Abstract
We present the hypothesis to the scientific community actively designing clinical trials and recommending public health guidelines to control the pandemic that – “Tetanus vaccination may be contributing to reduced severity of the COVID-19 infection” – and urge further research to validate or invalidate the effectiveness of the tetanus toxoid vaccine against COVID-19. This hypothesis was revealed by an explainable artificial intelligence system unleashed on open public biomedical datasets. As a foundation for scientific rigor, we describe the data and the artificial intelligence system, document the provenance and methodology used to derive the hypothesis and also gather potentially relevant data/evidence from recent studies. We conclude that while correlations may not be reason for causation, correlations from multiple sources is more than a serendipitous coincidence that is worthy of further and deeper investigation.
Introduction
Artificial intelligence (AI), when applied with explainable context is augmented intelligence – one that empowers human intelligence to excel at its best by doing what computers do best. This paper demonstrates such an example of AI augmenting subject-matter-expert intelligence on a hypothesis generation task of connecting a knowledge universe of medical facts/data to pose and address the question – “Is tetanus vaccination contributing to reduced severity of the COVID-19 infection?”.
In this paper, we describe the AI system instantiated for the task of hypothesis generation – the ability to reveal potentially innovative ideas/questions by mining meaningful implicit association between unlinked concepts. We demonstrate that a holistic approach that is able to associate and reason with insights from millions of publications, curated assay databases, and protein databanks can lead to hypotheses that are computationally feasible but impossible for humans to formulate. In application to the rapid response for the COVID-19 pandemic, our approach borrows several innovative concepts of creating [1], staging [2], [3] and reasoning [4] with medical facts represented as knowledge graphs. Our approach is differentiated in the following ways: (i) our approach does not limit to the body of knowledge with a siloed lens around the COVID-19 virus data, (ii) we extrapolate the search for hypotheses by associating protein sub-structures of disease-causing COVID-19 virus with other known “COVID-like” protein-structures in other viruses, bacteria and fungi etc., (iii) Using expert curated databases, we associate the “COVID-like” disease-causing-protein interactions with host-organism proteins in different organisms (rats, mice, rabbits, bats, humans, etc.), and (iv) we evaluate combinatorial drug-molecule interaction patterns between host-organism proteins and known/measured inhibitory activity of small drug molecules across multiple databases to score the strength of the hypothesis on the disease-causing protein. Computationally, this involves sifting through nearly 30 terabytes of data, comparing different protein-substructures of COVID-19 against 4+ million protein sequences and reasoning based on properties and interactions across millions of well-studied proteins and molecules in literature – an impossible task for humans to execute.
However, as computer science authors, we do not have the relevant medical resources to perform in-vivo animal studies with the potential for randomized clinical-trials and statistically validate the hypothesis. We also do not claim to have completely automated the hypothesis generation task with artificial intelligence algorithms. We admit to serendipitous discovery and will not argue that observed correlations are factors that contribute to causation. Our primary goal in this paper is to disseminate an insight if proven true may be valuable for public health amidst a global pandemic. Toward that goal, we present the provenance of connecting the dots across multiple public open-source data sources and linking multi-modal data - biological protein sequences, curated protein–protein interaction measurements - to known inhibitory protein-drug activity for drug repurposing which led to an insight that initially was considered as a serendipitous false positive. The insight appears to have gained supporting evidence recently [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]. Our goal is to present the insight and the emerging supporting evidence to stimulate discussion among experts that may be able to act on the insight. We present the facts that revealed the association between Tetanus vaccination and COVID-19 and the emerging evidence that appears to support the “discovered” association using strong correlations with vaccination rates, sequence similarity, and related symptoms.
