SUMMARY
Cell division axes during development are specified in different orientations to establish multicellular assemblies, but the mechanisms that generate division axis diversity remain unclear. We show here that patterns of cell contact provide cues that diversify cell division orientation by modulating cortical non-muscle myosin flow. We reconstituted in vivo contact patterns using beads or isolated cells to show two findings. First, we identified three contact-dependent cues that pattern cell division orientation and myosin flow: physical contact, contact asymmetry, and a Wnt signal. Second, we experimentally demonstrated that myosin flow generates forces that trigger plasma membrane movements and propose that their anisotropy drives cell division orientation. Our data suggest that contact-dependent control of myosin specifies the division axes of Caenorhabditis elegans AB, ABa, EMS cells, and the mouse AB cell. The contact pattern-dependent generation of myosin flows, in concert with known microtubule/dynein pathways, may greatly expand division axis diversity during development.
Graphical Abstract
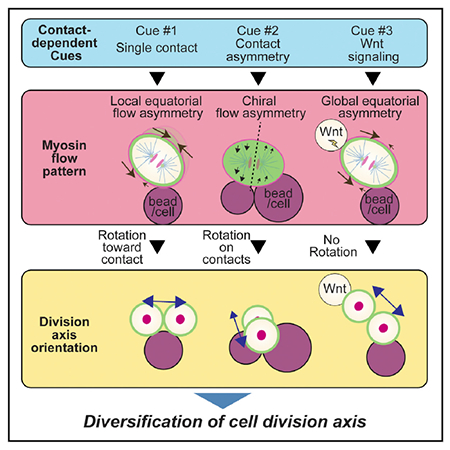
In Brief
Animal morphogenesis requires diverse orientations of cell division. However, how this division axis diversity is achieved remains to be elucidated. Sugioka et al. document cell contact-dependent mechanisms that diversify cell division axes by modulating cortical myosin flow and show that these mechanisms function in both C. elegans and mouse embryos.
INTRODUCTION
Cell division axes are arranged in different orientations during embryogenesis, stem cell division, and organogenesis (Gillies and Cabernard, 2011; Poulson and Lechler, 2012). Oriented divisions are critical for development as they contribute to both spatial cellular patterning and cell fate specification (Knoblich, 2010; Williams and Fuchs, 2013), and mutations in genes required for oriented cell division are associated with human diseases, including microcephaly, leukemia, and multiple cancers (Noatynska et al., 2012; Pease and Tirnauer, 2011). Although previous studies revealed the mechanisms that orient cell division in a specific axis, how cell division axes are arranged in different orientations in the course of development remains unclear. To understand the mechanisms that generate diversity in division axis orientation, three different regulatory layers should be considered: upstream developmental cues, downstream force generators that orient cell division, and cue-dependent spatial control of the force generators (Figure 1A, left). However, thus far only a few developmental cues and force generation mechanisms have been studied, limiting our knowledge of division axis regulation during multicellular assembly.
Figure 1. Oriented AB Cell Division during D-V Body Axis Establishment Does Not Require Microtubule-Pulling Forces.
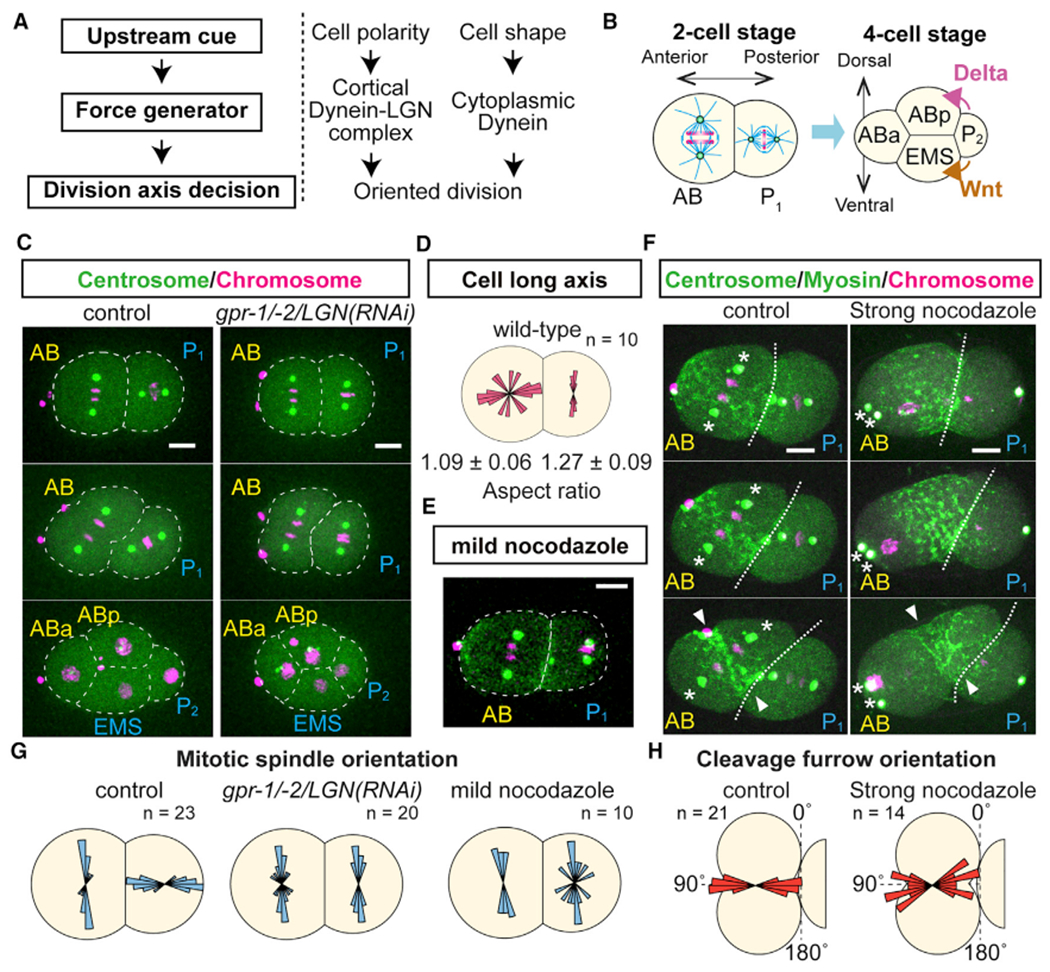
(A) General principle of cell division orientation mechanism (left) and known cell division orientation pathways (right).
(B) Oriented AB and P1 divisions at two-cell stage that precede establishment of the dorsal and ventral body axis.
(C) Orientation of AB cell division does not require cortical dynein recruiter LGN. Centrosomes (green), histone H2B (magenta), and cell outlines (white dotted line) are shown.
(D) Cell long axis does not dictate AB cell division orientation. Values at bottom are cellular aspect ratios.
(E) Mild nocodazole treatment (12.5 ng/mL) disrupted P1 but not AB division orientation.
(F) Cleavage furrow orientation is not affected after strong nocodazole treatment (20 μg/mL). Non-muscle myosin II (green), centrosomes (green; asterisks), histones (magenta), cell-cell boundary (white dotted line), and cleavage furrow position (arrowheads).
(G) Distributions of mitotic spindle orientations relative to the cell contact plane.
(H) Distributions of cleavage furrow orientations relative to the cell contact plane.
Scale bars, 10 μm.
For cell division axes to be oriented in a specific angle, cells need to employ force generation systems that move the division apparatus. Thus far, the microtubule motor protein dynein is the only known force generator. Dynein works at two different cellular locations: the cell cortex and the cytoplasm. At the cell cortex, upstream cues such as cell polarity (di Pietro et al., 2016), tricellular junctions (Bosveld et al., 2016), and mechanical forces (Fink et al., 2011) localize an evolutionarily conserved protein complex composed of Gα, LGN, and NuMA. The Gα/LGN/NuMA complex binds to dynein, which then generates microtubule pulling forces toward the cell cortex through minus-end-directed dynein movement in association with depolymerizing microtubules (Figure 1A, middle). In the cytoplasm, cell shape distortion serves as a cue that generates differences in astral microtubule length due to confinement by the cell cortex (Minc et al., 2011). Longer astral microtubules then bind more cytoplasmic dynein to generate greater pulling force and hence orient division along the longer cell axis (Minc et al., 2011), a phenomenon also known as Hertwig’s rule (Figure 1A, right). A mathematical model implementing microtubule-dependent force generation can predict early cell division orientations in fish, amphibian, echinoderm, and ascidian embryos (Pierre et al., 2016). However, it is unclear if these two microtubule-dependent force generation mechanisms are sufficient to create the diversity of division axes observed in vivo.
The actin cytoskeleton and its motor protein non-muscle myosin is an interesting candidate as an additional force generator for oriented cell division. Although recent reports suggest that F-actin and myosin participate in cell division orientation, they likely modulate microtubule-dependent pathways by controlling cell shape (Campinho et al., 2013), NuMA localization (Seldin et al., 2013), and microtubule dynamics (Kwon et al., 2015). However, one study coupling imaging and numerical simulation has suggested that cell autonomous myosin flow tilts the division plane by approximately 20° in the clockwise direction during establishment of the left-right body axis in Caenorhabditis elegans (Naganathan et al., 2014). We have therefore focused on cortical myosin flow; the concerted movement of a viscoelastic cell surface layer comprising F-actin, myosin, and cross-linking factors (Bray and White, 1988; Levayer and Lecuit, 2012). Myosin flow is generated by the anisotropy of cellular contractility and its velocity is further regulated by actin dynamics and non-muscle myosin II motor activity during cell division (Mayer et al., 2010; Reymann et al., 2016). Thus far, whether cortical myosin flow acts as a force generator during cell division orientation and is controlled by the developmental cues has not been experimentally shown.
We report here that patterns of cell contact diversify the choice of division axes by modulating cortical myosin flow. In multicellular contexts, a single cell is surrounded by multiple neighboring cells and thus can receive multiple physical and chemical cues, complicating the identification of cues required for cell division orientation. To overcome this predicament, we used isolated blastomeres and adhesive polystyrene beads to reconstitute and simplify contact-dependent cues. We thereby identified three contact-dependent cues (physical contact, asymmetry of two contacts, and a Wnt signal) that are sufficient to specify the oriented divisions of the C. elegans and mouse two-cell stage AB cells, the C. elegans four-cell stage ABa cell, and the C. elegans six-cell stage EMS cell, respectively. These three contact patterns distinctly modulate cortical myosin flows. Importantly, we experimentally demonstrated that myosin flow is a force generation mechanism that triggers cell surface movements and orients cell division. Our results suggest that the pattern of contact cues generates distinct myosin flows that specify diverse cell division axes during development.
RESULTS
Oriented Cell Division during C. elegans Dorsal-Ventral Axis Establishment Employs Microtubule-Independent Force Generation Mechanisms
To identify new mechanisms that orient cell division axes, we initially focused on the less studied oriented division of the AB cell in two-cell stage C. elegans embryos. The anterior AB cell divides before the posterior P1 cell, and their daughters always adopt a diamond shape at the four-cell stage, with the posterior daughter of AB (ABp) and anterior daughter of P1 (EMS) aligned perpendicularly to the anterior-posterior (A-P) axis to define the dorsal-ventral (D-V) body axis (Figure 1B) (Priess and Thomson, 1987; Sulston and Horvitz, 1977). This diamond shape is critical for later development, as signals from P2 activate Notch signaling in ABp and Wnt signaling in EMS to specify different cell fates (Figure 1B) (Priess, 2005; Sawa and Korswagen, 2013). During cell division, the AB and P1 mitotic spindles are oriented parallel and perpendicular to the plane of AB-P1 cell contact, respectively (Figures 1B, 1C, and 1G). For oriented P1 division, a midbody remnant of zygotic division appears to be an upstream cue (Singh and Pohl, 2014) and an LGN/dynein-dependent force generation mechanism is required to orient the division axis (Srinivasan et al., 2003). However, a previous report suggested that LGN knockdown did not affect AB division axes (Srinivasan et al., 2003). We confirmed that AB division axis oriented normally after LGN knockdown, suggesting that oriented AB division does not require cortical dynein-dependent microtubule pulling forces (Figures 1C and 1G). A second possibility is that cell shape distortion acts as a cue for oriented AB division through an anisotropy in cytoplasmic dynein-dependent microtubule pulling forces (Figure 1A, right). However, orientation of the long axis of the AB cell before division did not correlate with the later axis of division (Figure 1D), indicating that cell shape is not responsible for orienting AB division. To assess the potential roles of other microtubule-dependent mechanisms in oriented AB division, we treated embryos with a mild level of the microtubule-depolymerizing drug nocodazole (12.5 ng/mL, which still allowed mitotic spindle formation) and found that the AB division axes were unaffected while those of P1 were abnormal (Figures 1E and 1G). Furthermore, even when spindle formation was abolished by treatment with 20 μg/mL nocodazole, cleavage furrows oriented perpendicularly to the contact plane as in the control (Figure 1F; arrowheads, and 1H, Video S1). We conclude that a microtubule-independent force generation mechanism orients the AB division axis.
