/+ | aadA2 (995) |
| 2 | CK134 | C. jejuni | Chicken cloacal swab | AX, AM, SAM, AMC, KF, FOX, FEP, MEM, ATM, TOB, CN, E, AZM, CIP, NA, SXT, C, CT, LNZ, DA, DO | 21 | 14 | XDR | +/+ | dfrA15 (738) |
| 3 | CK110 | C. jejuni | Chicken thigh muscle | AX, AM, SAM, AMC, KF, FEP, ATM, S, TOB, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 21 | 13 | XDR | +/+ | dfrA15 (738); oxa1-aadA1a (2000) |
| 4 | H12 | C. jejuni | Human stool | AX, AM, KF, FOX, CEP, FEP, S, CN, E, AZM, CLR, CIP, SXT, C, LNZ, DA, DO | 17 | 10 | MDR | +/+ | dfrA17ab-aadA5 (1513); dfrA12-gcu-aadA2 (1864) |
| 5 | H19 | C. jejuni | Human stool | AX, AM, SAM, KF, FOX, FEP, ATM, S, TOB, CN, AK, E, AZM, CLR, NA, SXT, C, LNZ, DA, DO | 20 | 12 | XDR | +/+ | dfrA15 (738); dfrA17ab-aadA5 (1513); oxa1-aadA1a (2000) |
| 6 | CK139 | C. jejuni | Chicken breast muscle | AX,AM, SAM, AMC, KF, FOX, CEP, FEP, MEM, ATM, S, CN, TOB, E, AZM, CLR, CIP, NA, SXT, C, LNZ, DA, DO | 23 | 13 | XDR | +/+ | dfrA12-gcu-aadA2 (1864) |
| 7 | CK104 | C. jejuni | Chicken intestine | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, ATM, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 20 | 12 | XDR | +/+ | Phage tail TMP (349) |
| 8 | CK147 | C. jejuni | Chicken Liver | AX, AM, AMC, SAM, KF, FEP, CEP, FOX, MEM, ATM, S, TOB, CN, E, AZM, CLR, NA, SXT, C, CT, LNZ, DA, DO | 23 | 14 | XDR | +/+ | dfrA1-gcuC (1177) |
| 9 | C41 | C. jejuni | Raw milk | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, ATM, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 21 | 13 | XDR | +/+ | aacA4-cmlA4 (2600) |
| 10 | CK143 | C. coli | Chicken cloacal swab | AX, AM, SAM, AMC, KF, FOX, FEP, MEM, ATM, S, TOB, CN, AK, E, AZM, CIP, NA, SXT, C, CT, LNZ, DA, DO | 23 | 14 | XDR | +/+ | dfrA15 (738); dfrA17ab-aadA5 (1513) |
| 11 | H1 | C. coli | Human stool | AX, AM, KF, FOX, CEP, FEP, ATM, TOB, CN, AK, E, AZM, NA, SXT, CT | 15 | 8 | MDR | +/+ | dfrA15 (700); dfrA1ah-gcuCΔ (1146) |
| 12 | C52 | C. jejuni | Raw milk | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, ATM, S, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 22 | 13 | XDR | +/+ | dfrA17ab-aadA5 (1513); dfrA12-gcu-aadA2 (1864) |
| 13 | C8 | C. jejuni | Minced meat | AX, AM, AMC, SAM, KF, FEP, ATM, S, E, AZM, CLR, NA, SXT, C, CT, LNZ, DA, DO | 18 | 12 | XDR | +/+ | dfrA17-gcu5-aadA5 (1900) |
| 14 | H5 | C. jejuni | Human stool | AX, AM, AMC, KF, CEP, FEP, S, TOB, E, AZM, CLR, SXT, C, CT, DA, DO | 15 | 10 | MDR | +/+ | Phage tail TMP (349) |
| 15 | CK54 | C. coli | Chicken cloacal swab | AX, AM, AMC, KF, FEP, ATM, S, TOB, CN, E, CIP, NA, SXT, C, CT, LNZ, DA, DO | 18 | 13 | XDR | +/+ | aadA22 (947); dfrA17ab-aadA5 (1513) |
| 16 | CK80 | C. jejuni | Chicken breast muscle | AX, AM, SAM, AMC, KF, CEP, FEP, TOB, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DO | 19 | 11 | MDR | -/- | ND |
| 17 | CK125 | C. jejuni | Chicken Liver | AX, AM, SAM, AMC, KF, FEP, CEP, ATM, S, TOB, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 22 | 13 | XDR | +/+ | dfrA1-gcuC (1177); dfrA12-gcu-aadA2 (1864) |
| 18 | C12 | C. coli | Minced meat | AX, AM, AMC, KF, CEP, FEP, ATM, S, CN, E, AZM, CLR, CIP, NA, SXT, CT, LNZ, DA, DO | 19 | 12 | XDR | +/+ | dfrA15 (700); dfrA1-gcuC (1177); dfrA17ab-aadA5 (1513) |
| 19 | CK78 | C. jejuni | Chicken cloacal swab | AX, AM, AMC, KF, FEP, MEM, ATM, S, CN, E, AZM, CIP, NA, SXT, C, CT, LNZ, DA, DO | 19 | 14 | XDR | +/+ | dfrA15 (738); aadA22 (947); dfrA1ah-gcuCΔ (1146) |
| 20 | C46 | C. jejuni | Raw milk | AX, AM, AMC, KF, FOX, CEP, FEP, ATM, S, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 21 | 13 | XDR | +/+ | aadA2 (995); bla pse-1 (1200) |
| 21 | CK109 | C. jejuni | Chicken breast muscle | AX, AM, SAM, AMC, KF, CEP, FEP, ATM, S, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 21 | 13 | XDR | +/+ | dfrA15 (700); dfrA1-aadA1a (1600) |
| 22 | CK153 | C. jejuni | Chicken cloacal swab | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, MEM, ATM, S, TOB, CN, AK, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 25 | 14 | PDR | +/+ | dfrA15 (738); dfrA17-gcu5-aadA5 (1900) |
