Abstract
Background
Abnormal peripheral immunological features are associated with the progression of coronavirus disease 2019 (COVID-19).
Methods
Clinical and laboratory data were retrieved in a cohort of 146 laboratory-confirmed COVID-19 patients. Potential risk factors for the development of severe COVID-19 were evaluated.
Results
On admission, lymphocytes, CD3+, CD4+ and CD8+ T cells, eosinophils, and albumin and pre-albumin were dramatically lower, whereas neutrophils, and interleukin (IL)-10, C-reactive protein (CRP), aspartate aminotransferase (AST) and gamma-glutamyltransferase (GGT) were significantly higher in severe cases. By the second week after discharge, all variables improved to normal levels. Covariate logistic regression results showed that the CD8+ cell count and CRP level were independent risk factors for severe COVID-19.
Conclusion
Lower peripheral immune cell subsets in patients with severe disease recovered to normal levels as early as the second week after discharge. CD8+ T cell counts and CRP levels on admission are independent predictive factors for severe COVID-19.
Keywords: COVID-19, Immune cells, Severity, Predictive factor, Follow-up
1. Introduction
2019 novel coronavirus disease (COVID-19) has become a worldwide pandemic since its outbreak in December 2019. Unfortunately, both the confirmed cases and deaths related to COVID-19 continue to increase rapidly globally [1].
Several epidemiological features of the human-to-human transmission and clinical characteristics of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been reported. Phylogenetic analyses have revealed that SARS-CoV-2 is closely associated at 88%, 79%, and 50% identity with the bat-SL-CoVZXC21 and bat-SL-CoVZC45 SARS-like coronaviruses, SARS-CoV, and MERS-CoV, respectively [2]. The disease onset frequently manifests with cough and fever. In a cohort of 1099 patients with COVID-19, only 43.8% (473/1081) presented with fever which was much lower than seen in patients infected with SARS-CoV and MERS-CoV [3]. More importantly, confirmed asymptomatic patients with COVID-19 had similar viral transmission dynamics, in terms of the viral load, as the symptomatic patients [[4], [5], [6]]. Consequently, asymptomatic patients with COVID-19 may complicate the prevention, diagnosis, and control of this disease [7]. Clinical laboratory findings such as lymphopenia, lower counts of of CD4+ T and CD19+ B lymphocytes, and increased lactate dehydrogenase and C-reactive protein (CRP) levels are commonly observed in patients with COVID-19, and cases with advanced age, coexisting disorders, or dyspnoea were associated with the severity of the disease [[8], [9], [10]].
A follow-up study on the dynamics of the clinical laboratory features in patients with COVID-19 has not been carried out and the potential predictive factors for the severity of this disease remain unknown. In this study, we retrospectively assessed the clinical and laboratory data collected during and post-hospitalisation in patients diagnosed with COVID-19 at varying degrees of severity, to identify potential early predictive factors for the diagnosis of severe COVID-19 in affected patients.
2. Patients and methods
2.1. Patients and data collection
From January 19th, 2020 to March 11th, 2020, 146 laboratory-confirmed patients with COVID-19 (106 ordinary and 40 severe cases) were admitted to the Taizhou Hospital of Zhejiang Province and Taizhou Enze Hospital, Taizhou EnZe Medical Group (Center), the only officially designated medical center for COVID-19 in Taizhou City, Zhejiang, China. No death occurred and all cases were discharged by March 11th.
The medical histories of patients were reviewed, and the clinical classifications of COVID-19 were based on the 7th version of the Diagnosis and Treatment Guidance of Corona Virus Diseases 2019, National Health Commission (NHC), and National Administration of Traditional Chinese Medicine of the People's Republic of China [11]. Briefly, ordinary cases were categorised based on the presentation of fever, respiratory tract symptoms, and pneumonia in imaging results. Severe cases for adults were those with respiratory distress with a respiratory rate ≥ 30/min, or pulse oxygen saturation (SpO2) ≤ 93% at rest, or arterial blood oxygen partial pressure (PaO2)/oxygen concentration (FiO2) ≤300 mmHg (1 mmHg = 0.133 kPa), or with >50% pulmonary lesion progression within 24–48 h. Critical cases included respiratory failure requiring mechanical ventilation, shock, or complications of other organ failures.
Patients were clinically followed-up by the second and fourth week after discharge as recommended and were subjected to routine laboratory tests, including the real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for SARS-CoV-2 RNA screening. By May 11th, all available clinical laboratory data on the day of admission (145 patients), the day of discharge (115 out of 146 patients), and the second (1st follow-up, 113 out of 146 patients) and fourth week (2nd follow-up, 79 out of 146 patients) follow-ups after discharge were retrieved. After discharged, recovered COVID-19 patients were self-isolated in community quarantine facilities, and were subjected to RT-PCR re-testing during the second- (1st follow-up) and fourth-week (2nd follow-up) follow-ups as outlined by the guidelines [11].
The SARS-CoV-2 virus shedding duration was defined as the interval from the day of being confirmed positive for SARS-CoV-2 to the first day when SARS-CoV-2 testing returns a negative result (at least two consecutive negative RT-PCR results) during hospitalisation [12].
