Abstract
We are living through an unprecedented crisis with the rapid spread of the new coronavirus disease (COVID-19) worldwide within a short time. The timely availability of thousands of SARS-CoV-2 genomes has enabled the scientific community to study the origin, structures, and pathogenesis of the virus. The pandemic has spurred research publication and resulted in an unprecedented number of therapeutic proposals. Because the development of new drugs is time consuming, several strategies, including drug repurposing and repositioning, are being tested to treat patients with COVID-19. Researchers have developed several potential vaccine candidates that have shown promise in phase II and III trials. As of 12 November 2020, 164 candidate vaccines are in preclinical evaluation, and 48 vaccines are in clinical evaluation, of which four have cleared phase III trials (Pfizer/BioNTech's BNT162b2, Moderna's mRNA-1273, University of Oxford & AstraZeneca's AZD1222, and Gamaleya's Sputnik V vaccine). Despite the acquisition of a vast body of scientific information, treatment depends only on the clinical management of the disease through supportive care. At the pandemic's 1-year mark, we summarize current information on SARS-CoV-2 origin and biology, and advances in the development of therapeutics. The updated information presented here provides a comprehensive report on the scientific progress made in the past year in understanding of SARS-CoV-2 biology and therapeutics.
Keywords: SARS-CoV-2, Coronavirus, COVID-19, Drug repurposing, Vaccines, Therapeutics, Pandemic, Pathogenesis, Outbreak
1. Introduction
We have now been living with COronaVIrus Disease (COVID-19) for the past year. COVID-19 emerged in December 2019, and in March of 2020 was declared a pandemic by the World Health Organization. The devastating effect of the causative SARS-CoV-2 virus has infected millions of humans across 218 countries and terrotories and led to more than 1.4 million deaths globally as of 24 November 2020. The pandemic has significantly affected biomedical researchers, by first halting research and then resulting in the concentration of scientific resources toward better understanding the SARS-CoV-2 virus and developing vaccines and therapeutics (Palayew et al., 2020; Zamora-Ledezma et al., 2020). The advent of genomics technologies and computational approaches has accelerated scientific breakthroughs in the past year. There has been exponential growth in the number of scientific publications related to COVID-19. The genome sequence of the virus appeared online on January 10, and within weeks, the structures of several viral proteins were determined. Within months, clinical trials of vaccines and therapeutics began, and positive reports on vaccines are currently appearing. Moreover, an array of drugs approved for other viral infections are being studied for COVID-19 treatment in hundreds of clinical trials worldwide. Here, we review current knowledge related to SARS-CoV-2 gained in the past year, including its progression, pathology, prevention, and therapeutics. We discuss what is currently known about the virus and how far medicine has progressed in the fight against COVID-19.
2. Origin and diversification of human CoVs
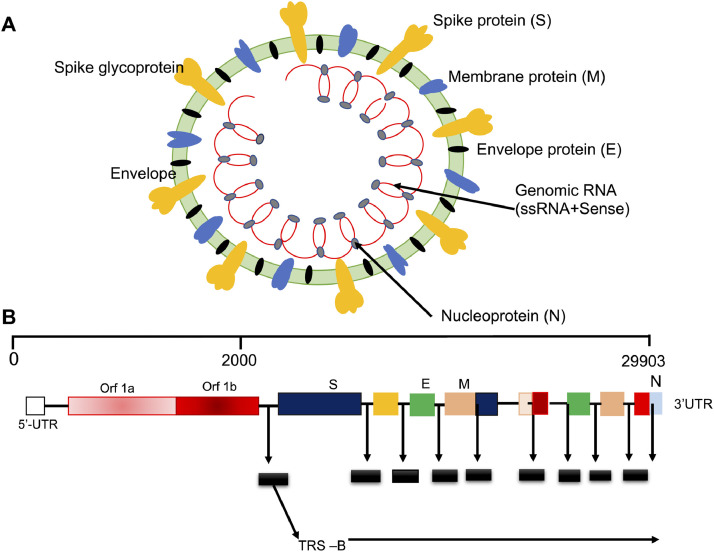
Coronaviruses (CoVs) are a large group of viruses that infect the upper respiratory tract in humans and cause common cold and flu-like infections. Their name originates from the presence of club-shaped glycoprotein projections (called spikes) that arise from the surface of the viral envelope and impart a crown-like appearance to the viral particles, similarly to the Sun's corona (Fig. 1 A). The CoVs belong to the order Nidovirales of the subfamily Orthocoronaviridae in the family Coronoviridae. All CoVs have zoonotic origin, and cause respiratory and intestinal infections in several animals, including humans. On the basis of genomic organization and phylogenetic relationships, CoVs are classified into four genera: α-CoV, β-CoV, γ-CoV, and δ- CoV. The α-CoVs and β-CoVs infect various mammals (such as bats, cattle, domestic animals, livestock, and humans), whereas the γ-CoVs and δ-CoVs infect avians and sometimes mammals (Woo et al., 2012).
