Mechanical force–repressed pectin degradation alters cell wall mechanics in control of cell growth and tissue morphogenesis.
Abstract
How mechanical forces regulate plant growth is a fascinating and long-standing question. After germination underground, buried seedlings have to dynamically adjust their growth to respond to mechanical stimulation from soil barriers. Here, we designed a lid touch assay and used atomic force microscopy to investigate the mechanical responses of seedlings during soil emergence. Touching seedlings induced increases in cell wall stiffness and decreases in cell elongation, which were correlated with pectin degradation. We revealed that PGX3, which encodes a polygalacturonase, mediates touch-imposed alterations in the pectin matrix and the mechanics of morphogenesis. Furthermore, we found that ethylene signaling is activated by touch, and the transcription factor EIN3 directly associates with PGX3 promoter and is required for touch-repressed PGX3 expression. By uncovering the link between mechanical forces and cell wall remodeling established via the EIN3-PGX3 module, this work represents a key step in understanding the molecular framework of touch-induced morphological changes.
INTRODUCTION
Plants continuously encounter diverse mechanical challenges, including wind, rainfall, and the physical barrier of soil. In adapting to these mechanical challenges, plants have evolved to respond and acclimate to mechanostimulation by triggering growth and developmental changes in a process termed thigmomorphogenesis (1–3). Among different plant species, the most common features of thigmomorphogenesis are a decrease in elongation growth and an increase in radial expansion, and both responses are thought to help plants withstand mechanical stresses (3, 4). Although repetitive-touch treatments over time are required to cause morphogenetic alterations, the intracellular responses to these treatments, including altered cytosolic calcium levels (5–7), phytohormone biosynthesis and catabolism (8–10), gene expression (1, 11), and protein phosphorylation and stabilization (12, 13), are very fast. The mechanisms by which mechanical forces alter cellular behavior, and thereby biological shapes, remain elusive.
Terrestrial seed plants often begin their life cycle under the soil. Germinating seedlings adopt an etiolation program when they are subjected to dark conditions, such as those in subterranean environments (14). At the same time, it is essential for buried seedlings to dynamically adjust their growth to respond to mechanical stimulation from soil barriers (11, 13, 15). Ethylene is a gaseous phytohormone that triggers marked morphological changes in dark-grown Arabidopsis seedlings, including inhibition of hypocotyl and root elongation, radial hypocotyl swelling, and exaggerated curvature of the apical hook (16, 17). This “triple response” morphology plays a critical role in protecting seedlings from mechanical injuries as they penetrate through the soil (11, 13, 15, 18). In Arabidopsis, perception of ethylene by a group of endoplasmic reticulum–localized receptors causes inactivation of CONSTITUTIVE TRIPLE RESPONSE1 (CTR1) (19–21) and stimulates cleavage and nuclear translocation of the C-terminal domain of ETHYLENE INSENSITIVE2 (EIN2) (22, 23). In the nucleus, EIN3 and EIN3 Like1 (EIL1) are stabilized by ethylene via EIN2 and two F-box proteins, EIN3-BINDING F-BOX PROTEIN1/2 (EBF1/2), allowing them to function as master transcription factors that direct transcriptional regulation to generate ethylene responses (24–26).
Plant cells are surrounded by cell walls whose mechanical properties must be regulated to adjust the growth behavior of individual cells and tissues (27–29). In land plants, the primary cell wall is mainly (approximately 90%) composed of polysaccharides, including cellulose, hemicellulose, and pectin (27, 30). Pectin polymers, defined as polysaccharides rich in galacturonic acid (GalA), form a hydrated gel-like matrix between cellulose/hemicellulose microfibrils (27, 31, 32). Structurally and functionally, pectins are the major constituents of cell walls and determine their physicochemical characteristics (33, 34). Degradation of pectin polymers by polygalacturonases (PGs) plays a crucial role in regulating cell and tissue growth in various developmental processes, including hypocotyl elongation, organ initiation, fruit ripening, and leaf or flower abscission (35–38).
Here, we describe a pathway underlying touch-induced morphological changes. Mechanical stimulation is linked to cell wall modifications via endogenous ethylene signaling in a process wherein pectin degradation controlled by PGX3 plays a central role. Moreover, we show that touch-regulated cell wall remodeling is transient and reversible, providing a molecular basis for the sensitive molecular responses and slow thigmomorphogenesis observed in plants. The mechanism described here allows plastic adjustments of plant cells in response to mechanical perturbations, ensuring the fitness of buried seedlings during growth and emergence from the soil.
RESULTS
Touch regulates cell elongation and cell wall stiffness of etiolated seedlings
To investigate the mechanical responses of seedlings emerging from the soil, we designed a lid touch assay in which seedlings were grown in darkness for 3 days on half-strength Murashige and Skoog (MS) medium in deep plate (D) or shallow plate (S) conditions (Fig. 1A). The distance between the medium surface and the plate lid was 12 mm for the D condition and 6 mm for the S condition (Fig. 1A). In the D condition, the etiolated seedlings grew upward without touching the lid. In the S condition, the etiolated seedlings touched the lid 2 days after germination and were grown under mechanical resistance from the plate lid for an additional day.
Fig. 1. Pectin degradation is involved in touch-regulated hypocotyl cell elongation and cell wall mechanics.
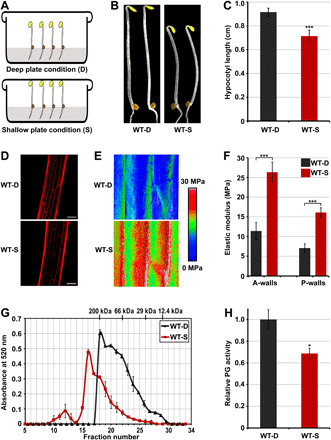
(A) Schematic diagram of the deep-shallow plate assay. (B and C) Images (B) and hypocotyl length (C) of 3-day-old etiolated seedlings grown in the D or S condition. Mean ± SD; n ≥ 20. (D) PI staining images of cells at the middle region of hypocotyls. Scale bar, 100 μm. (E and F) Elastic modulus map (E) and quantification (F) of epidermal cell walls at the middle region of hypocotyls. The elastic modulus on the A-walls and P-walls was obtained by AFM in the QNM mode. Mean ± SD; n ≥ 10. (G) Molecular mass profile of CDTA-soluble pectin in 3-day-old etiolated seedlings grown in the D or S condition. The mean ± SD was obtained from two technical replicates. (H) In vivo PG activity of etiolated seedlings in response to mechanical resistance stimulation. The relative PG activities of the samples were normalized to the absorbance of wild-type (WT) seedlings grown in the D condition. Mean ± SD; n = 3.
Previous studies have identified several touch-inducible genes (designated as TCHs) that are characteristically induced by touch treatments (1, 9). To determine whether the S condition stimulated these responses, we examined the expression levels of two representative touch-responsive genes: TCH3 (AT2G41100) and TCH4 (AT5G57560). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) and luciferase-reporter analysis showed that the expression levels of TCH3 and TCH4 were markedly up-regulated by the physical barrier of lid in the S condition (fig. S1). Next, we investigated developmental changes by plant seedlings in response to the lid touch stimulus. In comparison with the seedlings grown in the D condition, the hypocotyls of seedlings grown in the S condition were significantly shortened (Fig. 1, B and C). Analysis of hypocotyl cell length by propidium iodide (PI) staining showed that mechanical forces repressed cell elongation at the top, middle, and bottom regions of hypocotyls, with the greatest decrease and statistically high significance in the middle region (Fig. 1D and fig. S2). These results demonstrate that mechanical forces of the lid touch treatment used in this study inhibit hypocotyl cell elongation.
To determine the effects of the lid touch treatment on cell wall mechanics, we used a nanoindentation atomic force microscopy (AFM)–based method to measure the elastic modulus of the hypocotyl epidermal cell wall, which served as an indicator of its stiffness (39–43). After plasmolysis, the anticlinal cell walls (A-walls; perpendicular to the hypocotyl surface, transverse, and longitudinal) became evident. The AFM-based stiffness map revealed that the hypocotyl epidermal cell walls of seedlings grown in the S condition had an elastic modulus higher than that of seedlings grown in the D condition (Fig. 1E). The quantitative elastic modulus results from the A-walls and outer periclinal cell walls (P-walls) showed that the mechanical properties of cell walls changed in response to physical resistance, with the hypocotyl epidermal cell walls of seedlings grown in the S condition displaying an elastic modulus significantly higher than that of those grown in the D condition (Fig. 1F). Together, these findings demonstrate that mechanical touch represses hypocotyl cell elongation and increases cell wall stiffness.