Physical Contact Is a Sufficient Cue for Orienting the Second Embryonic Division in Both C. elegans and the Mouse
We next explored the origin of the upstream cue that orients the AB cell division. To test whether cell contact serves as a cue, we isolated early two-cell stage AB and P1 cells and recombined them in culture medium to randomize their contact sites (Figure 2A). The AB division axis was initially randomly oriented, due to the manipulation, but then rotated to become parallel to the AB-P1 contact plane by the end of telophase (hereafter referred to as parallel division) (Figure 2B and Video S2). Extrinsic cues transmitted through cell contact can be either mechanical or chemical in nature. To evaluate the former possibility, we reconstituted the physical contact by using carboxylate-modified polystyrene beads that nonspecifically bind to the amine groups of cell surface proteins. Attachment of a bead to isolated AB cells also resulted in parallel division (Figure 2B and Video S2), demonstrating that physical contact and adhesion are sufficient to orient AB cell division. Bead contact did not change cell shape; the ratio between the perpendicular and parallel cell diameters relative to the beads was approximately 1.0 (mean ± SD, 0.991 ± 0.021; n = 20). Consistent with the in vivo experiments, strong nocodazole treatment did not disrupt cleavage furrow orientation (Figure 2B). Thus, our results suggest that physical contact is an upstream cue that orients AB cell division through a microtubule-independent force generation system.
Figure 2. Physical Contact Is the Cue Sufficient for Oriented AB Cell Division in Both C. elegans and Mouse.
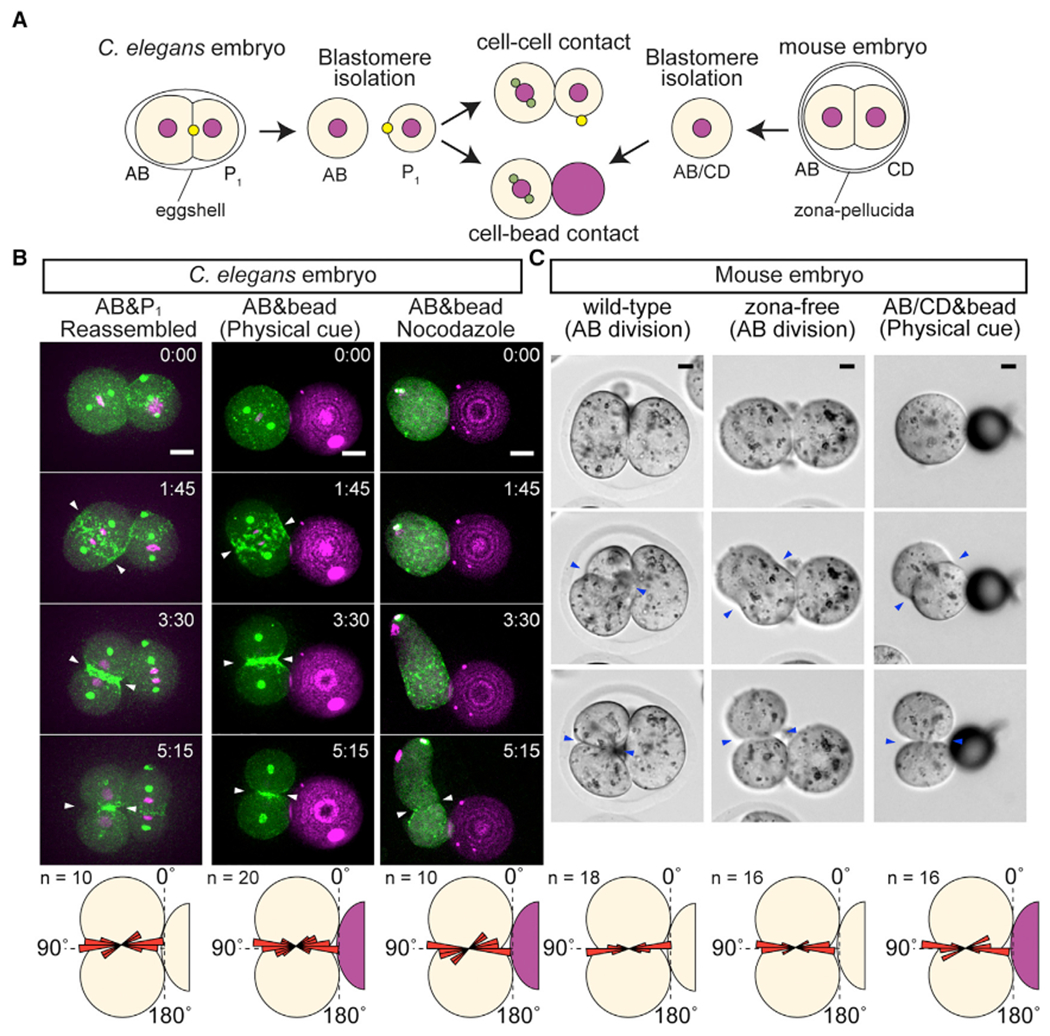
(A) Schematic illustration of contact reconstitution assay. Yellow dots with P1 cell indicate a midbody remnant.
(B) C. elegans AB division axis after the reconstitution of contact cues. Centrosomes (green), myosin (green), histone H2B (magenta), polystyrene bead (magenta), and cleavage furrow position (arrowheads) are shown along with the distribution of cleavage furrow orientations (bottom panel). Nocodazole 20 μg/mL was used.
(C) Mouse AB division axis in vivo and AB/CD division axis after contact reconstitution. Zona-free are embryos without zona pellucida. Cleavage furrow position (arrowheads) are shown along with the distribution of cleavage furrow orientation (bottom panel). Angle distributions are shown relative to the contact plane.
Scale bars, 10 μm. See also Figure S1.
We also tested whether this physical contact-dependent cue can orient cell division in mammalian embryos. As in C. elegans, the two-cell stage mouse embryo undergoes asynchronous divisions; the early dividing cell is called AB and the other CD (Kelly et al., 1978) (Figure 2A). Both AB and CD cells in intact two-cell stage embryos underwent division in parallel to the contact site (Figure 2C and Video S3). Previous studies showed that the mitotic spindle of the AB cell was randomly oriented at metaphase (Louvet-Vallee et al., 2005), and there are no data suggesting that microtubule pulling forces drive oriented AB division in mouse. While removal of the zona pellucida (a glycoprotein layer surrounding cells) was reported to result in abnormal cell division orientation (Graham and Deussen, 1978; Suzuki et al., 1995), we found that only CD cell divisions were affected and AB cells always underwent parallel division in zona-free embryos (Figures 2C and S1 and Video S3). We therefore assessed whether mouse AB cell division orientation is regulated by the physical contact cue. When attached to beads, isolated mouse two-cell stage blastomeres (either AB or CD) underwent parallel division, suggesting that physical contact acts as a cue for their oriented division (Figure 2C and Video S3). Taken together, physical contact is a sufficient cue that orients both the C. elegans and mouse second embryonic divisions.
Myosin Activity Is Required for Physical Contact-Dependent Division Axis Orientation
To investigate the force generation mechanism that underlies physical contact-dependent oriented division, we found in a C. elegans candidate screen that knockdown of the Cullin E3 ubiquitin ligase component CUL-3 resulted in abnormal AB division axes, with 27% of the four-cell stage embryos showing a linear cell arrangement and thus a severely disrupted D-V axis (Figures 3A and 3B). During the AB division in cul-3(RNAi) embryos, non-muscle myosin II/NMY-2 foci were abnormally distributed throughout the cell cortex; the ratio of myosin signal intensities between the polar and equatorial regions of the cell cortex became lower in cul-3(RNAi) than in control embryos (Figures 3C, 3E, S2A, and S2B). Although a functional contractile ring subsequently formed, orientation of the cleavage furrows was misoriented relative to the contact plane (Figures 3C and 3F). While we cannot rule out CUL-3 acting through the mitotic spindle, based on these results, we hypothesized that CUL-3 acts via a myosin-dependent pathway to orient AB cell division. To evaluate this possibility, we manipulated non-muscle myosin II activity by pharmacological treatment or RNAi. First, we knocked down the Rho GTPase-activating protein RGA-3 to activate myosin II; RGA-3 usually inactivates RhoA and hence inhibits downstream myosin II activity (Schonegg et al., 2007). We found that rga-3(RNAi) reduced the ratio of polar/equatorial myosin signal intensities and resulted in abnormal AB division axes, similar to cul-3(RNAi) (Figures 3C, 3E, and 3F). Second, we employed a myosin light-chain kinase inhibitor, ML-7, to inactivate myosin II (Saitoh et al., 1987), which is also known to inhibit myosin activity during C. elegans gastrulation (Lee and Goldstein, 2003). When early two-cell stage embryos were treated with ML-7, the AB cell did not form a cleavage furrow, and the mitotic spindle misoriented (Figures 3D and 3I). We obtained similar results after treatment with the actin depolymerizing drug Latrunculin A (Figure S2C). In addition, in the presence of cell-bead contact, ML-7 treatment also resulted in the abnormal AB division axis orientation (Figures 3H and 3I). Although cytokinesis was defective following ML-7 treatment, this was not the direct cause of abnormal division axis orientation, as ZEN-4/MKLP-1 mutants, which have normal myosin activity but fail to complete cytokinesis, oriented normally in parallel to the bead (Figure S2D). Consistent with the C. elegans results, ML-7 treatment disrupted mouse AB division axis orientation in zona-free embryos (Figures 3F and 3G). Taken together, these results indicate that these physical contact-dependent oriented divisions require myosin II activity instead of microtubule pulling forces.
Figure 3. Myosin Activity Is Required for Physical Contact-Dependent Oriented Division.
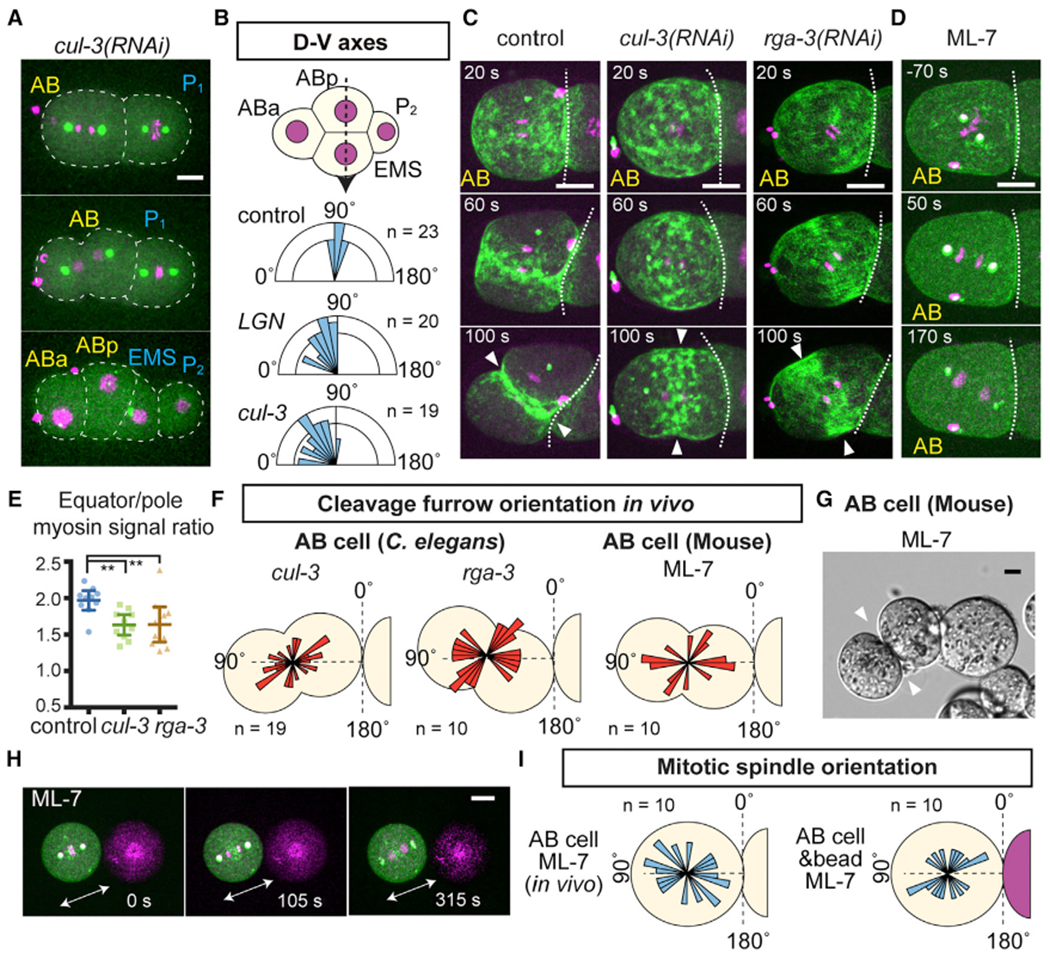
(A) Abnormal AB division axis in cul-3(RNAi). Centrosomes (green), histone H2B (magenta), and cell outlines (white dotted lines) are shown.