Fig. 1.
Schematic representation of SARS-CoV-2 (A) virus structure and (B) genome organization.
CoVs were first identified as infectious bronchitis viruses in 1937, infecting avian species and devastating poultry stocks. The strain of CoV responsible for 15–30% of cases of the common cold in humans was identified in 1965 (Bhargava 2020). To date, seven strains of human CoV have been identified as belonging to α-CoVs or β-CoVs. The four mild strains (HCoV-NL63, HCoV-229E, HCoV-OC43, and HKU1) induce mild respiratory disease of the upper respiratory tract in infants, older people, and immunocompromised individuals. In comparison, the three highly virulent pathogenic strains, severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome CoV (MERS-CoV), and SARS-CoV-2 cause acute respiratory distress syndrome, result in pulmonary failure and fatality in humans (Forni et al., 2016; Masters and Perlman, 2013; Su et al., 2016). The outbreak of SARS-CoV in 2002 in Guangdong province in South China spread to approximately 28 countries and infected nearly 8000 individuals, causing symptoms of fever, headache, and respiratory problems, such as shortness of breath, taking 774 human lives (Bhargava 2020; Zhong et al., 2003). Ten years later, in 2012, the MERS-CoV outbreak endemic in Middle Eastern countries, particularly Saudi Arabia, affected nearly 2500 people (Bhargava 2020; Wang et al., 2013; Zhong et al., 2003). Both these β-CoVs are genetically different and have been reported to originate from bats (Cui et al., 2019). These outbreaks indicate the potential for the emergence of super/lethal strains of human CoVs in the future with high transmission and mortality rates together with a long latency period between infection and detectable symptoms, thus, potentially resulting in devastating pandemics (Patrick). The viral pneumonia-like disease was first reported at the end of November 2019 near the wet seafood market in Wuhan Province, China. The CoV first reported in Wuhan was described to be a novel virus belonging to the β-CoV category and was designated as 2019 novel coronavirus (2019-nCOV) by Chinese researchers (Shereen et al., 2020). However, on 11 February 2020, the virus was renamed SARS-CoV-2, and the disease was named COVID-19 (Drosten et al., 2003; Fouchier et al., 2003; Zhong et al., 2003).
All human CoVs have zoonotic origin and are capable of transmission among mammalian hosts; however, most CoVs originate in bats and are transmitted to humans through domestic animals (Forni et al., 2016; Su et al., 2016). Thus, bats are considered the natural host and primary reservoir of human CoVs (Cui et al., 2019). For SARS-CoV, horseshoe bats (Rhinolophus) have been reported to be the natural host, and palm civets (Paguma larvata) have been reported to be the intermediate host, on the basis of the presence of antibodies in these organisms in China and European countries where the virus emerged during 2002–03. Later, several strains of related CoVs were identified in bats (Rhinolophus) that emerged through the recombination of existing strains of CoVs (Shi and Hu, 2008; Zheng et al., 2004). MERS-CoV had camels as the primary zoonotic host and was reported in bats of the genera Pipistrellus and Perimyotis, thus, suggesting that bats are the key reservoir and transmission agent (Cui et al., 2019; Huynh et al., 2012). The S-gene encoding spike protein is considered the hotspot of genetic recombination among different bat CoV strains (Hon et al., 2008; Huynh et al., 2012; Wu et al., 2016). Because of the genetic variability in different CoV strains and the frequent genetic recombination between strains, the emergence of novel variants of CoVs was predicted before 2002 (Nagy and Simon, 1997; Rowe et al., 1997). The progenitors of SARS-CoV evolved through recombination within Rhinolophus bats and then was transmitted to farm civets (Paguma larvata) through fecal-oral transmission; infected civets then transmitted it to market civets, in which the viruses underwent substantial mutation before spreading to the human population in Guangdong, China (Cui et al., 2019). SARS-CoV-2 is the seventh member of the human CoVs (Corman et al., 2018; Wu et al., 2020b; Zhou et al., 2020).