Touch alters the pectin matrix of cell walls in etiolated seedlings
Recently, pectin degradation has been shown to play critical roles in regulating cell wall mechanics and cell growth (37, 38, 44). We investigated whether mechanical stimulation modulates the molecular mass of pectin polymer and the PG activity of dark-grown seedlings. We performed fast protein liquid chromatography (FPLC) to examine the molecular mass distribution of pectin isolated from etiolated seedlings by 1,2-cyclohexylenedinitrilo-tetraacetic acid (CDTA) extraction. The CDTA-extracted pectins from seedlings grown in the D condition exhibited a polymer peak at fraction #18 with a molecular mass of 200 kDa, while seedlings grown in the S condition showed a polymer peak at fraction #16 with higher molecular weight (Fig. 1G), suggesting that the lid touch treatment resulted in larger average molecular masses of pectins. Consistent with this finding, the total PG activity of etiolated seedlings was significantly reduced by mechanical stimulation (Fig. 1H). These results suggest that touch-induced hypocotyl cell elongation repressing and cell wall stiffening are associated with inhibition of PG-mediated pectin degradation.
Touch-induced transcriptional inhibition of PGX3 alters cell wall mechanics
To explore the molecular mechanism underlying touch-inhibited cell wall pectin degradation, we analyzed the gene expression levels of PGs in response to mechanical stimulation. Three PG genes, POLYGALACTURONASE INVOLVED IN EXPANSION1–3 (PGX1, PGX2, and PGX3), were previously reported to be functional PGs involved in seedling growth and development (37, 38, 44). RT-qPCR results revealed that PGX3 (AT1G48100) was notably repressed by touch in wild-type (WT) seedlings grown in the S condition, but PGX1 (AT3G26610) and PGX2 (AT1G78400) were unaffected (Fig. 2A). To dissect the regulation of PGX3 proteins in response to touch stimuli, we generated PGX3p-PGX3-Myc/pgx3 (PGX3pc/pgx3) transgenic plants, in which the PGX3-Myc fusion gene driven by the native PGX3 promoter was expressed in pgx3 mutants. Two independent transgenic lines, in which the transcription levels of PGX3 were restored to levels comparable to those of WT, were used for further analyses (fig. S3A). Immunoblot analysis of PGX3 proteins in PGX3pc/pgx3 seedlings showed that PGX3 proteins accumulated much higher levels in seedlings grown in the D condition than those grown in the S condition (fig. S3B). However, the protein levels of constitutively expressed PGX3-Myc were not altered in 35S-PGX3-Myc/WT transgenic plants grown in the S condition (fig. S3C), indicating that touch did not regulate the stability of PGX3 protein. These results indicate that PGX3 is a touch-repressed PG in etiolated seedlings.
Fig. 2. PGX3 is a touch-responsive pectin-degrading enzyme that mediates seedling thigmomorphogenetic changes.
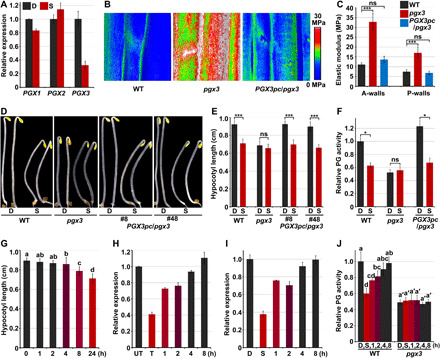
(A) RT-qPCR analysis of gene expression in response to mechanical resistance. Mean ± SD; n = 3. (B and C) Elastic modulus map (B) and quantification (C) of epidermal cell walls at the middle region of hypocotyls. The elastic modulus on the A-walls and P-walls was obtained by AFM in the QNM mode. Mean ± SD; n ≥ 10. ns, not significant. (D and E) Images (D) and hypocotyl length (E) of 3-day-old etiolated seedlings in response to mechanical resistance. Mean ± SD; n ≥ 20. (F) Mechanical forces inhibit PG activity via PGX3. Mean ± SD; n = 3. (G) Time-course analysis of changes in hypocotyl length in response to mechanical stimulation. Mean ± SD; n ≥ 20. (H) Reversible manner of PGX3 gene expression after 1 hour of touch treatment. Mean ± SD; n = 3. UT, untouched. (I and J) PGX3 gene expression (I) and PG activity (J) after removal of the touch stimulus. Seedlings were grown in the S condition (I) or in the low S condition (J) for 3 days, and then the lid was removed. Mean ± SD; n = 3. Lowercase letters above the bars in panels G and J represent significantly different groups, P < 0.05, one-way ANOVA and Tukey test.
Previous studies have shown that PGX3 is expressed in the hypocotyl, and pgx3-etiolated seedlings display a short hypocotyl phenotype (44). In this study, dark-grown pgx3 seedlings exhibited reduced hypocotyl cell length in comparison with WT seedlings, and complementation of PGX3 in the pgx3 background fully rescued the short hypocotyl cells of pgx3 seedlings (fig. S3, D and E). Moreover, mutation of PGX3 greatly increased cell wall stiffness, but stiffness was restored to a level comparable to that of WT in PGX3pc/pgx3 seedlings (Fig. 2, B and C). These results demonstrate that PGX3 plays a crucial role in regulating hypocotyl cell elongation and the cell wall mechanics of etiolated seedlings.
PGX3 is required for touch-induced morphological changes of etiolated seedlings
Our observation of transcriptional repression of PGX3 by touch and the similar effects of touch and PGX3 mutation on cell elongation and cell wall mechanics prompted us to investigate the involvement of PGX3 in touch-induced morphological changes. To avoid the possible variations on touch treatment caused by different hypocotyl lengths of WT and pgx3, seedlings were grown in the D condition for 2 days, followed by the lid touch treatment with a lower lid position (4 mm from the medium surface to the plate lid) for an additional day as low S condition. In the lid touch assay, we observed that the hypocotyl length of WT seedlings was significantly reduced, while pgx3 seedlings were insensitive to mechanical stimulation and displayed no significant hypocotyl growth alteration in the low S condition (Fig. 2, D and E). The shortened hypocotyls and reduced responsiveness to mechanical stimuli shown by pgx3 seedlings were fully restored in PGX3pc/pgx3 seedlings (Fig. 2, D and E). As PGX3 is a functional PG (44, 45), we examined whether PGX3 mediated touch-repressed PG activity. As mentioned above, WT etiolated seedlings displayed notably decreased PG activity when grown in the S condition (Figs. 1H and 2F). Compared with WT seedlings, pgx3 seedlings exhibited constitutively reduced PG activity in the D condition, which was not significantly different from that observed in the low S condition (Fig. 2F). Conversely, PGX3pc/pgx3 fully rescued the reduced PG activity of pgx3 seedlings and restored their ability to respond to mechanical stimulation (Fig. 2F). These results suggest that touch inhibits PG activity of seedlings through PGX3. The hypocotyl elongation of 35S-PGX3/pgx3 seedlings was found to be slightly repressed (fig. S4, A and B), but their PG activity was not altered when grown in the S condition (fig. S4C), suggesting that other factors independent of PG activity might be involved in touch-induced inhibition of hypocotyl growth. Collectively, we conclude that PGX3-mediated PG activity plays a primary role in regulating the thigmomorphogenetic responses of etiolated seedlings.
Touch represses PGX3 expression and PG activity in a rapid and reversible manner
To further dissect the characteristics of PGX3-mediated touch responses, we performed a short-term analysis. Seedlings were grown in the D condition for 3 days and then subjected to the S condition, in which they were exposed to the lid touch treatment, for the indicated periods. RT-qPCR results showed that PGX3 expression was effectively inhibited by the touch treatment and reached a steady level after 0.5 hours of treatment (fig. S5A). Consistent with changes in transcript levels, the PGX3 protein level in PGX3pc/pgx3 seedlings was rapidly decreased within 0.5 hours upon mechanical stimulation (fig. S5B). Given the rapid repression of PGX3 by the touch treatment, we determined whether the transient touch treatment caused morphogenetic changes. Two-day-old etiolated seedlings were exposed to mechanical stimulation by lid in the S condition for indicated periods of time (from 1 to 24 hours), after which they were grown in the D condition without touch for up to 3 days. In contrast with the shortened hypocotyl length induced by continuous 1-day-touch (24 hours) treatment in the S condition, 1 or 2 hours of touch treatment did not significantly affect hypocotyl length (Fig. 2G). However, after more than 4 hours of lid touch treatment, hypocotyl length started to decrease, and the reduction in hypocotyl length became more significant as the touch treatment was prolonged (Fig. 2G).