The protocol of this study was reviewed and approved by the Ethics Committee of Taizhou Hospital of Zhejiang Province (#K20200111), and written informed consent was obtained from all patients or guardians.
2.2. Statistical methods
General descriptive analyses of the variables were performed. Comparisons of variables were analysed using either t-tests or Mann–Whitney U tests for continuous variables. The χ2 test with Fisher's exact probability was performed for categorical variables. Covariate binary logistic regression analysis with the forward conditional method was performed for variables investigated as independent predictive factors for severe COVID-19. Receiver operating characteristics (ROC) curves for each variable were generated to evaluate the power of each variable in differentiating between severe and non-severe COVID-19. The cut-off value for the independent predictive factors was determined by Youden's index. Statistical analysis was performed using SPSS v.13.0 (SPSS, Inc., Chicago, IL, USA). A two-sided p value <0.05 was considered statistically significant.
3. Results
3.1. Patient clinical characteristics
Of the 146 patients, 77 were men and 69 were women, with a median age of 47 years (range, 4–86 years). A total of 76 (52.1%) patients were from Wuhan, and 70 (47.9%) were Taizhou residents.
Older patients were more commonly diagnosed with severe COVID-19 (median: 55.0 years vs. 45.0 years; p < 0.001). Body mass index values were significantly higher in severe cases (25.4 vs. 23.2; p = 0.001). Severe patients were more frequently accompanied by symptoms such as fever (p = 0.039), poor appetite (p = 0.009), and chest distress (p = 0.004) on admission, and had longer hospital stays (24 d vs. 17.5 d; p = 0.009), and viral shedding durations (15 d vs. 11 d; p < 0.001).
Among the 146 patients, following discharge, 13 (8.9%) patients without COVID-19 symptoms were re-admitted to the hospital after testing positive for SARS-CoV-2 RNA. 12 patients were confirmed to be SARS-CoV-2 repositivity by the 1st follow-up a one patients by the 2nd follow-up. However, no significant difference was observed in viral re-positivity after discharge between patients with severe and non-severe COVID-19 (p = 0.349). Details of the clinical characteristics of COVID-19 patients are shown in Table 1 .
Table 1.
Clinical characteristics of COVID-19 patients.
| Variables | All cases (n = 146) | Non-severe (n = 106) | Severe (n = 40) | p value |
|---|---|---|---|---|
| Gender (male/female) | 77/69 | 58/48 | 19/21 | 0.462 |
| Age (median, range) | 47 (4–86.) | 45 (4–81) | 55 (26–86) | <0.001 |
| Body Mass Index | 24.2 (16.0–31.3) | 23.2 (16.0–30.7) | 25.4 (19.8–31.3) | 0.001 |
| Wuhan Returned (yes/no) | 76/70 | 57/49 | 17/23 | 0.267 |
| On admission | ||||
| body temperature | 36.9 (36.0–39.0) | 36.9 (36.0–39.0) | 37.0 (36.0–38.7)) | 0.166 |
| respiratory rate/min | 19.0 (12.0–26.0) | 18.0 (16.0–22.0) | 19.0 (12.0–26.0) | 0.255 |
| heart rate/min | 83 (57–147) | 82 (57–115) | 84 (57–147) | 0.115 |
| systolic pressure (mmHg) | 127 (98–177) | 126 (101–166) | 129 (98–177) | 0.340 |
| diastolic pressure (mmHg) | 81 (59–110) | 81 (62–104) | 81 (59–110) | 0.589 |
| Symptoms (yes/no) | ||||
| fever | 105/41 | 71/35 | 34/6 | 0.039 |
| dry cough | 43/103 | 27/79 | 16/24 | 0.104 |
| fatigue | 37/109 | 29/77 | 8/32 | 0.402 |
| chills | 27/119 | 19/87 | 8/32 | 0.813 |
| sore throat | 22/124 | 14/92 | 8/32 | 0.310 |
| runny nose | 12/134 | 10/96 | 2/38 | 0.512 |
| sputum production | 47/99 | 33/73 | 14/26 | 0.694 |
| dizzy or headache | 28/118 | 18/88 | 10/30 | 0.346 |
| nausea or vomiting | 5/141 | 2/104 | 3/37 | 0.127 |
| myalgia | 10/136 | 5/101 | 5/35 | 0.137 |
| poor appetite | 47/99 | 27/79 | 20/20 | 0.009 |
| diarrhea | 14/132 | 7/99 | 7/33 | 0.060 |
| chest distress | 12/134 | 4/102 | 8/32 | 0.004 |
| Pre-existing disorders (yes/no) | 86/60 | 58/48 | 28/12 | 0.131 |
| chronic heart disease | 1/145 | 0/106 | 1/39 | / |
| diabetes | 18/128 | 9/97 | 9/31 | 0.044 |
| hypertension | 23/123 | 14/92 | 9/31 | 0.204 |
| chronic renal disease | 2 /144 | 1 /105 | 1/39 | 0.474 |
| cancer | 3/143 | 3/103 | 0/40 | / |
| chronic liver disease | 10/136 | 6/100 | 4/36 | 0.462 |
| HBV | 8/138 | 5/101 | 3/37 | 0.684 |
| HCV | 1/145 | 1/105 | 0/40 | / |
| chronic lung disease | 8/138 | 7/99 | 1/39 | 0.446 |
| other pre-existing disorders | 46/100 | 32/74 | 12/28 | 1.000 |
| Others | ||||
| hospital stay (days) | 20 (5–43) | 17.5 (5–43) | 24 (8–40) | <0.001 |
| virus shedding duration (days) | 12 (3–45) | 11 (3–43) | 15 (6–45) | 0.003 |
| hormone therapy | 47/99 | 12/94 | 35/5 | <0.001 |
| respiratory failure | 6/140 | 0/106 | 6/34 | / |
| shock occurs | 1/145 | 0/106 | 1/39 | / |
| viral re-positivity after discharge | 13/93 | 8/98 | 5/35 | 0.349 |
3.2. Laboratory dynamics across patients with differentially severe COVID-19
Detailed laboratory findings at different time points (admission, discharge, and follow-up at the second and fourth week) among COVID-19 patients, and comparison between severe and non-severe COVID-19 patients are shown in Table 2 .
