Abstract
Coronavirus disease 2019 (COVID-19) is a novel harmful respiratory disease that has rapidly spread worldwide. At the end of 2019, COVID-19 emerged as a previously unknown respiratory disease in Wuhan, Hubei Province, China. The world health organization (WHO) declared the coronavirus outbreak a pandemic in the second week of March 2020. Simultaneous deep learning detection and classification of COVID-19 based on the full resolution of digital X-ray images is the key to efficiently assisting patients by enabling physicians to reach a fast and accurate diagnosis decision. In this paper, a simultaneous deep learning computer-aided diagnosis (CAD) system based on the YOLO predictor is proposed that can detect and diagnose COVID-19, differentiating it from eight other respiratory diseases: atelectasis, infiltration, pneumothorax, masses, effusion, pneumonia, cardiomegaly, and nodules. The proposed CAD system was assessed via five-fold tests for the multi-class prediction problem using two different databases of chest X-ray images: COVID-19 and ChestX-ray8. The proposed CAD system was trained with an annotated training set of 50,490 chest X-ray images. The regions on the entire X-ray images with lesions suspected of being due to COVID-19 were simultaneously detected and classified end-to-end via the proposed CAD predictor, achieving overall detection and classification accuracies of 96.31% and 97.40%, respectively. Most test images from patients with confirmed COVID-19 and other respiratory diseases were correctly predicted, achieving average intersection over union (IoU) greater than 90%. Applying deep learning regularizers of data balancing and augmentation improved the COVID-19 diagnostic performance by 6.64% and 12.17% in terms of the overall accuracy and the F1-score, respectively. It is feasible to achieve a diagnosis based on individual chest X-ray images with the proposed CAD system within 0.0093 s. Thus, the CAD system presented in this paper can make a prediction at the rate of 108 frames/s (FPS), which is close to real-time. The proposed deep learning CAD system can reliably differentiate COVID-19 from other respiratory diseases. The proposed deep learning model seems to be a reliable tool that can be used to practically assist health care systems, patients, and physicians.
Keywords: COVID-19; Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); Respiratory diseases; Artificial intelligence (AI), Deep learning; Diagnosis
Introduction
Coronavirus disease 2019 (COVID-19) has recently become an unprecedented public health crisis worldwide [1]. At the end of December 2019, patients with a previously unknown respiratory disease were identified in Wuhan, Hubei Province, China [2]. By January 25, 2020, the diagnosis of COVID-19 had been confirmed in at least 1975 more patients since the first patient was hospitalized on December 12, 2019. COVID-19 caused by a new coronavirus named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [2, 3]. The typical symptoms of COVID-19 include fever, shortness of breath, dizziness, cough, headache, sore throat, fatigue, and muscle pain [2–4]. After the first case of COVID-19 was discovered in Wuhan, the virus has rapidly spread to 216 countries worldwide, largely due to human-to-human transmission of the virus early in the clinical course [1]. The COVID-19 pandemic has imposed substantial demands on the public health systems, health infrastructure, and economies of most countries worldwide [5]. Because the total number of people infected by SARS-CoV-2 has increased rapidly, the capacity of healthcare systems (i.e., beds, ventilators, care providers, masks, etc.) is insufficient to meet the demand. Due to the rapid transmission of SARS-CoV-2 from person to person, millions of people have been infected, more than four billion people have been instructed to remain at home, and many people have lost their jobs [1, 2, 5]. Severe COVID-19 has caused deaths worldwide [6]. As reported by the world health organization (WHO) on November 17, 2020 [6], the numbers of patients with confirmed cases of COVID-19, recovered COVID-19 patients, and non-surviving COVID-19 patients were 55.4M, 38.6M, and 1.3M, respectively. Moreover, education systems have been negatively affected by the COVID-19 pandemic, and schools and universities have switch to remote learning.
To date, the most widely used screening tool for the detection and diagnosis of COVID-19 has been real-time reverse-transcription polymerase chain reaction (RT-PCR) [7]. Radiological imaging techniques such as chest digital X-ray (CXR) and computed tomography (CT) are the standard screening tools used to detect and diagnose chest respiratory diseases early in the clinical course, including COVID-19 [1, 8]. Due to the low sensitivity of RT-PCR, radiological images are also used for diagnostic purposes in patients with symptoms of respiratory diseases. Although the CT is the gold standard, primary chest digital X-ray systems are still useful because they are faster, deliver a lower dose of radiation, are less expensive, and are more widely available [4, 8]. Indeed, CT scans or X-rays should be routinely obtained in addition to RT-PCR results to improve the accuracy of the diagnosis of COVID-19 [8]. However, the large number of patients who test positive for SARS-CoV-2 makes the use of regular screening on a daily basis challenging for physicians. Thus, on March 16, 2020,the United States administration encouraged experts and researchers to employ artificial intelligence (AI) techniques to combat the COVID-19 pandemic [1]. Currently, experts have started to use machine learning and deep learning technologies to develop CAD systems to assist physicians in increasing the accuracy of the diagnosis of COVID-19 [1, 8]. In the last few years, the use of deep learning methods as adjunct screening tools for physicians has attracted a great deal of interest. Deep learning CAD systems have been shown to be capable and reliable, and promising diagnostic performance has been achieved using the entire image without user intervention [9, 10]. The use of a deep learning CAD system could assist physicians and improve the accuracy of the diagnosis of COVID-19 [1]. Deep learning CAD systems have been successfully applied to predict different medical problems, such as breast cancer [9, 10], skin cancer [11, 12], and respiratory disease, using digital X-ray images [8]. The rapid spread of the COVID-19 pandemic and the consequent death of humans worldwide makes it necessary to apply deep learning technologies to develop CAD systems that can improve the diagnostic performance. This need was the motivation for developing a deep learning CAD system to diagnose COVID-19 based on entire digital X-ray images.
In this paper, our contributions to the diagnosis of COVID-19 based on digital X-ray images are as follows. First, a simultaneous deep learning CAD system that uses the YOLO predictor was adopted to detect and diagnose COVID-19 directly from entire chest X-ray images. Second, COVID-19 is differentiated from eight other respiratory diseases in a multiclass recognition problem. Third, deep learning regularizations of data balancing, augmentation, and transfer learning were also applied to improve the overall diagnostic performance for COVID-19. Finally, our proposed CAD system was trained and optimized with five-fold tests using data from two different digital X-ray datasets, COVID-19 [13, 14] and ChestX-ray8 [15]. The outcomes of this study can be used to guide other researchers when developing novel deep learning CAD frameworks to accurately diagnose COVID-19.
The objective of this work was to provide a practical and feasible CAD system based on AI that can help physicians, patients, healthcare systems, and hospitals by facilitating the faster and more accurate diagnosis of COVID-19.
The rest of this paper is organized as follows. A review of the relevant literature is presented in Section 2. The technical aspects of the deep learning CAD-based YOLO system are detailed in Section 3. The results of the experiment with COVID-19 are reported and discussed in Sections 4 and 5. Finally, the most important findings of this work are summarized in Section 6.