(B) D-V axis orientation measured by the angle of ABp-EMS nuclear axis (dotted arrow) relative to the A-P axis.
(C) Cleavage furrow misorientation after RNAi knockdowns. Myosin (green) and histones (magenta) are shown.
(D) Mitotic spindle misorientation after ML-7 treatment. Myosin (green), centrosome (green), and histones (magenta). White dotted lines in (C) and (D) are cell-cell boundaries.
(E) Ratio of equatorial to polar cortical myosin intensity at 100 s after anaphase onset. p values were calculated by one-way ANOVA with Holm-Sidak’s multiple comparison test. **p < 0.01.
(F) Distributions of cleavage furrow orientations. Schematics represent examples of abnormal orientations.
(G) Misorientation of AB division in zona-free mouse embryos after ML-7 treatment (see Method Details).
(H) Misorientation of C. elegans AB division in the presence of bead contact after ML-7 treatment. Arrows indicate mitotic spindle orientation.
(I) Distributions of C. elegans AB mitotic spindle orientations after ML-7 treatment. Angle distributions are shown relative to the contact plane.
Arrowheads show cleavage furrow position. Error bars indicate the mean ± 95% confidence interval. Scale bars, 10 μm. See also Figure S2.
Physical Contact Inhibits Myosin Regulatory Light-Chain Phosphorylation during Oriented Division
We next investigated how the physical contact cue controls myosin activity. For non-muscle myosin, each myosin II heavy chain dimer binds two essential light chains and two regulatory light chains (RLCs) to form a hexamer. RLC phosphorylation at the evolutionarily conserved Thr 18 and Ser 19 residues (Ser 17 and Thr 18 for C. elegans MLC-4/RLC) has central roles in myosin activation: RLC phosphorylations increase the myosin ATPase activity and trigger a conformational change from a closed to an open form that allows minifilament assembly and promotes downstream contraction (Figure 4A) (Vicente-Manzanares et al., 2009). Thus, we monitored myosin RLC phosphorylation as an indicator of myosin activity and tested the effects of physical contact on them by performing immunolabeling using anti-phospho-RLC antibody that recognizes RLC in both C. elegans and mouse embryos (Zonies et al., 2010; Maître et al., 2015). In C. elegans AB cells, phosphorylated RLC (p-RLC) signals were reduced at both cell and bead contact sites compared with the polar regions, while the myosin II signal ratios were not reduced (Figure 4B). In mouse AB/CD cells, p-RLC signals were also reduced at the cell or bead contact sites compared with the polar region (Figure 4C). Note that we performed the control experiment in mouse by comparing the signal intensity of p-RLC between polar region and sub-equator regions in the absence of contact, and their ratio was close to 1.0 (Figure 4C). To directly test the significance of RLC phosphorylation in cell division orientation, we expressed an RNAi-resistant version of MLC-4/RLC with Ser17A Thr18A phospho-deficient mutations introduced, with RNAi knockdown of endogenous MLC-4; strikingly, AB cells division axes were misoriented (Figure 4D). Thus, our results suggest that the contact-dependent local control of RLC phosphorylation is critical for AB cell division orientation.
Figure 4. Physical Contact Decreases Myosin Regulatory Light-Chain Phosphorylation.
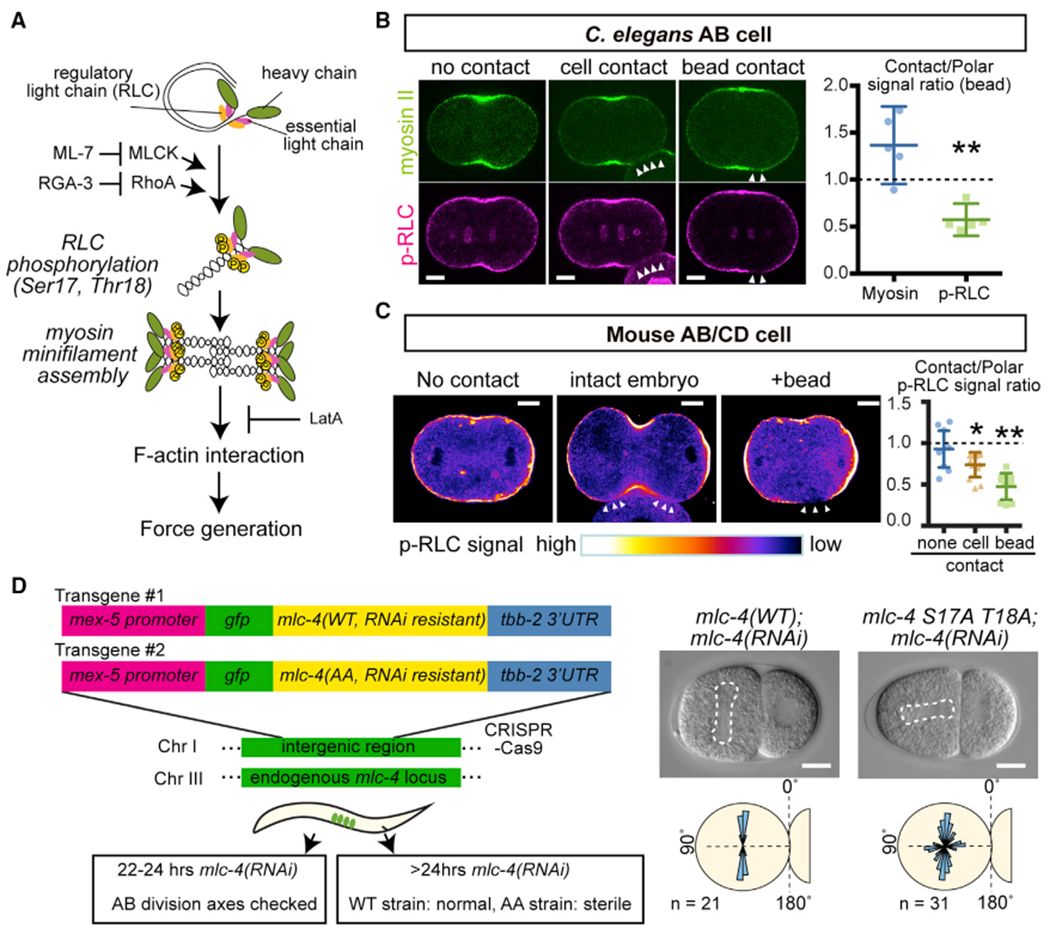
(A) Schematic representation of myosin activation process.
(B and C) Immunostaining of p-RLC individing C. elegans AB (B) or mouse AB/CD (C) cells. Arrowheads indicate the cell or bead contact site. In (C), p-RLC signal intensity is shown by the bottom color code. Right graph in (B) shows the ratio of signal intensities between bead contact and polar region. In (C), right graph shows the ratio of p-RLC signal intensities between contact and polar region. In the cell without contact, sub-equatorial region was used as “contact signal”. p values were calculated by the Welch’s t test in (B) and by the one-way ANOVA with Holm-Sidak’s multiple comparison test in (C). *p < 0.05 and **p < 0.01.
(D) RLC mutation leads to abnormal C. elegans AB division axis. Left schematics show the experimental design. Single copy transgenes were inserted into the intergenic region and endogenous MLC-4 was knocked-down by 22–24 hr RNAi treatments (see Method Details). Dotted lines in DIC images indicate mitotic spindle. Bottom panels are distributions of mitotic spindle orientations relative to the contact.
Error bars indicate the mean ± 95% confidence interval. Scale bars, 10 μm. See also Figure S3.
Physical Contact Induces Intracellular Myosin Flow Anisotropy
We hypothesized that the physical contact-dependent regulation of myosin activity resulted in force generation that orients AB division axis. To evaluate this possibility, we first analyzed the dynamics of cortical myosin II foci. We defined the x axis as passing through the spindle poles and the y axis as perpendicular to the x axis and within the plane of the cell and bead centers (Figure 5A). We also divided the AB cell surface into four quadrants (U1, U2, B1, and B2) with B2 attached to the bead (Figure 5A). Cell movements, induced by the orientation process or cellular drift in the culture media, were computationally removed to precisely analyze cortical myosin II dynamics. In isolated AB cells without physical contact, myosin foci flowed toward the cell equator along the x axis and exhibited no clear directionality along the y axis (Figures 5A, 5B, and S4A). However, upon bead attachment, x axis myosin movements were significantly reduced in B2, resulting in an asymmetric flow within B1 and B2 (Figures 5A and 5B and Video S4). On the other hand, y axis movements were oriented in the counter-clockwise direction around the division axis when viewed from the nearest pole, with myosin in U1 and B1 regions moving in the direction opposite to those in U2 and B2 (Figures 5A and S4A, and Video S4). Intact wild-type embryos exhibited similar myosin movements (Figures 5A–5C), suggesting that contact with either beads or cells induces anisotropic myosin II flow. Hereafter we refer to these x axis and y axis myosin flows as equatorial and chiral flows, respectively. In cul-3(RNAi) embryos, where division axes became abnormal, asymmetry in equatorial myosin flow was lost, whereas chiral flow remain normal (Figures 5A–5C). Moreover, although the direction of chiral flow was always counter-clockwise regardless of bead position, the equatorial flow was responsive to the contact location. These results suggest that the equatorial myosin flow asymmetry is in charge of the division axis orientation process.
Figure 5. Myosin Flow Is Tunable by Physical Contact and Triggers Cell Surface Movements.
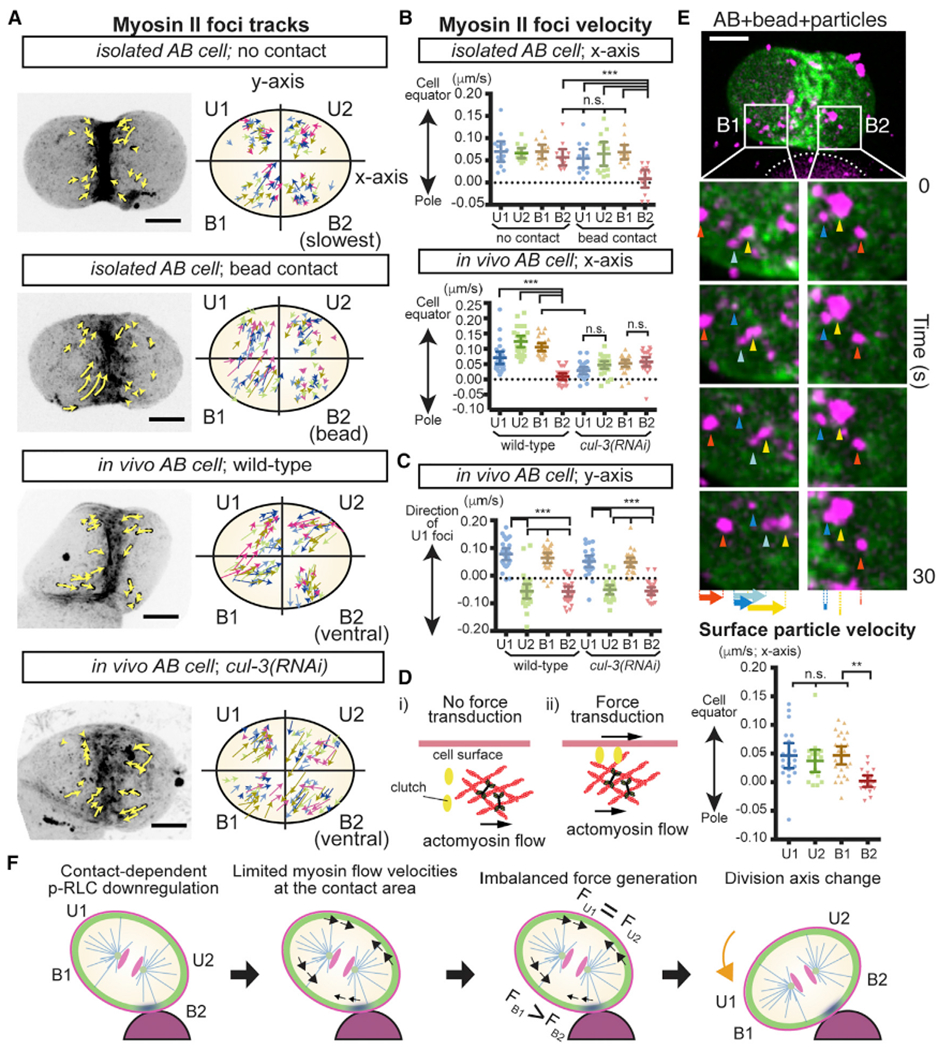
(A) Myosin foci movements during C. elegans AB cell division. Isolated AB cells with or without bead attachment and wild-type or cul-3(RNAi) embryos. Myosin foci tracks for 50 s are shown as yellow arrows in left panel. In the right panel, color of arrows indicates the tracks from different samples. The cell quadrant was defined as indicated. In the cells with no contact, regions exhibited slowest myosin velocities are B2 for convenience of comparison. In vivo, the quadrant of AB that showed the slowest myosin velocities always became the ventral side of the AB cell, but the direct cause of this asymmetry is unknown.