Table 2.
Comparison of laboratory data between non-severe and severe COVID-19 patients on different time points.
| Laboratory test | Normal reference | Day on admission |
Day on discharge |
||||
|---|---|---|---|---|---|---|---|
| Non-severe(n = 105) | Severe (n = 40) | p value | Non-severe (n = 84) | Severe (n = 31) | p value | ||
| WBC (109/L) | 3.5–9.5 | 5.2 (2.6–23.6) | 6.1 (3.6–22.5) | 0.073 | 5.8 (3.1–11.8) | 5.5 (3.2–14.4) | 0.609 |
| Neutrophil (109/L) | 1.8–6.3 | 3.35 (1.2–22.2) | 4.75 (2.0–21.4) | 0.005 | 3.3 (1.5–8.5) | 3.4 (1.5–11.6) | 0.123 |
| Lymphocyte (109/L) | 1.1–3.2 | 1.3 (0.3–3.0) | 0.80 (0.3–2.4) | <0.001 | 1.65 (0.6–3.5) | 1.25 (0.7–2.7) | <0.001 |
| CD3+ T cell (per μL) | 770–2041 | 745 (137–2012) | 424 (111–1684) | <0.001 | 1055 (377–2081) | 989 (463–2049) | 0.421 |
| CD4+ T cell (per μL) | 414–1123 | 450 (86–1236) | 239 (68–1177) | 0.001 | 589 (199–1253) | 520 (236–1348) | 0.538 |
| CD8+ T cell (per μL) | 238–874 | 269 (44–806) | 160 (41–561) | <0.001 | 433 (168–983) | 393 (151–738) | 0.476 |
| CD19+ B cell (per μL) | 90–560 | 142 (62–552) | 129 (28–551) | 0.427 | 157 (48–495) | 154 (456–351) | 0.642 |
| CD56+ NK cell (per μL) | 150–1100 | 209 (44–771) | 152(63–377) | 0.026 | 251 (122–656) | 154 (68–596) | 0.085 |
| Mononuclear (109/L) | 0.1–0.6 | 0.40 (0.2–1.2) | 0.4 (0.1–0.8) | 0.507 | 0.50 (0.2–0.9) | 0.50 (0.2–1.3) | 0.236 |
| Eosinophil (109/L) | 0.02–0.52 | 0.02 (0–0.34) | 0.0 (0–0.28) | 0.026 | 0.09 (0–0.55) | 0.11 (0–1.43) | 0.142 |
| Basophil (109/L) | 0.00–0.06 | 0.02 (0–0.07) | 0.01 (0–0.06) | 0.419 | 0.02 (0–0.09) | 0.02 (0–0.08) | 0.707 |
| IL-2 (pg/mL) | 1.1–9.8 | 1.35 (0.19–10.3) | 1.19 (0.33–2.81) | 0.205 | 1.32 (0.29–2.81) | 1.51 (0.49–2.67) | 0.120 |
| IL-4 (pg/mL) | 0.1–3.0 | 1.45 (0.10–5.96) | 1.54 (0.17–8.53) | 0.526 | 1.53 (0.13–3.86) | 1.71 (0.43–11.71) | 0.127 |
| IL-6 (pg/mL) | 1.7–16.6 | 5.93 (0.78–414.0) | 13.9 (2.76–251.8) | 0.217 | 3.24 (1.08–348) | 4.45 (1.76–105) | 0.712 |
| IL-10 (pg/mL) | 2.6–4.9 | 3.52 (0.19–22.0) | 4.37 (1.54–39.5) | 0.003 | 2.59 (0.57–7.19) | 2.83 (0.89–6.1) | 0.341 |
| TNF-α (pg/mL) | 0.1–5.2 | 1.20 (0–5.32) | 1.17 (0.09–2.93) | 0.982 | 1.04 (0.18–4.97) | 0.83 (0.15–2.50) | 0.039 |
| IFN-γ (pg/mL) | 1.6–17.3 | 1.85 (0.18–178.9) | 2.04 (0.3–13.55) | 0.632 | 1.60 (0.15–179) | 1.35 (0.45–2.98) | 0.503 |
| IgG (g/L) | 7.0–16.0 | 12.7 (8.5–28.5) | 12.2 (7.7–27.8) | 0.507 | 12.1 (9.5–14.63) | 12.5 (11.4–15.5) | 0.158 |
| IgA (g/L) | 0.70–4.0 | 2.35 (0.57–5.26) | 2.37 (0.79–3.98) | 0.592 | 2.18 (1.12–5.31) | 2.50 (1.2–3.89) | 0.543 |
| IgM (g/L) | 0.40–2.30 | 1.06 (0.38–4.41) | 1.02 (0.33–2.75) | 0.828 | 1.21 (0.81–2.30) | 1.11 (0.74–1.53) | 0.302 |
| CRP (mg/L) | <0.5 | 6.0 (0.1–89.9) | 26.5 (0.60–185) | <0.001 | 1.60 (0.20–22.0) | 3.00 (0.5–66.6) | 0.237 |