Related works
Starting in 2020, after the discovery of COVID-19, some artificial intelligence (AI) systems based on deep learning have been employed to detect COVID-19 on digital X-ray and CT images. In [16], Oh et al. presented a patch-based deep learning CAD system consisting of segmentation and classification stages that could identify COVID-19 based on CXR images. With regard to segmentation, FC-DenseNet103 was used to segment and extract the full lung regions from the entire CXR images. With regard to classification, multiple random patches (i.e., regions of interest) were extracted from the segmented lung regions for use as the input for the classification DL model. They used CXR images from multiple patients who were healthy and patients who were diagnosed with bacterial pneumonia, tuberculosis, and viral pneumonia associated with COVID-19. Diagnostic accuracies of 84.40% and 88.9% were achieved for the F1-score and overall accuracy, respectively. Ozturk et al. [8] proposed the deep learning DarkCovidNet that can automatically detect COVID-19 based on digital chest X-ray images. They developed their model using 17 convolutional layers with the aim of achieving binary classification (i.e., COVID-19 and no finding) and multinomial classification (i.e., COVID-19, no finding, and pneumonia) diagnoses. They achieved overall classification accuracies of 98.08% and 87.02% for the binary and multinomial classifications, respectively. Fan et al. [17] proposed a deep learning model called Inf-Net that can be used to identify or segment suspicious regions indicative of COVID-19 on chest CT images. They used a parallel partial decoder to generate the global representation of the final segmented maps. After that, they used implicit reverse attention and explicit edge attention to enhance the segmented boundaries. They achieved segmentation accuracies of 73.90% and 89.40% with regard to Dice and the enhanced-alignment index, respectively. In May 2020, Wang et al. [18] proposed COVID-Net, which was based on a deep learning model and could differentiate patients with COVID-19 from healthy individuals and those with pneumonia based on digital X-ray images. The classification performance of their model was compared with the those of VGG-19 and ResNet-50 using the same database of digital X-ray images [18]. The authors concluded that COVID-Net outperformed VGG-16 and ResNet-50, with positive predictive values (PPVs) of 90.50%, 91.30%, and 98.9% for healthy, pneumonia, and COVID-19, respectively. Hamdan et al. [19] presented a deep learning COVIDX-Net model that can be used to distinguish between COVID-19 patients and healthy individuals based on 50 digital chest X-ray images. They used seven well-established deep networks as feature extractors and compared their classification results. Compared with other deep learning models, VGG-19 and DensNet201 had the highest diagnostic performance value of 90%. Apostolopoulos et al. [20] tested the ability of five well-established deep learning networks to detect COVID-19 on digital X-ray images. They used three classifications, namely, normal, pneumonia, and COVID-19, and they achieved the best overall classification accuracy of 93.48% with VGG-19. Additionally, they tested all five deep learning models with regard to the binary classification problem (i.e., COVID-19 against non-COVID-19), and they achieved the highest accuracy of 98.75% with VGG-19. Sakshy et al. [21] proposed a three-phase deep learning detection model to detect COVID-19 on CT images with a binary classification task. They used data augmentation, transfer learning, and abnormality localization with different backend deep learning networks: ResNet18, ResNet50, ResNet101, and SqueezeNet. They concluded that the pre-trained ResNet18 using the transfer learning strategy achieved the best diagnostic results of 99.82%, 97.32%, and 99.40% in the training, validation, and test sets, respectively. Khan et al. [22] proposed a deep learning convolutional neural network (i.e., CoroNet) that could be used to diagnose COVID-19 as a multiclass problem based on whole chest X-ray images. They achieved an overall accuracy of 89.6% for the identification of COVID-19 from among bacterial pneumonia, viral pneumonia, and normal images. Narin et al. [23] compared the classification performances of three different deep learning convolutional neural networks (i.e., ResNet-50, InceptionV3, and InceptionResNetV2) using chest X-ray images. They evaluated the ability of those three models to differentiate patients with COVID-19 from individuals without COVID-19, and they achieved the best classification accuracy of 98% using ResNet-50. Ardakani et al. [24] evaluated ten different well-established DL models to diagnose COVID-19 on CT scans in routine clinical practice. They differentiated between COVID-19 and non-COVID-19 with a binary classification task, and they achieved the best diagnostic result using the ResNet-101 and Xception DL networks, with an overall accuracy of 99.40%. Pereira et al. [7] presented a classification scheme based on well-known texture descriptors and a convolutional neural network (CNN). They used a resampling algorithm to balance the training dataset for a multiclass classification problem. Their model achieved an average F1 score of 65%. Moreover, comprehensive survey studies on deep learning applications pertaining to COVID-19 are presented in [25, 26]. Such deep learning methods have been employed to diagnose COVID-19 on entire X-ray images. This is due to the lack of X-ray images with annotated regions of suspected lesions. However, it is not practical to use the entire X-ray image to achieve a reliable diagnosis of COVID-19 [27]. Thus, the detection of suspicious regions specific to individual respiratory diseases is critical for achieving a more accurate diagnosis because it could be used to derive more representative deep features of the abnormalities. To our knowledge, this is the regional convolutional deep learning CAD system developed to simultaneously detect COVID-19 and differentiate it from among other respiratory diseases based on chest X-ray images. The automatic detection of COVID-19 is a major challenge for researchers. Our previous promising diagnostic results from the breast cancer diagnosis CAD system using the YOLO predictor [9, 10] have encouraged us to employ a similar system to detect and classify COVID-19, with the aim of enhancing the diagnosis of COVID-19.
Material and methods
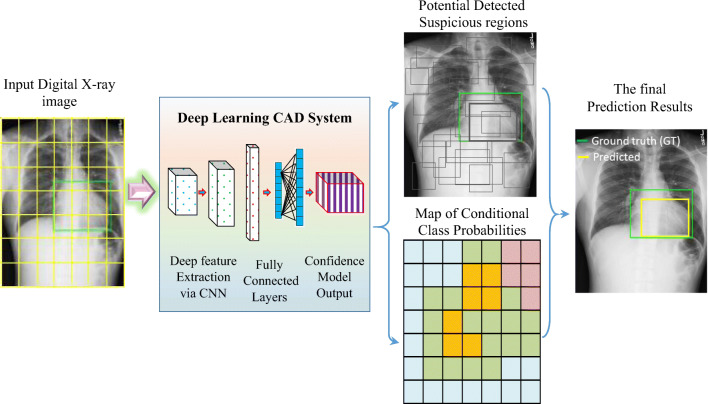
Deep learning computer-aided diagnosis (CAD) based on the YOLO predictor was used to simultaneously detect COVID-19 and differentiate it from eight other respiratory diseases: atelectasis, infiltration, pneumothorax, masses, effusion, pneumonia, cardiomegaly, and nodules. The CAD system presented in this paper has a unique deep learning framework structure; the system has been validated and can simultaneously detect and classify COVID-19. Figure 1 is a conceptual diagram of the proposed CAD system.
Fig. 1.
Schematic diagram of the proposed deep learning CAD system based on the YOLO predictor
Digital X-ray images dataset
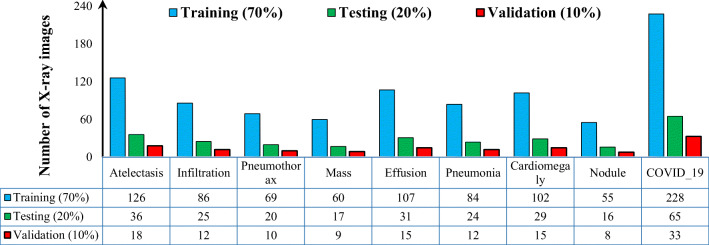
We used two different digital chest X-ray databases, namely, COVID-19 [13, 14] and ChestX-ray8 [15]. The data distributions for the two datasets are shown in Fig. 2.
Fig. 2.
Data distribution over all nine classes of respiratory diseases. The datasets for each classes were randomly split into 70%, 20%, and 10% for the training, testing, and validation sets, respectively
COVID-19 dataset
The COVID-19 dataset used in this study was collected from two different publicly available sources. First, we used the digital X-ray images from patients with COVID-19 collected by Cohen et al. [13] from different public sources, hospitals and radiologists. These images are publicly available to help expert researchers develop AI based on deep learning approaches to improve the diagnosis and understanding of COVID-19. Researchers from different countries try to constantly update these datasets and add more X-ray images. In this study, we used the available X-ray images acquired from 125 patients with COVID-19 (82 males and 43 females). Unfortunately, complete metadata were not yet available for all these patients. Age was provided for only 26 patients; the average age was 55 years. Second, we used digital X-ray images from patients with COVID-19 collected by a research team from Qatar University [14]. All these images are publicly available in portable network graphic (png) file format with a size of 1024 × 1024 pixels. This dataset is publicly provided for researchers to develop useful and impactful AI models with the aim of addressing the COVID-19 crisis. The metadata were not yet available for all patients with COVID-19. In this study, we used all available digital X-ray images from 201 patients with COVID-19. Thus, a total of 326 CXR images were collected and used to develop the proposed CAD system. The classification labels for these images are publicly available, but the information regarding the GT localization (i.e., bounding box) is not yet available for either COVID-19 dataset. This is because the CXR images are rapidly collected in the context of the pandemic. To locate the abnormalities, we asked two expert radiologists to annotate the abnormalities (i.e., lesions associated with COVID-19) localizations in a parallel manner. Since some CXR images were provided by the authors with some small white/black arrows, as shown in Figure 3a-c, showing the localization of the COVID-19 lesions, we compared the experts’ opinion with the existing annotations and marked the suspected lesions with a rectangle. Each bounding box GT was determined by the coordinates corresponding to the width (w), height (h), and center (x, y) of the abnormality. Figure. 3 shows some examples of COVID-19 lesions with the associated GT information.