| 23 | CK50 | C. jejuni | Chicken liver | AX, AM, SAM, AMC, KF, FEP, CEP, ATM, S, TOB, E, CLR, NA, SXT, CT, LNZ, DA, DO | 18 | 12 | XDR | +/+ | dfrA15 (738); bla pse-1 (1200); dfrA1-aadA1a (1600) |
| 24 | CK117 | C. jejuni | Chicken cloacal swab | AX, AM, SAM, KF, CEP, FEP, MEM, ATM, S, TOB, CN, AK, E, AZM, CIP, NA, SXT, C, CT, LNZ, DA | 21 | 13 | XDR | +/+ | aadA2 (995); dfrA17-gcu5-aadA5 (1900) |
| 25 | H21 | C. jejuni | Human stool | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, MEM, ATM, S, TOB, CN, AK, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 25 | 14 | PDR | +/+ | aadA22 (947); aacA4-cmlA4 (2600) |
| 26 | C32 | C. jejuni | Raw milk | AX, AM, AMC, KF, CEP, ATM, S, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 19 | 13 | XDR | +/+ | dfrA15 (700); aadA2 (995); bla pse-1 (1200); aacC5-aadA7Δ4 (1799) |
| 27 | CK44 | C. jejuni | Chicken thigh muscle | AX, AM, AMC, KF, S, TOB, CN, AK, E, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 18 | 12 | XDR | +/+ | aadA2 (1000) |
| 28 | H6 | C. jejuni | Human stool | AX, AM, AMC, KF,CEP, FEP, S, TOB, E, AZM, CLR, SXT, C, CT, DA, DO | 16 | 10 | MDR | +/+ | aadA2 (995) |
| 29 | CK112 | C. coli | Chicken Liver | AX, AM, AMC, KF, FEP, CEP, MEM, ATM, S, TOB, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, DA, DO | 21 | 13 | XDR | +/+ | dfrA1-gcuC (1177) |
| 30 | C17 | C. jejuni | Minced meat | AX, AM, AMC, KF, FOX, CEP, FEP, ATM, S, TOB, E, AZM, CLR, NA, SXT, C, CT, DA, DO | 19 | 12 | XDR | +/+ | dfrA15 (738); aadA2 (1000) |
| 31 | CK106 | C. coli | Chicken intestine | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, MEM, ATM, S, TOB, CN, E, AZM, CLR, NA, SXT, CT, DA, DO | 21 | 12 | XDR | +/+ | Phage tail TMP (349); aadA22 (947) |
| 32 | CK99 | C. coli | Chicken cloacal swab | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, ATM, S, CN, E, NA, SXT, C, CT, LNZ, DA, DO | 19 | 13 | XDR | +/+ | dfrA15 (738); aacC5-aadA7Δ4 (1799) |
| 33 | C53 | C. jejuni | Raw milk | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, ATM, S, CN, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 22 | 13 | XDR | +/+ | aadA2 (995) |
| 34 | C4 | C. coli | Minced meat | AX, AM, AMC, KF, CEP, ATM, S, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 18 | 13 | XDR | +/+ | aadA22 (947) |
| 35 | C21 | C. jejuni | Minced meat | AX, AM, AMC, KF, FOX, CEP, ATM, S, TOB, CN, E, AZM, CLR, NA, SXT, C, CT, LNZ, DA, DO | 20 | 13 | XDR | +/+ | oxa1-aadA1a (2000) |
| 36 | CK46 | C. jejuni | Chicken cloacal swab | AX, AM, AMC, KF, FEP, ATM, S, TOB, CN, E, AZM, CIP, NA, SXT, C, CT, DA, DO | 18 | 12 | XDR | +/+ | Phage tail TMP (349); dfrA1ah-gcuCΔ (1146); oxa1-aadA1a (2000) |
| 37 | CK151 | C. coli | Chicken cloacal swab | AX, AM, SAM, AMC, KF, FOX, FEP, MEM, ATM, S, TOB, CN, AK, E, AZM, CLR, CIP, NA, SXT, C, CT, LNZ, DA, DO | 24 | 14 | XDR | +/+ | dfrA15 (738); aadA22 (947); bla pse-1 (1200) |
| 38 | CK136 | C. jejuni | Chicken breast muscle | AX, AM, SAM, AMC, KF, FOX, CEP, FEP, ATM, S, CN, TOB, AK, E, AZM, CLR, CIP, NA, SXT, C, LNZ, DA, DO | 23 | 12 | XDR | +/+ | dfrA17ab-aadA5 (1513) |
AX, amoxicillin; AM, ampicillin; SAM, ampicillin-sulbactam; AMC, amoxycillin-clavulanic acid; KF, cephalothin; FOX, cefoxitin; CEP, cefoperazone; FEP, cefepime; MEM, meropenem; ATM, aztreonam; S, Streptomycin; TOB, tobramycin; CN, gentamicin; AK, amikacin; E, erythromycin; AZM, azithromycin; CLR, clarithromycin; NA, nalidixic acid; CIP, ciprofloxacin; SXT, sulfamethoxazole-trimethoprim; C, chloramphenicol; CT, colistin; LNZ, lenzolid; DA, clindamycin; DO, doxycycline; MDR, multiple drug-resistance; XDR, extensively drug-resistance; PDR, pan drug-resistance; TMP, tape measure protein; ND, not detected; CK, chicken; H, human; C, cattle; +, positive; -, negative. Bold gene cassette arrays were subjected to DNA sequencing, deposited in the GenBank database and assigned their accession numbers as depicted in the Material and Methods section.