Methodology
Data and system
The Cray Graph Engine (CGE) is an in-memory semantic graph database designed for the Cray XC supercomputer to support interactive querying of large datasets (~100 s of terabytes). A semantic graph is a collection of triples of the form < subject, predicate, object > . For this paper, the triples are medical facts such as {<Nexium, treats, heartburn>, <Malaria, causes, fever>, <Lipitor, is a, drug>…etc.}. Semantic graph databases differ from relational databases in that the underlying data structure is a graph, rather than a structured set of tables. The graph structure (with subject/object terms as vertices and the predicate terms as edges) makes semantic databases ideal for analyzing multi-modal unstructured data (sequences, signals, facts, properties) and reasoning based on relationships (semantic similarity, sequence similarity, image similarity etc.). The tabular form is preferred for hypothesis-driven research while knowledge graphs are preferred for hypothesis-generation [1], [2], [3], [4].
Motivated by the need to identify drug candidates for the novel COVID-19 virus, we used the existing capabilities of the Cray Graph Engine to provide an integrated life-science knowledge graph bringing together multiple multi-modal life-science databases. The scalability of CGE enables all of the relevant databases to be loaded in one environment, enabling seamless cross-database queries [5]. The performance and scalability of the database build process and the scalable load time enables updated data to be quickly pulled in and the database fully rebuilt in less than an hour. The integrated Life-Sciences knowledge graph assembled to study potential drug repurposing candidates for COVID-19 was generated from a collection of publicly available databases commonly used in life sciences and systems biology research, bringing together a knowledge graph with over 155 billion facts. To the best of our knowledge, no other graph database is able to host such a large dataset and offer interactive query capabilities.
| Dataset | Size (on disk) | Size (triples) | source |
|---|---|---|---|
| UniProt (Mar 2020) | 12.7 TeraBytes | 87.6 Billion | [26] |
| PubChemRDF (v1.6.3 beta) | 13.0 TeraBytes | 80.0 Billion | [27] |
| ChEMBL-RDF (27.0) | 81 Gigabytes | 539 Million | [28] |
| Bio2RDF (Release 4) | 2.4 TeraBytes | 11.5 Billion | [29] |
| OrthoDB (v10) | 275 Gigabytes | 2.2 Billion | [30] |
| Biomodels (r31) | 5.2 Gigabytes | 28 Million | [31] |
| Biosamples (v20191125) | 112.8 Gigabytes | 1.1 Billion | [32] |
| Reactome (r71) | 3.2 GigaBytes | 19 Million | [33] |
On this massive knowledge graph, our approach begins by first allowing researchers to understand the similarity/variability of the novel virus compared to other known viruses. If parts of the protein sequence that make up the novel virus have sequence overlaps of functional domains with other known viruses, the information in the integrated knowledge graph helps us extrapolate the search to identify potential drug candidates that are known to inhibit disease-causing activity of the known viruses. The typical workflow for researchers conducting hypothesis-driven research would have been to perform searches in one of the databases, then construct queries for another database, and iterate. The effort of manually mapping the ontologies of various data sources and piecing together results from multiple query end points (or using yet another database to perform this translation), is a cumbersome process. The performance scalability of CGE enables all of the relevant databases to be loaded in one environment, enabling seamless cross-database queries – and automated cross-database pattern search. Furthermore, CGE has the ability to compute similarity scores based on sequence and graph-theoretic properties implemented as domain-specific functions.