Fig. 3.
Example cases of COVID-19 in different patients. The ground-truth (GT) information of the bounding box (i.e., green) for each case is superimposed on the original chest X-ray (CXR) image. The GT information was determined by expert physicians
ChestX-ray8
The ChestX-ray8 [15] dataset is the most frequently used and widely accessible medical imaging examination dataset available for eight different respiratory diseases: atelectasis, infiltration, pneumothorax, masses, effusion, pneumonia, cardiomegaly, and nodules. In this study, we used all CXR images with ground truth (GT) information involving the disease class label and the disease localization information as a labeled bounding box. The information pertaining to the GT bounding box (i.e., the starting point of the box (x,y), width (w), and height (h)) for each image is publicly available in the XML file [15]. As shown in Fig. 2, a total of 984 frontal views of CXR images were used, which were representative of eight different respiratory diseases. These images were accurately converted from DICOM format into ‘.png’ file format with a size of 1024 × 1024 pixels. Figure. 4 shows an example of an X-ray image for each disease class with the associated GT information.
Fig. 4.
Example cases of eight common respiratory diseases in different patients from the ChestX-ray8 dataset [15]: a atelectasis, b infiltration, c pneumothorax, d mass, e effusion, f pneumonia, g cardiomegaly, and h nodule. The ground-truth (GT) information of the bounding box (i.e., green) for each case is superimposed on the original image
Data preparation: Training, validation, and testing
To fine-tune and evaluate the proposed CAD system, the COVID-19 [13, 14] and ChestX-ray8 [15] datasets were used. As shown in Fig. 2, the chest X-ray images for each disease class were randomly divided as follows: 70% in the training dataset, 20% in the evaluation dataset, and 10% in the validation dataset [9, 10]. The hypertrainable parameters of the proposed deep learning system were selected via the training process using the training and validation datasets. After that, the final performance of the proposed CAD system was assessed using the evaluation set. Meanwhile, our proposed CAD system was assessed using five-fold tests in the training, validation, and evaluation datasets. These sets were generated by stratified partitioning to ensure equal testing of each X-ray image and to avoid system bias error. It is important to use k-fold cross-validation to develop a robust, reliable, and efficient CAD system, especially given the small sizes of medical datasets [9–11]. In addition, to prevent the development of bias in the proposed prediction model during the learning process due to an unbalanced training set, we used the following techniques. First, the training set for each mini-batch was automatically shuffled. Second, a weighted cross-entropy was used as a loss function to optimize the deep learning trainable parameters [28].
Balancing and augmentation strategies for the training dataset
Data balancing and augmentation strategies were applied to enlarge the size of the training dataset, avoid overfitting, and accelerate the learning process [9, 10]. These practical solutions were successfully applied to address the challenge of small datasets of annotated medical images [9, 10]. During training, each mini-batch included an almost equal number of digital X-ray images for each disease class [29, 30]. This was to avoid overfitting and prevent the performance of the deep learning model from being biased towards the disease class with the largest number of images (i.e., COVID-19). To balance the training sets and avoid having a majority of images related to COVID-19, the training images from the eight disease classes in the ChestX-ray8 dataset were flipped twice (i.e., left-right and up-down), generating 1378 chest X-ray images. Thus, the total number of images in all disease classes in the training set after balancing was 2295 (i.e., 917 original images from all disease classes including COVID-19 and 1378 balanced images from eight disease classes from the ChestX-ray8 dataset).
After data balancing, an augmentation strategy was applied for all nine disease classes as follows. First, the original chest X-ray images were randomly scaled and translated ten times. Second, the X-ray images for each class were rotated around the origin center by 0°, 45°, 90°, 135°, 180°, 225°, 270°, and 315°. Finally, the rotated X-ray images for each class with θ = 0° and 270° were flipped left-right and up-down. This ensured that each X-ray image for each balanced class was augmented 22 times. Thus, a total of 50,490 X-ray images were generated and used to train our proposed CAD system. For each k-fold test, the same data balancing and augmentation strategy was utilized. In addition, transfer learning was applied to initialize the trainable parameters using ImageNet [9, 10]. Then, the deep learning CAD system was fine-tuned using our training set of chest X-ray images [31].
The concept of the deep learning CAD system
To simultaneously predict (detect and classify) COVID-19 from among the other respiratory diseases, a deep learning CAD system based on the YOLO predictor was adopted and used. With regard to object detection, previous studies have employed conventional image processing algorithms, machine learning classifiers, or complex deep learning pipelines [9, 10]. In contrast, our proposed CAD system is a regressor model that can simultaneously detect the localization of potential disease lesions and predict the probabilities of those lesions belonging to specific disease classes [10]. It has a robust ability to simultaneously learn the characteristics of the entire input X-ray image and the background. Thus, it can locate regions with lesions indicative of respiratory diseases with fewer background errors than other existing methods [30]. In addition, it has a unique deep learning structure allowing it to simultaneously optimize trainable parameters end-to-end to tune the training weights for the detection and classification tasks. Unlike the Faster R-CNN [32] and sliding window [33] methods, YOLO inspects the regions suspected of containing disease lesions directly in the context of the entire chest X-ray images. The conceptual diagram of the CAD-based YOLO predictor is shown in Fig. 1.
In fact, the YOLO predictor starts by dividing the input X-ray image into N × N grid cells, as shown in Fig. 1. If the lesion (i.e., the lesion associated with COVID-19 or any other respiratory diseases) center falls into any grid cell, that cell is responsible for predicting that disease. For each grid cell, five anchors (i.e., bounding boxes) are assigned and used to predict the disease class to which the lesion belongs (i.e., COVID-19, pneumonia, etc.). For each anchor, YOLO predicts the disease class of the lesions based on five prediction parameters: center location (x,y), width (w), height (h), and confidence score probability (Prconf.). The confidence score interprets the YOLO-based confidence that the predicted box contains a lesion and how accurate it expects the representation of the final output prediction by that box to be.
During the training process, the predicted confidence in each anchor is calculated by the multiplying the probability of the existing respiratory disease (i.e., lesion) by the value of the intersections over union (IoU) as follows:
| 1 |
If the grid cell does not contain any respiratory disease lesion, the confidence of all bounding boxes of that cell should be zero. In contrast, if any suspected disease lesion falls in that grid cell, Prob(Object ) should be greater than zero. Thus, the confidence of all bounding boxes of that cell should also be greater than zero. However, the network has been optimized to achieve the highest object probability and the highest object confidence. Based on both object probability and, the coordinates of all bounding boxes are simultaneously optimized and adjusted to fit the object that is falling in the specific grid cell. During the training process, each grid cell predicts the conditional class probabilities Prob(Classi| Object ) for all nine disease classes (i.e., COVID-19 and other respiratory diseases). During training, the confidence score for a detected bounding box is determined based on the conditional class probabilities as follows:
| 2 |
where
| 3 |
Then,
| 4 |
During testing, to obtain the confidence score when there is no GT, the conditional class probability is multiplied by the individual box confidence value. The detected bounding boxes with the highest confidence values indicate that COVID-19 or another respiratory disease is present, which should be considered the final prediction output. However, the confidence score probability for each detected bounding box encodes the probability for each disease class and how well each box fits the classes of respiratory diseases. The confidence score for each box is computed as follows:
| 5 |
For each bounding box, only one disease class is predicted and assigned (i.e., COVID-19, pneumonia, mass, etc.). As long as all bounding boxes are assigned to the same grid cell, the disease class for these boxes should be the same, but they can have different confidence values and conditional probabilities. Finally, the detected box that has the maximum confidence probability should be used to determine the final predicted output of the proposed CAD system. Moreover, all other detected bounding boxes have with lower confidence scores are suppressed using the algorithm of non-max suppression (NMS).