As shown in Table 4, 16 gene cassette arrays were identified among class1 integron-positive isolates. DNA sequence analysis revealed the predominance of aadA alleles (aadA1a, aadA2, aadA5, aadA7Δ4, and aadA22) in 30 out of 37 (81.08%) analyzed isolates, accounting resistance for aminoglycosides, particularly streptomycin. Other frequent gene cassettes reported herein were dfrA (25/37; 67.56%) alleles (dfrA1, dfrA1, dfrA12, dfrA15, and dfrA17), conferring resistance to the trimethoprim antimicrobial agent. Despite the high frequency of resistant Campylobacter isolates to B-lactams, the bla pse-1 and oxa1 gene cassettes were detected in only four isolates (4/37; 10.81% each). Likewise, the gene cassette aacA4-cmlA4 conferring resistance to chloramphenicol was found in only two Campylobacter isolates.
The most striking finding in the current study is the exclusive existence of a novel gene cassette array namely aacC5-aadA7Δ4 (In number in 1983) as a first report according to the INTEGRALL database in only two Campylobacter isolates (code Nos. 26 and 32). This conferred resistance to aminoglycosides in particular gentamicin and streptomycin, but not tobramycin, amikacin, nor kanamycin.
Two gene cassette arrays were reported within class 1 integrons of Campylobacter species, each one harbored triple genes. The dfrA17-gcu5-aadA5 (1900 bp) integron-borne cassette array existed in two XDR (code Nos. 13 and 24) and one PDR (code No. 22) Campylobacter isolates and dfrA12-gcu-aadA2 (1864 bp) gene cassette incorporated in four XDR Campylobacter isolates (code Nos. 4, 6, 12 and 17), both cassettes conferred resistance to aminoglycosides and trimethoprim antimicrobial agents (Table 4).
Of interest, all gene cassettes reported here were linked to antimicrobial resistance except one, whose product is a putative phage tail tape measure protein (349 bp; accession number MT612449). It was reported in three XDR Campylobacter isolates (code Nos. 7, 31, and 36) originated from chicken and one MDR C. jejuni isolate (code No. 14) of human origin, thus facilitates DNA transit to the cell cytoplasm during infection. To our knowledge, this is the first report of a putative phage tail protein associated with class 1 integrons in Campylobacter species.
3.4. Correlation between Class 1 Integrons and Antimicrobial Resistance Phenotypes in Campylobacter Isolates
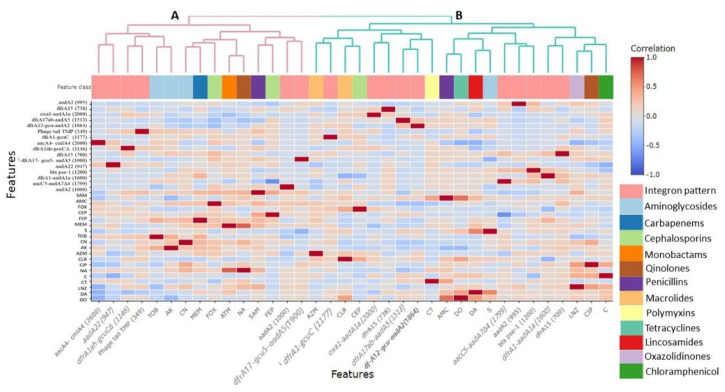
As depicted in Figure 2 and Table S2, PCR results and DNA sequence analysis were consistent with certain antimicrobial susceptibility phenotypes. It was noted that the existence of aadA and aacC5 genes positively correlated (r = 0.09–0.18) with streptomycin resistance. Moreover, class 1 integron-positive isolates carrying aacC5-aadA7Δ4 and aacA4-cmlA4 cassette arrays showed positive correlations with resistance to gentamicin (r = 0.11) and tobramycin (r = 0.17), respectively. The presence of bla pse-1 and oxa1 genes non-significantly (p > 0.05) associated with resistance to amoxicillin-clavulanate (r = 0.12 each) and cefoperazone (r = 0.21 and 0.01, respectively). However, both genes did not confer resistance to ampicillin-sulbactam (r = −0.07 and −0.25, respectively), cefoxitin (r = −0.21 and −0.04, respectively) or cefepime (r = −0.16 each).