Hypothesis generation
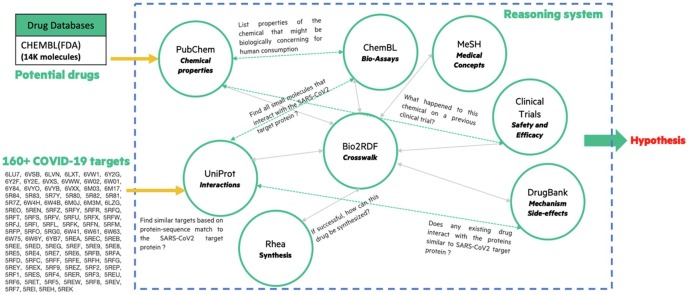
Leveraging the unique capabilities of CGE to interactively handle very large datasets [5], queries were written to search the life sciences knowledge graph for potential drugs that could be effectively repurposed against COVID-19. We have illustrated the approach in Fig. 1 below. The first query posed on the knowledge graph was to compute the similarity of all known protein sequences of known virus, bacteria and fungi in Uniprot to match protein-substructures of COVID-19. One potentially interesting result while computing and comparing the well-studied COVID-19 protein sub-structure called SPIKE_SARS2 [34] (also known as the Spike glycoprotein or simply Spike protein) was the tetanus toxin, which has the Uniprot identifier P04958 and mnemonic TETX_CLOTE. The similarity query against the SPIKE_SARS2 protein returns TETX_CLOTE as one of the highest non-coronavirus matches based on the similarity score across all available protein sequences in Uniprot.
Fig. 1.
An explainable AI system capable of hypothesis generation using knowledge graphs applied on COVID-19 protein targets. Our approach leverages a scalable graph database to host a knowledge graph of medically relevant facts integrated from multiple curated and open knowledge sources and also act as a computational engine capable of interactive in-database analytics to query/impute/infer new and novel relationships.
The next set of queries searched the existing published literature on proteins, compounds, chemical interactions and clinical trials as well as the protein structure information. For example, once we identified UniProt protein-substructures such as (P04958, P12821, P08183, P0A8V2, O43451, P47820, P12822, O00462, P35968, P47989…) bearing sequence similarity with COVID-19 protein structures we are able to associate proteins in a host organism that are known to interact with the disease-causing protein structure. For example, for the protein-substructures listed above, some of the host organism protein structures identified were the angiotensin-converting enzyme, ATP-dependent translocase ABCB1, DNA-directed RNA polymerase subunit beta, maltase-glucoamylase, etc. At this step, we did broaden the scope to include both animals and humans as host organisms. For these host-organism proteins, we then search for known small molecules in databases like CHEMBL, PubChem and/or Drugbank with documented evidence of protein-small molecule interaction. In our result set, several of the drugs currently under clinical trials [6] appear high in the list of compounds suggested by CGE-driven AI system. The AI system is able to look for patterns around prior evidence of efficacy and safety from open clinical trial datasets. Some of the drugs recommended by this AI system that were undergoing clinical trials include Baricitinib, Ribavirin, Ritonavir, Dexamethasone, Azithromycin and Lopinavir. The scores on sequence and functional overlap for these small-molecules (that experts expect to bind and modulate the COVID-19 SPIKE_SARS2 protein activity), which are undergoing clinical trials, are lower than the calculated score for TETX_CLOTE, which encouraged us to track the literature on our top match TETX_CLOTE. We present the evidence we have tracked since the outbreak in the Section below.
Observation and evidence of correlation to tetanus vaccination
Given the large rate of asymptomatic positive COVID-19 cases, which the Centers for Disease Control and Prevention (CDC) currently estimates to be 40% [7], the tetanus toxin result led to an unexpected hypothesis. The tetanus vaccine might be assuaging the symptoms of COVID-19 by enabling the immune system to mount a reasonable response to the virus thereby reducing the severity of symptoms, with a potential to even keep the patient asymptomatic. While tetanus is caused by bacteria and COVID-19 is caused by a virus, there are multiple examples of heterologous immunity between bacteria and viruses, which has been initially attributed to similarities in the protein sequences [8].
Vaccination rate correlations
According to the CDC, 63.4% of adults in the US have received the tetanus booster in the last 10 years, with a decrease in vaccination for adults older than 65 as well as minority groups [9], which correlates well with current COVID-19 asymptomatic rates around 40%. Overall, the United States has a mortality rate of approximately 2.76% [25]. By comparison, countries such as the United Kingdom do not routinely administer tetanus booster vaccines to adults [11] and the COVID-19 mortality rate in the United Kingdom is approximately 6.93% [25].