SARS-CoV-2 was transmitted to humans from infected animals in the market and then spread rapidly throughout the world via human-to-human contact involving exposure to respiratory droplets or aerosols through nosocomial transmission. Studies have indicated that bats are the natural reservoir of SARS-CoV-2, as with other human CoVs (Banerjee et al., 2019; Hampton, 2005; Li et al., 2005a; Wu et al., 2020b; Zhou et al., 2020). The first genome sequence of SARS-CoV-2 was compared with the bat CoV RaTG13 and human SARS-CoV, and the SARS-CoV-2 genome has been found to show a nucleotide sequence identity of 96.2% with the bat CoV RaTG13 and 79.5% with human SARS-CoV (Naqvi et al., 2020; Zhou et al., 2020), thus, indicating a close relationship between bat and human CoVs, and suggesting that both strains might have evolved from a common ancestor strain that diversified through genetic recombination, mutation, and natural selection during transmission. SARS-CoV-2 is thought to have originated in bats via genetic recombination of existing bat CoV strains and to have been transmitted from bats to humans either directly or through unknown intermediate hosts, similarly to the roles of civets and camels in SARS-CoV and MERS-CoV, respectively (Zhou et al., 2020). Recent data suggest that CoVs capable of infecting humans emerged in bats around 40 years ago and have been circulating undetected since then, thus suggesting bats as the primary reservoir of the SARS-CoV-2 lineage (Boni et al., 2020). Detailed analysis of the protein sequence data has also indicated the potential of pangolins, turtles, and snakes acting as intermediate hosts of SARS-CoV-2, owing to the presence of ACE2 receptors in the CoVs reported from these animals (Guan et al., 2020). A recent study has used protein mapping to characterize the protein interaction networks of the three CoVs (SARS-CoV-1, MERS-CoV, and SARS-CoV-2) in humans, thus revealing important molecular mechanisms and potential therapeutic interventions common to the three CoVs (Gordon et al., 2020a).
3. Structure and genomic organization of SARS-CoV-2
SARS-CoV-2 is an unsegmented single-stranded positive-sense RNA virus. Structurally, it is a spherical or pleomorphic enveloped virus. The genome size of SARS-CoV-2 is ∼29 Kb RNA, which is between the genome sizes of SARS-CoV (∼28 Kb) and MERS-CoV (∼30 Kb) (De Wit et al., 2016; Wu et al., 2020a; Wu et al., 2020b; Zehra et al., 2020; Zhou et al., 2020). The genome is organized as a 5′-leader-UTRs-replicase-S-E-M-N-3′UTR-poly (A) tail sequence and is characterized by the presence of a variable number (6–12) of open reading frames (ORFs) between conserved genes (ORF1ab, S, E, M, and N) and nine sub-genomic mRNAs, nine transcription regulatory elements and two terminal untranslated regions (UTRs) (Figure 1B) (Lu et al., 2020). The 5′ UTR and 3′ UTR are important for RNA-RNA interaction and binding of viral and cellular proteins. The sizes of UTRs differ among SARS-CoV-1, SARS-CoV-2, and MERS-CoV. Two-thirds of the viral genome located at the 5′ end constitutes the first ORF (ORF1a/b); ORF1a and ORF1b contain frameshifts encoding two long polypeptides (pp1a and pp1ab), which after priming and processing produce 16 non-structural proteins (nsp1–16), which are necessary for genome maintenance. These polypeptides are processed by viral encoded (chymotrypsin-like-3cLpro-/main protease-Mpro-nsp5/Papain like-nsp3) proteases. One-third of the genome near the 3′ end (ORFs 10 and 11) encodes four major structural proteins: the spike (S), membrane (M), envelope (E), and nucleocapsid (N). The spikes are club-shaped, halo-like membranous glycoprotein projections that constitute peplomers, induce neutralizing antibodies, and play the most significant role in the pathogenesis of SARS-CoV-2. The M-protein is the most abundant protein, which spans the membrane bilayer and maintains the shape of the virion. It plays a significant role during the budding of coronaviral particles from host cells. It is also a conserved viral protein, with 98% sequence similarity to bat and pangolin CoV M proteins (Bianchi et al., 2020). The E-protein is a valine and leucine rich hydropathic transmembrane protein essential for viral pathogenesis. It is a conserved protein of SARS-CoV-2 and has amino acid sequence similarity with pangolin CoV-MP798 and bat CoV-ZXC21, CoV-ZC45, and CoV-RaTG13, thus, indicating a close link with bat and pangolin CoVs (Bianchi et al., 2020). The proteins S, M, and E together constitute the viral envelope. The E and M proteins play significant roles in viral entry, replication, and particle assembly within human host cells during infection (Bianchi et al., 2020; EA and Jones, 2019; Schoeman and Fielding, 2019). During viral particle assembly, the M protein interacts with other structural proteins (S, E, and N) to constitute the complete virion (EA and Jones, 2019). The structural similarities and differences in the E and M proteins of SARS-CoV-2 proteins versus bat and pangolin CoVs are responsible for cross-species specificity and cross-species transmission of CoVs. The amino acid sequence variations in these structural proteins play significant roles in the evolution and diversification of CoVs. Similar variations arising either from mutation or genetic recombination may result in the zoonotic origin of a new viral strain with more severe virulence (Bianchi et al., 2020; EA and Jones, 2019). The N protein, in association with genomic RNA, forms the nucleocapsid, which maintains the genome structure inside the envelope and plays significant roles in viral assembly, budding, and the host cellular response to viral infection. The details on the various SARS-CoV-2 proteins and their roles are shown in Table 1 .