Figure 2.
Pairwise correlation (R) among different antimicrobial resistance phenotypes and integron gene cassette arrays. Red and blue colors indicate positive and negative correlations, respectively. The color key refers to the correlation coefficient (R). The darker colors imply stronger positive or negative correlations. The hierarchical clustering of the variables is shown as a dendrogram illustrating different clusters with different colors and letters (e.g., A and B). Variables that are identical among all strains are excluded, and thus not shown in this figure. Classes of antimicrobials are color-coded below the dendrogram.
The clustering pattern of class 1 integron-positive Campylobacter isolates is illustrated in Figure 2. The two variables (gene cassette arrays and antimicrobial resistance phenotypes) produced two distinct clusters (A and B). Notably, the aacA4-cmlA4 gene cassette gathered with tobramycin in cluster A. While, the aadA genes and bla pse-1 and oxa1-aadA1 cassette arrays, which confer resistance to streptomycin and amoxicillin-clavulanate and cefoperazone, respectively, were clustered together in cluster B.
3.5. Cluster Analysis of Gene Cassette Arrays in Campylobacter Isolates from Human and Animal Populations
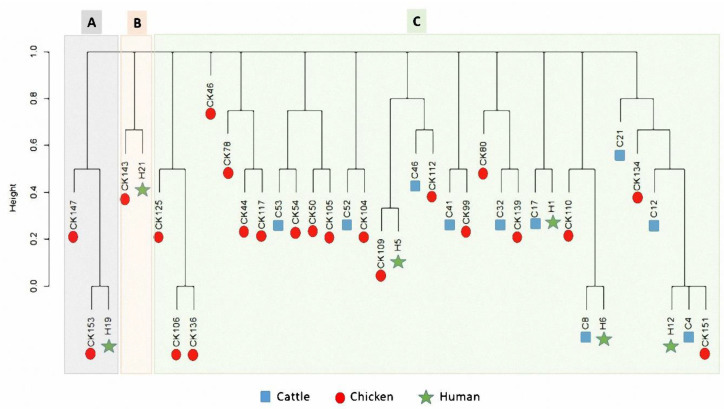
The dendrogram analysis (Figure 3) of class 1 integron-positive isolates (n = 38) simplified the existence of gene cassettes across livestock animals and humans. Three clusters were noticed in our dataset (A, B and, C). A close relatedness was observed among certain Campylobacter isolates of different sources. As exemplified, a Campylobacter isolate of the chicken source (code CK153) was closer to another one of human origin (code H19), both were gathered in cluster A. In addition, two Campylobacter isolates of human (H21) and chicken (CK143) sources clustered closely together in cluster B. Regarding the cluster C, several isolates of the three populations (cattle, chicken and human) clustered together.
Figure 3.
Dendrogram showing the binary distances among different Campylobacter isolates based on the integron patterns. The isolates are categorized based on their host, which are shown as different colors and symbols. The X-axis refers to the binary distance scale. Isolates codes are illustrated in Table 4.
4. Discussion
Thermophilic campylobacters as C. jejuni and C. coli are associated with infections in humans due to the consumption of undercooked meat, particularly poultry, and unpasteurized milk [2,3]. Currently, increasing resistance to major antibiotics in use among campylobacters is an emerging problem [34]. This is the first report to provide insights into the carriage of class 1 integrons by PDR, XDR, and MDR Campylobacter species isolated from livestock animals and humans in Egypt. In this study, the overall occurrence rate of C. jejuni (71.48%) was higher than that of C. coli (28.51%). Most Campylobacter isolates were detected in chicken samples (58.11%) represented as 67.53% C. jejuni and 32.47% C. coli, while previous studies reported varying rates of Campylobacter prevalence in chickens ranging from 24% to 62% [35,36].
Raw milk acts as a second main source of campylobacters [37]. The consumption of unpasteurized milk and milk products has been implicated in infections of 23% of human cases with campylobacteriosis in Egypt [38]. We reported a C. jejuni prevalence rate of 74.29% in raw milk samples, which have been previously documented with a lower prevalence rate (34%) [39]. Cross-contamination with Campylobacter species could occur during slaughter and milking of cattle. Herein, the prevalence of C. jejuni in fresh meat was 51.43%, which was lower than that reported in a previous study in Ethiopia (72%) [40]. Of note, milk and meat products were free from Campylobacter species, which was consistent with a previous study in India [41].
Poultry, milk, and meat are a reservoir for campylobacters; therefore, food processing with poor sanitation is an important source of transmission leading to increase the risk of human exposure, especially those in contact with food-producing animals. A higher prevalence of C. jejuni in the human stool (66.67%) was detected in this study when compared with previous studies conducted in Egypt with prevalence rates of 27.5% [35], 16.66% [42], and 4.07% [43].
Campylobacteriosis does not usually require antibiotic treatment; however, in some cases, antibiotics may be administered. Ciprofloxacin and erythromycin are considered drugs of choice for treating Campylobacter infections in humans [44]. However, the unregulated use of antibiotics in human and veterinary medicine, resulting in increasing their resistance [45].