Deep learning structure of the CAD system
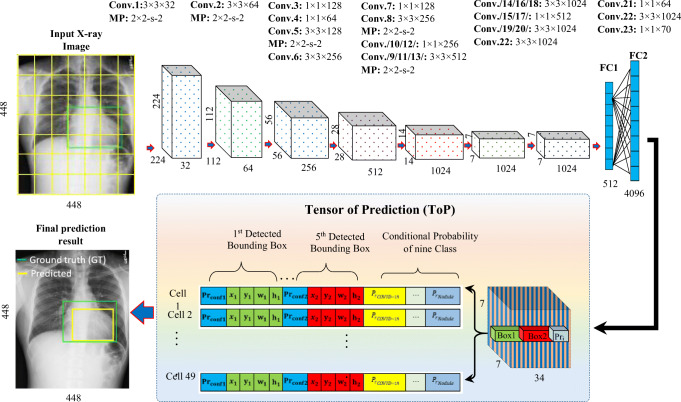
The structure of the proposed CAD system involves convolutional layers (Conv.), fully connected (FC) layers, and tensor of prediction (ToP), as shown in Fig. 5. Deep high-level features are extracted with 23 sequential convolutional layers, while the coordinates of the detected bounding boxes and the output probabilities are predicted with two FC layers. The total number of derived deep-feature maps depends mainly on the number of convolutional kernels that are used for each convolutional layer. Moreover, convolution reduction layers with a kernel size of 1 × 1 are added and utilized, followed by 3 × 3 convolutional layers, as shown in Fig. 5. This structure is used to reduce the size and compress the derived feature representations [9, 10]. In addition, batch normalization (BN) layer is used after each convolutional layer to reduce overfitting, accelerate convergence, and stabilize the training of the deep network [9, 10]. Down-sampling using max-pooling (MP) with a size of 2 × 2 is applied five times after the convolutional layers to minimize the dimensionality of the derived deep-feature maps and select the most appropriate deep features. The aggregated deep-feature maps from the last convolutional layer are concatenated and flattened using global average pooling (GAP) to feed directly into the fully connected layers. The numbers of nodes or neurons for the first and second dense layers are modified to 512 and 4096, respectively. The final output of the proposed model is called a tensor of prediction (ToP), which contains all detected predictors of the five anchors: coordinates (x, y, w, and h), confidence scores (Prconf.), and the conditional class probabilities of all nine disease classes (PrCOVID − 19, PrPneumonia, …etc). These predictors are encoded in the 3D matrix of the ToP with the size of N × N × (5 × B + C), where N, B, and C represent the number of grid cells, number of anchors, and number of classes, respectively [27]. As mentioned above, the input X-ray image is divided into 7 × 7 nonoverlapping grid cells, and each grid cell should detect any lesion (caused by COVID-19 or the other respiratory diseases) in that cell. The size of 7 × 7 was chosen to achieve the best performance, as shown in our previous studies. Meanwhile, five anchors or bounding boxes (i.e., B = 5) are used to detect the object in each grid cell. The proposed CAD system was built to detect and recognize nine classes of respiratory diseases (i.e., C = 9). Thus, the final output represents a 3D ToP with a size of 7 × 7 × 34. This means that the actual output layer of the fully connected layer has 7 × 7 × 34 or 1666 neurons. Each set of 34 neurons in the output FC layer is responsible for predicting all parameters of the five bounding boxes for each grid cell in the original chest X-ray image. Here, the key is that each grid cell can only make local predictions for its region of the input X-ray image. The proposed prediction model has the capability to detect and classify respiratory diseases faster than other recent detection methodologies. Moreover, the leaky rectified linear activation function is utilized in all the convolutional and fully connected layers, while the ReLU activation function, ϕ(x) = max (0, x), is only utilized in the final dense layer [27]. The leaky rectified linear activation function ϕ(θi) is expressed as the linear transformation of the input θi with a nonzero slope for the negative part of the activation function as follows:
| 6 |
Fig. 5.
Deep learning structure of the proposed CAD System
Experimental setting
The input digital CXR images were scaled using bilinear interpolation to a size of 448 × 448 pixels [9, 10]. In addition, the intensity of all CXR images was linearly normalized to a range of [0 ~ 1] as in [9, 10]. A multiscale training strategy was used to learn predictions across different resolutions of the input X-ray images [34]. Since the proposed network downsamples the derived deep-feature maps five times, the network randomly chose a new image dimension size for every 10 batches in multiplies of 32 (i.e., 320, 352, …, 608). Thus, the smallest input resolution was 320 × 320, and the largest input resolution was 608 × 608. Moreover, a mini-batch size of 24 and number of epochs of 120 were utilized to train and validate the proposed CAD system.
Implementation environment
To execute the experimental study, a PC with the following specifications was used. Intel® Core(TM) i7-6850K processor, RAM of 16.0 GB, 3.36 GHz, and four GPUs NVIDIA GeForce GTX1080.
Evaluation strategy
Our evaluation strategy used two conditions to determine whether the detected bounding boxes constituted a final true detection. First, the overlapping ratio (i.e., ) between the detected bounding box and its corresponding GT boxes had to be equal to or greater than an appropriate practical threshold. Second, the confidence score (i.e., Prconf) of the final detected box had to be equal to or greater than an appropriate threshold [27, 35]. Specifically, we always use the maximum confidence score to evaluate truly detected boxes [9, 10]. A high confidence score reflects a highly accurate prediction that the lesion exists in the detected bounding box [9, 10].
For the quantitative evaluation with each fold test, we used weighted objective metrics, including sensitivity (Sens.), specificity (Spec.), overall accuracy (Acc.), the F1-score or Dice, the Matthews correlation coefficient (Mcc.), the positive predictive value (PPV), and the negative predictive value (NPV) [9, 10]. To avoid having test sets that were unbalanced with regard to the nine disease classes, we used the weighted class strategy [27]. The weighted ratios for atelectasis, infiltration, pneumothorax, masses, effusion, pneumonia, cardiomegaly, nodules, and COVID-19 were 0.14, 0.10, 0.08, 0.06, 0.12, 0.09, 0.11, 0.06, and 0.25, respectively. All evaluation indices were computed using multiclass confusion matrices for each fold test [9, 10].
Experimental results
Detection results
The prober threshold of the IoU and confidence score
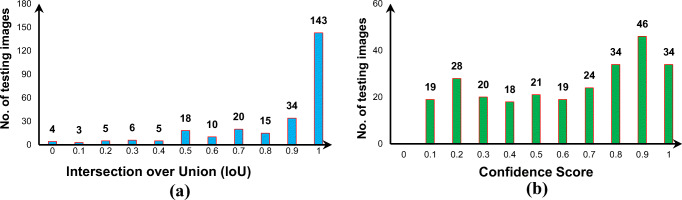
The presented CAD system is able to predict five anchors (i.e., bounding boxes) for each grid cell in entire X-ray images. To suppress undesirable detected boxes with very small confidence scores, the non-max suppression (NMS) technique was used [9, 34]. This algorithm required three consecutive stages during the testing phase. First, detected bounding boxes with confidence scores less than 0.005 were directly discarded. Second, among any remaining boxes, the box with the highest confidence score (i.e., Prconf.) was selected to represent the final predicted bounding box. Finally, any remaining boxes with IoUnms ≥ 50% with respect to the predicted box representing the final output identified in the second step were also discarded. Figure 6a shows the potential predicted boxes after applying NMS. During the evaluation phase, the overlapping ratio of the IoU between the final predicted box and its GT had to be greater than an appropriate threshold to ensure that the confidence that the predicted box includes the lesion is high. Experimentally, we found that the appropriate threshold for was greater than 45%, as shown in Fig. 7a. The majority of the final detected bounding boxes for the X-ray images in the test set had in IoU accuracy greater than 90%. The final detected boxes with were considered to be false detections. In addition to controlling the IoU, we also adjusted the appropriate threshold for the confidence score to ignore the undesirable detected boxes. Figure 6b-d show the detected bounding boxes stratified by different probability thresholds of the confidence score. Experimentally, we found that the appropriate confidence threshold was greater than 10%, as shown in Fig. 7b. This was for the detection of at least one suspected lesion in each test image for diagnostic purposes.
Fig. 6.
Effect of the confidence score (i.e., Prconf) threshold on the number of detected bounding boxes. The potential regions including suspected lesions (i.e., detected bounding boxes) caused by COVID-19 were detected using confidence score thresholds of a 0.005, b 0.02, c 0.10, and d 0.20
Fig. 7.
