Abstract
COVID-19, a novel identified coronavirus disease due to Severe Acute Respiratory Syndrome coronaviruses 2 (SARS-Cov-2) infection, has posed a significant threat to public health worldwide. It has been reported COVID-19 keeps substantial nucleotide similarity and shares common receptor, Angiotensin-converting enzyme 2 (ACE2) with Severe Acute Respiratory Syndrome coronaviruses (SARS-Cov). Here, we investigated the gene expression of ACE2 and identified associated pathways of SARS-Cov as a useful reference for a deepening understanding of COVID-19. The results indicated the ACE2 was overexpressed in human airway epithelial cells (HAEs), especially at 72 h after SARS-Cov infection. We found ACE2 might regulate immune response through immunological activation-associated pathways in the process of in both SARS-Cov and SARS-Cov-2 infection, where the activation of B cells, macrophages, helper T cells 1 (Th1 cells) and the inhibition of Foxp3 + regulatory T (Treg) cells and CD8 + T cells were found to be prominent. Finally, significant correlation between ACE2 and JAK-STAT signaling pathway was identified which indicate that JAK-STAT signaling pathway might involve in the downstream action of the overactivation of ACE2. These findings are expected to gain a further insight into the action mechanism of COVID-19 infection and provide a promising target for designing effective therapeutic strategies.
Abbreviations: COVID-19, Coronavirus disease 2019; ACE2, Angiotensin-converting enzyme 2; SARS-Cov, Severe Acute Respiratory Syndrome coronaviruses; HAEs, human airway epithelial cells; Th1 cells, helper T cells 1; Treg cells, Foxp3+ regulatory T; ARDS, Acute respiratory distress syndrome; HCoV-NL63, human coronavirus NL63; KNN, k-nearest neighboring; GSVA, gene set variation analysis; GSEA, Gene Set Enrichment Analysis
Keywords: COVID-19, ACE2, Signal-gene GSEA, Immune cell, JAK-STAT signaling pathway
1. Introduction
After several viral pneumonia cases were reported from Wuhan, China, in December 2019, a novel coronavirus, COVID-19 was identified based on the sequencing results of respiratory samples and named by the World Health Organization (WHO) on February 11, 2020 (Organization, 2020). With high infectiousness, COVID-19 spread rapidly and posed a high risk to the whole world. As of June 11, 2020, 84,652 definite patient cases have been reported (including 79,888 cured cases) and 4645 cases died from COVID-19 from 31 provinces in China according to the latest official report. Moreover, outside of China, 7,382,290 definite patient cases have been reported, including 413,583 deaths in 215 countries, most notably in America, Brazil, Britain and Spain. Acute respiratory distress syndrome (ARDS) and sepsis resulted from severe infection by COVID-19 were the major causes of death (Huang et al., 2020). It’s indicated by the phylogenetic analysis for the complete viral genome of COVID-19 that the new virus had 89.1% nucleotide similarity with SARS-like coronaviruses (Wu et al., 2020). Moreover, the SARS-CoV-2 has been identified as the pathogen of COVID-19 and the SARS-CoV-2 ribonucleic acid (RNA) has been used to detect COVID-19 (Cheng and Shan, 2020, Yu et al., 2020). However, the potential pathogenic mechanism of COVID-19 is still unclear and there is a lack of effective prevention or treatment for COVID-19 infection.
Only when combined with cell surface receptors, could this virus enter the target cells for further replication, which was the prerequisite of coronaviruses infection (Li, 2016). ACE2 had been known as a cell surface protein on cells in the heart, blood vessels, kidney, especially lung AT2 alveolar epithelial cells (Richardson et al., 2020). More importantly, it was identified as the receptor for SARS-Cov and human coronavirus NL63 (HCoV-NL63) (Li et al., 2007). Xu et al. also found ACE2 could serve as the receptor for COVID-19 through modeling the spike protein (Xu et al., 2020). In addition, Zhou et al. confirmed COVID-19 used ACE2 as the cell entry receptor from virus infectivity studies in HeLa cells from humans, pig, Chinese horseshoe bats and civets (Zhou et al., 2020). Hence, ACE2 played an essential role in the process of COVID-19 infection.
Based on the fact that COVID-19 remains substantial nucleotide similarity with SARS coronavirus and they share the same cell entry receptor ACE2, analysis of ACE2 expression and biological function in SARS-Cov would be of great reference value for the study of COVID-19 infection. In this study, we demonstrated the expression features of ACE2 in coronavirus infection and identified associated pathways of COVID-19 using gene expression profiles from public database using bioinformatics approaches. Moreover, all these results were further validated in other datasets of lung tissues and cells infected by SARS-Cov-2. We are the first that investigated the correlation between ACE2 and JAK-STAT signaling pathway to shed light on the potential pathogenesis of COVID-19 infection and provide hints to improve on therapeutic strategies.
2. Materials and methods
2.1. Data collection and preprocessing
Four eligible microarray datasets (GSE47960, GSE47961, GSE47962 and GSE47963) were downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/) with the following selection criteria: a) keywords of “SARS” or “coronavirus”; b) Using lung tissue or airway epithelial cells as research objects; c) Datasets contained a minimum of 10 samples and inclusion of >5,000 genes in the GEO platform. The microarray datasets comprised 174 HAEs samples which were divided into 85 samples dealt with mock infections as the control subgroup and 89 samples dealt with SARS coronavirus infection as the SARS subgroup. Moreover, each subgroup was further divided into 9 groups based on the time point at 0, 12, 24, 36 48, 60, 72, 84 and 96 h after infection (Supplementary Table 1). To increase the accuracy and reliability of the immune infiltration, we also downloaded another high-throughput sequencing datasets containing 16 SARS-Cov-2 infected and 5 healthy lung tissues from GSE150316 to conduct immune infiltration analysis. Further, another RNA sequencing dataset (including 14 SARS-Cov infected, 14 SARS-Cov-2 infected and 8 mock infected human Calu-3 cells) was acquired from GSE148729 to validate the conclusion.