Increasing resistance to the major antibiotics in use among campylobacters is an emerging problem [34]. Therefore, an investigation of the resistance rates and mechanisms is essential to prevent the spread of antibiotic-resistant campylobacters in livestock animals and humans. Herein, we provided better insight into different drug resistance patterns (n = 93-pattern) as well as an alarming increase of PDR, XDR, and MDR categories, while testing 25 antimicrobials among 14 antimicrobial categories.
Resistance to three or more antimicrobial classes (MDR) is a worldwide disturbing situation in C. jejuni [46]. In this study, 28.5% of Campylobacter isolates originating from livestock animals and humans exhibited MDR pattern. This level of resistance was less than that reported previously [47], while the resistance profile reaching 11 to 19 drugs is worrying compared with previous studies that recorded resistance to five to six [48] or three to four antimicrobial agents [49].
Resistance to all classes of antimicrobial agents except two or fewer is defined as XDR [26]. Of interest, this is the first report of XDR Campylobacter isolates (68.94%) among livestock animals and humans. In a previous study, an XDR C. jejuni CCARM 13,322 was recovered from a human case of diarrhea associated with international travel [17]. Moreover, 2.55% of Campylobacter isolates showed PDR (resistant to all antimicrobial agents among 14 categories), which was not reported in any previous study yet.
Class 1 integron is the main cause of multiple antibiotic resistance gene cassettes transmission in Gram-negative bacteria causing multidrug resistance [9,50]. Till date, few reports detected class 1 integrons in both C. jejuni and C. coli isolated from chicken house environment [12] and human sources [13,14] without showing the antimicrobial resistance profiles of isolates. However, no reports could detect class 1 integrons in PDR or XDR Campylobacter isolates [17].
In this study, the interested report of integrase gene and class 1 integrons (97.36% each) in C. jejuni and C. coli of chicken, cattle, and human origins representing MDR, XDR, and PDR patterns were documented for the first time, at least in Egypt. These integrons were associated with gene cassettes of different sizes ranging from 349 to 2600 bp. Considering, the previously published data, all Campylobacter isolates originating from a chicken house environment harbored a single cassette in the integron with 900 bp amplicon [12]. Additionally, those isolates originating from human and poultry sources had gene cassettes of molecular weights ranging from 300 to 1.4 kb [14].
Class 1 integrons detected in this study associated with 16 resistance gene cassettes. The most frequently reported were trimethoprim (dfrA1, dfrA1, dfrA12, dfrA15, and dfrA17) and aminoglycoside (aadA1a, aadA2, aadA5, aadA22, and aadA7Δ4) resistance gene cassette arrays. Consistently, previous studies detected class 1 integrons associated with aminoglycoside resistance genes (aadA2 and aacA4) in both C. jejuni and C. coli resulting from the sequencing of 1000 bp, and 900 bp amplicons, respectively [12,14]. Moreover, the trimethoprim resistance gene cassettes (dfr1 and dfr9) were carried by class 1 integrons in clinical isolates of C. jejuni following the sequencing of 399 bp, and 254 bp amplicons, respectively [13,15].
The most surprising points in the current study are the carriage of a novel gene cassette array, namely aacC5-aadA7Δ4 (in number in 1983) as a first report according to the INTEGRALL database. In addition, an unusual phage tail tape measure protein gene cassette was harbored by four Campylobacter isolates as a first record in campylobacters worldwide. It is widely assumed that Campylobacter–bacteriophage interactions may play a role in horizontal gene transfer. In accordance with previous reports, bacteriophages cause genomic instability in C. jejuni and mediate interstrain transfer of large DNA fragments [51,52].
Herein, the correlation between resistance to certain antimicrobials and the corresponding gene cassettes was shown similar to other studies as Campylobacter isolates harboring aminoglycoside resistance gene, aacA4, conferred higher tobramycin MICs but slightly increased resistance to gentamicin [12]. In addition, high-level resistance to trimethoprim in C. jejuni was associated with the acquisition of dfr genes [15].
Previous studies showed that the dfr cassette is mostly associated with the aadA gene cassette [53]. DNA sequence analysis of a 1513 bp amplicon of class 1 integron revealed dfrA17ab-aadA5 gene cassette in seven XDR Campylobacter isolates. Moreover, dfrA12-gcu-aadA2 gene cassette array was detected in four analyzed isolates. According to our results, the dfrA1-aadA1 gene cassette has been reported in MDR Citrobacter species with 1600 bp fragment size [54]. In addition, a previous study detected these gene cassettes of 2000 bp in MDR Salmonella isolates [55].
5. Conclusions
This is the first report, at least in Egypt, that showed the prevalence of PDR and XDR Campylobacter species in livestock animals and humans. Moreover, we demonstrated the existence of class 1 integrons and associated gene cassettes in analyzed isolates, which confer antimicrobial resistance and the possibility of Campylobacter–bacteriophage interactions by the carriage of an unusual phage tail protein as a first report.