| Alanine aminotransferase (U/L) | 7–40 | 20.0 (5.0–69.0) | 22.5 (6.0–152.0) | 0.236 | 25.0 (6.0–121.0) | 28.0 (9.0–98.0) | 0.054 |
| Aspartate aminotransferase (U/L) | 13–35 | 23.0 (11.0–57.0) | 28.0 (13.0–115.0) | 0.014 | 21.0 (12.0–68.0) | 22.0 (13.0–50.0) | 0.370 |
| Alkaline phosphatase (U/L) | 35–100 | 72.0 (35.0–376.0) | 68.0 (40.0–129.0) | 0.198 | 74.0 (40.0–356.0) | 73.0 (45.0–121.0) | 0.640 |
| gamma-glutamyltransferase (U/L) | 7–45 | 23.0 (10.0–109.0) | 32.0 (14.0–132.0) | 0.006 | 30.0 (11.0–293.0) | 48.0 (17.0–126.0) | <0.001 |
| Total bilirubin (mmol/L) | 5.0–21.0 | 12.7 (3.4–36.5) | 12.2 (5.1–36.4) | 0.862 | 12.3 (4.0–88.0) | 9.3 (4.3–31.5) | 0.018 |
| Total protein (g/L) | 65–85 | 68.9 (58.4–84.4) | 68.3 (54.6–83.6) | 0.465 | 66.2 (52.0–86.6) | 65.0 (55.0–80.4) | 0.050 |
| Albumin (g/L) | 40–55 | 39.9 (29.5–49.8) | 37.5 (27.6–47.7) | 0.001 | 40.6 (13.1–49.1) | 35.6 (28.6–44.3) | 0.000 |
| Globulin (g/L) | 20–40 | 29.3 (20.9–38.6) | 29.4 (23.0–47.3) | 0.115 | 26.7 (9.0–37.8) | 26.6 (19.7–46.6) | 0.451 |
| A/G ratio | 1.2–2.4 | 1.40 0.80–2.10) | 1.30 (0.70–1.70) | 0.001 | 1.50 (1.0–4.1) | 1.40 (0.70–1.90) | 0.009 |
| Pre- Albumin (mg/dL) | 20–45 | 17.7 (8.3–35.1) | 12.9 (4.5–30.6) | <0.001 | 23.6 (5.6–38.0) | 25.8 (20.2–46.8) | 0.063 |
| Laboratory tests | Normal reference |
Day on 1st follow-up |
Day on 2nd follow-up |
||||
|---|---|---|---|---|---|---|---|
| Non-severe (n = 84) | Severe (n = 29) | p value | Non-severe (n = 56) | Severe (n = 23) | p value | ||
| WBC (109/L) | 3.5–9.5 | 6.7 (2.4–11.0) | 6.1 (3.3–12.8) | 0.703 | 6.40 (2.4–10.6) | 6.00 (3.1–10.0) | 0.586 |
| Neutrophil (109/L) | 1.8–6.3 | 4.1 (1.1–8.9) | 4.0 (1.9–10.6) | 0.946 | 4.10 (1.1–7.5) | 3.70 (1.6–6.6) | 0.856 |
| Lymphocyte (109/L) | 1.1–3.2 | 1.70 (0.9–4.4) | 1.60 (0.70–2.7) | 0.093 | 1.70 (0.9–4.4) | 1.65 (1.0–3.5) | 0.551 |
| CD3+ T cell (per μL) | 770–2041 | 1163 (541–2738) | 1104 (645–2120) | 0.463 | 1185 (547–2700) | 1166 (725–1804) | 0.554 |
| CD4+ T cell (per μL) | 414–1123 | 620 (260–1497) | 539 (317–1083) | 0.240 | 620 (321–1497) | 555 (388–1108) | 0.712 |
| CD8+ T cell (per μL) | 238–874 | 477 (161–1243) | 466 (175–1150) | 0.661 | 475 (161–1243) | 495 (275–840) | 0.455 |
| CD19+ B cell (per μL) | 90–560 | 182 (57–586) | 108 (36.1–241.4) | <0.001 | 157 (42–541) | 119 (42.2–300) | 0.274 |
| CD56+ NK cell (per μL) | 150–1100 | 356 (77–1073) | 346 (131–766) | 0.396 | 367 (97–1043) | 444 (197–766) | 0.149 |
| Mononuclear (109/L) | 0.1–0.6 | 0.40 (0.20–0.70) | 0.50 (0.20–0.80) | 0.010 | 0.40 (0.2–0.9) | 0.40 (0.20–0.90) | 0.197 |
| Eosinophil (109/L) | 0.02–0.52 | 0.08 (0.00–0.39) | 0.08 (0.02–0.42) | 0.258 | 0.09 (0.01–0.41) | 0.12 (0.02–0.41) | 0.149 |