The reduced severity of COVID-19 in children has caused several researchers to consider whether a childhood vaccine is a contributing factor due to the immune response stimulated by the vaccine [13]. Some experts have reported that over 90% of children who test positive will be either asymptomatic or mildly symptomatic [14]. The World Health Organization (WHO) reports that 85% of infants worldwide receive the three recommended doses of the diphtheria-tetanus-pertussis (DTP) vaccine [35]. The high vaccination rate of DTP worldwide correlates well with the symptom severity of COVID-19 in children. Another childhood vaccine being considered for having a protective effect for COVID-19 is bacille Calmette-Guerin (BCG) for tuberculosis. However, several countries do not administer BCG to children and even in some countries that do, such as Brazil, COVID-19 is having a more significant impact than other countries [15], [16]. Brazil has reported poor vaccination coverage for DTP in a significant percentage of municipalities, which could correlate and explain the increased COVID-19 mortality rate in Brazil [15].
Other critical populations with high asymptomatic rates are pregnant women and prison inmates. A recent study of pregnant women admitted for delivery reported an asymptomatic rate of 87.9%, which is at least double the current estimated rate for the general population [17]. Pregnant women are advised to get two vaccines during the third trimester of pregnancy: the influenza vaccine and the Tetanus toxoid, reduced diphtheria toxoid and acellular pertussis (TDaP) vaccine [18]. In fact, for pregnant women, TDaP is the only childhood vaccine recommended. Additionally, prison inmates in Arkansas, Ohio, North Carolina and Virginia had an asymptomatic rate of 96% [7], and while the vaccination policies are not known for all of these states, North Carolina has a published policy to keep inmates current on vaccines such as TDaP [19]. While TDaP vaccination rates are not a proxy for a randomized case-control clinical trial, these observed correlations at international, national and cohort specific cannot be dismissed as random co-incidence.
Sequence similarities
A recent study [20] analyzed the protein sequence similarities of proteins in viruses targeted by childhood vaccines and the COVID-19 SPIKE_SARS2 protein. Based on the similarities they hypothesized that the measles-mumps-rubella (MMR) vaccine could be providing a protective effect. The Uniprot identifiers for the proteins in that study are Q786F3 (Measles virus strain Ichinose-B95a) and P08563 (Rubella virus strain M33) [26]. Using the Clustal Omega alignment program on the Uniprot site [26], the Q786F3 protein was found to have a sequence identity and similarity percentage with the SPIKE_SARS2 protein of 6.87% and 31%, respectively. The P08563 protein was found to have a sequence identity and similarity percentage of 10.68% and 24%, respectively. By comparison, the TETX_CLOTE protein (P04958) was found to have a sequence identity and similarity percentage of 12.78% and 30%, respectively.
To further strengthen the potential connection between tetanus and the COVID-19 spike protein, CGE was used to search the life science data to find any other connections between tetanus and other coronaviruses. These searches found a few other coronaviruses with the same similarity to the tetanus protein structure as was found between tetanus and the COVID-19 SPIKE_SARS2 protein. Two of those proteins, A0A023PTS3 and A0A023PUW9, are for strains of “Rhinolophus affinis coronavirus”. This is particularly interesting because one study found a SARS strain from 2013, SARs-Ra-BatCoV-RaTG13, that had a genomic identity of 96.1% to COVID-19, which was found in Rhinolophus affinis bats [24].
Tetanus related studies
Researchers have also discovered that COVID-19 is capable of infecting neurons and damaging the brain [21], though it is not yet clear why. One recent report stated that over 80% of hospitalized COVID-19 patients experience some neurological symptom [22]. Additionally, it has been reported that several recovering patients suffer from pain in the temporomandibular joint (TMJ) region, which some have suggested could be related to the increased difficulty breathing caused by COVID-19 [23]. Given the sequence similarity between the tetanus protein and the COVID-19 Spike protein, it seems possible that some of these neurological symptoms and TMJ could also be related/attributed to symptoms that patients suffer from a tetanus infection.