Table 1.
Details on the SARS-CoV-2 proteins and their roles.
| Proteins | Roles |
|---|---|
| Nsp1 | inhibits host innate immune response; increases proinflammatory chemokine production |
| Nsp2 | acts as an Nsp3 adaptor |
| PLpro/Nsp3 | interacts with Nsp4 and Nsp6 and forms a complex; strips ubiquitin and blocks the host innate immune response |
| Nsp4 | interacts with Nsp3 and Nsp6; anchors the replication complex to double-membrane vesicles |
| Nsp5/3CLPro | causes cleavage of viral polyproteins, thus decreasing individual Nsps |
| Nsp6 | interacts with Nsp3 and Nsp4; limits the expansion of autophagosome and lysosomal viral degradation |
| Nsp7/primase | forms the primase complex as part of the replication complex (Nsp7/8/12); can perform both de novo initiation and primer extension |
| Nsp8/primase | interacts with Nsp7 and the Nsp7/Nsp8 complex, thus forming the RNA transcriptase-replicase complex; the Nsp7/Nsp8 complex stabilizes the Nsp12 regions involved in RNA binding |
| Nsp9/RNA-binding protein | interacts with the replication complex (Nsp7/8/12) |
| Nsp10 | interacts with Nsp16, which is required for replication; stimulates Nsp16 methyltransferase activity; interacts with Nsp14, thus facilitating exoribonuclease and methyltransferase activities |
| Nsp12 (RNA-dependent RNA polymerase) | interacts with Nsp7 and Nsp8, thus forming an RNA transcriptase-replicase complex |
| Nsp13/NTPase/helicase | initiates the capping of viral mRNA (along with Nsp14 and Nsp16) and installs the cap structure onto viral mRNA |
| methyltransferase/exoribonuclease/NSP14 | repairs mutation errors during replication; involved in viral mRNA capping |
| uridylate-specific endoribonuclease/NSP15 | required for viral RNA synthesis |
| 2′-O-methyltransferase/NSP16 | forms a complex with NSP10; involved in the S-adenosyl-L-methionine cap methylation of mRNA |
| NSP11 | short peptide with unknown function |
| spike (S) protein | binds the ACE2 receptor on host cells and initiates viral fusion with the host cell membrane |
| envelope (E) protein | involved in viral assembly |
| membrane (M) protein | involved in viral assembly |
| nucleocapsid (N) protein | binds viral RNA |
| ORF3a | involved in the trafficking of S-protein and apoptosis |
| ORF3b | inhibits the activities of interferons |
| ORF6 | interferon I antagonist that binds karyopherins and decreases the interferon/antiviral response |
| ORF7a | involved in virus-induced apoptosis; inhibits CD317, which prevents the release of CoVs |
| ORF7b | unknown function |
| ORF8 | unknown function |
| ORF9b | involved in the degradation of signalosomes; limits host cell interferon responses |
| ORF9c | unknown |
| ORF10 | unknown |
4. Emergence of mutations in SARS-CoV-2
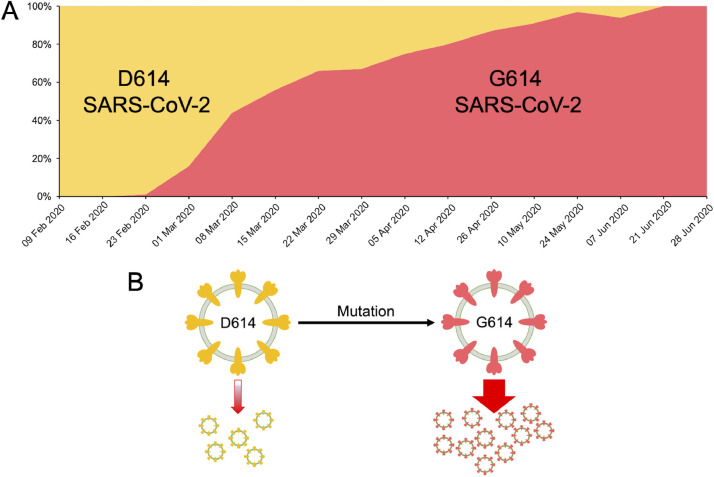
The global spread of SARS-CoV-2 has resulted in tens of thousands of mutations in the native strain within a short time. To rapidly share and disseminate the information related to virus genomes, mutations, and evolution GISAID took the initiative involving a public-private-partnership (Elbe and Buckland-Merrett, 2017). Subsequently, a CoV-GLUE database was developed to interpret and analyze the SARS-CoV-2 virus genome sequences, with a focus on amino acid sequence variation (http://cov-glue.cvr.gla.ac.uk/#/replacement) (Singer et al., 2020). The D614G mutation in the viral S-protein is now the most prevalent globally (Figure 2 ). However, it may be noted that the D614 prevalent epidemics are still prevalent in many locations when G614 first began to appear. The D614G mutant virus is not associated with increased mortality or clinical severity but is more transmissible among hosts and associated with a higher viral load (Korber et al., 2020; Volz et al., 2020). It has now been shown that the substitution D614G enhances viral replication in the respiratory tract and increases neutralization susceptibility (Plante et al., 2020). At present, the SARS-CoV-2 virus has a low mutation rate; however, as the pandemic progresses, it can acquire mutations with fitness advantages and immunological and drug resistance (Callaway, 2020b; Padhi et al., 2020c; Padhi and Tripathi, 2020).