| Basophil (109/L) | 0.00–0.06 | 0.02 (0.00–0.06) | 0.02 (0.01–0.09) | 0.069 | 0.02 (0.0–0.06) | 0.02 (0.01–0.06) | 0.075 |
| IL-2 (pg/mL) | 1.1–9.8 | 1.26 (0.30–2.45) | 1.26 (0.30–2.03) | 0.374 | 1.26(0.3–2.38) | 1.20 (0.59–1.63) | 0.353 |
| IL-4 (pg/mL) | 0.1–3.0 | 1.62 (0.26–6.42) | 1.62 (0.28–5.18) | 0.792 | 1.48 (0.28–6.42) | 1.62 (0.32–2.21) | 0.634 |
| IL-6 (pg/mL) | 1.7–16.6 | 2.57 (1.1–141.07) | 3.09 (1.33–20.25) | 0.789 | 2.45 (1.1–11.96) | 2.38 (1.43–4.17) | 0.618 |
| IL-10 (pg/mL) | 2.6–4.9 | 1.47 (0.18–8.50) | 1.58 (0.35–4.43) | 0.911 | 1.45 (0.18–8.50) | 1.15 (0.37–2.33) | 0.399 |
| TNF-α (pg/mL) | 0.1–5.2 | 0.89 (0.12–2.27) | 0.70 (0.19–1.92) | 0.126 | 0.78(0.15–2.27) | 0.47 (0.15–0.66) | 0.017 |
| IFN-γ (pg/mL) | 1.6–17.3 | 1.33 (0.15–122.3) | 0.93 (0.15–3.65) | 0.481 | 1.18 (0.15–122) | 1.03 (0.42–2.30) | 0.539 |
| IgG (g/L) | 7.0–16.0 | 12.8 (8.2–20.0) | 13.2 (9.44–17.1) | 0.336 | 12.8 (8.57–20.0) | 12.6 (9.5–14.7) | 0.684 |
| IgA (g/L) | 0.70–4.0 | 2.47 (0.58–6.36) | 2.20 (1.08–4.16) | 0.062 | 2.23 (0.58–6.36) | 2.17 (1.45–3.73) | 0.415 |
| IgM (g/L) | 0.40–2.30 | 1.10 (0.38–4.31) | 1.26 (0.45–2.78) | 0.864 | 1.19 (0.41–4.31) | 1.17 (0.41–2.43) | 0.600 |
| CRP (mg/L) | <0.5 | 0.81 (0.00–25.3) | 1.60 (0.20–17.5) | 0.054 | 0.88 (0.0–25.3) | 1.10 (0.00–3.90) | 0.681 |
| Alanine aminotransferase (U/L) | 7–40 | 31.5 (8.0–164.0) | 24.0 (5.0–120.0) | 0.215 | 21.0 (8.0–135.0) | 25.0 (5.0–104.0) | 0.556 |
| Aspartate aminotransferase (U/L) | 13–35 | 26.0 (12.0–103.0) | 25.0 (12.0–93.0) | 0.578 | 23.0 (14.0–103.0) | 24.5 (16.0–70.0) | 0.436 |
| Alkaline phosphatase (U/L) | 35–100 | 83.0 (46.0–201.0) | 87.0 (54.0–176.0) | 0.284 | 81.0 (48.0–127.0) | 79.0 (50.0–111.0) | 0.643 |
| gamma-glutamyltransferase (U/L) | 7–45 | 44.0 (12.3–229.0) | 36.0 (15.0–94.0) | 0.303 | 31.5 (15.0–137.0) | 32.0 (14.0–68.0) | 0.932 |
| Total bilirubin (mmol/L) | 5.0–21.0 | 12.3 (6.4–24.5) | 11.1 (3.9–33.4) | 0.083 | 13.8 (6.8–27.7) | 12.9 (3.7–33.2) | 0.480 |
| Total protein (g/L) | 65–85 | 75.3 (61.2–87.4) | 74.9 (66.1–83.3) | 0.821 | 75.0 (62.2–86.0) | 71.7 (65.3–78.4) | 0.022 |
| Albumin (g/L) | 40–55 | 49.2 (33.0–53.4) | 48.5 (38.0–53.4) | 0.304 | 49.8 (40.9–58.8) | 48.4 (44–52.5) | 0.143 |
| Globulin (g/L) | 20–40 | 26.7 (19.1–37.2) | 27.0 (18.6–35.1) | 0.354 | 25.1 (19.8–31.4) | 24.1 (20.4–29.5) | 0.081 |
| A/G ratio | 1.2–2.4 | 1.80 (1.0–2.4) | 1.80 (1.2–2.7) | 0.200 | 1.95 (1.5–2.6) | 2.00 (1.6–2.4) | 0.327 |
| Pre- Albumin (mg/dL) | 20–45 | 30.7 (23.7–47.7) | 33.7 (25.5–42.0) | 0.550 | 29.7 (21.3–37.2) | 30.5 (21.2–37.2) | 0.631 |
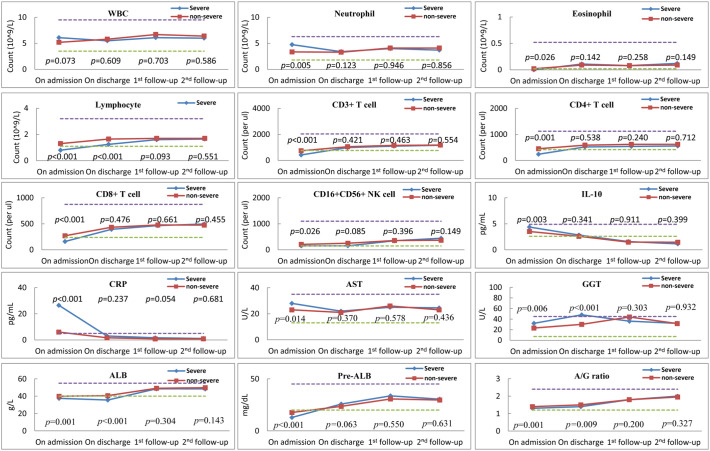