Two other studies [10], [12] have also suggested a possible protective effect from the TDaP vaccine against COVID-19. Both studies report the lower severity of symptoms in children and women and the overlap between these two groups of the DTP and TDaP vaccines. One study [12] focuses on the potential for the pertussis vaccine to provide a protective effect based upon the correlation of vaccination rates in children and pregnant women and the symptom severity for COVID-19 as well as the cytokine damping effects of the vaccine. The other study [10] again pointed out the reduced severity in children and suggested the TDaP vaccine could be helping by stimulating the immune system and reactivating CD4 + T cells. A recent study [36] also reported a potential protective effect against COVID-19 from the DTP childhood vaccine. The study noticed the reduced severity of COVID-19 in children to pose the question - Does a childhood vaccine provide cross-reactivite protective immunity against COVID-19? Investigating further, the study focused on the proteomes in 25 human pathogens and in vaccine antigens and concluded that only the DTP vaccine showed significant potential for cross-reactivity with COVID-19 Spike protein. This observaction significantly supports our theory that the tetanus toxoid vaccine could be providing a protective effect against COVID-19 in children as well as adults who are up-to-date on their tetanus vaccine.
Conclusion
In conclusion, we hypothesize that the tetanus toxoid vaccine could reduce the severity of COVID-19 symptoms. This hypothesis is based on the following:
-
-
Connections between the protein sequences for tetanus toxin and the COVID-19 Spike glycoprotein discovered using CGE on the life sciences knowledge graph
-
-
Similar connections between the tetanus toxin protein sequence and other coronaviruses, in particular, from the Rhinolophus affinis bat
-
-
Verification of connections between tetanus and the COVID-19 spike glycoprotein, as well as other coronaviruses, using the Uniprot Clustal Omega alignment tool to determine sequence similarities
-
-
Correlation between tetanus vaccination rates in the US and COVID-19 asymptomatic and mortality rates, especially when compared against countries with lower tetanus vaccination rates and significantly higher COVID-19 mortality rates
-
-
Correlation between DTP vaccination rates in children worldwide and COVID-19 symptom severity in children
-
-
Correlation between TDaP vaccination rates in pregnant women and asymptomatic COVID-19 rates
-
-
Potential for correlation between tetanus vaccination rates in prison inmates and asymptomatic COVID-19 rates
-
-
Neurological symptoms and TMJ that are typical symptoms of the tetanus disease are being reported in several COVID-19 patients
-
-
Additional studies also suggesting a protective effect from the DTP/TDaP vaccines against COVID-19 based on observed cross-reactivity
We present our preliminary findings on the correlation between tetanus vaccine and reduced symptoms severity of COVID-19 to the scientific community with the hope for an urgent accelerated pipeline to validate our findings in an in-vivo model organism like mice or rats given the potential to reverse/limit the far reaching debilitating impacts of the current pandemic situation. Our findings not only address the power of AI to potentially find a cure for novel illnesses with pre-existing compounds, it also aims to highlight the vulnerability of high-risk individuals in dire need of a cure in this race against time and why it is pivotal to pursue AI based analysis that can save time and billions of tax-payer dollars by searching for existing compounds that can potentially limit/cure disease progression.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The “Force for Good” pledge of intellectual property to fight COVID-19 brought into action Hewlett Packard Enterprise (HPE) products, resources and expertise to the problem of drug/vaccine discovery. Several scientists engaged in collaborations with HPE volunteers to accelerate efforts towards a cure. This paper is an example of the spirit of such a collaboration toward the true convergence of high-performance computing, artificial intelligence and data science to fight a pandemic. The authors (as HPE employees) are grateful to have leveraged a Cray XC40 supercomputer with 370 dual socket nodes (336 compute and 34 service nodes) with a mix of Skylake and Cascade Lake nodes ranging in frequency from 2.1-2.4 GHz. Each node had 192 + GB of memory. The supercomputer was mounted along with a Sonexion CS-L300N system with 8 object storage targets providing 655 TB of storage. The authors also thank Padmapriyadarshini Ravisankar (Cincinnati Children’s Hospital Medical Center), Dr. Ryan Yates (Principal Scientist at the Natural Center for Natural Products - University of Mississippi), Dr. Alex Madama (Chief Technologist for Healthcare and Life Sciences at HPE) and Dr. Eng-Lim Goh (Chief Technology Officer for AI at HPE) for encouraging this work and providing insightful feedback.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mehy.2020.110395.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Knowledge Graph Hub. https://github.com/Knowledge-Graph-Hub/kg-covid-19; 2020.