Prediction measurements in terms of a the intersection over union (IoU) and b the confidence score for the final predicted bounding box for all test sets for the nine disease classes
Detection results after 5-fold cross-validation
The presented deep learning CAD system can efficiently automatically predict suspected COVID-19 lesions and other respiratory disease lesions from entire X-ray images. Table 1 shows the overall detection performance according to 5-fold validation using the test images from all nine disease classes. For each k-fold test, the same deep learning structure, training, and testing parameters of the presented CAD system were applied. The detected regions of interest (ROIs) that involved COVID-19 or other respiratory diseases were considered to be correctly detected if and only if with Prconf. ≥ 10%. Otherwise, they were considered to be false detection cases even if.Prconf. ≥ 10%. Indeed, the most correct final detected bounding boxes had the maximum IoU and confidence scores as well. Based on the average of the 5-fold tests, the CAD-based YOLO was shown to be a reliable and feasible method of detecting COVID-19, with an overall detection accuracy of 96.31%. It failed to detect only 3.69% of COVID-19 cases in all the images. More generally, the presented CAD system has the capability to correctly detect respiratory diseases, with an overall detection accuracy of 90.67% for all nine disease classes. The true and false detection cases for the individual classes for the 5-fold validation are presented in Table 1.
Table 1.
Detection evaluation results for COVID-19 and eight other respiratory diseases over the 5-fold tests in the test set
| Fold Test | Atelectasis | Infiltration | Pneumothorax | Mass | Effusion | Pneumonia | Cardiomegaly | Nodule | COVID-19 | Total Classes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| True | False | True | False | True | False | True | False | True | False | True | False | True | False | True | False | True | False | True | False | |
| Fold1 | 32 | 4 | 24 | 1 | 12 | 8 | 15 | 2 | 28 | 3 | 21 | 3 | 29 | 0 | 12 | 4 | 62 | 3 | 235 | 28 |
| 88.89% | 11.11% | 96.0% | 4.0% | 60.0% | 40.0% | 88.24% | 11.76% | 90.32% | 9.68% | 87.5% | 12.5% | 100% | 0.0% | 75.0% | 25.0% | 95.38% | 4.62% | 89.35% | 10.65% | |
| Fold2 | 31 | 5 | 24 | 1 | 14 | 6 | 16 | 1 | 29 | 2 | 20 | 4 | 28 | 1 | 13 | 3 | 61 | 4 | 236 | 27 |
| 86.11% | 13.89% | 96.0% | 4.0% | 70.0% | 30.0% | 94.12% | 5.88% | 93.55% | 6.45% | 83.33% | 16.67% | 96.55% | 3.45% | 81.25% | 18.75% | 93.85% | 6.15% | 89.73% | 10.27% | |
| Fold3 | 31 | 5 | 25 | 0 | 13 | 7 | 16 | 1 | 27 | 4 | 22 | 2 | 27 | 2 | 12 | 4 | 63 | 2 | 236 | 27 |
| 86.11% | 13.89% | 100% | 0.0% | 65.0% | 35.0% | 94.12% | 5.88% | 87.10% | 12.90% | 91.67% | 8.33% | 93.10% | 6.90% | 75.00% | 25.0% | 96.92% | 3.08% | 89.73% | 10.27% | |
| Fold4 | 33 | 3 | 25 | 0 | 15 | 5 | 14 | 3 | 28 | 3 | 20 | 4 | 28 | 1 | 14 | 2 | 63 | 2 | 240 | 23 |
| 91.67% | 8.33% | 100% | 0.0% | 75.0% | 25.0% | 82.35% | 17.65% | 90.32% | 9.68% | 83.33% | 16.67% | 96.55% | 3.45% | 87.50% | 12.50% | 96.92% | 3.08% | 91.25% | 8.75% | |
| Fold5 | 33 | 3 | 24 | 1 | 15 | 5 | 16 | 1 | 30 | 1 | 21 | 3 | 29 | 0 | 12 | 4 | 64 | 1 | 264 | 19 |
| 91.67% | 8.33% | 96.00% | 4.00% | 75.0% | 12.50% | 94.12% | 5.88% | 96.77% | 3.23% | 87.50% | 12.50% | 100% | 0.0% | 75.00% | 25.0% | 98.46% | 1.54% | 93.29% | 6.71% | |
| Avg. (%) | 88.89 | 11.11 | 97.60 | 2.40 | 71.50 | 28.50 | 90.59 | 9.41 | 91.61 | 8.39 | 86.67 | 13.33 | 97.24 | 2.76 | 78.75 | 21.25 | 96.31 | 3.69 | 90.67 | 9.33 |
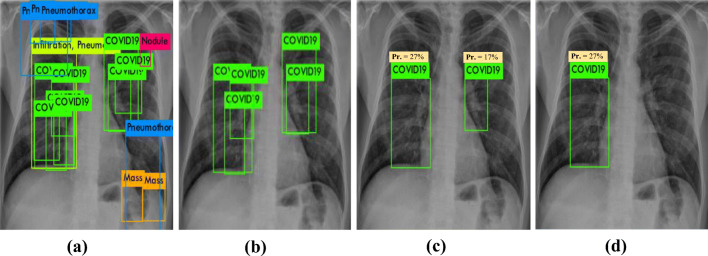
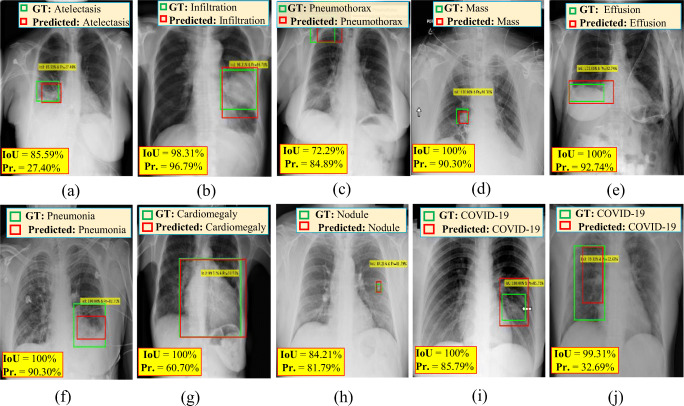
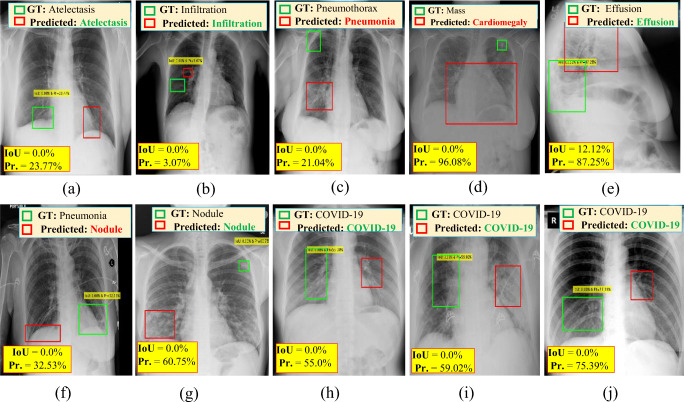
With regard to the qualitative evaluation, Fig. 8 shows examples of correctly detected suspicious lesions indicative of COVID-19 and all other disease classes. The overlapping ratios (i.e., IoU) for the resulting bounding boxes beside their corresponding confidence scores from each case are also presented. The detected boxes of these cases have acceptable IoU ratios and high confidence scores, indicating that the lesions have been accurately detected. Figure. 9 shows some examples of falsely detected cases of all nine disease classes. The final detected boxes of these cases have undesirable overlapping ratios with their GTs. Therefore, they were considered incorrect detection cases even if they satisfied the confidence score condition.
Fig. 8.
Examples of correctly predicted cases of COVID-19 and other respiratory diseases from chest X-ray (CXR) images: a atelectasis, b infiltration, c pneumothorax, d mass, e effusion, f pneumonia, g cardiomegaly, h nodule, and i & j COVID-19. The GT information (green), detected bounding box (red), IoU, and probability or confidence score (Pr.) for each case are superimposed on the original chest X-ray images
Fig. 9.