The discrepancy between the rapid responses of PGX3 mRNA/protein levels and the requirement for continuous touch stimulation to achieve hypocotyl growth inhibition implies that the repressive effect of touch on PGX3 might be reversible. To test this assumption, we exposed 2-day-old etiolated seedlings to short-term lid touch treatment for 1 hour and then removed the lid to let the seedlings continue to grow untouched. Touch-inhibited PGX3 expression was partially restored within 1 hour after the cessation of touch treatment and was fully restored to its original level in 4 hours (Fig. 2H). Moreover, seedlings were grown in the S condition with 1 day of touch treatment, after which the lid was removed to allow the seedlings to grow untouched for the indicated periods of time. PGX3 expression increased within 1 hour and reached a level comparable to that of untouched seedlings within 4 hours (Fig. 2I). Consistently, assessment of in vivo PG activity showed that touch-repressed PG activity in WT seedlings grown in the low S condition gradually increased after cessation of touch stimulation (Fig. 2J). In contrast, the PG activity of pgx3 mutant was not altered by either application or removal of the lid touch treatment (Fig. 2J). Thus, touch rapidly and reversibly repressed the action of PGX3 in etiolated seedlings.
Ethylene is associated with touch responses and enhances cell wall stiffness
Next, we explored the molecular link between mechanical stimulation of seedlings and PGX3-mediated morphological regulation. Mechanical impedance has been reported to increase phytohormone ethylene production in many plant species (8, 46, 47), while recent studies have demonstrated that ethylene signaling promotes seedling soil emergence (11, 13, 15, 18). By using the lid touch assay, we found that seedlings grown in the S condition released more ethylene gas than those grown in the D condition (Fig. 3A), confirming mechanical stimulation–induced ethylene production in etiolated seedlings. Since, in the S condition, 1-day-touch treatment was imposed onto the 2-day-old seedlings, we grew seedlings in the D condition for 2 days, after which they were exposed to ethylene gas for an additional day to assess the effects of ethylene in plant growth. Ethylene suppressed hypocotyl elongation of seedlings grown in the D condition in a dose-dependent manner, and treatment of these seedlings with 0.1–parts per million (ppm) ethylene generated a hypocotyl phenotype similar to that of seedlings grown in the S condition (Fig. 3B and fig. S6A). Moreover, ethylene markedly enhanced the hypocotyl cell wall stiffness of seedlings grown in the D condition (fig. S6, B and C). These results suggest that ethylene is implicated in the responses of seedlings to mechanical stimulation.
Fig. 3. Ethylene signaling is activated by touch and is required for touch-induced changes in morphogenesis mechanics.
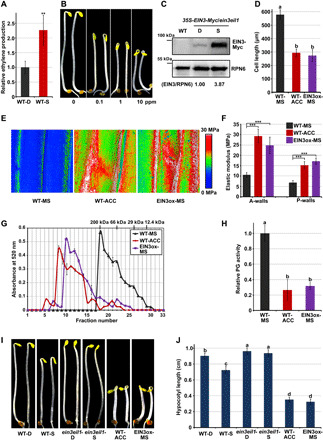
(A) Ethylene production in seedlings upon mechanical stimulation. Mean ± SE; n = 3. (B) Hypocotyl length of 2-day-old seedlings in response to the indicated (air: 0.1-, 1-, and 10-ppm ethylene) treatments for an additional day. (C) Immunoblot analysis of EIN3 protein levels. EIN3/RPN6 indicates the relative band intensities of EIN3-Myc normalized to RPN6 and is presented relative to that in the D condition set at unity. (D) Cell length of cells at the middle region of hypocotyls. Three-day-old seedlings were grown on 1/2 MS medium (MS) or supplemented with ACC (ACC). Mean ± SD; n ≥ 15. (E and F) Elastic modulus map (E) and quantification (F) of epidermal cell walls at the middle region of hypocotyls. The elastic modulus on the A-walls and P-walls was obtained by AFM in the QNM mode. Mean ± SD; n ≥ 10. (G) Molecular mass profile of CDTA-soluble pectin in seedlings. Mean ± SD was obtained from two technical replicates. (H) In vivo PG activity of seedlings. Mean ± SD; n = 3. (I and J) Images (I) and hypocotyl length (J) of seedlings. Mean ± SD; n ≥ 20. Lowercase letters above the bars in panels D, H, and J represent significantly different groups, P < 0.05, one-way ANOVA and Tukey test.
EIN3 influences the thigmomorphogenetic responses of seedlings to touch
Consistent with ethylene production, protein levels of EIN3, the master transcription factor in the ethylene signaling pathway, were notably elevated by mechanical resistance in seedlings grown in the S condition (Fig. 3C). Microscopic observation of hypocotyl cells using PI staining revealed that the cell length of WT seedlings was markedly reduced by treatment with ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC), and overexpression of EIN3 (EIN3ox) resulted in constitutively shortened hypocotyl cells (Fig. 3D and fig. S6D). AFM results showed that the elastic modulus was greatly increased by ACC treatment or EIN3 overexpression (Fig. 3, E and F). Moreover, the molecular mass profile of cell wall pectin matrix revealed that the average molecular mass of pectin from seedlings treated with ACC or overexpressing EIN3 was much larger than that of untreated WT seedlings (Fig. 3G), and seedlings grown in the D condition and subjected to ACC treatment or EIN3 overexpression displayed constitutively reduced total PG activity (Fig. 3H).
These thigmomorphogenetic-like alterations prompted us to investigate whether EIN3 is involved in the responses of etiolated seedlings to mechanostimulus. By examining the phenotypes of ethylene-related mutants, we found that repression of hypocotyl elongation by mechanical resistance in the S condition was mostly abolished by mutation of EIN3 and its homolog EIL1 (ein3eil1), while ACC application or EIN3 overexpression induced exaggerated hypocotyl inhibition (Fig. 3, I and J), indicating that EIN3 is required for touch-inhibited hypocotyl cell elongation.
EIN3 influences touch-repressed PGX3 expression
To dissect the regulatory effect of ethylene on PGX3, we performed a time-course experiment with PGX3p-LUC/WT transgenic seedlings to monitor dynamic changes of PGX3 gene expression in response to ethylene application. The bioluminescence images showed that the expression levels of PGX3 in two independent lines were visibly reduced within 15 min by ethylene treatment and were progressively decreased as the ethylene treatment was prolonged (Fig. 4A), implying that ethylene repressed PGX3 transcription. To explore tissue-specific regulation of PGX3, we generated transgenic plants in which the β-glucuronidase (GUS) reporter gene was driven by the PGX3 promoter. GUS staining indicated that PGX3 was expressed in the cotyledon and hypocotyl regions of etiolated seedlings (fig. S7A). Moreover, the expression level of PGX3p-GUS was notably increased in the hypocotyl by treatment with silver ions (Ag+), an inhibitor of ethylene signaling, but was mostly eliminated by ACC treatment (fig. S7A). Next, changes in PGX3 protein abundance in response to ethylene signaling activation and inhibition were assessed. Immunoblots showed that the level of PGX3 protein in etiolated PGX3pc/pgx3 seedlings was decreased by ACC but increased by Ag+ (Fig. 4B). However, the abundance of constitutively expressed PGX3 protein in 35S-PGX3-Myc/WT seedlings was not affected by Ag+ or ACC (fig. S7B). Collectively, these results demonstrate that ethylene reduces PGX3 protein abundance in etiolated seedlings through transcriptional repression.
Fig. 4. EIN3 directly binds to the promoter of PGX3 and represses PGX3 gene expression.