Acknowledgments
We thank Taif University researchers supporting project number TURSP-2020/13, Taif University, Taif, Saudi Arabia. We thank the staff members of Faculty of Veterinary Medicine, Zagazig University, Egypt for their technical support during this study.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2615/10/11/2067/s1, Table S1. Oligonucleotide primer sequences used in the study, Table S2: Antibiotypes, MAR-indices, and detection of PDR, XDR, and MDR Campylobacter isolates originated from different sources. Figure S1: Distribution and clustering of studied C. jejuni isolates (n = 168) and the patterns of their antimicrobial resistance. Different hosts, sample types, antimicrobial classes, and resistance categories are shown for each isolate as color codes. The heat map represents the hierarchical clustering of the isolates and the antimicrobial classes, Figure S2: Distribution and clustering of studied C. coli isolates (n = 67) and the patterns of their antimicrobial resistance. Different hosts, sample types, antimicrobial classes, and resistance categories are shown for each isolate as color codes. The heat map represents the hierarchical clustering of the isolates and the antimicrobial classes.
Author Contributions
N.K.A.E.-A. and A.M.A. contributed equally in the conception and design of the study. M.M.H. performed the classical microbiological techniques. A.A.G. and E.A. conceived the study and participated in the design. A.A.G., E.A., A.H.S. and M.M.H. participated in acquisition of data and analysis and interpretation of data. N.K.A.E.-A. carried out the sequence analysis and participated in the data analysis. N.K.A.E.-A. and A.H.S. wrote the initial draft of the manuscript and revised it critically for important intellectual content. A.A.G. and E.A. are responsible for the project administration and funding resources. All authors revised the manuscript and gave the final approval of the version to be published. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Moore J.E., Corcoran D., Dooley J.S., Fanning S., Lucey B., Matsuda M., McDowell D.A., Mégraud F., Millar B.C., O’Mahony R. Campylobacter. Vet. Res. 2005;36:351–382. doi: 10.1051/vetres:2005012. [DOI] [PubMed] [Google Scholar]
- 2.Friedman C.R., Hoekstra R.M., Samuel M., Marcus R., Bender J., Shiferaw B., Reddy S., Ahuja S.D., Helfrick D.L., Hardnett F. Risk factors for sporadic Campylobacter infection in the United States: A case-control study in FoodNet sites. Clin. Infect. Dis. 2004;38:S285–S296. doi: 10.1086/381598. [DOI] [PubMed] [Google Scholar]
- 3.Neimann J., Engberg J., Mølbak K., Wegener H.C. A case–control study of risk factors for sporadic Campylobacter infections in Denmark. Epidemiol. Infect. 2003;130:353–366. doi: 10.1017/S0950268803008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.European Food Safety Authority. European Centre for Disease Prevention and Control EU Summary Report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2013. EFSA J. 2015;13:4036. doi: 10.2903/j.efsa.2017.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luangtongkum T., Jeon B., Han J., Plummer P., Logue C.M., Zhang Q. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Future Microbiol. 2009;4:189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laroche E., Pawlak B., Berthe T., Skurnik D., Petit F. Occurrence of antibiotic resistance and class 1, 2 and 3 integrons in Escherichia coli isolated from a densely populated estuary (Seine, France) FEMS Microbiol. Ecol. 2009;68:118–130. doi: 10.1111/j.1574-6941.2009.00655.x. [DOI] [PubMed] [Google Scholar]
- 7.Brink A., Feldman C., Richards G., Moolman J., Senekal M. Emergence of extensive drug resistance (XDR) among Gram-negative bacilli in South Africa looms nearer. S. Afr. Med. J. 2008;98:586–592. [PubMed] [Google Scholar]
- 8.Park Y.K., Peck K.R., Cheong H.S., Chung D.-R., Song J.-H., Ko K.S. Extreme drug resistance in Acinetobacter baumannii infections in intensive care units, South Korea. Emerg. Infect. Dis. 2009;15:1325. doi: 10.3201/eid1508.080772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mazel D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006;4:608–620. doi: 10.1038/nrmicro1462. [DOI] [PubMed] [Google Scholar]
- 10.Labbate M., Case R.J., Stokes H.W. Horizontal Gene Transfer. Springer; Berlin/Heidelberg, Germany: 2009. The integron/gene cassette system: An active player in bacterial adaptation; pp. 103–125. [DOI] [PubMed] [Google Scholar]