The series matrix files of four gene expression profiles were downloaded from GEO and the k-nearest neighboring (KNN) imputation algorithm was applied to impute the few missing values through the “Impute. Knn” function of “impute” package (Suyundikov et al., 2015). Then we deleted the probes without a corresponding gene symbol and calculated the average value as the final expression value for genes corresponding to more than one probe. Subsequently, probes with zero (the lowest expression) was eliminated by a filtering process and the “ComBat” function of R package “sva” was used to remove known batch effects from microarray data (Leek et al., 2012). Finally, quantile normalization within and between arrays on all samples was conducted using “normalizeWithinArrays” and “normalizeBetweenArrays” function and the probe IDs were converted into gene symbols based on the annotation file for probes of the platform. In addition, the counts files of lung tissues and human Calu-3 cells were filtered with low expression and normalized by the DEseq2 package (Love et al., 2014).
2.2. Signal-gene GSEA and GSVA
To investigate the potential role of ACE2 in SARS-Cov infection, we divided the SARS subgroups into two groups with high or low expression levels of ACE2 based on Candidate-Gene Scores, calculated as described in other studies (Kirou et al., 2004, Kirou et al., 2005, Feng et al., 2006). The Mean and SD levels of ACE2 in the control (Mean-control and SD-control) were calculated for the standardization of expression levels of ACE2 for each infected sample. Then the standardized expression levels of each sample were reckoned as following calculation formula:
Candidate-Gene Scores (ACE2) i= (ACE2 i SARS – Mean-control)/(SD-control), where i = number of the SARS-Cov samples, ACE2 i SARS = expression levels of ACE2 in each SARS-Cov sample.
Subsequently, the threshold of Candidate-Gene Scores was identified through double normal distribution model from R packages “mixtools”. The GESA v4.0.3 software was used for Gene Set Enrichment Analysis (GSEA) of ACE2 in SARS-Cov groups [37]. The parameters were set as following: number of permutations = 1000, min size for excluding sets = 15, max size for excluding sets = 500, the pathways with p value < 0.05 were identified significant. We then conducted gene set variation analysis (GSVA) by “GSVA” package (Hanzelmann et al., 2013) and used annotation gmt file as the reference gene sets from MSigDB v7.0 (https://www.gsea-msigdb.org/gsea/msigdb/) (Liberzon et al., 2011).
2.3. Immune infiltration analysis
To further evaluate the immune cell infiltration features of normal lung tissues and SARS-Cov-2 infected samples, we used the “ssGSEA” method of “GSVA” package to transform the gene expression profiles into immune infiltration files based on immune annotation gmt file including 29 immune cell types or functions (Barbie et al., 2009, He et al., 2018). Then the samples were redeployed into immune-associated groups through average-neighbor clustering. The heatmap of immune infiltration was constructed using “pheatmap” R package and the comparison of ACE2 and the components of immune cells among subgroups with different extent of immune response was performed using Kruskal-Wallis test.
2.4. Correlation analysis
For validating the mechanism of ACE2 modulating immune response in SARS-Cov infection, we performed Pearson correlation between ACE2 expression and key genes of JAK-STAT signaling pathway in SARS-Cov and SARS-Cov-2 infected samples and control, and used an R package “ggstatsplot” to perform the data visualization (https://CRAN.R-project.org/package=ggstatsplot). Moreover, Finally, those genes with significant positively correlated with the expression of ACE2 were compared between SARS-Cov and SARS-Cov-2 infected samples and control again using Wilcoxon test.
3. Results
3.1. Expression features of ACE2 in coronavirus infection
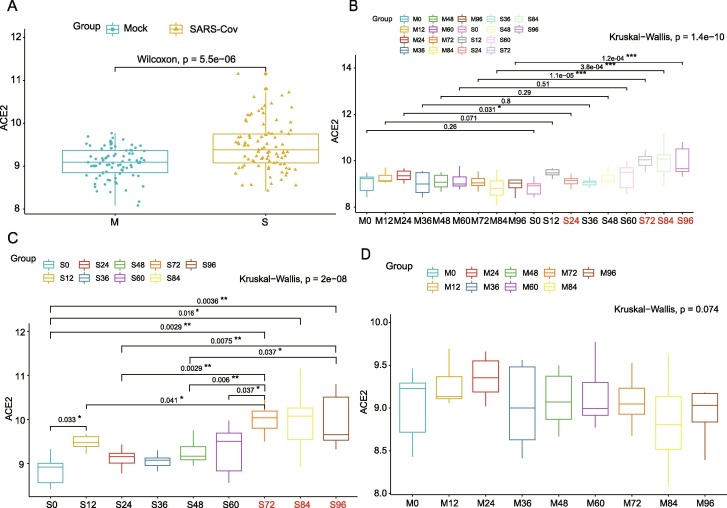
To evaluate the temporal feature of ACE2 expression after coronavirus infection, we analyzed the dynamic change of expression levels of ACE2 in distinct infection time phases. The expression of ACE2 in SARS-Cov groups, as expected, was significantly elevated compared with the control (Fig. 1 A, p < 0.001). Interestingly, the expression level of ACE2 was not significantly altered at early stage (<72 h) of infection but markedly upregulated at 72, 84 and 96 h after infection (Fig. 1B, p < 0.001). We further analyzed the expression levels of ACE2 in different time groups in control and SARS-Cov group respectively. Notably, the expression of ACE2 didn’t significantly increased at any time in the control while dramatically increased at 72 h after SARS-Cov infection compared to previous times and reminded a high level after 72 h (Fig. 1C, p < 0.001; Fig. 1D, p > 0.05).