Fig. 2.
The D614G mutation. (A) Graph showing the increasing frequency of the D614G variant over time. (B) The virus with D614G mutation is associated with increased transmissibility and higher viral loads in COVID-19 patients.
5. Spike protein and its role in the pathogenesis of CoVs
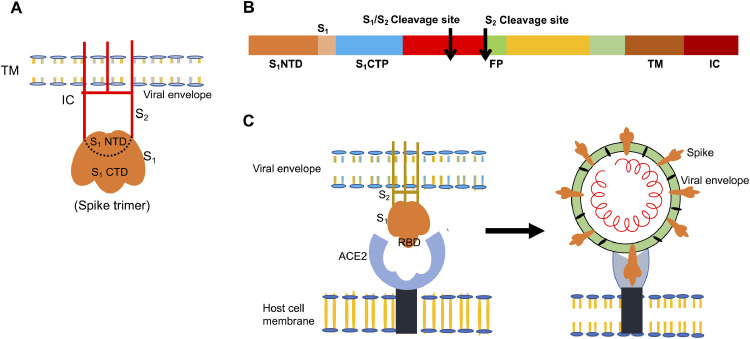
The S-protein of CoVs forms a trimeric clover-shaped structure that binds a range of hosts and initiates pathogenesis (Fig. 3 A). The S-protein is highly glycosylated and is made up of two subunits, the S1 head and the S2 filament, which protrudes as a club-shaped projection from the viral envelope (Towler et al., 2004). The receptor-binding domain (RBD) of the trimeric S-protein binds the human angiotensin-converting enzyme 2 (ACE2) receptor, thus, initiating conformational changes that drive membrane fusion (Figure 3B) (Tortorici and Veesler, 2019). The RBD is responsible for determining the cellular tropism and host range of human CoVs, whereas the S2 subunit, with the specific tandem domain heptads HR1 and HR2, mediates the fusion of the viral envelope with the host cell membrane (Xia et al., 2020; Yu et al., 2020). The S1 subunit is differentiated into an N-terminal and a C-terminal domain designated S1-NTP and S1-CTP, respectively. The RBD is present in the C-terminal domain of the S-protein and mediates specific binding of the S1 subunit to the ACE2 receptor. In contrast, the N-terminal domain plays a role in transmission between hosts and modulates the host cell endoplasmic reticulum, thus, inhibiting the interferon responses in the host cell (Babcock et al., 2004; Li et al., 2003; Li et al., 2005b; Qu et al., 2005).
Fig. 3.
(A) Schematic structure of a single CoV spike-protein, showing the receptor binding S1 subunit, membrane fusion S2 subunit, and transmembrane anchor (TM) emerging from the viral envelope. (B) The domain structure of S-protein, containing the S1N-terminal transmembrane domain (S1NTD) S1-C terminal (S1CTD) fusion peptide, heptad repeats (HRN and HRC), and protective cleavage sites (S1/S2 and S2). (C) Binding of CoV to the ACE2 receptor on the host cell through the S-protein RBD.
The phylogenetic and genomic similarities among various CoVs, specifically in the gene encoding RBD, render CoVs more amenable to human transmission (Guo et al., 2020). Strains of CoV differ in binding affinity for human ACE2, thus, resulting in variation in their infection ability, transmission rate, and pathogenicity. In the past few months, SARS-CoV-2-RBD has undergone several mutations conferring stability to the viral particle (Andersen et al., 2020; Benvenuto et al., 2020; Zhou et al., 2020). The binding affinity of the S-protein of SARS-CoV-2 for the human receptor is much higher (∼4.7 nM) than that of SARS-CoV (∼31 nM) and MERS-CoV (∼16.7 nM) (Wang et al., 2020c). A detailed understanding of the mechanism and specificity of the RBD-ACE2 interaction may aid in predicting the generation of potential strains with greater transmission and pathogenicity, and in monitoring future disease outbreaks. In addition, the information should aid in disease management and potentially in designing specific RBD-based therapeutics (vaccines, antibodies, or drugs). Knowledge of the direction of evolution and prediction of the residues whose mutation increases the affinity of RBD/ACE2 interactions may aid in the identification of favorable and unfavorable target residues in the design of new therapeutic strategies (Padhi et al., 2020b).