Examples of the incorrectly predicted cases of COVID-19 and other respiratory diseases from chest X-ray images: a atelectasis, b infiltration, c pneumothorax, d mass, e effusion, f pneumonia, g nodule, and (h, i, & j) COVID-19. The GT information (green), detected bounding box (red), IoU, and probability or confidence score (Pr.)for each case are superimposed on the original chest X-ray images
Classification results
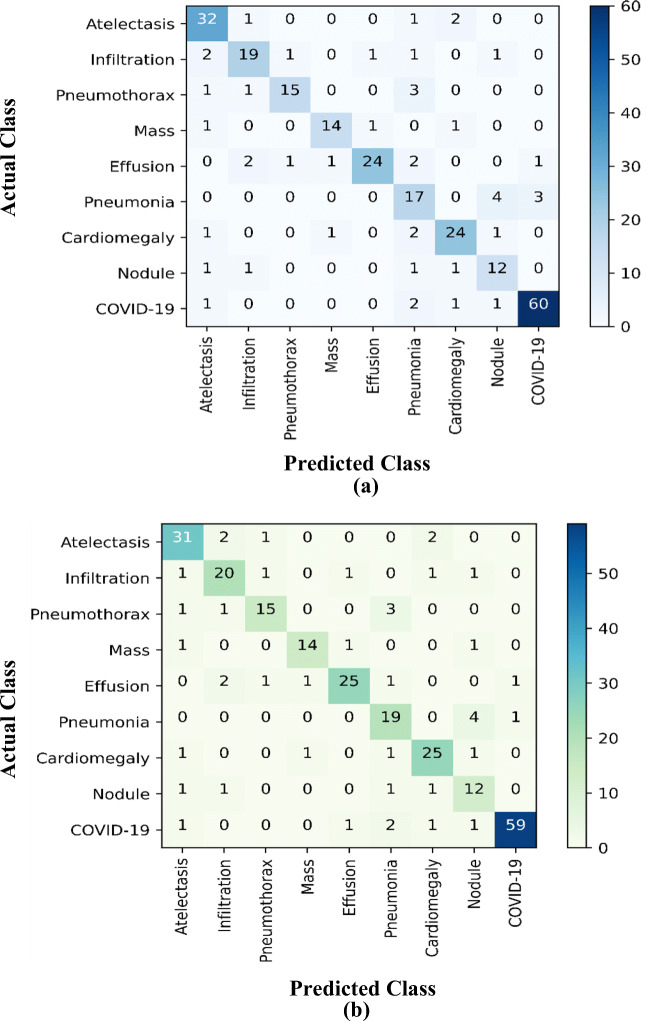
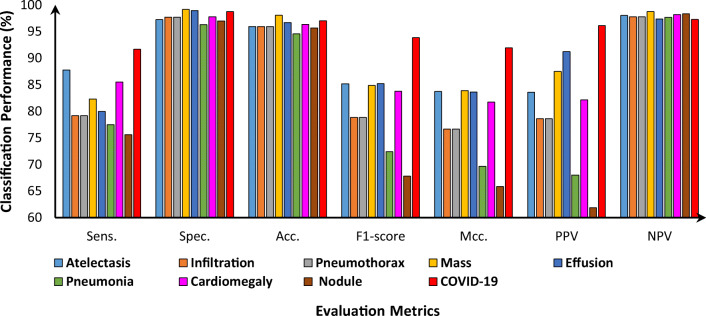
The presented CAD-based YOLO predictor has the capability to simultaneously detect ad classify end-to-end the detected ROIs as COVID-19 or other respiratory diseases. As shown in Figs. 8 and 9, the presented CAD system detects the final regions with suspected lesions of respiratory diseases and classifies them at the same time. In fact, this is the key characteristic that makes the YOLO predictor faster and more accurate than other techniques, such as Faster R-CNN [10, 27, 34]. All final detected bounding boxes are classified even if they have been incorrectly detected. With regard to classification, it is important to know the final diagnosis status of each X-ray image (i.e., COVID-19 or another disease) since its GT label is available. The classification evaluation results are derived based on the multiclass confusion matrices for all nine classes over each fold test. Figure 10 shows an example of the confusion matrices for all disease classes from the 3-fold and 5-fold tests. Indeed, most of the COVID-19 cases were correctly distinguished from other respiratory diseases. Due to the high degree of similarity between COVID-19 and other respiratory diseases, some cases of COVID-19 were misclassified as pneumonia and vice versa. The weighted recognition evaluation metrics obtained via the five-fold test for all classes are reported in Table 2. Specifically, the classification evaluation results for each individual disease class as an average of the tests are shown in Fig. 11. It is clear that the proposed CAD system achieved an average overall accuracy of classification between 94.60% for pneumonia and 97.40% for COVID-19. The sensitivity was 91.69%, the specificity was 98.79%, and the Mcc. was 91.96% for differentiating COVID-19 from the other respiratory diseases. The classification performance of the system for COVID-19 as represented by the F1-score was 93.86%. We can conclude that the CAD system achieved satisfactory and promising classification performance with regard to the problem of the multiclass recognition of respiratory diseases.
Fig. 10.
The derived multiclass confusion matrices of COVID-19 against other lung diseases from the test sets over a 3-fold and b 5-fold tests
Table 2.
Weighted classification measurements (%) for COVID-19 among the other lung diseases as an average over the 5-fold tests in the test set
| Fold Test | Sens. | Spec. | Acc. | F1-score | Mcc. | PPV | NPV |
|---|---|---|---|---|---|---|---|
| Fold1 | 88.25 | 99.16 | 97.59 | 85.47 | 83.60 | 86.0 | 98.90 |
| Fold2 | 84.47 | 99.09 | 97.37 | 84.75 | 82.75 | 85.33 | 98.72 |
| Fold3 | 83.33 | 98.78 | 97.08 | 83.63 | 81.50 | 84.40 | 98.68 |
| Fold4 | 85.25 | 99.16 | 97.59 | 85.47 | 83.60 | 86.0 | 98.90 |
| Fold5 | 84.47 | 99.09 | 97.37 | 84.75 | 82.75 | 85.33 | 98.72 |
| Average (%) | 85.15 | 99.056 | 97.40 | 84.81 | 82.84 | 85.412 | 98.784 |
Fig. 11.
Classification evaluation measurements (%) for each individual class of lung diseases as an average over the 5-fold tests in the test set
Effects of the regularization strategies
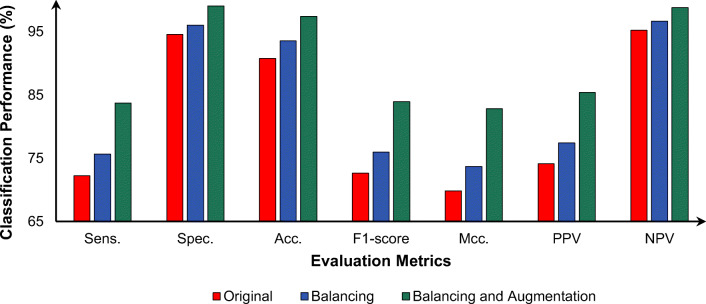
To improve the diagnostic performance for COVID-19 and the differentiation of COVID-19 from other respiratory diseases, data balancing and augmentation strategies were used. In this regard, the presented CAD system was trained and fine-tuned over 5-fold tests using the original, balanced, and augmented datasets in three separate scenarios. In each scenario, the same deep learning structure and learning settings were used. Figure 12 shows the weighted classification performance as an average of the 5-fold tests for each scenario. The balancing strategy improved the diagnostic performance by 3.43%, 1.47%, 2.79%, 3.35%, 3.86%, 3.28%, and 1.43% in terms of the sens., spec., Acc., F1-score, Mcc., PPV, and NPV, respectively. The major improvement was achieved through data augmentation after balancing. After applying the augmentation strategy, the classification performance was improved by 12.91%, 4.49%, 6.64%, 12.17%, 12.99%, 11.72%, and 3.56% in terms of the sens., spec., Acc., F1-score, Mcc., PPV, and NPV, respectively.
Fig. 12.
Effect of enlarging the training set sizes using different deep learning regularization strategies on the overall classification performance of the proposed CAD system. The evaluation results are presented as the average of the 5-fold tests in the test sets for all disease classes
The cost of the prediction time
The training time depends on the deep learning structure, training settings (i.e., number of epochs and mini-batch size), number of training sets, and specifications of the PC. For each fold test, the presented CAD system required almost 18 h for training. To make predictions for all test images, the proposed CAD system required 2.44 s. Since we had 263 test images across all disease classes, the predication time for an individual X-ray image was 0.0093 s. Our CAD system can make reliable preditctions in real time by 108 FPS. The rapid global spread of COVID-19 is challenging for physicians. The accurate and fast detection of COVID-19 based on entire chest X-ray image can help physicians, patients, and health care systems.