(B and C) Velocities of myosin foci in x (B) and y (C) axes of the cells summarized in (A).
(D) Clutch enables force transduction from actomyosin cortex to cell surface; identity of clutch unknown.
(E) Cell surface x axis movements during oriented AB division. Movements of 0.35-μm particles attached to the membrane (arrowheads) and their velocities are shown along with myosin (green) and beads (white dotted line).
(F) Model for cell division orientation in which equatorial myosin flow asymmetry generates a cell surface torque(FB1 > FB2). Myosin cortex (green), inactive myosin (blue), myosin flow (black arrow), and cell rotation (orange arrow) are shown.
p values were calculated by one-way ANOVA with Holm-Sidak’s multiple comparisons tests. **p < 0.01; ***p < 0.0001; n.s., not significant (p > 0.05). Error bars indicate the mean ± 95% confidence interval. Scale bars, 10 μm. See also Figure S4.
Myosin Flow Generates Forces to Trigger Cell Surface Movements during Oriented Division
The anisotropy of myosin flow has been proposed to control division axes in C. elegans six-cell stage embryos, when the ABa and ABp cell division axes rotate 20° in a clockwise manner. A chiral myosin flow was proposed to provide the driving force for the rotation of these two division axes rotation (Naganathan et al., 2014), but it has not been experimentally tested whether myosin flow can generate forces to orient cell division. If myosin flow can trigger cell surface movements, then the flow forces cause the cell to move and might orient the axis of cell division. However, it is also unclear as a state of physical linkage between actomyosin cortex and cell surface affects the outcome of force transduction (Figure 5D) (Rho-Johnson et al., 2012; Case and Waterman, 2015). To determine whether actomyosin flow generates forces that trigger cell surface movement, AB cells attached to a 30-μm bead were coated with fluorescently labeled carboxylate-modified particles (0.35 μm in diameter) to track cell surface movements. We found that the surface particles exhibited equatorial flows that were similar to those of myosin II and also were limited near the contact site (Figure 5E and Video S5). These results suggest that myosin flow is a force generation mechanism that triggers cell surface movements and can be controlled by the position of physical contact. Consistent with the physical contact-dependent myosin inhibition, ML-7 treatment limited flow velocities (Figure S4B).
Based on our results, we propose the following model for cell division orientation (Figure 5F). First, contact-dependent p-RLC downregulation leads to the inhibition of myosin activity and flow at the contact area (B2). Second, the myosin flow forces are transmitted to the cell surface. Third, the cell surface flow forces are balanced in the area distal to the contact site (U1 and U2) but asymmetric in the proximal (B1 and B2), resulting in a force anisotropy. Third, the greater equatorial force in B1 generates a directional torque that orients cell division (similar to how a basketball spins in response to a hand stroke) such that it becomes parallel to the contact plane.
Second Contact Cue: Contact Asymmetry Orients Left-Right Division Axis at the Four-Cell Stage
Given that a physical contact cue suffices to orient the two-cell stage AB cell division through the generation of myosin flow, we looked for other contact-dependent cues in different developmental contexts. We focused on the four-cell stage ABa cell that divides along the left-right body axis (Figure 6A) (Sulston et al., 1983). ML-7 treatment disrupted ABa division axis orientation, suggesting that this process also requires myosin activity (Figure 6B). Before cell division, ABa is adjacent to an equally sized ABp cell and a smaller EMS cell, creating a contact asymmetry. To determine the causal link between this contact pattern and the subsequent ABa division axis orientation, isolated ABa cells were attached to differently sized beads. When ABa cells were attached to symmetrically sized (two 30-μm or two 20-μm diameter) beads, the division pattern did not recapitulate the in vivo situation (Figure 6B). However, when attached to asymmetrically sized beads (one 30-μm and one 20-μm), 82% of the ABa cells underwent a normal division pattern analogous to the left-right oriented division in wild-type embryos (Figure 6B and Video S6). AB cells, the mother of ABa and larger in size, showed similar behavior (Figure 6B). These results suggest that asymmetry in the sizes of contacting cells is another contact-dependent cue that orients cell division, which in an intact embryo specifies the left-right oriented ABa cell division.
Figure 6. Asymmetry in Contacting Cell Sizes Is a Sufficient Cue for ABa Division Axis.
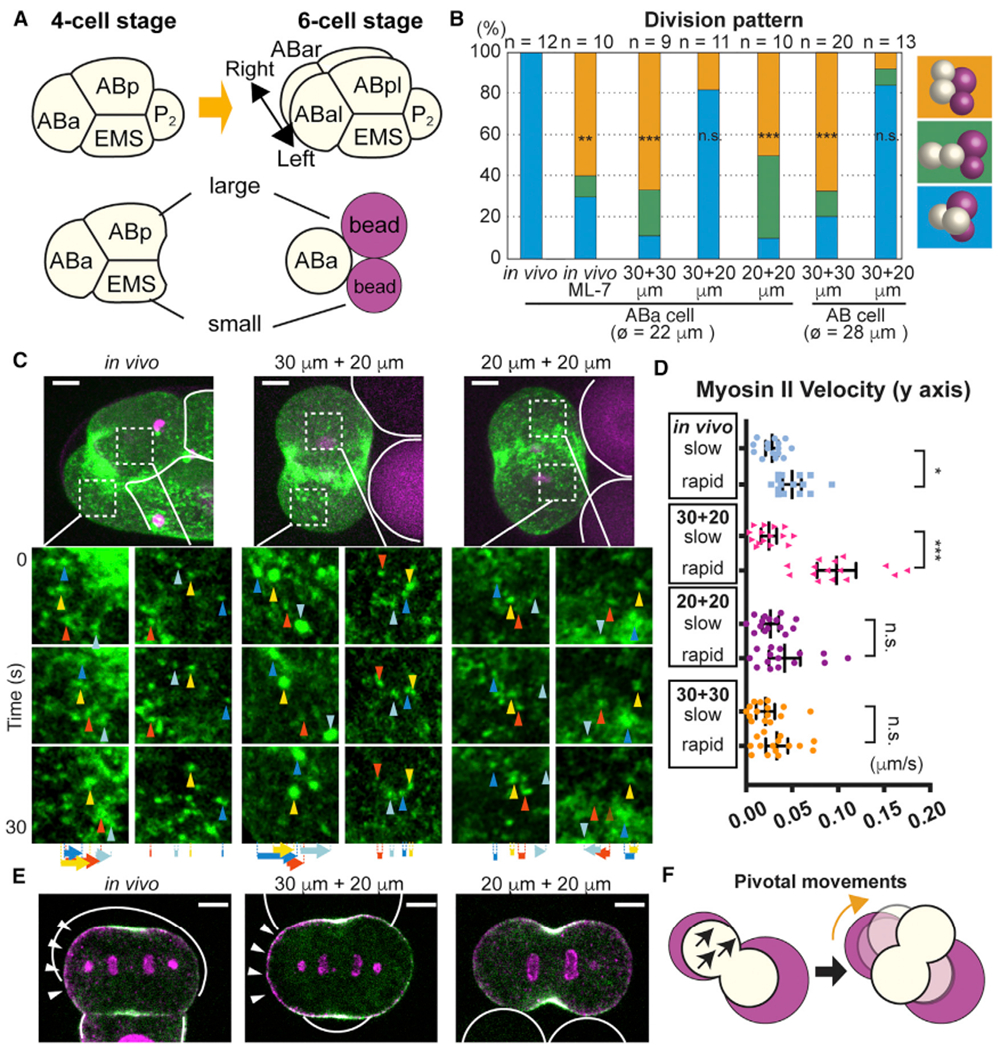
(A) ABa cell division at the four-cell stage and recapitulation of contact asymmetry using beads.
(B) Effects of bead size asymmetry on ABa and AB cell division axes. Color represents different cell division pattern as shown in right; ø = cell diameter.
(C and D) Myosin foci movements during ABa cell division. Myosin foci in the white dotted box were magnified in the bottom panels. Solid white lines indicate contact boundary. Arrowheads and arrows in (C) indicate myosin foci and their total displacement, respectively. Velocities of three myosin foci from each dividing cell half are quantified for five embryos and shown in (D) with the slow or rapid group determined by the average velocities in single embryos.
(E) Polarized p-RLC localization in response to in vivo and in vitro cell/bead contacts. Myosin (green), p-RLC (magenta), cell or bead outlines (white solid lines), and polarized p-RLC (arrowheads) are shown.
(F) Schematic model of ABa division axis orientation (see text). Myosin flow (black arrows) and cell rotation (orange arrows).
Scale bars, 5 μm. p values in (B) were determined with the Fisher’s exact test and those in (D) were calculated by one-way ANOVA with Holm-Sidak’s multiple comparisons test. *p < 0.05; **p < 0.01; ***p < 0.0001; n.s., not significant (p > 0.05). Error bars indicate the mean ± 95% confidence interval. See also Figure S5.
As myosin activity is required for ABa cell division orientation, we analyzed myosin flow during this process. In vivo, both x axis and y axis myosin flows exhibited asymmetric velocities between the different cell halves, consistent with a previous report (Naganathan et al., 2014) (Figures 6C, 6D, and S5A). On the other hand, when isolated ABa cells were attached to asymmetrically sized beads, only the y axis myosin flow was asymmetric (Figures 6C, 6D, and S5A). Given that ABa cells attached to asymmetrically sized beads can recapitulate their in vivo division axes, the y axis myosin flow asymmetry may be sufficient for ABa division axis orientation. Conversely, myosin flow asymmetries were undetectable when isolated ABa cells were attached to symmetrically sized beads, suggesting that contact asymmetry is the cue that activates y axis myosin flow asymmetry (Figures 6C and 6D). Consistent with the myosin flow asymmetry, asymmetrically sized cell or bead contacts induced a polarized localization of p-RLC both in vivo and in vitro (Figures 6E and S5C). These results indicate that asymmetry in the sizes of the contacting cells is a cue that polarizes myosin activity and flow to specify left-right oriented ABa cell division. Although we do not have strong causal evidence, we speculate that greatery axis myosin flow in one cell half results in a pivotal cellular movement on the contact plane, and hence division axis rotation (Figure 6F). Note that the previously reported left-right skew of the ABa and ABp division axes (Naganathan et al., 2014) occurs after the initial specification of left-right oriented division that we address here.