The optimization of the binding affinity of the SARS-CoV-2 S-protein for the human ACE2 receptor may also be attributed to the presence of functional polybasic cleavage sites for proteases, such as furin. These cleavage sites, also known as S1–S2 cleavage sites, occur at the interface of the S1 and S2 subunits through the insertion of 12 nucleotides, thus resulting in the acquisition of O-linked glycans at the region flanking the cleavage site (Walls et al., 2020). The RBD is the most variable part of the genome of CoVs. A seven-residue stretch critical for delimiting the host according to the specific affinity toward the ACE2 receptor differs in various CoVs, and natural selection has promoted optimal binding affinity during the course of host shifting (Wan et al., 2020; Wrapp et al., 2020; Zhou et al., 2020). The furin cleavage site at the junction of the S1 and S2 subunits allows for furin-mediated cleavage of the two subunits of S-protein, and enables the RBD to specifically bind the ACE2 receptor and facilitate the fusion of host cells and subsequent entry of the virus into host respiratory tract cells (Follis et al., 2006; Jia et al., 2005; Zhou et al., 2020). Moreover, O-linked glycans have a mucin-like domain that hides the antigenic determinant sites of the S-protein of SARS-CoV-2, thus, resulting in immunoevasion (Bagdonaite and Wandall, 2018).
6. SARS-CoV-2 pathogenesis
The specific RBD-ACE2 interaction facilitates the attachment and subsequent entry of viral particles into host cells. The entry of the CoVs is caused by furin-mediated cleavage and triggered by cellular proteases, such as transmembrane protease serine 2 (TMPRSS2) and cathepsins present on the host cell surface (Bertram et al., 2011; Glowacka et al., 2011). A Gln residue in the RBD (Gln479 in SARS-CoV and Gln394 in SARS-CoV-2) interacts with the Lys31 residue of the human ACE2 receptor, thus, forming a strong interaction between the RBD and ACE2 (Wan et al., 2020; Wu et al., 2012). After the formation of the RBD-ACE2 ligand-receptor complex, the S2 protein activates the fusion of the viral envelope with the host cell membrane. HR1 and HR2 interact, forming a six-helical bundle that juxtaposes the viral S2 protein with the C-terminus of the ectodomain of the ACE2 receptor (Xia et al., 2020; Yu et al., 2020). Hypertension drugs (such as losartan) that act as ACE2 inhibitors or angiotensin receptor blockers can competitively inhibit the RBD-ACE2 interaction and prevent the entry of the virus into the host cell; therefore, these drugs may be used as therapeutic agents against SARS-CoV-2 (Gurwitz, 2020). After membrane fusion, the virus uses the clathrin-dependent or independent endocytic pathway and autophagy via lysosomes to enter host cells (Pelkmans and Helenius, 2003; Sieczkarski and Whittaker, 2002). The endocytic pathway used by SARS-COV-2 is pH-dependent. Lysosomes are characterized by acidic pH, and acid hydrolases play a crucial role in the degradation of the viral envelope inside the autophagosome-lysosome complex, thus, releasing the viral genome for replication, transcription, and translation. Because the degradation of the autophagosome-lysosome complex is pH-dependent, lysosomotropic drugs such as chloroquine and hydroxychloroquine (which neutralizes the pH inside the lysosomal cavity by blocking proton pumps), chlorpromazine (an inhibitor of clathrin-dependent endocytosis), and inhibitors of cathepsins (endosomal/lysosomal cysteine proteases that act in protein degradation) could aid in controlling the entry of viral particles and subsequent infection (Al Bari, 2017; Degtyarev et al., 2008; Inoue et al., 2007; Schrezenmeier and Dorner, 2020; Wang et al., 2008; Yang and Shen, 2020).
The acidic pH of the autophagosomes aids in the dissolution of the viral envelope and the release of the viral genomic RNA inside the host cell. Because the viral RNA is positive-sense, it can directly enter the replication and transcription cycle; these processes occur at the cytoplasmic membrane and involve coordinated synthesis of continuous and discontinuous RNA. Two-thirds of the 5′-end of the SARS-CoV-2 RNA, constituting the first open reading frame (ORF 1a/b), has a frameshift between ORF 1a and ORF1a/b that guides the production of two large polypeptides, pp1a and pp1b (de Wilde et al., 2018; Guo et al., 2020). These polypeptides are then processed by the virally encoded chymotrypsin-like-3CLpro, main protease-Mpro-nsp5, and papain-like-nsp3 proteases into 16 nonstructural proteins (nsps) that constitute the viral replicase-transcriptase complex. This complex consists of helicase (nsp13), RNA dependent RNA polymerase (RdRp, also known as nsp12), and other nsps that are packed in membrane vesicles (Sawicki and Sawicki, 2005). After ORF1, a series of multiple ORFs are preceded by a short repeated sequence called the transcription regulatory sequence (TRS). Replication and transcription are performed by the replicase-transcriptase complex, which organizes itself in double-membrane vesicles (ER). Transcription termination occurs at transcription regulatory sequences located between ORFs that serve as a template for the production of RNA. The replicase-transcriptase complex, similarly to those in other positive-sense RNA viruses, has RNA-dependent RNA polymerase, RNA helicase, and protease activities. The transcription catalyzed by RdRP toward the 3′-end of genome results in the production of a nested series of many sub-genomic RNAs with leader sequences at the 5′-end. This region produces the structural proteins S, E, M, and N, which are needed for virion assembly; other accessory proteins required for maintenance and replication of the viral genome; and several additional proteins that aid in blocking the host innate immune response (Hussain et al., 2005; Perrier et al., 2019).