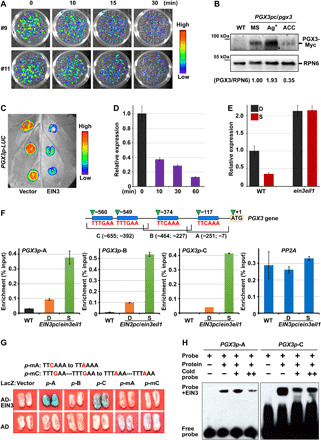
(A) Bioluminescent luciferase images showing the gene expression levels of PGX3 with 10-ppm ethylene treatment for the indicated periods. (B) Immunoblot analysis of PGX3 protein levels. PGX3/RPN6 indicates the relative band intensities of PGX3-Myc normalized to RPN6 and is presented relative to that grown on 1/2 MS medium set at unity. (C) Luciferase bioluminescence images of PGX3p-LUC transiently coexpressed with the 35S vector (Vector, left) or 35S-EIN3 (EIN3, right) in tobacco leaves. (D) RT-qPCR analysis of regulation of PGX3 gene expression by β-estradiol–induced EIN3 proteins. Mean ± SD; n = 3. (E) RT-qPCR analysis of PGX3 gene expression in response to mechanical resistance. Mean ± SD; n = 3. (F) ChIP-qPCR analysis of the interactions between EIN3 protein and PGX3 promoter. The top diagram shows the PGX3 promoter fragments, which contain putative EBSs (blue rectangles). Mean ± SD; n = 3. (G) Yeast one-hybrid assay showing that specific elements are required for physical binding of EIN3 to PGX3 promoter. The top diagram shows mutated PGX3 promoter fragments A and C with mutations located in the EBSs. (H) EMSA indicating physical binding between EIN3 and PGX3 promoter.
To investigate whether ethylene represses PGX3 via EIN3, we first examined the effects of EIN3 on PGX3 expression in a tobacco transient expression system, which revealed that coexpression with EIN3 strongly decreased PGX3-driven luciferase activity in comparison with the coexpression with the empty vector control (Fig. 4C and fig. S7C). Next, we used β-estradiol to induce EIN3 protein abundance in pER8-EIN3-Myc/ein3eil1ebf1ebf2 Arabidopsis seedlings to assess the inhibitory role of EIN3 on PGX3 expression. In pER8-EIN3-Myc/ein3eil1ebf1ebf2 seedlings, EIN3 protein visibly accumulated after 10 min of induction and promptly inhibited PGX3 expression (Fig. 4D and fig. S7D). As the EIN3 protein level was increased by prolonging the induction period, the expression level of PGX3 was further decreased (Fig. 4D and fig. S7D). RT-qPCR analysis showed that ethylene repressed PGX3 expression in a manner that was dependent on EIN3, with the inhibitory effect of ethylene on PGX3 expression being abolished in the ein3eil1 mutants (fig. S7E). These results indicate that EIN3 plays a primary role in repressing PGX3 transcription.
After showing that touch specifically and rapidly represses PGX3 transcription, we assessed the functional importance of EIN3 in touch-induced inhibition of PGX3 expression. The PGX3 expression levels of etiolated WT seedlings grown in the S condition were decreased (Fig. 4E). In contrast, PGX3 transcription was constitutively stimulated in ein3eil1 mutant seedlings and failed to respond to mechanical stimuli (Fig. 4E). Collectively, these results demonstrate that touch stimulation represses PGX3 transcription via the ethylene-EIN3 pathway.
EIN3 directly binds to EIN3-binding site motifs in the PGX3 promoter
Given the rapid and dose-dependent regulation of PGX3 expression by EIN3, we analyzed the promoter sequence of PGX3 and identified four known conserved EIN3-binding site (EBS) motifs (Fig. 4F). We then carried out chromatin immunoprecipitation (ChIP)–qPCR assays using EIN3p-EIN3-Myc/ein3eil1 (EIN3pc/ein3eil1)–etiolated seedlings to examine interactions between EIN3 proteins and PGX3 promoter fragments in vivo. We divided the PGX3 promoter region into three fragments (fragment A, B, and C), each of which contained at least one putative EBS motif (Fig. 4F). In comparison with WT seedlings, all three PGX3 promoter fragments, but not the coding region of control gene PP2A, were notably enriched in the EIN3pc/ein3eil1 seedlings (Fig. 4F). Furthermore, the enrichment of PGX3 promoter fragments by EIN3 was largely enhanced by mechanical stimulation of seedlings in the S condition (Fig. 4F). These results indicate that EIN3 specifically associates with the PGX3 promoter in etiolated seedlings.
To illustrate the physical interaction between EIN3 proteins and PGX3 promoter fragments in vitro, we performed yeast one-hybrid assays. We found that Activation domain (AD)–EIN3 fusion proteins, but not AD alone, bound to promoter fragments A and C, exhibiting strong LacZ reporter gene expression (Fig. 4G). Moreover, point mutations of critical C/G nucleotides in the EBS motifs fully abolished the LacZ activation of fragments A and C (Fig. 4G), revealing the essential role of the EBS motifs in the physical interaction between EIN3 and the PGX3 promoter. Electrophoretic mobility shift assay (EMSA) results further confirmed the direct interaction between EIN3 protein and fragments A and C of the PGX3 promoter (Fig. 4H). By combining our in vivo and in vitro data, we conclude that EIN3 specifically and directly binds to EBS motifs in the PGX3 promoter.
PGX3 acts downstream of EIN3 to regulate PG activity and cell elongation
Since biochemical and molecular evidence has shown that PGX3 is a primary target of EIN3, we next investigated the effects of ethylene on PG activity in relevant mutants. Two-day-old etiolated seedlings grown in the D condition were subjected to a time course of ethylene treatment. The total PG activity of WT seedlings was found to progressively decrease in response to prolonged ethylene application (Fig. 5A). Mutation of EIN3 and EIL1 (ein3eil1) resulted in elevated PG activity and fully abolished the capacity of seedlings to respond to ethylene (Fig. 5A), suggesting that EIN3 mediates ethylene-induced inhibition of PG activity. Intriguingly, the pgx3 mutant displayed constitutively decreased PG activity and failed to respond to ethylene treatment (Fig. 5A). The ethylene response defects of pgx3 seedlings were restored in PGX3pc/pgx3 seedlings (Fig. 5A), confirming that ethylene-induced inhibition of PG activity required PGX3. These results demonstrate that ethylene-induced inhibition of PG activity requires both EIN3 and PGX3.
Fig. 5. PGX3 functions genetically downstream of EIN3 to regulate the morphology and soil emergence of etiolated seedlings.
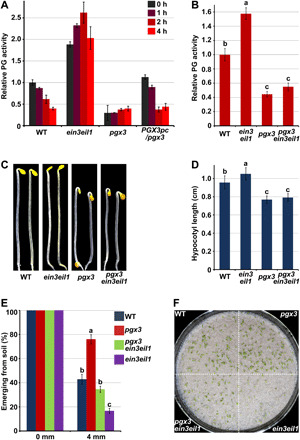
(A) Time-course analysis of the in vivo PG activity of etiolated seedlings in response to ethylene treatment. Three-day-old etiolated seedlings were treated with 10-ppm ethylene for the indicated periods. Relative PG activity was normalized to the absorbance of the WT–0 hour sample. Mean ± SD; n = 3. (B) In vivo relative PG activity of 3-day-old etiolated seedlings. The relative PG activity of the samples was normalized to the absorbance of samples from WT seedlings. Mean ± SD; n = 3. (C and D) Images (C) and hypocotyl length (D) of 3-day-old etiolated seedlings. Mean ± SD; n ≥ 20. (E and F) Soil emergence rate (E) and phenotype (F) of seedlings with soil cover. Seedlings were grown in the dark for 7 days on 1/2 MS medium without (0 mm) or with 4 mm sand cover, and the emergence rate was counted (E). Mean ± SE; n = 3. Soil emergence phenotypes were imaged following additional 2 days of light irradiation (F). Photo credit: Q. Wu, Peking University. Lowercase letters above the bars in panels B, D and E represent significantly different groups, P < 0.05, one-way ANOVA and Tukey test.
Next, we generated triple-mutant pgx3ein3eil1 seedlings by crossing the pgx3 and ein3eil1 backgrounds to dissect the genetic relationship between EIN3 and PGX3. We found that ein3eil1 seedlings exhibited increased PG activity in comparison with that of WT seedlings, whereas pgx3 seedlings displayed reduced PG activity (Fig. 5B). Mutation of PGX3 in the ein3eil1 background markedly reduced PG activity to a level comparable with that of pgx3 seedlings (Fig. 5B). Moreover, the hypocotyl cell length of pgx3ein3eil1 seedlings was notably shortened, resulting in a hypocotyl elongation phenotype similar to that of pgx3 seedlings but different from that of ein3eil1 seedlings (Fig. 5, C and D, and fig. S8). These findings suggest that PGX3 functions genetically downstream of EIN3 to regulate hypocotyl cell elongation and PG activity.