Third Contact Cue: An Extrinsic Wnt Signal Overrides the Physical Contact Cue at the Six-Cell Stage
Following the ABa and ABp cell divisions at the six-cell stage, the EMS cell undergoes an A-P oriented division (Figure 7A). The oriented EMS division is regulated by a Wnt signal from the posteriorly located P2 cell and is critical for the asymmetric specification of anterior mesoderm and posterior endoderm precursor cell fates (Figure 7A) (Rocheleau et al., 1997; Thorpe et al., 1997; Schlesinger et al., 1999; Goldstein et al., 2006). Although it was reported that cortical dynein pathways are involved in EMS division axis orientation (Liro and Rose, 2016; Tsou et al., 2003), cortical dynein is uniformly localized throughout EMS and thus the mechanism of its division axis orientation remains unclear (Heppert et al., 2018). Moreover, the EMS cell contacts not only the P2 cell but also the ABal, ABar, ABpl, and ABpr cells. Given that physical contact is a cue that controls the AB division axis at the two-cell stage (see above), multiple contact cues may compete with each other for control of the EMS division axis. We therefore investigated how contact cues from P2 and other early embryonic cells interact. When attached to a single bead, isolated EMS cells underwent parallel division (Figure S6A). When attached to a bead on one side and to ABxx cells (i.e., a pair of ABa or ABp daughters that do not induce Wnt signaling; Goldstein, 1992) on the other, EMS cells again underwent parallel division relative to the plane of bead contact (Figure 7B). However, when attached to a bead on one side and P2 on the other, EMS division axes were misoriented relative to the bead and oriented exclusively toward the P2 cell (Figure 7B and Video S7). Furthermore, EMS cells attached to a bead on one side and P2 isolated from Wnt mutants on the other restored the parallel division orientation relative to the cell-bead contact plane (Figure 7B and Video S7). These results suggest that a contact-dependent chemical cue, Wnt from P2, cancels the physical contact cues during EMS division axis orientation.
Figure 7. Extracellular Wnt Signal Overrides Physical Contact Cues to Control EMS Division Axis.
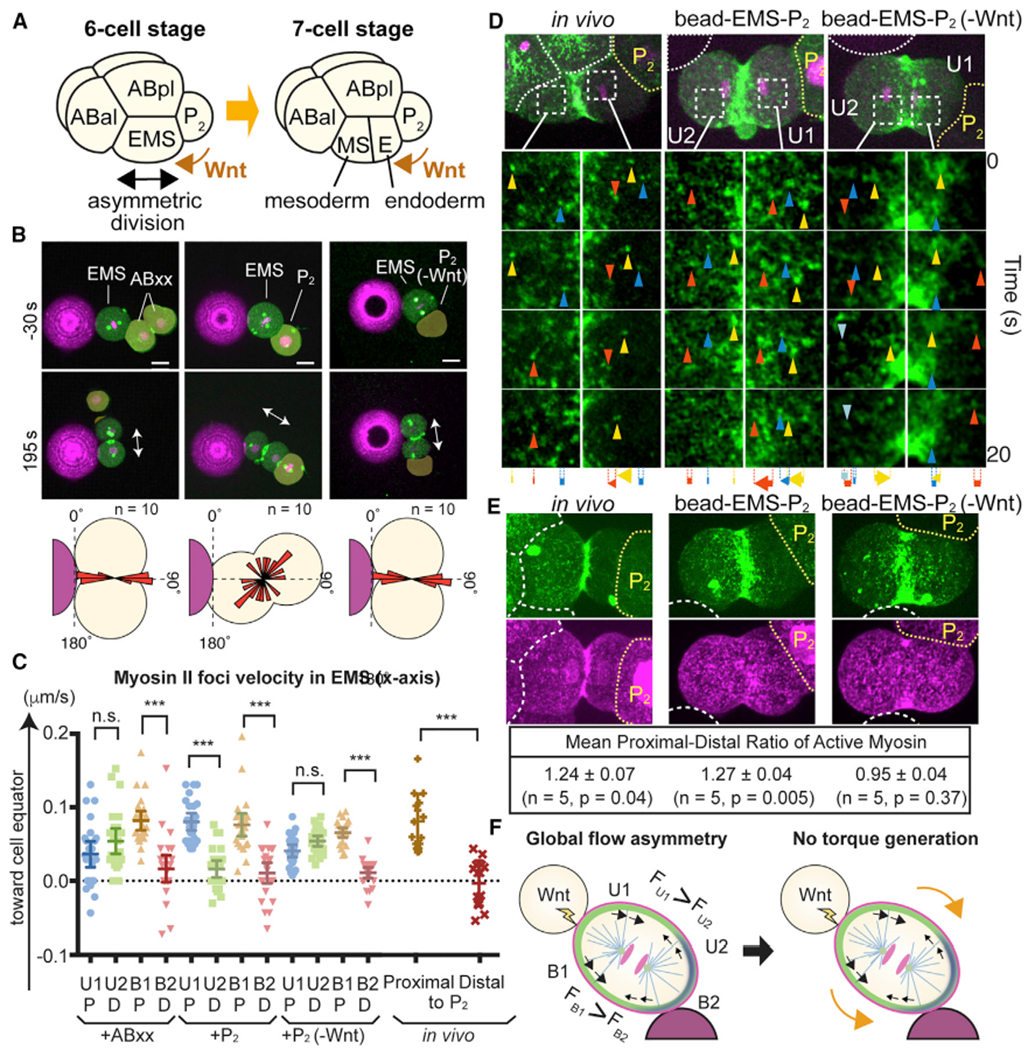
(A) Asymmetric EMS division at the six-cell stage is oriented toward P2 cells despite the presence of multiple physical contact cues.
(B) Bead contact cue blocked by Wnt signal from P2 cell. Myosin (green), centrosomes (green), histones (magenta), beads (magenta), and EMS division axis (arrows) are shown. P2(-Wnt) indicates P2 cells isolated from Wnt(−) mutants. Bottom panel shows the distributions of cleavage furrow orientations.
(C and D) Myosin foci velocities (C) and movements (D) during EMS division. Arrowheads and arrows indicate myosin foci and their total displacement, respectively. Cell quadrants were determined by bead contact site as in Figure 5. P, cell halves proximal to P2 cell; D, cell halves distal to P2 cell.
(E) Wnt-dependent in vivo and in vitro polarization of active myosin localization. Myosin (green), p-RLC (magenta), cell or bead boundaries (white dotted lines), and P2 cells (yellow dotted lines) are shown. Active myosin level was determined as the ratio of p-RLC to myosin intensity.
(F) Model of Wnt-dependent overriding of physical contact cue (see text). Myosin (black arrows) and cell surface (orange arrows) movements are shown.
p values were determined by one-way ANOVA with Holm-Sidak’s multiple comparisons test in (C) and by a paired t test in (E). ***p < 0.0001; n.s., not significant (p > 0.05). Error bars indicate the mean ± 95% confidence interval. Scale bars, 10 μm. See also Figure S6.
We next analyzed the influence of Wnt signaling on cortical myosin flow dynamics during EMS division. We again divided the cell surface area into four quadrants (U1, U2, B1, and B2), with B2 attached to the bead. Upon attachment to a bead and ABxx or P2 from Wnt mutants, myosin equatorial flows became asymmetric between B1 and B2 regions but not in U1 and U2 regions, recapitulating the contact-dependent local myosin flow asymmetry we observed for two-cell stage AB cells (Figures 7C, 7D, and 7G and Video S7). However, attachment to a bead and a wild-type P2 cell yielded asymmetric myosin velocities for both U1-U2 and B1-B2, generating a global flow asymmetry (Figures 7C, 7D, and 7G, and Video S7). Such a global flow asymmetry also exists in vivo during EMS cell division (Figures 7C and 7D, and Video S7). In both cases, myosin flows were always faster in the cell halves proximal to the P2 cell compared with those in distal halves (Figures 7C and 7D).
To determine if Wnt activates myosin to generate the global myosin flow asymmetry, we analyzed the RLC phosphorylation state. In both in vivo and in vitro experiments, the localization of myosin II was decreased in the region proximal to P2, whereas that of p-RLC was more symmetric (Figures 7E and S6B). Consequently, the level of p-RLC per myosin II, indicative of active myosin, was higher in the proximal cortex (Figure 7E). Attachment to P2 cells from a Wnt mutant abolished the myosin II asymmetry, causing myosin activity to become symmetric (Figures 7E and S6B). Taken together, these results demonstrate that the Wnt signal is a cue that induces asymmetric myosin activation along an axis proximal-distal to Wnt-expressing cells. In contrast to the local myosin flow asymmetry induced by the physical contact (Figure 5F), we propose that the Wnt-dependent global myosin flow asymmetry does not generate torque toward the physical contact. If we consider force generation between U1 and U2, greater forces toward the equator from U1 would result in outward cell rotation, while asymmetric force generation between B1 and B2 would rotate the cell in the opposite direction, with these asymmetries thus canceling each other (Figure 7F). This model explains why EMS ignores physical contact cues in the presence of Wnt signaling and thus ensures robust division axis orientation, although the mechanism that orients the EMS division axis along the A-P axis, while possibly requiring dynein, remains unclear.
DISCUSSION
In this study, we have shown that the patterns of cell contact diversify division axis orientations. All three contact-dependent cell division orientation mechanisms involve cortical non-muscle myosin flow control, and at least one of these cell division orientations that is regulated by the physical contact cue occurs independently of known microtubule-dynein pathways. We have shown that physical contact with a cell or a bead acts as a sufficient upstream cue for orienting the division axes of the two-cell stage C. elegans and mouse AB cells. In both the C. elegans and mouse AB cells, physical contact downregulated myosin regulatory light-chain phosphorylation. As a consequence, in the C. elegans AB cell, myosin equatorial flow became asymmetric in the region proximal to the contact site, while symmetric in the distal region, creating a local myosin flow asymmetry. By tracking cell surface movements, we experimentally showed that myosin flow is a force generator that triggers cell surface movements. Based on these data, we proposed a model in which locally asymmetric cell surface movements generate a directional torque that orients cell division in parallel to the contact site. In addition, we identified two additional contact cues, contact asymmetry and Wnt signaling, that orient the ABa and EMS cell division axes, respectively. Although requirements for the myosin flow forces during ABa and EMS division orientation remain to be tested, our study demonstrates that cortical myosin flow is a force generator that can drive cell division orientation and can be tuned by the patterns of cell contact. In combination with microtubule pulling forces, tunable myosin flow forces may allow for a greater diversity of division axis specification during development.
Our results provide the first examples of extrinsic control of actomyosin flow in dividing cells. The molecular mechanism of contact-dependent myosin flow regulation remains to be elucidated. For the physical contact cue, there are two potential scenarios of mechanosensitive myosin inhibition, depending on the nature of the changes in mechanical properties induced by cell contact. First, if physical contact increases cortical tension, it may directly inhibit myosin by exerting resistive forces; a previous single-molecule study showed that purified myosin II molecules prematurely detached from F-actin and exhibited smaller working strokes when resistive forces were exerted (Capitanio et al., 2012). Second, if physical contact reduces cortical tension, it may inhibit myosin light-chain kinase; another single-molecule study showed that myosin light-chain kinase can be activated by force application and proposed that an auto-inhibitory domain that blocks kinase-substrate interaction is released upon force application (Baumann et al., 2017). Future characterization of the mechanical properties at the cell-cell contact site may sort out these possibilities. Furthermore, how contact-dependent regulation affects myosin flow in the contact-adjacent area is unknown. Notably, we showed that myosin lost the ability to respond to the contact site after knockdown of CUL-3 E3 ubiquitin ligase. Thus CUL-3 substrates are interesting candidate proteins that might shed light on the nature of both physical contact-dependent and asymmetric contact-dependent myosin regulation. For the Wnt-dependent control of myosin activity, we speculate that the noncanonical Wnt pathway may regulate myosin flow. In vertebrates, Rho activity is regulated by the Frizzled and Dishevelled during morphogenesis (Sokol, 2015). Future analysis should reveal the regulatory networks that influence myosin flow in response to different cues.
We have shown that physical contact-dependent cell division orientation employs cortical myosin activity as a force generation system, and our data strongly suggest that myosin flow generates force to trigger cell surface movements independently of the microtubule cytoskeleton. Studies in other organisms suggest that this mechanism may be widely conserved. In Drosophila neuroblasts, it was reported that a cell polarity pathway can specify cleavage furrow positioning independently of microtubules (Cabernard et al., 2010). Polarized myosin can also regulate the unequal division of a C. elegans neuroblast (Ou et al., 2010). Although it has not been experimentally tested, it is possible that myosin flow-dependent force generation contributes to cleavage furrow positioning in these systems. Furthermore, it also is interesting to consider how physical contact cues might affect polarity-dependent myosin control in these systems.