The replicase component of the replicase/transcriptase complex system encoded by ORF 1a recognizes the single-stranded positive-sense genomic RNA and copies it into a complementary strand (genomic minus-strand), which is used as a template to produce multiple copies of the positive-sense genomic RNA through continuous RNA synthesis. The transcriptase produces a nested series of subgenomic messenger RNAs via discontinuous transcription, which are translated into structural and non-structural viral proteins. The viral proteins are assembled at the surfaces of the ER membrane along with the genomic RNA, thus, constituting a mature virion that is enclosed in the vesicles at the ERGIC face of the Golgi complex. The vesicles enclosing viral particles are fused with the internal plasma membrane, bud out by exocytosis, and release vesicle-coated virus particles from infected cells (Perrier et al., 2019). The released viral particles trigger the host immune system and result in the production of aggressive inflammatory responses, thus releasing an excessive amount of proinflammatory cytokines in a process called cytokine storm (Braciale and Hahn, 2013; Ragab et al., 2020). The overactivated and abnormal host immune response during SARS-CoV-2 pathogenesis results in the host attacking its own cells and tissues rather than targeting the viral particles (Asrani and Hassan, 2020). The production of proinflammatory cytokines and the resulting excessive inflammatory reactions lead to acute respiratory distress syndrome (ARDS), which can result in lung injury, decreased oxygen saturation, and widespread tissue damage, thus, causing multiorgan failure, unfavorable prognosis and instantaneous death in patients with COVID-19 (Huang et al., 2020; Ragab et al., 2020; Shimizu, 2019; Thompson et al., 2011). The increased levels of cytokines facilitate the influx of immune cells, process such as macrophages, neutrophils, and T-cells, from the circulation toward the site of infection; this process has a destructive effect on human tissues by destabilizing endothelial cell to cell interactions and causing lung injury and severe ARDS, thereby leading to low oxygen saturation levels, which are a significant cause of mortality in patients with COVID-19 (Shimizu, 2019). The patients show lymphopenia with or without leucopenia; the lymphocyte counts decline, thus resulting in high neutrophil to lymphocyte ratios, which are typically associated with the severity of the disease (Singhal, 2020).
7. SARS-CoV-2 pathogenesis associated clinical complications and symptoms
The inoculum of COVID-19 is nosocomial infection, which spreads primarily through respiratory droplets, secretions, and direct contact with the upper respiratory tract (Li et al., 2020b). SARS-CoV-2 isolated from fecal and blood samples from patients with pneumonia suggests the possibility of multiple transmission routes of the virus (Zhang et al., 2020). The abundance of ACE2 receptor proteins on the epithelia of lung alveoli and enterocytes of the small intestine supports the existence of multiple transmission routes (Hamming et al., 2004; Zhang et al., 2020). Epidemiological reports have revealed an incubation period of 1–14 days with a peak at 3–7 days; during the latency period, the virus becomes severely contagious (Jin et al., 2020). Patients show symptoms of influenza, sore throat, fever, cough, fatigue, and shortness of breath; in some cases, patients develop gastrointestinal problems, such as diarrhea and vomiting, whereas in severe cases, patients develop ARDS, thus resulting in multiorgan failure and death (Chen et al., 2020b; Huang et al., 2020). Older people and those with co-morbidities, such as diabetes, hypertension, pulmonary disease, asthma, bronchitis, and cardiovascular disorders, are prone to developing ARDS and death (Huang et al., 2020). Most people have good prognosis, but those older in age and/or with critical chronic disorders are adversely affected by the disease. Recently, a connection between the virus and neurological problems has been reported (Marshall, 2020; Paterson et al., 2020). Some people who become ill with COVID-19 develop neurological symptoms including confusion, disorientation, agitation, and even psychosis; however, the underlying mechanisms are unclear (Paterson et al., 2020; Varatharaj et al., 2020).