- 11.Recchia G.D., Hall R.M. Gene cassettes: A new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 12.Lee M.D., Sanchez S., Zimmer M., Idris U., Berrang M.E., McDermott P.F. Class 1 integron-associated tobramycin-gentamicin resistance in Campylobacter jejuni isolated from the broiler chicken house environment. Antimicrob. Agents Chemother. 2002;46:3660–3664. doi: 10.1128/AAC.46.11.3660-3664.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gibreel A., Sköld O. An integron cassette carrying dfr1 with 90-bp repeat sequences located on the chromosome of trimethoprim-resistant isolates of Campylobacter jejuni. Microb. Drug Resist. 2000;6:91–98. doi: 10.1089/107662900419384. [DOI] [PubMed] [Google Scholar]
- 14.O’Halloran F., Lucey B., Cryan B., Buckley T., Fanning S. Molecular characterization of class 1 integrons from Irish thermophilic Campylobacter spp. J. Antimicrob. Chemother. 2004;53:952–957. doi: 10.1093/jac/dkh193. [DOI] [PubMed] [Google Scholar]
- 15.Gibreel A., Sköld O. High-level resistance to trimethoprim in clinical isolates of Campylobacter jejuni by acquisition of foreign genes (dfr1 and dfr9) expressing drug-insensitive dihydrofolate reductases. Antimicrob. Agents Chemother. 1998;42:3059–3064. doi: 10.1128/AAC.42.12.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Y.-C., Tien N., Yang J.-S., Lu C.-C., Tsai F.-J., Huang T.-J., Wang I.-K. Class 1 integrons and plasmid-mediated multiple resistance genes of the Campylobacter species from pediatric patient of a university hospital in Taiwan. Gut Pathog. 2017;9:1–8. doi: 10.1186/s13099-017-0199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin E., Hong H., Oh Y., Lee Y. First report and molecular characterization of a Campylobacter jejuni isolate with extensive drug resistance from a travel-associated human case. Antimicrob. Agents Chemother. 2015;59:6670–6672. doi: 10.1128/AAC.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandepitte J., Verhaegen J., Engbaek K., Rohner P., Piot P., Heuck C., Heuck C. Basic Laboratory Procedures in Clinical Bacteriology. World Health Organization; Geneva, Switzerland: 2003. [Google Scholar]
- 19.Quinn P.J., Markey B., Carter M.E. Clinical Veterinary Microbiology. 1st ed. Wolfe Publishing; London, UK: 1994. Campylobacter spp. pp. 268–272. [Google Scholar]
- 20.Wang G., Clark C.G., Taylor T.M., Pucknell C., Barton C., Price L., Woodward D.L., Rodgers F.G. Colony multiplex PCR assay for identification and differentiation of Campylobacter jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus subsp. fetus. J. Clin. Microbiol. 2002;40:4744–4747. doi: 10.1128/JCM.40.12.4744-4747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin E., Lee Y. Comparison of three different methods for Campylobacter isolation from porcine intestines. J. Microbiol. Biotechnol. 2009;19:647–650. [PubMed] [Google Scholar]
- 22.Bauer A., Kirby W., Sherris J., Turck M. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966;45:493–496. doi: 10.1093/ajcp/45.4_ts.493. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Susceptibility Testing. Twenty-First Informational Supplement. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2011. CLSI document M100-S21. [Google Scholar]
- 24.European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of mics and zone diameters. Version 7.1, Valid from 2017-03-10. [(accessed on 20 August 2019)]; Available online: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf.
- 25.Tambekar D., Dhanorkar D., Gulhane S., Khandelwal V., Dudhane M. Antibacterial susceptibility of some urinary tract pathogens to commonly used antibiotics. Afr. J. Biotechnol. 2006;5:1562–1565. [Google Scholar]
- 26.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 27.Kashif J., Buriro R., Memon J., Yaqoob M., Soomro J., Dongxue D., Jinhu H., Liping W. Detection of class 1 and 2 integrons, β-lactamase genes and molecular characterization of sulfonamide resistance in Escherichia coli isolates recovered from poultry in China. Pak. Vet. J. 2013;33:321–324. [Google Scholar]
- 28.White P.A., McIver C.J., Deng Y.-M., Rawlinson W.D. Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 2000;182:265–269. doi: 10.1111/j.1574-6968.2000.tb08906.x. [DOI] [PubMed] [Google Scholar]
- 29.Sow A.G., Wane A.A., Diallo M.H., Boye C.S.-B., Aïdara-Kane A. Genotypic characterization of antibiotic-resistant Salmonella Enteritidis isolates in Dakar, Senegal. J. Infect. Dev. Ctries. 2007;1:284–288. [PubMed] [Google Scholar]
- 30.Kolde R. Package ‘pheatmap’: Pretty Heat Map. [(accessed on 12 May 2020)];2018 Available online: https://rdrr.io/cran/pheatmap/
- 31.Friendly M. Corrgrams. Am. Stat. 2002;56:316–324. doi: 10.1198/000313002533. [DOI] [Google Scholar]
- 32.Galili T., O’Callaghan A., Sidi J., Sievert C. Heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2018;34:1600–1602. doi: 10.1093/bioinformatics/btx657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrell F.E., Jr. Package ‘Hmisc’. [(accessed on 20 May 2020)]; Available online: https://cran.r-project.org/web/packages/Hmisc/Hmisc.pdf.