The EIN3-PGX3 regulatory module facilitates seedling soil emergence
To verify the regulatory role of the EIN3-PGX3 module in seedling emergence from the soil, we examined the soil penetration capacity of etiolated seedlings. Seeds were plated on agar medium and evenly covered with a layer of soil. The emergence rate was counted after 7 days of growth in the dark, and the emergence phenotype was photographed when the emerged seedlings were exposed to dim light for an additional 2 days. All the mutant seedlings grew upward in a manner similar to WT seedlings when there is no soil covering (Fig. 5E and fig. S9A). With a soil covering depth of 4 mm, approximately 40% of WT seedlings emerged from the soil (Fig. 5, E and F, and fig. S9A). In comparison with WT seedlings, pgx3 mutant seedlings exhibited enhanced seedling emergence, with an emergence rate of approximately 80% (Fig. 5, E and F, and fig. S9A), and the emergence rate of PGX3pc/pgx3 was comparable to that of WT seedlings (fig. S9A). EIN3 was previously reported to promote seedling soil emergence (15). Consistently, more than 80% of ein3eil1 mutant seedlings failed to emerge from the soil (Fig. 5, E and F). Mutating PGX3 in ein3eil1 seedlings largely rescued these severe defects and restored the emergence frequency of ein3eil1 to a level comparable to that of WT seedlings (Fig. 5, E and F). Therefore, inhibition of PGX3 via EIN3 played a crucial role in the emergence of seedlings from the soil.
DISCUSSION
Mechanical signals are important factors that shape the architecture of plants and animals (3, 12, 43, 48–50). However, the molecular and cellular mechanisms of thigmomorphogenesis remain poorly understood. In this study, we demonstrate that the physical resistance of a barrier induces thigmomorphogenetic characteristics in dark-grown seedlings, and this process requires suppression of the key cell wall remodeling enzyme PGX3. In addition, we show that PGX3 expression responds sensitively to touch. PGX3 expression is rapidly down-regulated in response to touch and is restored when the stimulus is removed. We further find that ethylene production is stimulated to suppress pectin degradation and strengthen cell walls in response to touch. Ethylene represses PGX3 transcription and PG activity promptly and in a dose-dependent manner through the transcription factor EIN3. EIN3 protein directly binds to the PGX3 promoter to mediate touch-induced inhibition of PGX3 expression and facilitate emergence from the soil. Our results reveal that buried seedlings transduce the impedance of the soil into activation of ethylene signaling and further use an EIN3-PGX3 regulatory module to optimize cell wall stiffness and cell growth, providing a signaling transduction pathway for plant seedling thigmomorphogenesis (fig. S9B).
Pectins have been shown to largely determine plant cell wall stiffness, in which pectin methylesterification is a key control point (27, 29). Our study reveals that touch represses pectin degradation through PGX3 and increases the elastic modulus of hypocotyl cell walls, illustrating the regulatory role of PG in cell wall mechanics. Pectin degradation might disrupt the polysaccharide matrix and increase cellulose microfibril sliding, leading to cell wall loosening (29, 36). In addition, with reduced PG activity, demethylesterified pectin tends to undergo Ca2+ cross-linking, which is associated with cell wall stiffening (34, 44, 51). In the Arabidopsis hypocotyl, asymmetric pectin demethylesterification selectively loosens longitudinal cell walls to initiate symmetry breaking, following the cortical microtubule–mediated orientation of cellulose microfibrils to restrict increases in girth and allow longitudinal cell growth (28, 40). In this way, touch evenly stiffens hypocotyl cell walls but produces inhibition of longitudinal growth. This regulation mode enables rapid adjustment of cell growth rate in response to external mechanical cues.
In contrast with the touch-induced rapid leaf movement of Mimosa pudica and the Venus flytrap, only repetitive touches over a long period of time are able to produce visible morphogenetic changes in most plants (3). However, touch was shown to alter gene expression within 0.5 hours in Arabidopsis (1). In this study, we found that touch regulates cell wall remodeling in a transient and reversible manner. Therefore, although plants respond to touch sensitively, the effects of touch, including changes in gene expression and enzyme activity, are rapidly reversed when the touch stimulus is removed. Constant or repeated touches can maintain or revive the effects of touch on plants, resulting in gradual changes in cell wall mechanics over time and eventually leading to thigmomorphogenetic responses. The mechanism revealed by our study allows dynamic adjustments by plant cells in response to mechanical information, accomplishing plasticity of morphology to acclimate to ever-changing environments. Intriguingly, mechanical stimulus treatments, such as purposefully stepping on wheat and barley seedlings or releasing ducklings onto rice fields, have been used by farmers to strengthen plant stems for centuries (52). Tissue- and developmental stage–specific regulation of PGX3 orthologs may be used in the future to improve crop yield and stress resistance.
As critical endogenous regulators, phytohormones play pivotal roles in regulating plant morphogenesis in response to environmental changes. In adult plants, touch increases accumulation of jasmonate, which is required for touch-induced delayed flowering, reduced rosette radial growth, and enhanced biotic resistance (9). On the other hand, touch reduces plant height and delays flowering by down-regulating bioactive gibberellin levels (10). Whether ethylene is involved in regulating plant touch responses has long been debated. Ethylene was previously suspected to be involved in regulating thigmomorphogenesis (46, 53). However, in studies of ethylene-insensitive mutants etr1-3 and ein2-1, the response of the mutant plants to wind was found to be similar to that of WT plants (54). In this study, we demonstrate that ethylene is required for and promotes thigmomorphogenesis in etiolated seedlings. It is likely that plants adopt flexible mechanisms to optimize their growth and development in diverse conditions during different developmental stages. Our data also show that external signals, such as mechanical forces, use endogenous hormones to alter cell wall mechanics. Because the physical properties of cell walls dominate cellular behaviors and biological shapes, this finding is critical for deciphering the general principles underlying the integration of external cues and cell activities in regulating plant morphogenesis.
MATERIALS AND METHODS
Plant materials and growth conditions
The WT Arabidopsis thaliana ecotype used in this study was Columbia-0 (Col-0). Mutants and transgenic plants, including EIN3ox, ein3eil1, pER8-EIN3-Myc/ein3eil1ebf1ebf2, EIN3p-EIN3-Myc/ein3eil1, and 35S-EIN3-Myc/ein3eil1, have been described previously (13, 55–57). The transferred DNA insertion mutant pgx3 (Salk_010192) was obtained from the Arabidopsis Biological Resource Center (44, 58). Multiple mutants were generated by crossing, and homozygous lines were genotyped. The 752-, 907-, and 2000-bp promoter fragments upstream from the predicted translational initiation sites of TCH3 (AT2G41100), TCH4 (AT5G57560), and PGX3 (AT1G48100) were cloned into pGW435 (59) to generate the TCH3p-LUC, TCH4p-LUC, and PGX3p-LUC constructs, respectively (59). The PGX3 promoter fragments were cloned into pGW433 (59) to generate the PGX3p-GUS construct. The full-length coding sequence (CDS) of PGX3 amplified from Arabidopsis complementary DNA (cDNA) by PCR was cloned into pGWB617 to generate the 35S-PGX3-Myc construct (59). The PGX3 promoter fused upstream of the PGX3 CDS was cloned into pGWB616 to generate the PGX3pc-Myc construct (59). The constructs were transformed into WT or pgx3 mutants as indicated, and transgenic plants were selected by phosphinothricin (20 μg/ml) or kanamycin (50 μg/ml; Sigma-Aldrich). #8 and #48 are two independent PGX3pc/pgx3 transgenic lines, and #8 was used in the study unless specified otherwise.
Seeds were sterilized with 75% (v/v) ethanol containing 0.1% (v/v) Triton X-100 for 10 min, after which they were washed four times with sterile water. Surface-sterilized seeds were sown on half-strength (1/2) MS medium [MS salts (2.2 g/liter), sucrose (5 g/liter), and agar (8 g/liter; pH 5.7)]. For chemical treatment, 1/2 MS medium was supplemented with 10 μM ACC (Sigma-Aldrich) or 100 μM AgNO3. For β-estradiol induction, seedlings were immerged in 100 μM β-estradiol (Sigma-Aldrich) solution for the indicated periods.