We showed that physical contact is a sufficient cue for orienting both the C. elegans and mouse second embryonic divisions. As zona pellucida-free human two-cell stage embryos also appear to undergo cell division in parallel to cell contact (Bodri et al., 2015), this oriented division mechanism may also explain the geometry of the human second embryonic division. The two-cell stage division axes in these different animals affect four-cell stage architectures that are significant for further development: the diamond shape in C. elegans establishes the D-V axis, while the tetrahedron shape in mouse and human embryos promotes more successful in vitro fertilization outcomes than do other patterns (Cauffman et al., 2014; Ebner et al., 2012; Graham and Deussen, 1978; Suzuki et al., 1995). Moreover, the tetrahedron and diamond shapes in mouse embryos are associated with distinct pluripotency factor activities and gene expression profiles among blastomeres (Goolam et al., 2016; Torres-Padilla et al., 2007; White et al., 2016).
We also have shown that physical contact inhibits myosin activity at the contact site in both C. elegans and mouse. While extrinsic control has yet to be documented, actomyosin regulation at the four-cell stage is associated with developmental delay or arrest in human embryos (Wong et al., 2010) and with specification of left-right body asymmetry in C. elegans, snail, and frog embryos (Pohl and Bao, 2010; Naganathan et al., 2014; Davison et al., 2016). Thus, actomyosin regulation plays critical roles during the early development of Bilateria, and further studies of myosin flow regulation during cell division should contribute to our understanding of multicellular assembly processes in diverse organisms.
In a broader perspective, our results indicate that different patterns of cell contact between cells of distinct sizes and fates arising during embryogenesis function as unique physical and chemical cues to control cell division and produce diverse axisorientations. Indeed, C. elegans is remarkable in having an invariant lineage of division axis orientation (Sulston et al., 1983), and reproducible patterns of cell contact cues, along with cell shape cues, may help to explain how this remarkable invariance is achieved. By examining the influence of cell-cell contact on the orientations of unexplored cell divisions, future studies may uncover new patterns of cell contact cues, and determine if sequences of cell contact patterns regulate microtubule and myosin-dependent force generation systems to specify the assembly of multicellular architecture throughout development.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kenji Sugioka (sugioka@zoology.ubc.ca).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mus musculus (mouse) and Caenorhabditis elegans strains were used in this study. To obtain mouse embryos, female C57BL/6J in 3-12 week old were superovulated by intraperitoneal injections of 5 international unit (IU) of Pregnant Mare Serum Gonadotropin (PMSG) followed by 5 IU of Human Chorionic Gonadotropin (hCG) 48 hours later. Each female was then placed with a C57BL/6J or B6.Cg-Tg(HIST1H2BB/EGFP)1Pa/J male (The Jackson Laboratory) overnight and all females were checked for a copulation plug the following morning. Two-cell stage embryos were collected by flushing oviducts with FHM medium and cultured in EmbryoMax Advanced KSOM medium (EMD Millipore) under 37°C, 5% CO2, and 5% O2. The experiments have been approved by the University of Oregon Animal Care and Use Committee (IACUC). The housing and husbandry of animals are University of Oregon standard methods and complied with the guideline of the IACUC.
All C. elegans strains except for temperature sensitive mutants were cultured at 25°C as described (Brenner, 1974). zen-4(or153ts) temperature sensitive mutants were cultured at 15°C before experiments. The following transgenes were used: ddIs299 (YFP::SPD-5, centrosome marker, a gift from Tony Hyman), itIs37 (mCherry::histone H2B), zuIs45 (NMY-2::GFP, non-muscle myosin II), cp13[nmy-2::GFP + LoxP] (non-muscle myosin II), orSi1[mex-5p::GFP:mlc-4WT::tbb-2 3’UTR], and orSi3[mex-5p::GFP::mlc-4AA::tbb-2 3UTR]. orSi1 and orSi3 were made by CRISPR/Cas9 as described later. Some strains carry the viable α-tubulin and β-tubulin gene mutations tba-2(sb25) and tbb-2(sb26) (Lu and Mains, 2005), respectively (gifts from Paul Mains), to suppress cul-3(RNAi) defects in early stage (see Figure S7).
METHOD DETAILS
Suppression of Early Cul-3 Knockdown Defects Caused by MEI-1
The CUL-3/Cullin 3 E3 ubiquitin ligase is required for the post-meiotic degradation of the meiosis-specific microtubule severing protein MEI-1/katanin. cul-3(RNAi) results in a failure to degrade MEI-1/katanin after the completion of oocyte meiosis and hence abnormal mitotic spindles with short microtubules and associated early embryonic cell division defects, and embryonic lethality (Figure S7A) (Kurz et al., 2002; Pintard et al., 2003). However, the tubulin suppressor mutations tba-2(sb25) and tbb-2(sb26) almost fully rescue the lethality and cell division defects of the dominant mei-1 mutation ct46, which encodes a degradation-defective form of MEI-1 (Figures S7A and S7B) (Lu and Mains, 2005). tba-2(sb25) and tbb-2(sb26) confer resistance to MEI-1-dependent microtubule severing without affecting normal mitosis (Lu and Mains, 2005). We therefore used cul-3(RNAi) in a tba-2(sb25);tbb-2(sb26) mutant background to suppress early cul-3(RNAi) defects caused by ectopic MEI-1 function. Importantly, we confirmed that AB cleavage furrow orientation was normal in mei-1(ct46); tba-2(sb25);tbb-2(sb26) mutants, indicating that AB spindle orientation is regulated by the CUL-3 E3 ligase dependent degradation of unknown substrate(s) other than MEI-1 (Figure S7C).
C. elegans RNAi
Feeding RNAi was performed for control, cul-3, gpr-1/-2, mlc-4 and perm-1 at 25°C. For control RNAi, a bacterial strain with empty vector (L4440) was used. For control and gpr-1/-2 RNAi, L2 larvae were grown on the feeding RNAi plates and embryos were analyzed. For cul-3 RNAi, L1 larvae were grown on the feeding RNAi plates and sterile F1 adults were crossed with cul-3(RNAi) males to obtain embryos for analysis. The sterile F1 adult worms have oocytes that appear normal but abnormally large sperm that probably failed to fully differentiate. For perm-1 and mlc-4 RNAi, L4 larvae were grown on the feeding RNAi plates and allowed to grow for 10-14 hrs and 22-24hrs for perm-1 and mlc-4, respectively.
Generation of Mlc-4/RLC Phosphorylation Deficient Strain
We first designed the gBlock synthetic DNA oligos that encode mlc-4 cDNA either with wild-type Ser 17 Thr 18 or mutant Ser17A Thr18A fused with GFP at the N-terminus. Codon usage of mlc-4 cDNA was modified by the C. elegans Codon Adapter program so that it is resistant to the RNAi against original gene (Redemann et al., 2011). Also, the codon adaptation index that might affect transgene expression was adjusted to 0.36 for both transgenes (Redemann et al., 2011). Each transgene was inserted between the mex-5 promoter and the tbb-2 3′UTR of in house plasmid that contains ~1 kb homology arms around Chr I: 6504486 intergenic regions to make a repair template. Injection solution was prepared according to the previously reported method (Paix et al., 2015). Before injection, 40 μM crispr RNA (crRNA) targeting Chr I: 6504486 (gtcgcatcgctaaaaacgag), 10 μM dpy-10 crRNA, 50 μM transactivating crispr RNA (tracrRNA) were mixed and incubated at 95°C for 5 min and allowed to cool down at 22°C for 5 min. We then added 3.3 μg/μl Alt-R S.p. Cas9 protein with 3xNLS (Integrated DNA technology) and incubated at room temperature for 5 min and then added 30 ng/μl mlc-4 repair template and 3.75 μM dpy-10 single stranded DNA repair template to make an injection solution. Young adults were injected with the injection solution and progeny of animals that carry dpy-10 repair (Dumpy phenotype) were transferred to new plates 3–4 days post injection and screened for GFP fluorescence using a compound fluorescence microscope.
Polystyrene Beads Preparation
10 mg carboxyl-modified polystyrene beads of 30 μm diameter (KISKER BIOTECH GmbH &Co.) and of 20 μm, 10 μm, and 0.35 μm diameter (Polysciences) were washed twice with 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 6.5) and incubated with the 1 mL MES buffer containing 10 mg 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) for 15 minutes at room temperature. The beads were washed twice with phosphate buffered saline (PBS) and incubated with the 0.5 mL PBS containing 0.05 μg Rhodamine Red-X succinimidyl ester (ThermoFisher Scientific) for 5 minutes. The beads were washed twice and stored with PBS at 4°C before use. Cells adhere to the beads probably due to the interactions between carboxyl-modified bead and extracellular proteins or the electrostatic interactions between the negative charge of plasma membrane and positive charge of Rhodamine.
Blastomere Isolation
For C. elegans embryos, embryonic cells were isolated as described before (Edgar, 1995; Park and Priess, 2003), with the following modifications. After cutting the adult worms in egg salt buffer, embryos were placed in the freshly made hypochlorite solution [75% Clorox (Clorox) and 2.5 N KOH] for 50 seconds. After washing with Shelton’s growth medium twice (Shelton and Bowerman, 1996), embryos were put into Shelton’s medium on the coverslip with metallic holds. Eggshell and permeabilization barrier were removed by repeated mouth pipetting with hand-drawn glass microcapillary tubes (10 microliters, Kimble Glass Inc.) to obtain eggshell and permeabilization barrier-free embryos. We call these embryos eggshell-free embryos for simplicity. For blastomere isolation, 2-cell stage eggshell-free embryos were further drawn to separate the two cells. ABx, ABxx, EMS and P2 cells were isolated by separating daughter cells after each cell division.
Two-cell stage mouse embryos were briefly placed in M2 medium containing Tyrode’s acid (Sigma-Aldrich) to remove the zona-pellucida. They were used as zona-free embryos or further separated by repeated mouth pipetting with hand-drawn glass microcapillary tubes to obtain individual blastomeres.
Cell-Beads Interaction Assay
Cells isolated prior to prometaphase were attached to either one or two polystyrene beads before imaging. For 1 cell-2 bead assays, 2 beads were combined initially and then attached to the isolated cell. For surface particle tracking, cells were first coated with 0.35 μm beads and then attached to a 30 μm bead. All manipulations were done with a mouth pipette. For zen-4 temperature sensitive mutants, isolated AB cells were prepared at 15°C and incubated at 25°C using a temperature controlled stage and a Leica DMi8 microscope (Leica) described below.
Drug Treatments
For the drug treatments of C. elegans embryos surrounded by the eggshell and permeability barrier (intact embryos), we performed perm-1 RNAi to permeabilize embryos for drug treatment as previously described (Carvalho et al., 2011). Early 2-cell stage embryos were transferred to Shelton’s growth medium containing 2% DMSO (control), 20 μM Latrunculin A (Sigma-Aldrich), 200 μM ML-7, 12.5 ng/mL Nocodazole (Sigma-Aldrich) or 20 μg/ml Nocodazole.
For drug treatments of isolated C. elegans blastomeres, Shelton’s growth medium containing 200 μM ML-7 (Sigma-Aldrich) or 20 μg/ml Nocodazole (Sigma-Aldrich) were used.
For drug treatments of mouse blastomeres, EmbryoMax Advanced KSOM medium containing 50 μM ML-7 was used.
Specificity of ML-7
Although ML-7 is an inhibitor of myosin light chain kinase that disrupted AB division orientation and cytokinesis in our experiments, there is a report claiming that C. elegans myosin light chain kinases are not essential for cytokinesis, casting the doubt for the specificity of ML-7 inhibitor (Batchelder et al., 2007). However, our data confirm the role of myosin RLC phosphorylation in cell division orientation. First, ML-7 treatment resulted in the loss of myosin foci without affecting cell cycle progression (Figures 3D and S3), which is consistent with studies of mammalian epithelial cell division (Murthy and Wadsworth, 2005), and with the biochemical requirement of RLC phosphorylation for myosin filament assembly (Scholey et al., 1980). Second, expression of an RNAi-resistant MLC-4/RLC with Ser17A Thr18A phospho-deficient mutations, after partial RNAi knockdown of endogenous MLC-4, resulted in the abnormal AB cell division orientation (Figure 4D). About 1/3 of these partial RNAi knockdown embryos were defective in the first embryonic mitosis; we analyzed AB cell division orientation in those embryos that completed the first embryonic cytokinesis. Third, C. elegans has seven myosin light chain kinase homologs, and a deletion mutation of one homolog, ttn-1, exhibited sterile phenotype (n = 50), which phenocopied mlc-4(RNAi) (n = 50). Previous report used RNAi to knock-down ttn-1 and found no defects (Batchelder et al., 2007), but in our experiment ttn-1(RNAi) animals were completely viable and did not phenocopy ttn-1 deletion mutants (n > 100), suggesting that RNAi was not working in the previous report. Thus, while myosin light chain kinase actively regulates oriented division at the contact site remains unknown, our results indicate that the contact-dependent control of RLC-phosphorylation is critical for AB cell division orientation.