Fig. 1.
Expression features of ACE2 in HAEs after SARS-Cov infection. A. the expression of ACE2 in SARS-Cov and control groups; B. Different expression of ACE2 between SARS-Cov and control groups based on time phases. C. Different expression of ACE2 in SARS-Cov groups based on time phases. D. Different expression of ACE2 in the control group based on time phases.
3.2. Functional enrichment analysis of ACE2 in coronavirus infection
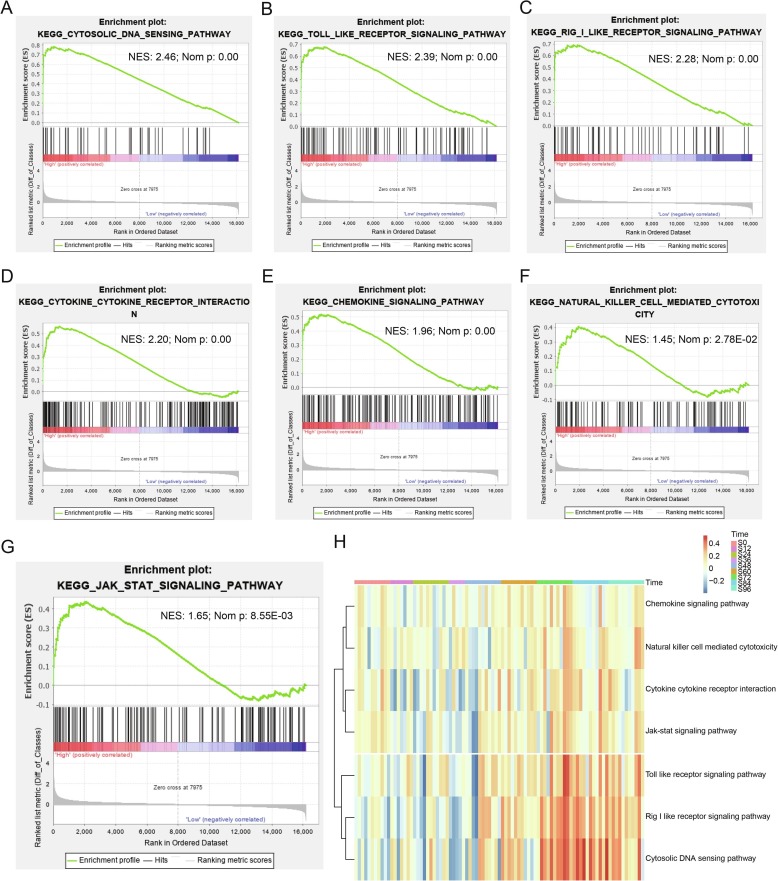
To further investigate the biological potential role of ACE2 in SARS-Cov infection, we divided the SARS-Cov subgroups into two groups with high or low expression levels of ACE2 based on Candidate-Gene Scores. As shown in Supplementary Fig. 1A, the Candidate-Gene Score 2.4 was set as the threshold for division based on double normal distribution model. As expected, single-gene GSEA analysis indicated high-expression ACE2 was mainly enriched in pathways of immunological activation including cytosolic DNA sensing pathway, toll like receptor signaling pathway, rig i like receptor signaling pathway, cytokine-cytokine receptor interaction, chemokine signaling pathway, JAK-STAT signaling pathway and natural killer cell mediated cytotoxicity (Table 1 , Fig. 2 A–G). Moreover, ACE2 was significantly associated with process of virus replication such as cell cycle, homologous recombination and oocyte meiosis (Table 1). Subsequently, gene set enrichment score analyzed by GSVA and the heatmap of pathways in SARS-Cov showed enrichment scores increased over infection time, maintaining with a high level after 72 h.
Table 1.
Results of GSEA of high-expression ACE2 in SARS groups.
| KEGG pathway | NES | NOM p value |
|---|---|---|
| Cytosolic DNA sensing pathway | 2.46 | 0.00 |
| Toll like receptor signaling pathway | 2.39 | 0.00 |
| Rig I like receptor signaling pathway | 2.28 | 0.00 |
| Cytokine-Cytokine receptor interaction | 2.20 | 0.00 |
| Chemokine signaling pathway | 1.96 | 0.00 |
| Cell cycle | 1.58 | 8.49E-03 |
| JAK-STAT signaling pathway | 1.60 | 8.55E-03 |
| Nicotinate and nicotinamide metabolism | 1.65 | 2.18E-02 |
| Natural killer cell mediated cytotoxicity | 1.45 | 2.78E-02 |
| Homologous recombination | 1.64 | 2.93E-02 |
| Oocyte meiosis | 1.42 | 4.49E-02 |
Fig. 2.
Results of GSEA and GSVA for high-expression ACE2 in SARS-Cov infection. A-G. GSEA showing the related biological potential role of ACE2 in SRAS, including (A) Cytosolic DNA sensing pathway, (B) Toll like receptor signaling pathway, (C) Rig I like receptor signaling pathway, (D) Cytokine-Cytokine receptor interaction, (E) Chemokine signaling pathway, (F) JAK-STAT signaling pathway and (G) Natural killer cell mediated cytotoxicity. H. The heatmap showing enrichment of associated-pathways through GSVA in SARS-Cov groups among different time phases.