D and S conditions
For the deep and shallow plates, the distances from the medium surface to the plate lid were 12 and 6 mm, respectively. Surface-sterilized seeds were plated on 1/2 MS agar medium, and the plates were incubated in the dark at 4°C for 3 days. After stratification, the plates were illuminated with white light for 6 hours and incubated in the dark for 3 days. In the S condition, growing seedlings encountered the physical barrier of the lid and were studied as touch-treated samples, while the seedlings grown in the D condition did not reach the lid and were used as untouched controls.
Ethylene treatment assay
Surface-sterilized seeds were plated on 1/2 MS agar medium in a deep plate and incubated in the dark at 4°C for 3 days. Next, the plates were illuminated with white light for 6 hours and cultivated in the dark for 2 days. Two-day-old dark grown seedlings were placed into a beaker sealed with plastic wrap. The indicated concentrations of ethylene (Beijing Hua Yuan Gas Chemical Industry Co. Ltd) were injected into the beaker. The ethylene concentration was 10 ppm unless specified otherwise.
Soil emergence assay
Soil emergence assays were performed as previously described with minor modifications (15). Briefly, 200 seeds were surface-sterilized and plated on 1/2 MS medium with an area of 50 cm2. Next, 20 ml of 50- to 70-mesh particle size sand (Sigma-Aldrich) was spread evenly onto the medium, covering the seeds to a depth of 4 mm. After stratification in the dark for 3 days at 4°C, the plates were illuminated under white light for 6 hours. Next, the plates were incubated in darkness for the indicated time periods to determine the percentage of dark-grown seedlings that emerged from the soil. To allow phenotypes to be recorded, the plates were transferred into dim white light for an additional 2 days. Three independent biological replicates were used for statistical analysis.
Ethylene measurement
Eight hundred seeds were plated on four plates of the D or S condition, respectively, and sealed in a 1000-ml glass beaker with plastic wrap. Three replicates were placed in three individual beakers for each sample. The seedlings were incubated in darkness for 3 days. A 100-μl headspace gas sample was withdrawn from each beaker using a gas-tight syringe and injected into a gas chromatograph. A gas chromatograph (6890N Network GC System; Agilent Technologies) equipped with a flame ionization detector with a HP-PLOTQ column (40 m by 530 μm by 40 μm) was used to measure the amount of ethylene produced by the seedlings. Using N2 as the carrier gas, separations were carried out at 60°C. The ethylene peak area was integrated with Agilent ChemStation, and results were calculated as the average relative ethylene production (%) for seedlings subjected to the D or S treatment. Two independent experiments were performed with comparable results, and representative results are shown.
PI and histochemical GUS staining
For PI staining, 3-day-old dark-grown seedlings were submerged in PI (10 μg/ml) solution (Sigma-Aldrich; dissolved in 1× PBS) for 10 min and washed three times with sterile water. Fluorescence was detected at an excitation wavelength of 543 nm using an LSM 710 Meta confocal laser scanning microscope (Zeiss).
For GUS staining, 3-day-old etiolated seedlings were submerged in 0.1 M sodium phosphate buffer (pH 7.0) containing 1 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 1 mM EDTA, 1% Triton X-100, and X-gluc (1 mg/ml), after which they were incubated in the dark at 37°C. Stained tissues were observed using a Leica Microsystems DFC295 microscope.
Hypocotyl length and cell length measurement
For hypocotyl length measurement, at least 20 seedlings per sample were measured using ImageJ software (https://imagej.nih.gov/ij/). The error bars represent the average value ± SD from n ≥ 20 seedlings. For cell length measurement, at least 15 cells from 10 seedlings per sample were measured using ImageJ software. The error bars represent the average value ± SD from n ≥ 15 cells per sample.
AFM measurements
The seedling hypocotyls treated with 0.55 M mannitol for 0.5 hours were immobilized on a glass slide using nail polish, and the middle parts of the hypocotyls were measured under 0.55 M mannitol at room temperature (40). The samples were scanned using a BioScope Resolve atomic force microscope (Bruker, Billerica, MA, USA) with a “PeakForce QNM in fluid” operation mode (39, 41–43). The “PeakForce QNM in fluid” operation mode records the force-distance curve at every pixel and thereby maps the distribution of nanomechanical properties: height, elastic modulus, adhesion, and deformation. Images were collected over a 40 μm by 40 μm image size with a resolution of 256 × 256 pixels. A 0.2-Hz scanning rate was used.
The probe was a standard pyramidal silicon nitride ScanAsyst-Fluid (Bruker) probe, containing a triangular silicon nitride cantilever with a thickness of 0.59 to 0.61 μm. The spring constant of the probe was estimated to be 0.7 N/m (frequency = 150 kHz), and the silicon point probe tip had a radius of 20 nm. The deflection sensitivity was calibrated on a hard glass sample. The spring constant was calibrated for a ramp mode by thermal tuning before each measurement. Cell wall elasticity was measured using an indentation depth of 150 to 250 nm and an indentation force of 30 nN.
Quantification of elasticity (elastic modulus) was obtained by AFM using the Quantitative Nanoscale Mechanical (QNM) mode and calculated on the basis of the Derjaguin-Muller-Toporov modulus. Data were analyzed with NanoScope Analysis version 1.8. The elastic modulus of at least 10 hypocotyl cells from four seedlings per sample was measured.
RNA extraction and RT-qPCR analysis
Seedlings were harvested in liquid nitrogen under dim green light. Total RNA was extracted using the Spectrum Plant Total RNA Kit (Sigma-Aldrich) along with On-Column DNase I Digestion treatment. Gel electrophoretic and spectrophotometric analyses (NanoDrop 2000c) were performed to assess RNA quality and determine RNA concentrations. Total RNA (1 μg) was used to synthesize first-strand cDNA using the ReverTra Ace qPCR RT Kit (TOYOBO). RT-qPCR was performed using cDNA as the template with SYBR Green Mix (TaKaRa) on the ABI Fast 7500 Real-Time System. All gene expression results were normalized with respect to the values obtained for housekeeping genes PP2A (AT1g13320) and SAND (AT2G28390). All RT-qPCR experiments were independently performed at least three times using different pools of seedlings under the indicated conditions, and representative results are presented.
Immunoblot assay
Seedlings were harvested in liquid nitrogen and ground to powder under dim green light. Total proteins were extracted in buffer [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× one Roche cOmplete EDTA-free Protease Inhibitor Cocktail]. The proteins in SDS-loading buffer were separated on 8 or 10% SDS–polyacrylamide gel electrophoresis gels. Anti-Myc (Sigma-Aldrich; M4439, 1:5000 dilution) and anti-RPN6 (Abmart; X-Q9LP45-N, 1:1000 dilution) antibodies were used for the immunoblotting analysis. The intensity of immunoblot bands was quantified using ImageJ software (https://imagej.nih.gov/ij/).
ChIP assay
For the ChIP assay, EIN3p-EIN3-Myc/ein3eil1 seedlings were grown in the S or D condition for 3 days. Approximately 2 g of whole seedlings was harvested and cross-linked in 1% formaldehyde solution for 30 min. The ChIP procedure was performed in a dark room with dim green light as previously described (60). Anti-Myc Affinity Gel (Sigma-Aldrich) was used to perform the immunoprecipitation. After the ChIP procedure, qPCR was performed to examine the enrichment level of selected DNA fragments. The enrichment of each target site was indicated by the percentage of coimmunoprecipitated DNA relative to the corresponding input DNA. WT seedlings were subjected to the same experimental procedures as a negative control group. A fragment of the PP2A coding region was used as an internal control.
Yeast one-hybrid assay
Yeast one-hybrid assays were performed as previously described (55). Briefly, LacZ reporter genes driven by the PGX3 WT or mutant promoter fragments were cotransformed with the pB42AD empty vector or pB42AD-EIN3 construct into yeast strain EGY48. Yeast transformation was conducted according to the instructions provided in the Yeast Protocols Handbook (Clontech). Transformants were grown on SD-Ura-Trp dropout plates containing X–β-gal overnight for blue color development.
Transient expression assays in Nicotiana benthamiana
Agrobacterium bacteria carrying PGX3p-LUC and the 35S-EIN3 or 35S-Vector were prepared in infiltration buffer [10 mM MES (pH 5.6), 10 mM MgCl2, and 0.2 mM acetosyringone] at an optical density of 0.4. PGX3p-LUC was infiltrated into tobacco leaves with the 35S-EIN3 or 35S-Vector at a ratio of 1:1. After 2 days, 1 mM d-luciferin potassium salt solution (Biotium) was infiltrated, and luciferase bioluminescence images were obtained using a Berthold Technologies Multimode Reader LB942. For each combination, at least five infiltration spots were assessed. The bioluminescence intensity was quantified by Indigo software.