Discussion
Recently, researchers have been encouraged to apply artificial intelligence (AI) methodologies to help physicians in hospitals diagnose COVID-19. Indeed, deep learning based on CNN has been shown to achieve promising classification results with different applications. To date, a few studies based on machine learning and deep learning models have been designed and presented. Such studies employed deep learning models to classify entire input X-ray images. However, it is neither efficient nor accurate to base a diagnosis on an entire X-ray image [12, 27]. Thus, the detection by the CAD system of regions containing suspected lesions related to a respiratory disease (i.e., COVID-19 or another disease) represents a crucial prerequisite for achieving a more accurate diagnosis. Table 3 compares the prediction compression performance of our proposed CAD system with the performance of the latest deep learning models. Ozturk et al. [8] presented the deep learning model of DarkCovidNet that can be used to differentiate COVID-19 cases from pneumonia and normal cases. They achieved an overall diagnostic performance of 87.02%. Wand et al. developed the COVID-Net deep learning model to differentiate COVID-19 cases from normal and pneumonia cases. They achieved an overall diagnotic performance of 92.40%. Meanwhile, Khan et al. [22] presented the deep learning model of CoroNet, which can be used to differentiate COVID-19 cases from bacterial pneumonia, viral pneumonia, and normal cases. A diagnostic performance of 89.60% was achieved for the multiclass recognition problem.
Table 3.
Prediction performance comparison against the latest deep learning models for the diagnosis of COVID-19 based on chest x-ray images
| Reference | Method | Prediction Classes: No. of images | Diagnosis Accuracy (%) |
|---|---|---|---|
| Ozturk et al. [8] | DarkCovidNet | COVID-19: 125, Pneumonia: 500, and Normal: 500 | 87.02 |
| Wang et al. [18] | COVID-Net | COVID-19: 53, Pneumonia: 5526, and Normal: 8066 | 92.40 |
| Apostolopoulos et al. [20] | VGG19, Mobile Net, Inception, Xception, and InceptionResNet v2 | COVID-19: 224, Pneumonia: 700, and Normal: 504 | 93.48 ➔ for VGG19 |
| Khan et al. [22] | CoroNet | COVID-19: 284, Pneumonia bacterial: 330, Pneumonia viral: 327, and Normal: 310 | 89.60 |
| The Presented CAD system | CAD-based YOLO Predictor | COVID-19: 326 and the number of images from other eight classes are shown in Fig. 2. |
Detection: 90.67 Classification: 97.40 |
In this study, the proposed CAD system could effectively differentiate COVID-19 from eight other respiratory diseases. The detection accuracies for all nine disease classes ranged from 71.50% for pneumothorax to 97.60% for infiltration. The overall performance for the correct detection of regions with suspicious lesions was 90.67%. With regard to the detection of COVID-19, an overall detection accuracy of 96.31% was achieved. The results of the evaluation of the detection capability of the model for each individual disease class are reported in Table 1. The proposed CAD system could simultaneously predict the diagnosis (i.e., COVID-19 or not) for each detected ROI to determine the final diagnosis of the input X-ray image. As shown in Table 2, a promising classification accuracy of 97.40% was achieved over 5-fold tests. The simultaneous detection and classification of COVID-19 or other respiratory diseases in a single assessment of an entire X-ray image is helpful for physicians, especially when the number of patients is large. This will directly help support health care systems in hospitals as well. By controlling the confidence score threshold for the detected bounding boxes, we can select the desired number of boxes that should be used for the final real-time diagnosis. As shown in Fig. 6c, after adjusting the confidence threshold to be greater than 10%, two detected boxes were finally assigned two different regions with lesions suspected of being related to COVID-19. These results are logical and acceptable because COVID-19 and other respiratory diseases can affect both lungs in the same patient. Meanwhile, it is important to consider the final detected regions with suspicious lesions for classification even if they have been incorrectly detected. As shown in Fig. 9, most falsely detected cases were correctly classified. Additionally, it may help physicians focus on regions with suspicious lesions other than those with GTs. Figure 9h-j show the incorrectly detected ROIs according to the annotated position of the GT, but the final diagnosis was accurate. Meanwhile, deep learning regularizations for data balancing and augmentation were applied to improve the final diagnostic performance of the proposed CAD system. As shown in Fig. 12, these regularizers obviously improved the diagnostic performance as reflected in all evaluation indices. The average of the five-fold tests for the overlap class problem showed that the classification performance increased from 90.76% to 97.40% and from 72.64% to 84.81% with regard to the Acc. and F1-score, respectively. Generally, CAD systems could support physicians by providing a second opinion that could be used when making the final decision regarding the diagnosis. The fast and accurate diagnosis of COVID-19 based on entire X-ray images is key to helping physicians, patients, and health care systems.
The proposed CAD system has some advantages. First, the model has promising predictive accuracy for differentiating COVID-19 from other respiratory diseases is achieved. Second, the model can rapidly predict the presence of COVID-19 and other respiratory diseases based on entire X-ray images. Finally, user interventions are not required to detect and classify COVID-19 because the proposed CAD system has a unique end-to-end deep learning structure.
Despite the encouraging and rapid diagnostic performance for COVID-19, some drawbacks and limitations need to be addressed. Annotated digital X-ray images from COVID-19 patients are still unavailable. Considerable time and effort on the part of physicians is needed to label and localize the exact regions containing lesions associated with COVID-19.
In the future, when the annotated chest X-ray images become available, we plan to validate the presented CAD system. For increase the reliability of the diagnosis, we will expand our proposed CAD system to diagnose COVID-19 based on digital CT images. Additionally, we plan to locally collect digital X-ray and CT images for further validation. To achieve more accurate pre-training of deep learning models, a generative adversarial network (GAN) could be used to synthesize images [27].
Conclusion
In this work, a deep learning CAD system is proposed that can simultaneously detect and diagnose COVID-19 based on chest X-ray images. Our presented deep learning system was built in a unique deep learning structure and can rapidly predict the regions containing suspicious lesions likely associated with COVID-19 on entire X-ray images. The proposed CAD system was validated with regard to the multiclass recognition problem, achieving a promising diagnostic accuracy of 97.40% over 5-fold tests. Highly accurate and rapid information extraction from entire CXR images is a key for developing a comprehensive and useful patient triage system in hospitals and healthcare systems. The promising diagnostic performance and the rapid prediction time make this proposed CAD system practical and reliable as a means of assisting physicians, patients, and health care systems.
Acknowledgement
This research was supported by the MSIT(Ministry of Science and ICT), Korea, under the ITRC(Information Technology Research Center) support program(IITP-2017-0-01629) supervised by the IITP(Institute for Information & communications Technology Promotion)”, by Institute for Information & communications Technology Promotion(IITP) grant funded by the Korea government(MSIT) (No.2017–0-00655), by the MSIT(Ministry of Science and ICT), Korea, under the Grand Information Technology Research Center support program(IITP-2020-0-01489) supervised by the IITP(Institute for Information & communications Technology Planning & Evaluation) NRF-2016K1A3A7A03951968 and NRF-2019R1A2C2090504.
Biographies
Mugahed A. Al-antari
received his Ph.D. degree from the Department of Biomedical Engineering, Kyung Hee University, Republic of Korea, in 2019. Now, Al-antari is a post-doctoral researcher in the Ubiquitous Computing Laboratory (UCLab), Department of Computer Science and Engineering, Kyung Hee University, Republic of Korea. Al-antari is an official lecturer since 2010 in the Department of Biomedical Engineering at Sana’a Community College, Republic of Yemen. Al-antari has professional industrial experience (Executive Engineering Director) for more than 5 years for developing mini- and whole body DXA machines using X-ray pencil and fan beams at YOZMA BMTech Group, Seoul, Republic of Korea. He is a member of IEEE EMBS since 2014. His current research interests include deep learning, machine learning, artificial intelligence (AI), AI-based medical platforms, AI-based energy analysis, pattern recognition, medical signal and image processing, and medical imaging of dual-energy X-ray absorptiometry (DXA). His recent AI-based publications have earned a lot of attention from researchers as well as the international journal editorials and been selected to represent a cornerstone for modern medicine.
Cam-Hao Hua
received the B.S. degree in Electrical and Electronic Engineering from Bach Khoa University, Ho Chi Minh City, Vietnam, in 2016. He is currently pursuing the Ph.D. degree with the Department of Computer Science and Engineering, Kyung Hee University, Gyeonggi, Republic of Korea. His research interests are computer vision and deep Learning.
Jaehun Bang
received his Ph.D. degree from the Department of Computer Science and Engineering, Kyung Hee University, Republic of Korea, in 2019. Now, Jaehun Bang is a post-doctoral researcher in the Ubiquitous Computing Laboratory (UCLab) and Intelligent Medical Platform Research Center (IMPRC), Department of Computer Science and Engineering, Kyung Hee University, Republic of Korea. His current research interests include ubiquitous computing and applications, context awareness, signal processing, pattern recognition, machine learning, and activity & emotion recognition.