3.3. Immune infiltration characterization in normal lung and coronavirus infection
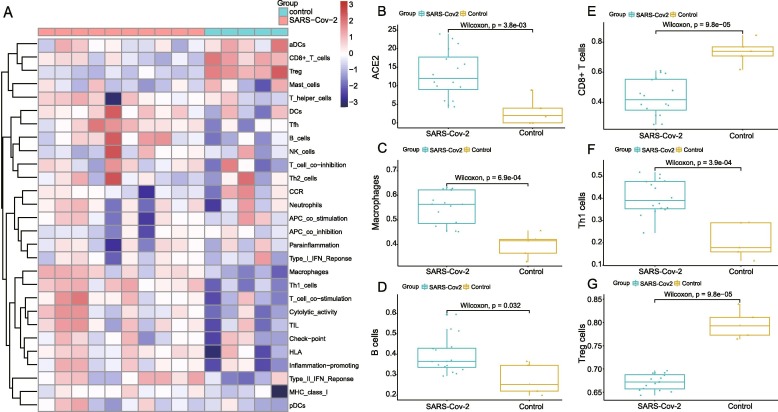
In order to further present immune infiltration features, we conducted ssGSEA analysis between SARS-Cov-2 infected and healthy lung tissues (Supplementary Table 3). The heatmap of immune infiltration is exhibited in Fig. 3 A and the different degree of immune cell infiltration was clearly displayed between SARS-Cov-2 and control group. Furthermore, the high-expression of ACE2 was identified in SARS-Cov-2 infected groups (Fig. 3B). To further demonstrate the role of different immune cells in SARS-Cov-2 infection, main immune cells of adaptive immune response were compared between infected and control subgroups. After SARS-Cov-2 infection, high immune cell infiltration was predominantly linked with macrophages, B cells and Th1 cells while low-level infiltration was mainly associated with CD8 + T cells and Treg cells (Fig. 3C–G).
Fig. 3.
Results of ACE2′s immune infiltration characterization in normal lung and SARS-Cov infection. A The heatmap showing the degree of immune infiltration between SARS-Cov-2 infected and control lung tissue including 29 immune cells and responses. B. Different expression of ACE2 based on groups of SARS-Cov-2 infection and control (B). C-G. Infiltration of immune cells between SARS-Cov-2 and control group including macrophages (C), B cells (D), CD8 + T cells (E), Th1 cells (F) and Treg cells (G).
3.4. Correlation analysis between ACE2 and JAK-STAT signaling pathway
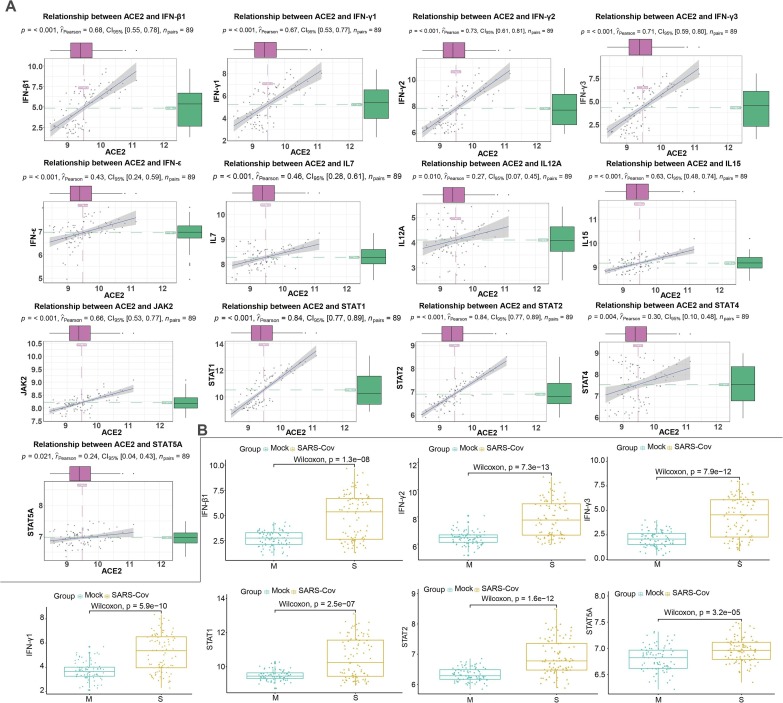
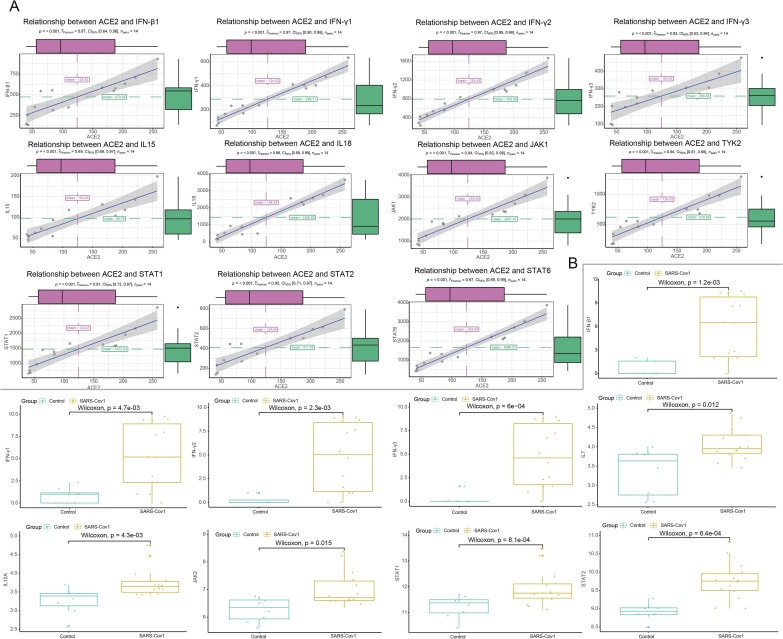
Considering the JAK-STAT signaling pathway was instrumental in inflammatory networks (Chen et al., 2011), we further explored the correlation analysis between ACE2 and key genes related to JAK-STAT pathway in airway epithelial cells with or without infection respectively. Interestingly, cytokines including I/II interferon family (including IFN-β1, IFN-γ1, IFN-γ2, IFN-γ3 and IFN-ε) and interleukin family members (such as IL7, IL 12A and IL15) manifested significantly positively correlation with ACE2 in SARS-Cov groups (Fig. 4 A, Table 2 ) while uncorrelated or less correlated with ACE2 in the control (Supplementary Fig. 1C). As for main components in JAK-STAT pathway, we found JAK2, STAT1, STAT2, STAT4 and STAT5A were significantly positively correlated with ACE2 in the SARS-Cov infetion while other factors (JAK1, JAK3, STAT3, STAT5B and STAT6) were uncorrelated or negatively correlated with ACE2. In addition, we also conducted comparison of the expression of these components between the infected and control. In accordance with the results of correlation analysis, IFN-β1, IFN-γ1, IFN-γ2, IFN-γ3, STAT1, STAT2 and STAT5A were still up-regulated in SARS-Cov group (Fig. 4B). However, the expression of JAK1, JAK2 and JAK3 were not significantly altered between healthy and SARS-Cov (Supplementary Fig. 1B). Further, members of the I/II interferon family, interleukin family and JAK-STAT signaling members also demonstrated similar positive correlation with ACE2 in human Calu-3 cells after SARS-Cov and SARS-Cov- 