On the day of admission, the neutrophil count (median: 4.75 × 109/L vs. 3.75 × 109/L; p = 0.005), levels of IL-10 (4.37 pg/mL vs. 3.52 pg/mL; p = 0.003), CRP (26.9 mg/L vs. 6. 0 mg/L; p < 0.001), aspartate aminotransferase (AST; 28.0. vs. 23.0 U/L; p = 0.014), and gamma-glutamyltransferase (GGT; 32.0 U/L vs. 23.0 U/L; p = 0.006) were markedly higher in patient with severe COVID-19 than in the non-severe group. Contrastingly, counts of lymphocytes (0.80 × 109/L vs. 1.30 × 109/L; p < 0.001), and subsets of CD3+ T cells (424/μL vs. 745/μL; p < 0.001), CD4+ T cells (239/μL vs. 450/μL; p = 0.001), CD8+ T cells (160/μL vs. 269/μL; p < 0.001), CD56+ NK cells (152/μL vs. 209/μL; p = 0.042), eosinophils (0.00 × 109/L vs. 0.02 × 109/L; p < 0.001), and albumin (37.5 g/L vs. 39.9 g/L; p = 0.001) and pre-albumin (12.9 mg/dL vs. 17.7 mg/dL; p < 0.001) were significantly lower in patients with severe COVID-19 than that of the non-severe group. Immunoglobulin (Ig)-G, IgA, and IgM were within the normal range and comparable between the two groups.
On the day of discharge, counts of lymphocytes (1.25 × 109/L vs. 1.65 × 109/L; p < 0.001), levels of cytokine TNF-α (0.83 pg/mL vs. 1.04 pg/mL; p = 0.039) and albumin (35.6 g/L vs. 40.6 g/L; p = 0.001) were much lower, and GGT (48.0 U/L vs. 30.0 U/L; p < 0.001) remained markedly higher in patients with severe COVID-19.
On the day of the first follow-up, variables were comparable across the two groups, except for a marked decrease in B cells (108/μL vs. 182/μL; p < 0.001) in severe cases. Similarly, on the day of 2nd follow-up, variables were comparable and recovered to normal levels across the two groups, except for markedly lower levels of TNF-α (0.47 pg/mL vs. 0.78 pg/mL; p = 0.017) and total protein (71.7 g/L vs. 75.0 g/L; p = 0.022) in severe cases. All laboratory findings at different time points are shown in Fig. 1 .
Fig. 1.
The dynamics and comparison of peripheral immunological variables at admission, discharge, 1st follow-up (second week) and 2nd follow-up (fourth week) for patients with severe and non-severe COVID-19. Blue and red dashes indicate the lower and upper values of the normal reference interval. Squares and diamonds on the lines represent median. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Early predictive factors for severe COVID-19
To assay the predictive value of the above-mentioned variables with a p value less than 0.05 on admission (patient age, BMI, WBC count, neutrophil, eosinophil, lymphocyte, and its subsets CD3+ T cell, CD4+ and CD8+ T cell and NK cell, IL-10, CRP, AST, GGT, albumin, pre-albumin, and albumin/globulin (A/G) ratio) in differentiating between severe and non-severe COVID-19, we used covariate binary logistic regression analysis with the forward conditional method. Results showed that CD8+ T cells (HR = 0.995; p = 0.056) and CRP levels (HR = 1.040; p = 0.002) on admission were independent predictive factors for severe COVID-19 (Table 3 ).
Table 3.