Electrophoretic mobility shift assay
EMSAs were performed using biotin-labeled probes and the LightShift Chemiluminescent EMSA Kit as described previously (55, 60). Biotin-labeled probes (10 nM) were incubated with EIN3N-His protein in a 20-μl reaction mixture [10 mM tris-HCl, 150 mM KCl, poly–deoxyinosine-deoxycytidine (50 ng/ml), 1 mM dithiothreitol (DTT), 0.05% NP-40, 2.5% glycerol, 100 μM ZnCl2, and bovine serum albumin (0.5 μg/ml)] for 20 min at 30°C. For the cold competitor, unlabeled probes [0.5 μM (+) or 2 μM (++)] were added to the reaction mixture. After incubation, all mixtures were separated on 6% native polyacrylamide gels.
PG activity assay
For the PG activity assay, plant proteins were extracted according to previously reported methods with minor modifications (37, 44). Briefly, 2 g of 3-day-old dark-grown seedlings was harvested under dim green light and ground into powder in liquid nitrogen. The sample powder was homogenized in 5 ml of precooled extraction buffer [50 mM tris-HCl (pH 7.5), 2.5 mM DTT, 3 mM EDTA, 2 mM PMSF, 1 M NaCl, and 10% (v/v) glycerol] and centrifuged at 14,000g for 40 min at 4°C. The supernatant was dialyzed against 50 mM Na-acetate buffer (pH 5.0) using 8000- to 14,000–molecular weight cutoff D membrane MD55 (Solarbio). To measure the PG activity, the protein samples were added to the substrate solution [50 mM Na-acetate (pH 5.0) and 0.2% (w/v) polygalacturonic acid (Sigma-Aldrich)] and incubated at 30°C for 3 hours. Next, 200 μl of released reducing-end group solution was added to 1 ml of 100 mM borate (pH 9.0) and 200 μl of 1% (w/v) 2-cyanoacetamide (Sigma-Aldrich), and the resulting mixture was boiled for 10 min. The samples were cooled to room temperature, after which the absorbance was measured at 276 nm (NanoDrop 2000c). The experiment was repeated at least two times using independent sample pools, and in each experiment, three equal-sized aliquots from one sample were used for substrate reaction and absorbance measurement as technical replicates.
Preparation of alcohol-insoluble residue extracts and alcohol-insoluble residue fractionation
Alcohol-insoluble residue (AIR) extraction and fractionation were performed according to previously reported methods with minor modifications (37, 44). Briefly, 2 g of 3-day-old dark-grown seedlings harvested under dim green light was ground into a powder in liquid nitrogen. The powder was suspended in 30-ml chloroform/methanol (1:1, v/v) and rotated at room temperature for 1 hour. The cell wall residue was collected by centrifugation, resuspended in 100% acetone, and dried under a vacuum at room temperature for 2 hours. Starch was removed by treatment with 30 ml of 90% dimethyl sulfoxide (DMSO; Sigma-Aldrich) overnight with rotation. The residue was rinsed with 30 ml of 90% DMSO and washed six times with 70% ethanol. The final residue was dried with 100% acetone. AIR was sequentially extracted with 50 mM sodium acetate buffer (pH 6.0) containing 50 mM CDTA (Sigma-Aldrich) by rotation at room temperature for 24 hours. The CDTA-insoluble residues were discarded by centrifugation at 3800g for 20 min. Last, the CDTA fraction was precipitated from the soluble supernatant in 70% ethanol and lyophilized under a vacuum.
FPLC and uronic acid assays
A sample of the CDTA-soluble lyophilized powder (5 mg) was dissolved in 1 ml of 0.1 M sodium acetate buffer on a vortex mixer and filtered through a 0.22-μm filter. Fractionation was performed using a Superdex 75 10/300 GL (GE Healthcare) gel filtration column. Gel media were equilibrated with 0.1 M sodium acetate buffer. Each sample (500 μl) was loaded into a 24-ml column and eluted at a flow rate of 0.5 ml/min. Forty fractions (500-μl elution per fraction) were collected and analyzed for uronic acid (UA) content. A size standard kit (Sigma-Aldrich, MWGF200) was used to determine molecular mass.
UA content was determined by the m-hydroxydiphenyl (Sigma-Aldrich) method as previously described with minor modifications (37). Briefly, 250 μl of each fraction was mixed with 1 ml of 0.0125-M sodium tetraborate (Sigma-Aldrich) sulfuric acid solution, boiled for 5 min, and cooled to room temperature. The absorbance of the sample at 520 nm was measured as the background absorbance. Next, each sample was mixed with 20 μl of 0.15% (w/v) m-hydroxydiphenyl (Sigma-Aldrich), dissolved in 0.5% (w/v) NaOH, and incubated at room temperature for 10 min. The absorbance of each sample was measured at 520 nm and recorded as the UA absorbance. The experiment was repeated at least two times using independent sample pools, and in each experiment, two equal-sized aliquots of each fraction were used for UA reaction and absorbance measurement as technical replicates.
Statistical analysis
Microsoft Excel was used to perform Student’s t tests for two independent groups, with significant differences indicated as *P < 0.05, **P < 0.01, and ***P < 0.001. R 3.5.1 was used to perform one-way ANOVA and Tukey’s honest significant difference post hoc test with a confidence level of 0.95 for more than two groups; for significantly different groups from one-way ANOVA and Tukey test, P < 0.05. The sample sizes for each experiment are given in the figure legends.
Supplementary Material
Acknowledgments
We thank the National Center for Protein Sciences at Peking University in Beijing, China, particularly C. Li and S. Qin for technical help with AFM, and G. Li for professional technical assistance with FPLC. We thank K. Kuang (Capital Normal University) and Y. Zhao (Peking University) for experimental assistance. Funding: This work was supported by grants from the National Key R&D Program of China (2016YFA0502900) and the National Science Foundation of China (31822004, 31621001). Q.W. was supported by a China Postdoctoral Science Foundation grant (2019 M650325). H.S. was supported by the Support Project of High-level Teachers in Beijing Municipal Universities in the Period of 13th Five-year Plan (CIT&TCD20190331). Author contributions: S.Z. and H.S. designed the research. Q.W., Y.Li, M.L., and Y.Luo performed the experiments. S.Z., H.S., and Q.W. analyzed the data. S.Z., H.S., and Q.W. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/48/eabc9294/DC1
REFERENCES AND NOTES
- 1.Braam J., Davis R. W., Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 60, 357–364 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Jaffe M. J., Forbes S., Thigmomorphogenesis: The effect of mechanical perturbation on plants. Plant Growth Regul. 12, 313–324 (1993). [DOI] [PubMed] [Google Scholar]
- 3.Chehab E. W., Eich E., Braam J., Thigmomorphogenesis: A complex plant response to mechano-stimulation. J. Exp. Bot. 60, 43–56 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Monshausen G. B., Gilroy S., Feeling green: Mechanosensing in plants. Trends Cell Biol. 19, 228–235 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa Y., Katagiri T., Shinozaki K., Qi Z., Tatsumi H., Furuichi T., Kishigami A., Sokabe M., Kojima I., Sato S., Kato T., Tabata S., Iida K., Terashima A., Nakano M., Ikeda M., Yamanaka T., Iida H., Arabidopsis plasma membrane protein crucial for Ca2+ influx and touch sensing in roots. Proc. Natl. Acad. Sci. U.S.A. 104, 3639–3644 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monshausen G. B., Bibikova T. N., Weisenseel M. H., Gilroy S., Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu D., Haswell E. S., Plant mechanosensitive ion channels: An ocean of possibilities. Curr. Opin. Plant Biol. 40, 43–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goeschl J. D., Rappaport L., Pratt H. K., Ethylene as a factor regulating the growth of pea epicotyls subjected to physical stress. Plant Physiol. 41, 877–884 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chehab E. W., Yao C., Henderson Z., Kim S., Braam J., Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Curr. Biol. 22, 701–706 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Lange M. J. P., Lange T., Touch-induced changes in Arabidopsis morphology dependent on gibberellin breakdown. Nat. Plants 1, 14025 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Shen X., Li Y., Pan Y., Zhong S., Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front. Plant Sci. 7, 1571 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K., Yang Z., Qing D., Ren F., Liu S., Zheng Q., Liu J., Zhang W., Dai C., Wu M., Chehab E. W., Braam J., Li N., Quantitative and functional posttranslational modification proteomics reveals that TREPH1 plays a role in plant touch-delayed bolting. Proc. Natl. Acad. Sci. U.S.A. 115, E10265–E10274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi H., Liu R., Xue C., Shen X., Wei N., Deng X. W., Zhong S., Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and Ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 26, 139–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H., Lyu M., Luo Y., Liu S., Li Y., He H., Wei N., Deng X. W., Zhong S., Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. U.S.A. 115, 6482–6487 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong S., Shi H., Xue C., Wei N., Guo H., Deng X. W., Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc. Natl. Acad. Sci. U.S.A. 111, 3913–3920 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bleecker A. B., Estelle M. A., Somerville C., Kende H., Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science 241, 1086–1089 (1988). [DOI] [PubMed] [Google Scholar]