2 infection (including IFN-β1, IFN-γ1, IFN-γ2, IFN-γ3, IL18, JAK1, TYK2, STAT1, STAT2 and STAT6), consistent with the results of SARS-Cov infection of HAEs (Fig. 5 A; Fig. 6 A). In addition, the expression of several essential roles in this pathway (such as IFN-β1, IFN-γ1, IFN-γ2, IFN-γ3, STAT1 and STAT2) also dramatically increased in human Calu-3 cells with SARS-Cov and SARS-Cov-2 infection (Fig. 5B; Fig. 6B). Notably, the expression of JAk1, IL7 and IL12A was also up-regulated in Calu-3 cells after infection, which is different from that in HAEs. To better understand the relationship of these components, a schematic model of coronavirus infection was exhibited in Fig. 7 .
Fig. 4.
Correlation analysis between ACE2 and JAK-STAT signaling pathway in SARS-Cov infected HAEs. A. The scatter diagrams showing significant positively-associated factors with ACE2 in JAK-STAT signaling pathway. B Expression of associated factors of JAK-STAT signaling pathway in SARS-Cov infection.
Table 2.
Results of Correlation analysis between ACE2 and JAK-STAT pathway in SARS-Cov infected HAEs.
| Group | Symbol | Correlation | p value | Group | Symbol | Correlation | p value |
|---|---|---|---|---|---|---|---|
| SARS | IL15 | 0.63 | 4.16E-11 | Control | IL15 | 0.60 | 1.10E-09 |
| SARS | IL29 | 0.67 | 8.30E-13 | Control | IL29 | −0.38 | 3.18E-04 |
| SARS | IL6 | −0.14 | 2.06E-01 | Control | IL6 | 0.05 | 6.78E-01 |
| SARS | IL23A | 0.05 | 6.50E-01 | Control | IL23A | 0.67 | 2.12E-12 |
| SARS | IL7 | 0.46 | 6.61E-06 | Control | IL7 | 0.39 | 2.55E-04 |
| SARS | IL12A | 0.27 | 9.88E-03 | Control | IL12A | 0.31 | 4.48E-03 |
| SARS | IL28B | 0.71 | 4.03E-15 | Control | IL28B | 0.05 | 6.67E-01 |
| SARS | IL28A | 0.73 | 6.31E-16 | Control | IL28A | −0.02 | 8.50E-01 |
| SARS | IFNA16 | 0.08 | 4.51E-01 | Control | IFNA16 | 0.27 | 1.16E-02 |
| SARS | IFNA10 | −0.11 | 3.12E-01 | Control | IFNA10 | −0.05 | 6.62E-01 |
| SARS | IFNE | 0.43 | 2.49E-05 | Control | IFNE | −0.26 | 1.70E-02 |
| SARS | IFNA6 | −0.14 | 1.93E-01 | Control | IFNA6 | 0.19 | 8.10E-02 |
| SARS | IFNB1 | 0.68 | 3.04E-13 | Control | IFNB1 | −0.34 | 1.68E-03 |
| SARS | IFNA5 | −0.03 | 8.07E-01 | Control | IFNA5 | −0.27 | 1.33E-02 |
| SARS | IFNA21 | −0.07 | 5.26E-01 | Control | IFNA21 | 0.26 | 1.66E-02 |
| SARS | IFNA4 | −0.15 | 1.71E-01 | Control | IFNA4 | −0.51 | 5.19E-07 |
| SARS | JAK2 | 0.66 | 1.39E-12 | Control | JAK2 | 0.69 | 3.93E-13 |
| SARS | JAK3 | −0.40 | 1.22E-04 | Control | JAK3 | −0.21 | 5.93E-02 |
| SARS | JAK1 | −0.31 | 3.47E-03 | Control | JAK1 | −0.39 | 2.11E-04 |
| SARS | TYK2 | −0.16 | 1.38E-01 | Control | TYK2 | −0.22 | 4.46E-02 |
| SARS | STAT1 | 0.84 | 6.22E-25 | Control | STAT1 | 0.52 | 2.55E-07 |
| SARS | STAT2 | 0.84 | 4.14E-25 | Control | STAT2 | 0.20 | 6.10E-02 |
| SARS | STAT3 | −0.13 | 2.37E-01 | Control | STAT3 | −0.47 | 4.79E-06 |
| SARS | STAT4 | 0.30 | 3.99E-03 | Control | STAT4 | 0.42 | 5.63E-05 |
| SARS | STAT5A | 0.24 | 2.15E-02 | Control | STAT5A | −0.01 | 9.22E-01 |
| SARS | STAT5B | −0.29 | 6.21E-03 | Control | STAT5B | −0.40 | 1.42E-04 |
| SARS | STAT6 | −0.13 | 2.40E-01 | Control | STAT6 | −0.49 | 1.93E-06 |
Fig. 5.