Live Imaging
For worm cells with eggshell removed or permeabilized, metallic slides with a hole in the center were sandwiched between two coverslips, and the cells were mounted on one of the coverslips with Shelton’s medium to avoid compression and dehydration. For other C. elegans experiments, embryos were mounted on 4% agar pads on glass slides and sealed with petroleum jelly (Vaselline) after overlaying with a coverslip. For mouse cells, embryos in a drop of EmbryoMax Advanced KSOM medium, covered with mineral oil on a cell culture dish with a glass bottom (FluoroDish, World Precision Instruments), were used for the imaging. For worm DIC videos, imaging was performed with generic CCD cameras mounted on either AxioPlan or AxioSkop compound microscopes (Zeiss). For worm fluorescence imaging, a confocal unit CSU10 (Yokogawa electric) and an EMCCD camera Image EM (Hamamatsu photonics) mounted on an inverted microscope Leica DMI 4000 (Leica), or a confocal unit CSU-W with Borealis (Andor) and an EMCCD camera iXon Ultra 897 (Andor) mounted on an inverted microscope Leica DMi8 (Leica), were used. Both imaging systems were controlled by Metamorph (Molecular Devices). Samples were illuminated by diode-pumped lasers with 488 nm and 561nm wavelength. NMY-2::GFP, YFP::SPD-5 and mCherry::histone H2B signals were obtained every 15 sec with 2 μm Z spacing for most experiments. NMY-2::GFP, mCherry::histone H2B and 0.35 μm particle signals were obtained every 5 sec with 1.5 μm Z spacing for the tracking of cortical movements in cell-bead experiments. NMY-2::GFP, mCherry::histone H2B signals were obtained every 10 sec with 1.5 μm Z spacing for the tracking of cortical NMY-2 movements in intact embryos. For the live-imaging of mouse embryo, Nikon Eclipse Ti inverted fluorescence microscope (Nikon) equipped with Live Cell stage top incubation system (Pathology Devices) controlled by NIS Elements Advanced Research software was used. Embryos were maintained at 37°C, 5% CO2, and 5% O2 condition.
Immunofluorescence
Mouse or C. elegans embryos were fixed with 4% Paraformaldehyde in PBS for 15 mins. Fixed embryos were washed three times with PBS containing 1% Tween-20. Embryos were then incubated with anti-phospho-myosin light chain (Ser19) antibody (1:50 dilution, #3671, Cell Signaling Technology) in PBS containing 1% Bovine Serum Albumin (BSA) and 0.1% Tween-20 at 4°C for overnight. After washing three times, samples were incubated with 1:500 anti-rabbit Rhodamine Red-X (Jackson ImmunoResearch) for 2hrs at room temperatures. Mouse embryos were imaged at 0.5 μm Z spacing while those of worms were at 0.25 μm.
Image Analysis
All images were analyzed with Fiji (Schindelin et al., 2012). Division axis orientations were measured in two ways. First, cleavage furrow orientations were measured relative to the cell or bead contact plane using myosin signal and furrow formation as a marker. Second, mitotic spindle orientations were measured relative to the cell or bead contact plane using either fluorescent or DIC image of centrosome. D-V axes were measured from the angle between (i) the line that passes the centroids of ABp and EMS nuclei at 4-cell stage and (ii) the A-P axis, using DIC videos. Except DIC videos, division axis orientations relative to the plane of cell-bead or cell-cell contact were measured using the 3D reconstructed fluorescence images. All angular data were analyzed with PAST software (Hammer et al., 2001). For the quantification of myosin foci movements in the cell, cell position and orientation were corrected by the image J plug-in StackReg, to eliminate the movements of myosin foci caused by the cell movements or rotation. Supplemental Videos were made with Adobe Premiere Elements 15 (Adobe) or Imaris (Bitplane).
QUANTIFICATION AND STATISTICAL ANALYSIS
Error bars indicate the mean ± 95% confidence interval. Statistical methods were described in the Figure legends. Statistical analysis was performed using PAST software, Microsoft Excel (Microsoft) and Prism 6 (GraphPad Software). *P < 0.05, **P < 0.01, ***P < 0.0001; n.s., not significant (P > 0.05).
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-myosin light chain 2 (Ser19) antibody (rabbit) | Cell Signaling Technology | #3671; RRID: AB_330248 |
| Anti-rabbit Rhodamine Red-X | Jackson ImmunoResearch | 111-295-003; RRID: AB_2338022 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Carboxyl-modified polystyrene beads (30 μm) | Kisker Biotech GmbH & Co. | PPS-30.0COOHP |
| Carboxyl-modified polystyrene beads (20 μm) | Polysciences, Inc. | 24811-2 |
| Carboxyl-modified polystyrene beads (0.35 μm) | Polysciences, Inc. | 21753-15 |
| 2-(N-morpholino)ethanesulfonic acid (MES) | Sigma-Aldrich | M8250 |
| 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDAC) | Sigma-Aldrich | E1769 |
| Rhodamine Red-X succinimidyl ester | ThermoFisher Scientific | R6160 |
| Clorox | Clorox | N/A |
| Microcapillary tubes (10 μL) | Kimble Glass Inc. | 71900-10 |
| ML-7 | Sigma-Aldrich | I2764 |
| Cytochalasin D | Sigma-Aldrich | C8273 |
| Latrunculin A | Sigma-Aldrich | L5163 |
| Experimental Models: Organisms/Strains | ||
| C. elegans: Strain TH102: unc-119(ed3)III; ddIs299[pie-1p::YFP::F54C4.3; unc-119(+)] | Laboratory of Tony Hyman | N/A |
| C. elegans: Strain EU2988: tba-2(sb25)I; tbb-2(sb26)III; itIs37[pie-1p::mCherry::his-58 + unc-119(+)]IV; zuIs45[nmy-2p::NMY-2::GFP + unc-119(+)]V; ddIs299[pie-1p::YFP::SPD-5 + unc-119(+)] | This paper | N/A |
| C. elegans: Strain EU2987: tba-2(sb25)I; tbb-2(sb26)III; itIs37[pie-1p::mCherry::his-58 + unc-119(+)]IV; zuIs45[nmy-2p::NMY-2::GFP + unc-119(+)]V | This paper | N/A |
| C. elegans: Strain HR1044: mei-1(ct46) unc-29(e1072) tba-2(sb25)I; tbb-2(sb26)III | Laboratory of Paul Mains | N/A |
| C. elegans: Strain EU3028: zen-4(or153ts)IV; [nmy-2p::NMY-2::GFP + unc-119(+)]V; ddIs299[pie-1p::YFP::SPD-5 + unc-119(+)] | This paper | N/A |
| C. elegans : Strain LP162: nmy-2(cp13[nmy-2::GFP + LoxP]) I | Caenorhabditis Genetics Center | LP162 |
| C. elegans: Strain EU3131: orSi2[mex-5p::GFP::mlc-4(S17A T18A; RNAi resistant)::tbb-2 3′UTR] I; itIs37 | This paper | |
| C. elegans: Strain EU3130: orSi1[mex-5p::GFP::mlc-4(WT; RNAi resistant)::tbb-2 3′UTR] I; itIs37 | This paper | |
| Mus musculus: Strain C57BL/6J | The Jackson Laboratory | 000664 |
| Mus musculus: Strain B6.Cg-Tg(HIST1H2BB/EGFP)1Pa/J | The Jackson Laboratory | 006069 |
| Oligonucleotides | ||
| Forward primer for cul-3(RNAi): ACCGGCAGATCTGATATCATCGATGAATTCTTAGCAAATTTTGGCGCTTTtccg | This paper | N/A |
| Reverse primer for cul-3(RNAi): CCGGTACTGTCACCGAATCT | This paper | N/A |
| Forward primer for gpr-1/-2(RNAi): GAACGAGGAACCCGTAGACA | This paper | N/A |
| Reverse primer for gpr-1/-2(RNAi): CGACTGACAGCCTGAAAACA | This paper | N/A |
| Synthetic oligo for mlc-4(RNAi) (inserted into L4440 feeding RNAi vector): ACCGGCAGATCTGATATCATCGATGAATTCGCCTCCCGCAAAACCGTAAACCGCCGTCAACGGCCACAAAGAGCCACTTCCAATGTGTTCGCCATGTTCGATCAGGCTCAAATTCAAGAGTTCAAAGAGGCATTCAACATGATCGACCAAAACCGCGATGGATTCATCGATCAAGAGGATCTGAAGGATATGTTCGCTTCCCTTGGAAAAGAAGTCACCGAACAATTCATCGATTCGATGATTAACGAGGCTCCCGGAGCACAGCCGATCAATTTCACAATGTTCCTGACACTTTTCGGTGAAAAATTGACTGGAACGGATCCGGAGGAAGTCATACGGAATGCGTTCCAATGCTTCGATGAGGATAATTCGGGAAAACTCAATGAGGAGCATCTTCGCGAGTTGCTCACCACGATGGGCGAGAGATATTCCGAGGAACAGGTTGATGAGCTCTTTAGAGATGCTCCGATTAAGGGCGGACAATTCGACTACGTGGAGTTCACTCGTATGCTCAAACACGGAACCAAGGACAAGGATGAGGCTTAAGGAATTCGATATCAAGCTTATCGATACCGT | This paper | N/A |
| Crispr RNA (crRNA) targeting ChrI: 6504486: gtcgcatcgctaaaaacgag | This paper | N/A |
| Crispr RNA (crRNA) targeting dpy-10: GCTACCATAGGCACCACGAG | Paix et al., 2015 | N/A |
| Transactivating crispr RNA (tracrRNA) | IDT | 1072532 |
| Cas9 Nuclease 3NLS | IDT | 1074181 |
| Software and Algorithms | ||
| Metamorph | Molecular Devices | N/A |
| Fiji | Schindelin et al., 2012 | http://fiji.sc |
| PAST software | Hammer et al., 2001 | http://folk.uio.no/ohammer/past/ |
| Prism 6 | Graphpad Software | N/A |
| Imaris | Bitplane | N/A |
| Adobe Premiere Elements 15 | Adobe | N/A |
| Microsoft Excel | Microsoft | N/A |
| Other | ||
| Feeding RNAi E. coli for perm-1(RNAi) | Source BioScience | II-5J22 |
Highlights.