- 2.Sadegh S., Matschinske J.O., Blumenthal D.B., Galindez G., Kacprowski T., List M., Nasirigerdeh R., Oubounyt M., Pichlmair A., Rose T., Salgado-Albarran M., Spath J., Stukalov A., Wenke N.K., Yuan K., Pauling J., Baumbach J. Exploring the sars-cov-2 virus host-drug interactome for drug repurposing. Nat Commun. 2020;11 doi: 10.1038/s41467-020-17189-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Networkbased drug repurposing for novel coronavirus 2019-ncov/sars-cov-2. Cell Discovery. 2020;6 doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gysi DM, Valle ID, Zitnik M, Ameli A, Gan X, Varol O, Sanchez H, Baron R, Ghiassian D, Loscalzo J, Barabasi A-L. Network medicine framework for identifying drug repurposing opportunities for covid-19. ArXiv, 2020. [DOI] [PMC free article] [PubMed]
- 5.Rickett C., Haus U., Maltby J., Maschhoff K. Stockholm; Sweden: 2018. Loading and Querying a Trillion RDF Triples with Cray Graph Engine on the Cray XC, in Cray User Group Conference (CUG ’18) [Google Scholar]
- 6.“Clinicaltrials.gov,” 2020, accessed 6/3/2020. [Online]. Available: https://clinicaltrials.gov/ct2/results?cond=COVID-19.
- 7.Cha AE. Forty percent of people with coronavirus infections have no symptoms. might they be the key to ending the pandemic? Aug. 2020. [Online]. Available: https://www.washingtonpost.com/health/2020/08/08/asymptomatic-coronavirus- covid/.
- 8.Agrawal B. Heterologous immunity: role in natural and vaccine-induced resistance to infections. Front Immunol. 2019 doi: 10.3389/fimmu.2019.02631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaccination coverage among adults in the united states, national health interview survey, 2017. CDC, Tech. Rep.; 2018. [Online]. Available: https://www.cdc.gov/vaccines/imz-managers/coverage/adultvaxview/pubs- resources/NHIS- 2017.html.
- 10.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7175879/, accessed 10/02/20.
- 11.https://patient.info/doctor/immunisation-schedule-uk, accessed 10/02/20.
- 12.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7365096/, accessed 10/02/20.
- 13.Miri S.M., Noorbakhsh F., Mohebbi S.R., Ghaemi A. Higher prevalence of asymptomatic or mild covid-19 in children, claims and clues. J Med Virol. 2020 doi: 10.1002/jmv.26069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michaud J, Kates J. What do we know about children and coronavirus transmission? Jul. 2020. [Online]. Available: https://www.kff.org/coronavirus-covid-19/issue-brief/what-do-we-know-about-children-and-coronavirus-transmission/.
- 15.Sato A.P.S. What is the importance of vaccine hesitancy in the drop of vaccination coverage in brazil? Rev Saude Publica. 2018;52(96) doi: 10.11606/S1518-8787.2018052001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coronavirus: Brazil passes 100,000 deaths as outbreak shows no sign of easing, Aug. 2020. [Online]. Available: https://www.bbc.com/news/ world-latin-america-53712087.