Correlation analysis between ACE2 and JAK-STAT signaling pathway in SARS-Cov infected human Calu-3 cells. A. The scatter diagrams showing significant positively-associated factors with ACE2 in JAK-STAT signaling pathway. B Expression of associated factors of JAK-STAT signaling pathway in SARS-Cov infection.
Fig. 6.
Correlation analysis between ACE2 and JAK-STAT signaling pathway in SARS-Cov-2 infected human Calu-3 cells. A. The scatter diagrams showing significant positively-associated factors with ACE2 in JAK-STAT signaling pathway. B Expression of associated factors of JAK-STAT signaling pathway in SARS-Cov-2 infection.
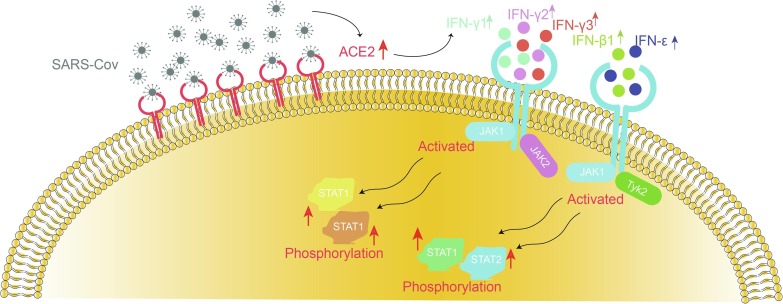
Fig. 7.
The schematic model showing the hypothesis for COVID-19 infection.
4. Discussion
Although substantial clinical retrospective studies have clarified the clinical features of COVID-19, there is still lack of explicit molecular mechanism and effective treatment for COVID-19 infection. In this study, a total of six serial gene expression files of HAEs, human Calu-3 cells and lung tissues were conjointly analyzed and we validated the high-expression levels of ACE2 in SARS-Cov and SARS-Cov-2 infection, coincided with previous studies (Gui et al., 2017). Moreover, comparison among time phases indicated the expression of ACE2 dramatically increased and remained a high level at 72 h after SARS-Cov infection, suggesting that there was a comparable incubation period before ACE2 drastically upregulated after the infection of COVID-19 and its relationship with clinical latency needs further investigation.
It has been reported that ACE2 was associated with adaptive immune responses and participated in regulating the produce of cytokines associated with ARDS which induced by coronavirus infection (Rockx et al., 2009, Fischer et al., 2017). Similarly, in our study, results of single-gene GSEA and GSVA also revealed ACE2 was related to the pathways of immunological activation such as cytokine-cytokine receptor interaction, chemokine signaling pathway, JAK-STAT signaling pathway and natural killer cell mediated cytotoxicity, consistent with above studies. In addition, we also found the elevated ACE2 was associated with virus replication through the pathway of cell cycle, homologous recombination and oocyte meiosis (He et al., 2010, Gillespie et al., 2012), also reported in He’s study(He et al., 2020).
Previous researches have certified coronavirus could activate innate immunity system (Frieman et al., 2008) and cause T-cell response irregular through stimulating T-cell apoptosis(Zhou et al., 2014). In this study, the comparison of immune cells between SARS-Cov-2 infection and control showed the increase of antigen presenting cells (macrophages and B cells) and the decrease of CD8 + T cells, suggesting activation of innate immunity response by enhancing the capacity of presenting antigen and releasing inflammatory factors. Moreover, Treg cells were exert an essential role to maintain immune homeostasis by inhibiting various inflammatory responses (Belkaid and Tarbell, 2009) and Treg cell ablation could lead to multiple autoimmune syndrome (Kim et al., 2007). Notably, experimental evidence from animal models indicated lack or dysfunction of Treg cells was accompanied with sharply augmented Th1 responses and activation of IFN-γl-dependent immune responses (Siebler et al., 2003, Lu et al., 2010). Interestingly, the expression of Treg cells significantly decreased while Th1 cells dramatically increased in SARS-Cov-2 infection, which in consistent with an adjusted immunity and overactivated inflammation during the process of SARS-Cov-2 infection. However, whether the overexpression of ACE2 is related to the infiltration of immune cells needs further investigation.