- 17.Ecker J. R., The ethylene signal transduction pathway in plants. Science 268, 667–675 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Harpham N. V. J., Berry A. W., Knee E. M., Roveda-Hoyos G., Raskin I., Sanders I. O., Smith A. R., Wood C. K., Hall M. A., The effect of ethylene on the growth and development of wild-type and mutant Arabidopsis thaliana (L.) Heynh. Ann. Bot. 68, 55–61 (1991). [Google Scholar]
- 19.Schaller G. E., Bleecker A. B., Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science 270, 1809–1811 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R., CTR1, a negative regulator of the ethylene response pathway in arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441 (1993). [DOI] [PubMed] [Google Scholar]
- 21.Chang C., Kwok S. F., Bleecker A. B., Meyerowitz E. M., Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262, 539–544 (1993). [DOI] [PubMed] [Google Scholar]
- 22.Alonso J. M., Hirayama T., Roman G., Nourizadeh S., Ecker J. R., EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Ji Y., Guo H., From endoplasmic reticulum (ER) to nucleus: EIN2 bridges the gap in ethylene signaling. Mol. Plant 6, 11–14 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Chao Q. M., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J. R., Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89, 1133–1144 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Chen Y.-F., Etheridge N., Schaller G. E., Ethylene signal transduction. Ann. Bot. 95, 901–915 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang K. N., Zhong S., Weirauch M. T., Hon G., Pelizzola M., Li H., Huang S.-s., Schmitz R. J., Urich M. A., Kuo D., Nery J. R., Qiao H., Yang A., Jamali A., Chen H., Ideker T., Ren B., Bar-Joseph Z., Hughes T. R., Ecker J. R., Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife 2, e00675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolf S., Hématy K., Höfte H., Growth control and cell wall signaling in plants. Annu. Rev. Plant Biol. 63, 381–407 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Bidhendi A. J., Geitmann A., Relating the mechanics of the primary plant cell wall to morphogenesis. J. Exp. Bot. 67, 449–461 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Chebli Y., Geitmann A., Cellular growth in plants requires regulation of cell wall biochemistry. Curr. Opin. Cell Biol. 44, 28–35 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Charrier B., Rabillé H., Billoud B., Gazing at cell wall expansion under a golden light. Trends Plant Sci. 24, 130–141 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Peaucelle A., Braybrook S. A., Le Guillou L., Bron E., Kuhlemeier C., Höfte H., Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21, 1720–1726 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Szymanski D. B., Cosgrove D. J., Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 19, R800–R811 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H., Toward a systems approach to understanding plant cell walls. Science 306, 2206–2211 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Wolf S., Greiner S., Growth control by cell wall pectins. Protoplasma 249 ( Suppl. 2), S169–S175 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Kim J., Shiu S.-H., Thoma S., Li W.-H., Patterson S. E., Patterns of expansion and expression divergence in the plant polygalacturonase gene family. Genome Biol. 7, R87 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babu Y., Bayer M., Plant polygalacturonases involved in cell elongation and separation—The same but different? Plants (Basel) 3, 613–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao C., Somerville C., Anderson C. T., POLYGALACTURONASE INVOLVED IN EXPANSION1 functions in cell elongation and flower development in Arabidopsis. Plant Cell 26, 1018–1035 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao C., Barnes W. J., Zamil M. S., Yi H., Puri V. M., Anderson C. T., Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf expansion, stem lignification, mechanical stiffening, and lodging. Plant J. 89, 1159–1173 (2017). [DOI] [PubMed] [Google Scholar]
- 39.Braybrook S. A., Measuring the elasticity of plant cells with atomic force microscopy. Methods Cell Biol. 125, 237–254 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Peaucelle A., Wightman R., Höfte H., The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr. Biol. 25, 1746–1752 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Milani P., Gholamirad M., Traas J., Arnéodo A., Boudaoud A., Argoul F., Hamant O., In vivo analysis of local wall stiffness at the shoot apical meristem in Arabidopsis using atomic force microscopy. Plant J. 67, 1116–1123 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Sampathkumar A., Krupinski P., Wightman R., Milani P., Berquand A., Boudaoud A., Hamant O., Jönsson H., Meyerowitz E. M., Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife 3, e01967 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qi J., Wu B., Feng S., Lü S., Guan C., Zhang X., Qiu D., Hu Y., Zhou Y., Li C., Long M., Jiao Y., Mechanical regulation of organ asymmetry in leaves. Nat. Plants 3, 724–733 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Rui Y., Xiao C., Yi H., Kandemir B., Wang J. Z., Puri V. M., Anderson C. T., POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 29, 2413–2432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogawa M., Kay P., Wilson S., Swain S. M., ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 21, 216–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi H., Jaffe M. J., Thigmomorphogenesis: The relationship of mechanical perturbation to elicitor-like activity and ethylene production. Physiol. Plant. 61, 405–411 (1984). [DOI] [PubMed] [Google Scholar]
- 47.Sarquis J. I., Jordan W. R., Morgan P. W., Ethylene evolution from maize (Zea mays L.) seedling roots and shoots in response to mechanical impedance. Plant Physiol. 96, 1171–1177 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sampathkumar A., Yan A., Krupinski P., Meyerowitz E. M., Physical forces regulate plant development and morphogenesis. Curr. Biol. 24, R475–R483 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lecuit T., Lenne P.-F., Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Monshausen G. B., Haswell E. S., A force of nature: Molecular mechanisms of mechanoperception in plants. J. Exp. Bot. 64, 4663–4680 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hocq L., Pelloux J., Lefebvre V., Connecting homogalacturonan-type pectin remodeling to acid growth. Trends Plant Sci. 22, 20–29 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Braam J., Chehab E. W., Thigmomorphogenesis. Curr. Biol. 27, R863–R864 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Telewski F. W., Jaffe M. J., Thigmomorphogenesis: The role of ethylene in the response of Pinus taeda and Abies fraseri to mechanical perturbation. Physiol. Plant. 66, 227–233 (1986). [DOI] [PubMed] [Google Scholar]
- 54.Johnson K. A., Sistrunk M. L., Polisensky D. H., Braam J., Arabidopsis thaliana responses to mechanical stimulation do not require ETR1 or EIN2. Plant Physiol. 116, 643–649 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong S., Shi H., Xue C., Wang L., Xi Y., Li J., Quail P. H., Deng X. W., Guo H., A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr. Biol. 22, 1530–1535 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lyu M., Shi H., Li Y., Kuang K., Yang Z., Li J., Chen D., Li Y., Kou X., Zhong S., Oligomerization and photo-deoligomerization of HOOKLESS1 controls plant differential cell growth. Dev. Cell 51, 78–88.e3 (2019). [DOI] [PubMed] [Google Scholar]
- 57.Shi H., Shen X., Liu R., Xue C., Wei N., Deng X. W., Zhong S., The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev. Cell 39, 597–610 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., Shinn P., Stevenson D. K., Zimmerman J., Barajas P., Cheuk R., Gadrinab C., Heller C., Jeske A., Koesema E., Meyers C. C., Parker H., Prednis L., Ansari Y., Choy N., Deen H., Geralt M., Hazari N., Hom E., Karnes M., Mulholland C., Ndubaku R., Schmidt I., Guzman P., Aguilar-Henonin L., Schmid M., Weigel D., Carter D. E., Marchand T., Risseeuw E., Brogden D., Zeko A., Crosby W. L., Berry C. C., Ecker J. R., Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T., Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104, 34–41 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Shi H., Zhong S., Mo X., Liu N., Nezames C. D., Deng X. W., HFR1 sequesters PIF1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25, 3770–3784 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/48/eabc9294/DC1