Covariate logistic regression analysis of clinical variables between severe and non-severe COVID-19 patients.
| Laboratory variables | Covariate logistic regression |
p |
|---|---|---|
| Exp(B) (95% CI) | ||
| Age (years) | / | 0.208 |
| BMI | / | 0.119 |
| White blood cell count | / | 0.265 |
| Neutrophil count | / | 0.296 |
| Eosinophil count | / | 0.399 |
| Lymphocyte count | / | 0.985 |
| CD3+ T cell | / | 0.805 |
| CD4+ T cell | / | 0.974 |
| CD8+ T cell | 0.995 (0.990–1.000) | 0.056 |
| CD16 + CD56+ NK cell | / | 0.252 |
| IL-10 | / | 0.945 |
| C-reactive protein | 1.040 (1.015–1.066) | 0.002 |
| Aspartate aminotransferase | / | 0.134 |
| gamma-glutamyltransferase | / | 0.268 |
| Albumin | / | 0.484 |
| A/G ratio | / | 0.405 |
| Pre- Albumin | / | 0.320 |
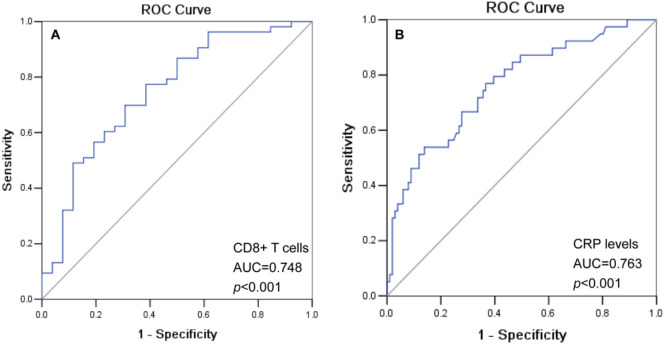
Fig. 2 shows the ROCs for the CD8+ T cells [area under curve (AUC) = 0.748; 95% CI: 0.631–0.866; p < 0.001], and CRP levels [AUC = 0.763 (95% CI: 0.674–0.853; p < 0.001)]. The optimal cut-off was determined by the Youden's index for CD8+ T cells as 232/μL (sensitivity: 0.698; specificity: 0.692), and CRP as 10.30 mg/L (sensitivity: 0.769; specificity: 0.634; data not shown).
Fig. 2.
ROC curve for (A) CD8+ T cells (AUC = 0.748; 95% CI: 0.631–0.866; p < 0.001), and (B) CRP levels (AUC = 0.763; 95% CI: 0.674–0.853; p < 0.001) to distinguish patients with severe and non-severe COVID-19.
4. Discussion
COVID-19 outbreaks continue at staggering rates globally since December 2019 and have become a serious public health concern [13]. Patients with advanced age and underlying diseases are associated with acquiring more severe forms of COVID-19. Laboratory abnormalities such as lymphopenia, leukopenia, and pneumonia imaging are more frequently observed in severe cases, as they are prone to poor clinical outcomes [3,14]. However, most information regarding laboratory findings for the disease was obtained during the initial stages of the disease in previous studies. A full spectrum of the peripheral immunological features from the disease onset, patient hospitalisation, and recovery remains to be investigated.
In this study, with a cohort of 146 laboratory-confirmed patients with COVID-19, clinical and laboratory data during hospitalisation and follow-up in the second and fourth week after discharge were retrieved and compared between the severe and non-severe COVID-19 patients. Our findings revealed that, on admission, the counts of lymphocytes and subsets of CD3+, CD4+, and CD8+ T cells, NK cells and eosinophils, and levels of albumin, and pre-albumin were dramatically lower, but neutrophil counts, IL-10, CRP, AST, and GGT levels were significantly higher in severe COVID-19 patients. These immunological abnormalities can favour virus immune escape from host anti-viral immune responses [15,16]. Fortunately, along with the disease convalesce, most of these variables are improved, reaching a normal and comparable level as early as the second week after discharge among both severe and non-severe COVID-19 patients.
Notably, 13 (8.9%) asymptomatic patients tested as positive for the SARS-CoV-2 virus during the follow-ups. Our findings revealed that the SARS-CoV-2 re-positivity was not related to the severity of COVID-19. Re-positivity of SARS-CoV-2 in patients with COVID-19 following discharge has been reported and raised concerns in recovery management. However, the viral transmission potential of the patients re-positive for SARS-CoV-2 remains unknown. Wang et al. [17] showed that eight (6.10%) of 131 patients with COVID-19 were confirmed to be re-positive for SARS-CoV-2. Additionally, 292 (3.3%) patients that were re-positive out of 8922 were released by the South Korean Centres for Disease Control and Prevention [18]. The human-to-human transmission risks by asymptomatic patients with COVID-19 have been emphasised in previous studies [19,20]. In this scenario, virus isolation in patients re-positive for SARS-CoV-2 is extremely necessary, to testify whether virus are reactivated and replicated, or only genetic material of the ‘dead virus’ in patients with SARS-CoV-2 re-positivity after discharge.
Various clinical laboratory findings during early stages of COVID-19 have helped the evaluation of disease severity and have been highly related to mortality [[21], [22], [23], [24]]. However, the predictive significance of these variables in differentiating patients with severe and non-severe COVID-19 is unknown. Using covariate binary logistic regression analysis, our data showed that the CRP levels on admission were an independent predictive factor for patients with severe COVID-19. This might help develop more proactive management for severe COVID-19. T cell depletion and dysfunction are associated with COVID-19 progression [10,23]. Subpopulations of T cells such as CD4+ and CD8+ T lymphocytes were delicately balanced for host cellular immune responses against viral infection. The CD8+ cytotoxic T lymphocytes orchestrate cytotoxic anti-virus responses and kill virus-infected cells directly, whereas the CD4+ helper T lymphocytes are essential for B lymphocyte maturation, which can produce virus-specific antibodies to neutralise viral antigens [16]. In this context, the decreased CD8+ T cells observed are associated with longer SARS-COV-2 virus shedding duration [12]. Although the counts of the peripheral immune cells were recovered during the convalesce as our findings indicated, whether immune functions of these immune cells restored remain unknown.