The JAK-STAT signaling pathway was reported widely involved in the regulation of massive inflammatory responses and promotes cell migration and apoptosis (Chen et al., 2011). Besides, The JAK-STAT signaling pathway was the major downstream pathway of the activation type I IFNs after virus infection and dysfunction of JAK-STAT pathway was also major patterns for immune escape of virus from normal immune surveillance (Ma and Suthar, 2015, Nelemans and Kikkert, 2019, Li et al., 2020). However, the relation between high expression of ACE2 and JAK-STAT pathway in coronavirus infection has few been reported. In our study, we found significant association between ACE2 and components (including JAK2, STAT1, STAT2, STAT4 and STAT5A), which indicating ACE2 activation in SARS-Cov infection was associated with high levels of type I and II IFNs and STAT family. These results indicated the elevated ACE2 expression might activate JAK-STAT signaling pathway to modulate the immune response after SARS-Cov infection. More importantly, above relation between ACE2 and IFN-JAK-STAT signaling was further validated in another different cell line with SARS-Cov and SARS-Cov-2 infection, directly indicating the close connection of ACE2 and JAK-STAT signaling during the process of COVID-19 infection.
However, there still are some limitations in our study. Despite of the common cell surface receptors ACE2 and 89.1% nucleotide similarity with SARS-Cov, there might be different in potential mechanism of COVID-19 virus infection. Besides, the sample size in our study is still limited and need enlarged studies to support our results. Furthermore, concrete relation between COVID-19 and JAK-STAT signaling pathway in pathogenesis of COVID-19 infection remains to be verified through subsequent animal models or cell experiments.
In conclusion, we observed the expression of ACE2 dramatically increased and remained a high level at 72 h after SARS-Cov infection. The activation of innate and adaptive immune responses might participate in the progression of SARS-Cov-2 infection, which accompanied with marked infiltrated immune cells, including B cells, macrophages and Th1 cells and reduced present of Treg T cells and CD8 + T cells. JAK-STAT signaling pathway, in particular, the downstream of type I/II IFNs might to be involved in the related action mechanism of COVID-19 infection. These findings are expected to shed light on the potential pathogenesis and provide hints to improve on therapeutic strategies for COVID-19 infection.
CRediT authorship contribution statement
Jing Luo: Conceptualization, Formal analysis, Writing - original draft, Writing - review & editing. Saisai Lu: Formal analysis, Writing - original draft. Mengjiao Yu: Software, Visualization. Lixia Zhu: Data curation, Formal analysis. Chengwei Zhu: Data curation, Validation. Chenlu Li: Data curation. Jinxia Fang: Visualization. Xiaochun Zhu: Project administration. Xiaobing Wang: Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81700062) and the Natural Science Foundation of Zhejiang Province grants (LQ16H010003), Science and Technology Project of Zhejiang Provincial Health Commission (2019RC050), Zhejiang Xinmiao Talents Program (2019R413082) and the General scientific projects of Zhejiang Education Department (Y201942208).
Ethics Statement
There is no animal or experimental ethics involved in this study.
Data Availability Statement
The public datasets supporting the conclusions of this article are available in the.
GEO database with GSE47960, GSE47961, GSE47962 and GSE47963 (https://www.ncbi.nlm.nih.gov/geo/). All data of GSEA and immune infiltration supporting this paper have been uploaded as electronic supplementary material.
Author contributions
Jing Luo and Saisai Lu contributed to the drafting of the manuscript. Mengjiao Yu, Lixia Zhu and Chengwei Zhu contributed to data analysis, Chenlu Li and Jinxia Fang contributed to data acquisition and analysis, Xiaochun Zhu and Xiaobing Wang contributed to design of the study and revision of the manuscript. All authors have read and approved the final version of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2020.145325.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., Frohling S., Chan E.M., Sos M.L., Michel K., Mermel C., Silver S.J., Weir B.A., Reiling J.H., Sheng Q., Gupta P.B., Wadlow R.C., Le H., Hoersch S., Wittner B.S., Ramaswamy S., Livingston D.M., Sabatini D.M., Meyerson M., Thomas R.K., Lander E.S., Mesirov J.P., Root D.E., Gilliland D.G., Jacks T., Hahn W.C. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y., Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annu. Rev. Immunol. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- Chen P., Huang L., Zhang Y., Qiao M., Yao W., Yuan Y. The antagonist of the JAK-1/STAT-1 signaling pathway improves the severity of cerulein-stimulated pancreatic injury via inhibition of NF-kappaB activity. Int. J. Mol. Med. 2011;27:731–738. doi: 10.3892/ijmm.2011.632. [DOI] [PubMed] [Google Scholar]
- Cheng Z.J., Shan J. 2019 Novel coronavirus: where we are and what we know. Infection. 2020 doi: 10.1007/s15010-020-01401-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Wu H., Grossman J.M., Hanvivadhanakul P., FitzGerald J.D., Park G.S., Dong X., Chen W., Kim M.H., Weng H.H., Furst D.E., Gorn A., McMahon M., Taylor M., Brahn E., Hahn B.H., Tsao B.P. Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54:2951–2962. doi: 10.1002/art.22044. [DOI] [PubMed] [Google Scholar]
- Fischer, D.D., Kandasamy, S., Paim, F.C., Langel, S.N., Alhamo, M.A., Shao, L., Chepngeno, J., Miyazaki, A., Huang, H.C., Kumar, A., Rajashekara, G., Saif, L.J., Vlasova, A.N., 2017. Protein malnutrition alters tryptophan and angiotensin-converting enzyme 2 homeostasis and adaptive immune responses in human rotavirus-infected gnotobiotic pigs with human infant fecal microbiota transplant. Clin. Vaccine Immunol. 24. [DOI] [PMC free article] [PubMed]
- Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133:101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie K.A., Mehta K.P., Laimins L.A., Moody C.A. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol. 2012;86:9520–9526. doi: 10.1128/JVI.00247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27:119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., Zhang, L., Ran, Q., Xiong, A., Wang, J., Wu, D., Chen, F., Li, G., 2020. Integrative bioinformatics analysis provides insight into the molecular mechanisms of 2019-nCoV. MedRxiv.