- 34.Mshana S., Joloba M., Kakooza A., Kaddu-Mulindwa D. Campylobacter spp among Children with acute diarrhea attending Mulago hospital in Kampala-Uganda. Afr. Health Sci. 2009;9:201. [PMC free article] [PubMed] [Google Scholar]
- 35.Abushahba M.F., Ahmed S.O., Ibrahim A.A., Mosa H.A. Prevalence of zoonotic species of Campylobacter in broiler chicken and humans in Assiut governorate, Egypt. Approaches Poult. Dairy Vet. Sci. 2018;3:1–9. doi: 10.31031/APDV.2018.03.000568. [DOI] [Google Scholar]
- 36.Jouahri M., Asehraou A., Karib H., Hakkou A., Touhami M. Prevalence and control of thermo-tolerant Campylobacter species in raw poultry meat in Morocco. MESO Prvi Hrvat. Časopis Mesu. 2007;9:262–267. [Google Scholar]
- 37.Leedom J.M. Milk of nonhuman origin and infectious diseases in humans. Clin. Infect. Dis. 2006;43:610–615. doi: 10.1086/507035. [DOI] [PubMed] [Google Scholar]
- 38.Jun W., Guo Y.C., Ning L. Prevalence and risk assessment of Campylobacter jejuni in chicken in China. Biomed. Environ. Sci. 2013;26:243–248. doi: 10.3967/0895-3988.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Wicker C., Giordano M., Rouger S., Sorin M.-L., Arbault P. Campylobacter detection in food using an ELISA based method. Int. J. Med. Microbiol. 2001;291:1–12. [Google Scholar]
- 40.Woldemariam T., Asrat D., Zewde G. Prevalence of thermophilic Campylobacter species in carcasses from sheep and goats in an abattoir in Debre Zeit area, Ethiopia. Ethiop. J. Health Dev. 2009;23 doi: 10.4314/ejhd.v23i3.53245. [DOI] [Google Scholar]
- 41.Singh H., Rathore R.S., Singh S., Cheema P.S. Comparative analysis of cultural isolation and PCR based assay for detection of Campylobacter jejuni in food and faecal samples. Braz. J. Microbiol. 2009;42:181–186. doi: 10.1590/S1517-83822011000100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hassanain N.A. Antimicrobial resistant Campylobacter jejuni isolated from humans and animals in Egypt. Glob. Vet. 2011;6:195–200. [Google Scholar]
- 43.El-Hamid M.I., El-Aziz A., Khairy N., Samir M., Remela A., Etab M., El-Naenaeey E.-s.Y., Bendary M.M., Mosbah R.A. Genetic diversity of Campylobacter jejuni isolated from Avian and human sources in Egypt. Front. Microbial. 2019;10:2353. doi: 10.3389/fmicb.2019.02353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghunaim H., Behnke J.M., Aigha I., Sharma A., Doiphode S.H., Deshmukh A., Abu-Madi M.M. Analysis of resistance to antimicrobials and presence of virulence/stress response genes in Campylobacter isolates from patients with severe diarrhoea. PLoS ONE. 2015;10:e0119268. doi: 10.1371/journal.pone.0119268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Vries S.P., Vurayai M., Holmes M., Gupta S., Bateman M., Goldfarb D., Maskell D.J., Matsheka M.I., Grant A.J. Phylogenetic analyses and antimicrobial resistance profiles of Campylobacter spp. from diarrhoeal patients and chickens in Botswana. PLoS ONE. 2018;13 doi: 10.1371/journal.pone.0194481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mdegela R., Nonga H., Ngowi H., Kazwala R. Prevalence of thermophilic Campylobacter infections in humans, chickens and crows in Morogoro, Tanzania. J. Vet. Med. Ser. B. 2006;53:116–121. doi: 10.1111/j.1439-0450.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- 47.Bouhamed R., Bouayad L., Messad S., Zenia S., Naïm M., Hamdi T.-M. Sources of contamination, prevalence, and antimicrobial resistance of thermophilic Campylobacter isolated from turkeys. Vet. World. 2018;11:1074. doi: 10.14202/vetworld.2018.1074-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marotta F., Garofolo G., di Marcantonio L., Di Serafino G., Neri D., Romantini R., Sacchini L., Alessiani A., Di Donato G., Nuvoloni R. Antimicrobial resistance genotypes and phenotypes of Campylobacter jejuni isolated in Italy from humans, birds from wild and urban habitats, and poultry. PLoS ONE. 2019;14:e0223804. doi: 10.1371/journal.pone.0223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Di Giannatale E., Calistri P., Di Donato G., Decastelli L., Goffredo E., Adriano D., Mancini M.E., Galleggiante A., Neri D., Antoci S. Thermotolerant Campylobacter spp. in chicken and bovine meat in Italy: Prevalence, level of contamination and molecular characterization of isolates. PLoS ONE. 2019;14:e0225957. doi: 10.1371/journal.pone.0225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hall M.A.L.-v., Blok H.E.M., Donders T.R., Paauw A., Fluit A.C., Verhoef J. Multidrug resistance among Enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infec. Dis. 2003;187:251–259. doi: 10.1086/345880. [DOI] [PubMed] [Google Scholar]
- 51.Scott A.E., Timms A.R., Connerton P.L., Carrillo C.L., Radzum K.A., Connerton I.F. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 2007;3 doi: 10.1371/journal.ppat.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott A.E., Timms A.R., Connerton P.L., El-Shibiny A., Connerton I.F. Bacteriophage influence Campylobacter jejuni types populating broiler chickens. Environ. Microbiol. 2007;9:2341–2353. doi: 10.1111/j.1462-2920.2007.01351.x. [DOI] [PubMed] [Google Scholar]
- 53.Yan H., Li L., Zong M., Alam M.J., Shinoda S., Shi L. Occurrence and characteristics of Class 1 and 2 integrons in clinical bacterial isolates from patients in south China. J. Health Sci. 2010;56:442–450. doi: 10.1248/jhs.56.442. [DOI] [Google Scholar]
- 54.Lorestani R.C., Akya A., Elahi A., Hamzavi Y. Gene cassettes of class 1 integron-associated with antimicrobial resistance in isolates of Citrobacter spp. with multidrug resistance. Iran. J. Microbiol. 2018;10:22. [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao X., Yang J., Zhang B., Sun S., Chang W. Characterization of integrons and resistance genes in Salmonella isolates from farm animals in Shandong Province, China. Front. Microbiol. 2017;8:1300. doi: 10.3389/fmicb.2017.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.