Cortical myosin flow is a force generation mechanism that orients cell division
Three contact-dependent cues specify distinct patterns of cell division orientation
Contact-dependent oriented division regulates C. elegans and mouse embryogenesis
ACKNOWLEDGMENTS
We thank Ute Hostick and the University of Oregon transgenic mouse facility for providing mouse embryos; Paul Mains, Tony Hyman, Chien-Hui Chuang, and the Caenorhabditis Genetics Center (funded by the NIH Office of Research Infrastructure Programs; P40 OD010440) for C. elegans strains; Akatsuki Kimura for helpful discussion; Kryn Stankunas, Diana Libuda, and Chris Doe for sharing laboratory equipment; and Chris Doe for critical reading of the manuscript. This work was supported by NIH grant R01GM049869 to B.B. and Human Frontier Science Program (LT000345/2012-L) to K.S.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and seven videos and can be found with this article online at https://doi.org/10.1016/j.devcel.2018.06.020.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Batchelder EL, Thomas-Virnig CL, Hardin JD, and White JG (2007). Cytokinesis is not controlled by calmodulin or myosin light chain kinase in the Caenorhabditis elegans early embryo. FEBS Lett. 581, 4337–4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann F, Bauer MS, Rees M, Alexandrovich A, Gautel M, Pippig DA, and Gaub HE (2017). Increasing evidence of mechanical force as a functional regulator in smooth muscle myosin light chain kinase. Elife 6, e26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodri D, Kato R, Kondo M, Hosomi N, Katsumata Y, Kawachiya S, and Matsumoto T (2015). Time-lapse monitoring of zona pellucida-free embryos obtained through in vitro fertilization: a retrospective case series. Fertil. Steril. 103, e35. [DOI] [PubMed] [Google Scholar]
- Bosveld F, Markova O, Guirao B, Martin C, Wang Z, Pierre A, Balakireva M, Gaugue I, Ainslie A, Christophorou N, et al. (2016). Epithelial tricellular junctions act as interphase cell shape sensors to orient mitosis. Nature 530, 495–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D, and White JG (1988). Cortical flow in animal cells. Science 239, 883–888. [DOI] [PubMed] [Google Scholar]
- Brenner S (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabernard C, Prehoda KE, and Doe CQ (2010). A spindle-independent cleavage furrow positioning pathway. Nature 467, 91–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campinho P, Behrndt M, Ranft J, Risler T, Minc N, and Heisenberg CP (2013). Tension-oriented cell divisions limit anisotropic tissue tension in epithelial spreading during zebrafish epiboly. Nat. Cell Biol. 15, 1405–1411. [DOI] [PubMed] [Google Scholar]
- Capitanio M, Canepari M, Maffei M, Beneventi D, Monico C, Vanzi F, Bottinelli R, and Pavone FS (2012). Ultrafast force-clamp spectroscopy of single molecules reveals load dependence of myosin working stroke. Nat. Methods 9, 1013–1019. [DOI] [PubMed] [Google Scholar]
- Carvalho A, Olson SK, Gutierrez E, Zhang K, Noble LB, Zanin E, Desai A, Groisman A, and Oegema K (2011). Acute drug treatment in the early C. elegans embryo. PLoS One 6, e24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, and Waterman CM (2015). Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat. Cell Biol. 17, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauffman G, Verheyen G, Tournaye H, and Van de Velde H (2014). Developmental capacity and pregnancy rate of tetrahedral- versus non-tetrahedral-shaped 4-cell stage human embryos. J. Assist. Reprod. Genet. 31, 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A, McDowell GS, Holden JM, Johnson HF, Koutsovoulos GD, Liu MM, Hulpiau P, Van Roy F, Wade CM, Banerjee R, et al. (2016). Formin is associated with left-right asymmetry in the pond snail and the frog. Curr. Biol. 26, 654–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Pietro F, Echard A, and Morin X (2016). Regulation of mitotic spindle orientation: an integrated view. EMBO Rep. 17, 1106–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner T, Maurer M, Shebl O, Moser M, Mayer RB, Duba HC, and Tews G (2012). Planar embryos have poor prognosis in terms of blastocyst formation and implantation. Reprod. Biomed. Online 25, 267–272. [DOI] [PubMed] [Google Scholar]
- Edgar LG (1995). Blastomere culture and analysis. Methods Cell Biol. 48, 303–321. [DOI] [PubMed] [Google Scholar]
- Fink J, Carpi N, Betz T, Bétard A, Chebah M, Azioune A, Bornens M, Sykes C, Fetler L, Cuvelier D, and Piel M (2011). External forces control mitotic spindle positioning. Nat. Cell Biol. 13, 771–778. [DOI] [PubMed] [Google Scholar]
- Gillies TE, and Cabernard C (2011). Cell division orientation in animals. Curr. Biol. 21, R599–R609. [DOI] [PubMed] [Google Scholar]
- Goolam M, Scialdone A, Graham SJ, Macaulay IC, Jedrusik A, Hupalowska A, Voet T, Marioni JC, and Zernicka-Goetz M (2016). Heterogeneity in Oct4 and Sox2 targets biases cell fate in 4-cell mouse embryos. Cell 165, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein B (1992). Induction of gut in Caenorhabditis elegans embryos. Nature 357, 255–257. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Takeshita H, Mizumoto K, and Sawa H (2006). Wnt signals can function as positional cues in establishing cell polarity. Dev. Cell 10, 391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CF, and Deussen ZA (1978). Features of cell lineage in preimplantation mouse development. J. Embryol. Exp. Morphol. 48, 53–72. [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, and Ryan PD (2001). PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9. [Google Scholar]
- Heppert JK, Pani AM, Roberts AM, Dickinson DJ, and Goldstein B (2018). A CRISPR tagging-based screen reveals localized players in Wntdirected asymmetric cell division. Genetics 208, 1147–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly SJ, Mulnard JG, and Graham CF (1978). Cell division and cell allocation in early mouse development. J. Embryol. Exp. Morphol. 48, 37–51. [PubMed] [Google Scholar]
- Knoblich J (2010). Asymmetric cell division: recent developments and their implications for tumour biology. Nat. Rev. Mol. Cell Biol. 11, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Pintard L, Willis JH, Hamill DR, Gönczy P, Peter M, and Bowerman B (2002). Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295, 1294–1298. [DOI] [PubMed] [Google Scholar]
- Kwon M, Bagonis M, Danuser G, and Pellman D (2015). Direct microtubule-binding by myosin-10 orients centrosomes toward retraction fibers and subcortical actin clouds. Dev. Cell 34, 323–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, and Goldstein B (2003). Mechanisms of cell positioning during C. elegans gastrulation. Development 130, 307–320. [DOI] [PubMed] [Google Scholar]
- Levayer R, and Lecuit T (2012). Biomechanical regulation of contractility: spatial control and dynamics. Trends Cell Biol. 22, 61–81. [DOI] [PubMed] [Google Scholar]
- Liro MJ, and Rose LS (2016). Mitotic spindle positioning in the EMS cell of Caenorhabditis elegans requires LET-99 and LIN-5/NuMA. Genetics 204, 1177–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvet-Vallee S, Vinot S, and Maro B (2005). Mitotic spindles and cleavage planes are oriented randomly in the two-cell mouse embryo. Curr. Biol. 15, 464–469. [DOI] [PubMed] [Google Scholar]
- Lu C, and Mains PE (2005). Mutations of a redundant α-tubulin gene affect Caenorhabditis elegans early embryonic cleavage via MEI-1/katanin-dependent and -independent pathways. Genetics 170, 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maître JL, Niwayama R, Turlier H, Nédélec F, and Hiiragi T (2015). Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat. Cell Biol. 17, 849–855. [DOI] [PubMed] [Google Scholar]
- Mayer M, Depken M, Bois JS, Jülicher F, and Grill SW (2010). Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature 467, 617–621. [DOI] [PubMed] [Google Scholar]
- Minc N, Burgess D, and Chang F (2011). Influence of cell geometry on division-plane positioning. Cell 144, 414–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy K, and Wadsworth P (2005). Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 15, 724–731. [DOI] [PubMed] [Google Scholar]
- Naganathan SR, Furthauer S, Nishikawa M, Julicher F, and Grill SW (2014). Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. Elife 3, e04165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noatynska A, Gotta M, and Meraldi P (2012). Mitotic spindle (DIS) orientation and DISease: cause or consequence? J. Cell Biol. 199, 1025–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Stuurman N, D’Ambrosio M, and Vale RD (2010). Polarized myosin produces unequal-size daughters during asymmetric cell division. Science 330, 677–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, and Seydoux G (2015). High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park FD, and Priess JR (2003). Establishment of POP-1 asymmetry in early C. elegans embryos. Development 130, 3547–3556. [DOI] [PubMed] [Google Scholar]
- Pease JC, and Tirnauer JS (2011). Mitotic spindle misorientation in cancer—out of alignment and into the fire. J. Cell Sci. 124, 1007–1016. [DOI] [PubMed] [Google Scholar]
- Pierre A, Sallé J, Wühr M, and Minc N (2016). Generic theoretical models to predict division patterns of cleaving embryos. Dev. Cell 39, 667–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Willis JH, Willems A, Johnson JL, Srayko N, Kurz T, Glaser S, Mains PE, Tyers M, Bowerman B, and Peter M (2003). The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425, 311–316. [DOI] [PubMed] [Google Scholar]
- Pohl C, and Bao Z (2010). Chiral forces organize left-right patterning in C. elegans by uncoupling midline and anteroposterior axis. Dev. Cell 19, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulson ND, and Lechler T (2012). Asymmetric cell divisions in the epidermis. Int. Rev. Cell Mol. Biol. 295, 199–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priess JR, and Thomson JN (1987). Cellular interactions in early C. elegans embryos. Cell 48, 241–250. [DOI] [PubMed] [Google Scholar]
- Priess JR (2005). Notch signaling in the C. elegans embryo. In WormBook, The C. elegans Research Community., ed. (WormBook) 10.1895/wormbook.1.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redemann S, Schloissnig S, Ernst S, Pozniakowsky A, Ayloo S, Hyman AA, and Bringmann H (2011). Codon adaptation–based control of protein expression in C. elegans. Nat. Methods 8, 250–252. [DOI] [PubMed] [Google Scholar]
- Reymann AC, Staniscia F, Erzberger A, Salbreux G, and Grill SW (2016). Cortical flow aligns actin filaments to form a furrow. Elife 5, e17807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau CE, Downs WD, Lin R, Wittmann C, Bei Y, Cha YH, Ali M, Priess JR, and Mello CC (1997). Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90, 707–716. [DOI] [PubMed] [Google Scholar]
- Roh-Johnson M, Shemer G, Higgins CD, McClellan JH, Werts AD, Tulu US, Gao L, Betzig E, Kiehart DP, and Goldstein B (2012). Triggering a cell shape change by exploiting preexisting actomyosin contractions. Science 335, 1232–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh M, Ishikawa T, Matsushima S, Naka M, and Hidaka H (1987). Selective inhibition of catalytic activity of smooth muscle myosin light chain kinase. J. Biol. Chem. 262, 7796–7801. [PubMed] [Google Scholar]
- Sawa H, and Korswagen HC (2013). Wnt signaling in C. elegans. In WormBook, The C. elegans Research Community., ed. (WormBook) 10.1895/wormbook.1.7.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger A, Shelton C, Maloof J, Meneghini M, and Bowerman B (1999). Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev. 13, 2028–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey JM, Taylor KA, and Kendrick-Jones J (1980). Regulation of non-muscle myosin assembly by calmodulin-dependent light chain kinase. Nature 287, 233–235. [DOI] [PubMed] [Google Scholar]
- Schonegg S, Constantinescu AT, Hoege C, and Hyman AA (2007). The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc. Natl. Acad. Sci. USA 104, 14976–14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin L, Poulson ND, Foote HP, and Lechler T (2013). NuMA localization, stability, and function in spindle orientation involve 4.1 and Cdk1 interactions. Mol. Biol. Cell 24, 3651–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton CA, and Bowerman B (1996). Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development 122, 2043–2050. [DOI] [PubMed] [Google Scholar]
- Singh D, and Pohl C (2014). Coupling of rotational cortical flow, asymmetric midbody positioning, and spindle rotation mediates dorsoventral axis formation in C. elegans. Dev. Cell 28, 253–267. [DOI] [PubMed] [Google Scholar]
- Sokol SY (2015). Spatial and temporal aspects of Wnt signaling and planar cell polarity during vertebrate embryonic development. Semin. Cell Dev. Biol. 42, 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, and van den Heuvel S (2003). A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C. elegans. Genes Dev. 17, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, and Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, and Thomson JN (1983). The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Togashi M, Adachi J, and Toyoda Y (1995). Developmental ability of zona-free mouse embryos is influenced by cell association at the 4-cell stage. Biol. Reprod. 53, 78–83. [DOI] [PubMed] [Google Scholar]
- Thorpe CJ, Schlesinger A, Carter JC, and Bowerman B (1997). Wnt signaling polarizes an early C. elegans blastomere to distinguish endoderm from mesoderm. Cell 90, 695–705. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, and Zernicka-Goetz M (2007). Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature 445, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou MFB, Hayashi A, and Rose LS (2003). LET-99 opposes Gα/GPR signaling to generate asymmetry for spindle positioning in response to PAR and MES-1/SRC-1 signaling. Development 130, 5717–5730. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, and Horwitz AR (2009). Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E, Bruno L, Bissiere S, Levi V, and Plachta N (2016). Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 165, 75–87. [DOI] [PubMed] [Google Scholar]
- Williams SE, and Fuchs E (2013). Oriented divisions, fate decisions. Curr. Opin. Cell Biol. 25, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Loewke KE, Bossert NL, Behr B, De Jonge CJ, Baer TM, and Pera RAR (2010). Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 28, 1115–1121. [DOI] [PubMed] [Google Scholar]
- Zonies S, Motegi F, Hao Y, and Seydoux G (2010). Symmetry breaking and polarization of the C. elegans zygote by the polarity protein PAR-2. Development 137, 1669–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