- 17.Sutton D., Fuchs K., D’Alton M., Goffman D. Universal screening for sars-cov-2 in women admitted for delivery. New England J Med. 2020;382(22):2163–2164. doi: 10.1056/NEJMc2009316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobah YB. Which vaccines during pregnancy are recommended and which ones should i avoid? Feb. 2020. [Online]. Available: https://www.mayoclinic.org/healthy-lifestyle/pregnancy-week-by-week/expert-answers/vaccines-during- pregnancy/faq-20057799.
- 19.Health Services Policy Procedure Manual. Available: https://files.nc.gov/ncdps/documents/IC- 8\%20Immunizations.pdf.
- 20.Sidiq K.R., Sabir D.K., Ali S.M., Kodzius R. Does early childhood vaccination protect against covid-19? Front Mol Biosci. 2020 doi: 10.3389/fmolb.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.https://www.nature.com/articles/d41586-020-02599-5, accessed 10/06/20.
- 22.https://www.foxnews.com/health/hospitalized-coronavirus-patients-neurological-symptoms-study, accessed 10/06/20.
- 23.https://www.foxnews.com/health/jaw-pain-coronavirus-patient-breathing-issues, accessed 10/06/20.
- 24.Lau S.K., Luk H.K., Wong A.C., Li K.S., Zhu L., He Z. Possible bat origin of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. Jul. 2020;26(7) doi: 10.3201/eid2607.200092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.https://coronavirus.jhu.edu/map.html, accessed 10/12/20.
- 26.Consortium U. Uniprot: the universal protein knowledgebase. Nucleic Acids Res. 2019;47:D506. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., Zaslavsky L., Zhang J., Bolton E.E. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 10 2018.;47(D1):D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Félix E., Magariños M., Mosquera J., Mutowo P., Nowotka M., Gordillo- Marañón M., Hunter F., Junco L., Mugumbate G., Rodriguez-Lopez M., Atkinson F., Bosc N., Radoux C., Segura-Cabrera A., Hersey A., Leach A. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2018;47(D1):D930–D940. doi: 10.1093/nar/gky1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belleau F., Nolin M.-A., Tourigny N., Rigault P., Morissette J. Bio2RDF: Towards a mashup to build bioinformatics knowledge systems. J. Biomed. Inform. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. http://www.sciencedirect.com/science/article/pii/S1532046408000415. [DOI] [PubMed] [Google Scholar]
- 30.Zdobnov E.M., Tegenfeldt F., Kuznetsov D., Waterhouse R.M., Simão F.A., Ioannidis P., Seppey M., Loetscher A., Kriventseva E.V. OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs. Nucleic Acids Res. 2019;47(D1):D807–D811. doi: 10.1093/nar/gky1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chelliah V., Juty N., Ajmera I., Ali R., Dumousseau M., Glont M. BioModels: ten-year anniversary. Nucl Acids Res. 2015;43:D542–D548. doi: 10.1093/nar/gku1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jupp S., Malone J., Bolleman J., Brandizi M., Davies M., Garcia L., Gaulton A., Gehant S., Laibe C., Redaschi N., Wimalaratne S.M., Martin M., Le Novere N., Parkinson H., Birney E., Jenkinson A.M. The ebi rdf platform: linked open data for the life sciences. Bioinformatics. 2014;30(9):1338–1339. doi: 10.1093/bioinformatics/btt765. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3998127/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P. The reactome pathway knowledgebase. Nucleic Acids Res. Jan 2018;46(D1):D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang Y., Yang C., Xu X. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.“Immunization coverage,” Jul. 2020. [Online]. Available: https://www.who.int/news-room/fact-sheets/detail/immunization-coverage.
- 36.Reche P.A. Potential cross-reactive immunity to SARS-CoV-2 from common human pathogens and vaccines. Front Immunol. 2020;11:2694. doi: 10.3389/fimmu.2020.586984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.