In summary, our findings provided evidence that peripheral immune cells recovered during the course of the disease, and that the CRP levels on admission were the only early predictive factors for patients with severe COVID-19. These findings will help develop more appropriate management protocols for patients diagnosed with COVID-19. A major limitation of our study was that the retrospective analyses were based on a small cohort from our single medical center. A multiple center-based study with a larger cohort is necessary to identify generalised findings and will help to further evaluate the significance of early predictive factors for COVID-19 severity.
Author contributions
AL and W—H Y participated in the following: study design, data analysis, literature search, and writing of the manuscript. X-H Jin, H-L Zhou, L-L Chen, G-F Wang, J-G Z, XZ, Q-Y Han, Q-Y C and Y—H Y participated in the following: medical history review, data collection, and data interpretation.
Fundings
This work was supported by grants from the Science and Technology Bureau of Taizhou (1901ky01; 1901ky04).
Declaration of Competing Interest
The authors declare no conflict of interest. All work was conducted in the absence of any commercial or financial relationships.
References
- 1.WHO Coronavirus disease 2019 (COVID-19) Situation Report – 63. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200323-sitrep-63-covid-19.pdf?sfvrsn=d97cb6dd_2
- 2.Lu R., Zhao X., Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X. Et al; China medical treatment expert Group for Covid-19. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382(18):1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothe C., Schunk M., Sothmann P. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N. Engl. J. Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Liu Y., Liu L., Wang X., Luo N., Ling L. Clinical outcomes in 55 patients with severe acute respiratory syndrome coronavirus 2 who were asymptomatic at hospital admission in Shenzhen. China. J Infect Dis. 2020;221(11):1770–1774. doi: 10.1093/infdis/jiaa119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou L., Ruan F., Huang M. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai C.C., Liu Y.H., Wang C.Y. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J Microbiol Immunol Infect. 2020;53(3):404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Z., McGoogan J.M. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72 314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323(13):1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 9.Li L.Q., Huang T., Wang Y.Q. 2019 novel coronavirus patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J. Med. Virol. 2020;92(6):577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giamarellos-Bourboulis E.J., Netea M.G., Rovina N. Complex Immune Dysregulation in COVID-19 Patients with Severe Respiratory Failure. Cell Host Microbe. 2020;27(6) doi: 10.1016/j.chom.2020.04.009. 992-1000.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palacios Cruz M., Santos E., Velázquez Cervantes M.A., León Juárez M. COVID-19, a worldwide public health emergency. Rev. Clin. Esp. 2020;S0014-2565(20):30092–30098. doi: 10.1016/j.rce.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou F., Yu T., Du R. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kikkert M. Innate immune evasion by human respiratory RNA viruses. J Innate Immun. 2020;12(1):4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiappelli F., Khakshooy A., Greenberg G. CoViD-19 immunopathology and immunotherapy. Bioinformation. 2020;16(3):219–222. doi: 10.6026/97320630016219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., Xu H., Jiang H. The clinical features and outcomes of discharged coronavirus disease 2019 patients:a prospective cohort study. QJM. 2020;113(9):657–665. doi: 10.1093/qjmed/hcaa178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang Y.J. South Korea’s COVID-19 infection status: from the perspective of re-positive after viral clearance by negative testing. Disaster Med Public Health Prep. 2020:1–3. doi: 10.1017/dmp.2020.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C., Ji F., Wang L. Asymptomatic and human-to-human transmission of SARS-CoV-2 in a 2-family cluster, Xuzhou. China. Emerg Infect Dis. 2020;26(7):1626–1628. doi: 10.3201/eid2607.200718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huff H.V., Singh A. Asymptomatic transmission during the COVID-19 pandemic and implications for public health strategies. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian S., Hu N., Lou J. Characteristics of COVID-19 infection in Beijing. J. Inf. Secur. 2020;80(4):401–406. doi: 10.1016/j.jinf.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan S., Xiang Y., Fang W. Clinical features and treatment of COVID-19 patients in Northeast Chongqing. J. Med. Virol. 2020;92(7):797–806. doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin C., Zhou L., Hu Z. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin Infect Dis. 2020;71(15):762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Health Commission (NHC), National Administration of Traditional Chinese Medicine of the People's Republic of China Diagnosis and Treatment Guidance of Corona Virus Diseases 2019 (Tentative 7th Edition) http://www.gov.cn/zhengce/zhengceku/2020-03/04/content_5486705.htm
- 24.Lin A., He Z.B., Zhang S. Early risk factors for the duration of SARS-CoV-2 viral positivity in COVID-19 patients. Clin. Infect. Dis. 2020;71(16):2061–2065. doi: 10.1093/cid/ciaa490. [DOI] [PMC free article] [PubMed] [Google Scholar]