Sungyoung Lee
(M’89) received the B.S. degree from Korea University, Seoul, Republic of Korea, and the M.S. and Ph.D. degrees in computer science from the Illinois Institute of Technology, Chicago, IL, USA, in 1987 and 1991, respectively. He was an Assistant Professor with the Department of Computer Science, Governors State University, University Park, IL, USA, from 1992 to 1993. He has been a Professor with the Department of Computer Engineering, Kyung Hee University, Republic of Korea, since 1993, where he has been the Director of the Neo Medical ubiquitous-Life Care Information Technology Research Center since 2006. He is currently the Founding Director of the Ubiquitous Computing Laboratory. His current research interests include ubiquitous computing and applications, wireless ad hoc and sensor networks, context aware middle-ware, sensor operating systems, real-time systems and embedded systems, and activity and emotion recognition. He is a member of ACM.
Compliance with ethical standards
Conflict of interest
None.
Declarations
Not applicable.
Footnotes
Highlights of the Article
A fast deep learning computer-aided diagnosis (CAD) based on the YOLO predictor is proposed to simultaneously detect and diagnose COVID-19 respiratory disease from the entire chest X-ray (CXR) images.
The COVID-19 respiratory disease is automatically detected and classified end-to-end with overall detection and classification accuracies of 96.31% and 97.40%, respectively.
The proposed deep learning CAD system is able to detect and classify COVID-19 or other respiratory diseases in a single X-ray image within 0.0093 seconds.
The presented CAD system is able to predict at least 108 frames/sec at the real-time of prediction.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mugahed A. Al-antari, Email: en.mualshz@oslab.khu.ac.kr
Cam-Hao Hua, Email: hao.hua@oslab.khu.ac.kr.
Jaehun Bang, Email: jhb@oslab.khu.ac.kr.
Sungyoung Lee, Email: sylee@oslab.khu.ac.kr.
References
- 1.Alimadadi A, Aryal S, Manandhar I, Munroe PB, Joe B, Cheng X (2020) Artificial intelligence and machine learning to fight COVID-19. American Physiological Society Bethesda, MD [DOI] [PMC free article] [PubMed]
- 2.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinazzi M, Davis JT, Ajelli M, Gioannini C, Litvinova M, Merler S, y Piontti AP, Mu K, Rossi L, Sun K (2020) The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science 368 (6489):395–400 [DOI] [PMC free article] [PubMed]
- 4.Robson B. Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput Biol Med. 2020;119:103670. doi: 10.1016/j.compbiomed.2020.103670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao S, Lin Q, Ran J, Musa SS, Yang G, Wang W, Lou Y, Gao D, Yang L, He D. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coronavirus disease (COVID-19) (2020) World Health Organization. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed November 2020
- 7.Pereira RM, Bertolini D, Teixeira LO, Silla CN, Jr, Costa YM. COVID-19 identification in chest X-ray images on flat and hierarchical classification scenarios. Comput Methods Prog Biomed. 2020;194:105532. doi: 10.1016/j.cmpb.2020.105532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput Biol Med. 2020;121:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Antari MA, Al-Masni MA, Choi MT, Han SM, Kim TS. A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification. Int J Med Inform. 2018;117:44–54. doi: 10.1016/j.ijmedinf.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 10.Al-Masni MA, Al-Antari MA, Park JM, Gi G, Kim TY, Rivera P, Valarezo E, Choi MT, Han SM, Kim TS. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput Methods Prog Biomed. 2018;157:85–94. doi: 10.1016/j.cmpb.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 11.Al-Masni MA, Al-Antari MA, Choi MT, Han SM, Kim TS. Skin lesion segmentation in dermoscopy images via deep full resolution convolutional networks. Comput Methods Prog Biomed. 2018;162:221–231. doi: 10.1016/j.cmpb.2018.05.027. [DOI] [PubMed] [Google Scholar]
- 12.Al-Masni MA, Kim D-H, Kim T-S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput Methods Prog Biomed. 2020;190:105351. doi: 10.1016/j.cmpb.2020.105351. [DOI] [PubMed] [Google Scholar]
- 13.Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection. https://www.githubcom/ieee8023/covid-chestxray-dataset. Accessed June [online] 2020
- 14.Chowdhury M, Rahman T, Khandakar A, Kadir R, Mahbub Z, Islam K, Khan M, Iqbal A, Emadi N, Reaz M (2020) Can AI help in screening Viral and COVID-19 pneumonia? https://www.kagglecom/tawsifurrahman/covid19-radiography-database. . Accessed June [online] 2020
- 15.Wang X, Peng Y, Lu L, Lu Z, Bagheri M (2017) Summers RM Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 2097–2106
- 16.Oh Y, Park S, Ye JC. Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans Med Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- 17.Fan D-P, Zhou T, Ji G-P, Zhou Y, Chen G, Fu H, Shen J, Shao L. Inf-net: automatic COVID-19 lung infection segmentation from CT images. IEEE Trans Med Imaging. 2020;39(8):2626–2637. doi: 10.1109/TMI.2020.2996645. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Wong A (2020) COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv preprint arXiv:200309871 [DOI] [PMC free article] [PubMed]
- 19.Hemdan EE-D, Shouman MA, Karar ME (2020) Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images: arXiv preprint arXiv:2003.11055
- 20.Apostolopoulos I, Mpesiana T. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahuja S, Panigrahi BK, Dey N, Rajinikanth V, Gandhi TK (2020) Deep transfer learning-based automated detection of COVID-19 from lung CT scan slices. Appl Intell. 10.1007/s10489-020-01826-w [DOI] [PMC free article] [PubMed]
- 22.Khan AI, Shah JL, Bhat M. Coronet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Prog Biomed. 2020;196:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks: arXiv preprint arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- 24.Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med. 2020;121:103795. doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohamadou Y, Halidou A, Kapen PT (2020) A review of mathematical modeling, artificial intelligence and datasets used in the study, prediction and management of COVID-19. Appl Intell 50(11):3913–3925 [DOI] [PMC free article] [PubMed]
- 26.Shoeibi A, Khodatars M, Alizadehsani R, Ghassemi N, Jafari M, Moridian P, Khadem A, Sadeghi D, Hussain S, Zare A (2020) Automated detection and forecasting of COVID-19 using deep learning techniques: A review. arXiv preprint arXiv:2007.10785
- 27.Al-Antari MA, Kim T-S. Evaluation of deep learning detection and classification towards computer-aided diagnosis of breast lesions in digital X-ray mammograms. Comput Methods Prog Biomed. 2020;196:105584. doi: 10.1016/j.cmpb.2020.105584. [DOI] [PubMed] [Google Scholar]
- 28.Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In: 25th International Conference on Neural Information Processing Systems, USA, pp. 1097–1105. pp 1097–1105
- 29.Takahashi R, Kajikawa Y. Computer-aided diagnosis: a survey with bibliometric analysis. Int J Med Inform. 2017;101:58–67. doi: 10.1016/j.ijmedinf.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 30.Chougrad H, Zouaki H, Alheyane O. Deep convolutional neural networks for breast cancer screening. Comput Methods Prog Biomed. 2018;157:19–30. doi: 10.1016/j.cmpb.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Celik Y, Talo M, Yildirim O, Karabatak M, Acharya UR. Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images. Pattern Recogn Lett. 2020;133:232–239. doi: 10.1016/j.patrec.2020.03.011. [DOI] [Google Scholar]
- 32.Girshick R (2015) Fast r-cnn. In: Proceedings of the IEEE international conference on computer vision, pp. 1440–1448
- 33.Felzenszwalb PF, Girshick RB, McAllester D, Ramanan D. Object detection with discriminatively trained part-based models. IEEE Trans Pattern Anal Mach Intell. 2010;32(9):1627–1645. doi: 10.1109/TPAMI.2009.167. [DOI] [PubMed] [Google Scholar]
- 34.Redmon J, Farhadi A (2017) YOLO9000: better, faster, stronger. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7263–7271
- 35.Al-Antari MA, Al-Masni MA, Kim T-S (2020) Deep learning computer-aided diagnosis for breast lesion in digital mammogram. In: Deep Learning in Medical Image Analysis. Springer, Cham, pp 59–72 [DOI] [PubMed]