- He Y., Jiang Z., Chen C., Wang X. Classification of triple-negative breast cancers based on immunogenomic profiling. J. Exp. Clin. Cancer Res. 2018;37:327. doi: 10.1186/s13046-018-1002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Xu K., Keiner B., Zhou J., Czudai V., Li T., Chen Z., Liu J., Klenk H.D., Shu Y.L., Sun B. Influenza A virus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 2010;84:12832–12840. doi: 10.1128/JVI.01216-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Rasmussen J.P., Rudensky A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Kirou K.A., Lee C., George S., Louca K., Papagiannis I.G., Peterson M.G., Ly N., Woodward R.N., Fry K.E., Lau A.Y., Prentice J.G., Wohlgemuth J.G., Crow M.K. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- Kirou K.A., Lee C., George S., Louca K., Peterson M.G., Crow M.K. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–1503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Sui J., Huang I.C., Kuhn J.H., Radoshitzky S.R., Marasco W.A., Choe H., Farzan M. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. 2007;367:367–374. doi: 10.1016/j.virol.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L.F., Boldin M.P., Chaudhry A., Lin L.L., Taganov K.D., Hanada T., Yoshimura A., Baltimore D., Rudensky A.Y. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–929. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D.Y., Suthar M.S. Mechanisms of innate immune evasion in re-emerging RNA viruses. Curr. Opin. Virol. 2015;12:26–37. doi: 10.1016/j.coviro.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelemans T., Kikkert M. Viral innate immune evasion and the pathogenesis of emerging RNA virus infections. Viruses. 2019;11 doi: 10.3390/v11100961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Organization, W.H., 2020. Director-General's remarks at the media briefing on 2019-nCoV on 11 February 2020.
- Richardson P., Griffin I., Tucker C., Smith D., Oechsle O., Phelan A., Stebbing J. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet. 2020;395:e30–e31. doi: 10.1016/S0140-6736(20)30304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Baas T., Zornetzer G.A., Haagmans B., Sheahan T., Frieman M., Dyer M.D., Teal T.H., Proll S., van den Brand J., Baric R., Katze M.G. Early upregulation of acute respiratory distress syndrome-associated cytokines promotes lethal disease in an aged-mouse model of severe acute respiratory syndrome coronavirus infection. J. Virol. 2009;83:7062–7074. doi: 10.1128/JVI.00127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebler J., Wirtz S., Klein S., Protschka M., Blessing M., Galle P.R., Neurath M.F. A key pathogenic role for the STAT1/T-bet signaling pathway in T-cell-mediated liver inflammation. Hepatology. 2003;38:1573–1580. doi: 10.1016/j.hep.2003.09.020. [DOI] [PubMed] [Google Scholar]
- Suyundikov A., Stevens J.R., Corcoran C., Herrick J., Wolff R.K., Slattery M.L. Accounting for dependence induced by weighted KNN imputation in paired samples, motivated by a colorectal cancer study. PLoS ONE. 2015;10:e0119876. doi: 10.1371/journal.pone.0119876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F., Zhao, S., Yu, B., Chen, Y.-M., Wang, W., Hu, Y., Song, Z.-G., Tao, Z.-W., Tian, J.-H., Pei, Y.-Y., Yuan, M.-L., Zhang, Y.-L., Dai, F.-H., Liu, Y., Wang, Q.-M., Zheng, J.-J., Xu, L., Holmes, E.C., Zhang, Y.-Z., 2020. Complete genome characterisation of a novel coronavirus associated with severe human respiratory disease in Wuhan, China. bioRxiv.
- Xu, X., Chen, P., Wang, J., Feng, J., Zhou, H., Li, X., Zhong, W., Hao, P., 2020. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. [DOI] [PMC free article] [PubMed]
- Yu F., Du L., Ojcius D.M., Pan C., Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22:74–79. doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Chu H., Li C., Wong B.H., Cheng Z.S., Poon V.K., Sun T., Lau C.C., Wong K.K., Chan J.Y., Chan J.F., To K.K., Chan K.H., Zheng B.J., Yuen K.Y. Active replication of Middle East respiratory syndrome coronavirus and aberrant induction of inflammatory cytokines and chemokines in human macrophages: implications for pathogenesis. J. Infect. Dis. 2014;209:1331–1342. doi: 10.1093/infdis/jit504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P., Yang, X.-L., Wang, X.-G., Hu, B., Zhang, L., Zhang, W., Si, H.-R., Zhu, Y., Li, B., Huang, C.-L., Chen, H.-D., Chen, J., Luo, Y., Guo, H., Jiang, R.-D., Liu, M.-Q., Chen, Y., Shen, X.-R., Wang, X., Zheng, X.-S., Zhao, K., Chen, Q.-J., Deng, F., Liu, L.-L., Yan, B., Zhan, F.-X., Wang, Y.-Y., Xiao, G.-F., Shi, Z.-L., 2020. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in 2 humans and its potential bat origin. bioRxiv.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The public datasets supporting the conclusions of this article are available in the.
GEO database with GSE47960, GSE47961, GSE47962 and GSE47963 (https://www.ncbi.nlm.nih.gov/geo/). All data of GSEA and immune infiltration supporting this paper have been uploaded as electronic supplementary material.