Growing evidence suggests that about one in five people infected with COVID-19 experience no symptoms. Such asymptomatic individuals can also transmit the virus as symptomatic people, albeit at a far lesser rate (Nogrady, 2020). Though estimating their contribution to outbreaks is challenging, it is crucial for managing the disease spread (Bi et al., 2020).
8. Control and management of COVID-19
8.1. Preventive approach
Immunization through vaccination is a preferred protection method in public health. Vaccines are developed to enhance the passive immunity in individuals who do not exhibit the symptoms of the disease against the targeted pathogen, to prevent the occurrence of the disease after exposure to the specific pathogen (Han, 2015). The development of an effective vaccine is a complex process that requires a long time for successful optimization, production, and clinical trials to establish the vaccine's purity, capability, and efficacy in vaccinated individuals (Black, 2015). The most challenging issue in developing an anti-SARS-CoV-2 vaccine is achieving efficacy and clinical safety. An effective vaccine promotes the development of the innate immune system with specific instructions for recognizing the antigenic determinant of the causative pathogen (Calina et al., 2020). In classical viral vaccines, the antigenic determinant epitope or attenuated viral particles are used as vaccine antigens to activate the host immune system to produce appropriate antibodies against the antigenic vaccine, thus, aiding in recognition of the specific viral pathogen upon future exposure. However, vaccine development is a time-consuming and expensive process with high failure rates. To produce a successful commercial vaccine, multiple candidates must be tested, and several years of high-quality research are typically required. Another challenge in the vaccine development process is the rapid development of mutations in RNA viruses in the genes encoding surface glycoproteins, which trigger the antigenic immune response and lessen the effectiveness of vaccines (Kramps and Probst, 2013). Moreover, the CoVs appear to have a specific mechanism of being enclosed in vesicles in the host cells that lack the receptors. This process aids in recognition of the invading virus; in the absence of the receptors, the viruses are not recognized by the antibodies developed post-vaccination (Kikkert, 2020). If the vaccine development process proceeds smoothly from conception to market, an effective and successful vaccine can be ready in approximately 12–18 months (Amanat and Krammer, 2020). Vaccines will be essential to decreasing morbidity and mortality if the SARS-CoV-2 establishes itself in the population.
Serious efforts are being made worldwide to develop and launch effective vaccines against SARS-CoV-2 through advanced molecular biology and biotechnological approaches. To support the development of vaccines against SARS-CoV-2, the Coalition for Epidemic Preparedness Innovations (CEPI) is working with global health authorities and scientists. On 11 August 2020, the first (and only) vaccine against SARS-CoV-2, Sputnik V, was approved by Russia; however, the safety and efficacy of the vaccine has been questioned, because it has not yet entered phase III clinical trials. As of 12 November 2020, 164 vaccines are in different stages of clinical trials worldwide, and 48 vaccines have already reached the crucial phase of human trials. More than 280,000 participants from at least 470 sites in 34 different countries are enrolled. Details of the various vaccine platforms and candidate SARS-CoV-2 vaccines are shown in Table 2 . The four major vaccine programs in the final stages of clinical trials are Moderna's mRNA-1273, The University of Oxford and AstraZeneca's AZD1222, Gamaleya Research Institute's Sputnik V, and Pfizer & BioNTech's BNT162; these trials are being conducted at hundreds of sites worldwide. Moderna's mRNA-based vaccine candidate, mRNA-1273, has shown great promise and is in phase III clinical trials. mRNA-1273 results in synthesis of the prefused form of S-protein as the antigenic determinant that is recognized by host immune cells and generates a specific immune response against SARS-CoV-2 (Kramps and Probst, 2013). The second vaccine candidate, AZD1222 (also known as ChAdOx1 nCoV-19 and Covisheild), developed by the University of Oxford, has also reached the phase III of clinical trials. AZD1222 is a DNA-based adenoviral vector vaccine that uses a weakened chimpanzee adenovirus, which causes the common cold in chimpanzees. The virus has been modified so that it does not multiply in humans. The DNA encoding the SARS-CoV-2 S-protein was fused to the viral vector and introduced as recombinant DNA. When the vaccine enters a cell, S-protein is produced and induces an immune response against SARS-CoV-2. The third is Pfizer's BNT162 program evaluating four mRNA-based vaccines, all of which are combined with a lipid nanoparticle formulation. Two vaccine candidates include a nucleoside modified mRNA (BNT162b1 and BNT162b2), one includes mRNA of optimized SARS-CoV-2 RBD (BNT162b1), and one uses mRNA of SARS-CoV-2 full-length S-protein (BNT162b2). Other than these nanoparticle-based vaccines, ankara vector-based vaccines, adjuvanted vaccines, inactivated vaccines, fusion-protein based vaccines, recombinant protein, and live-attenuated vaccines are also being developed; however, all these vaccines appear to be many months away from reaching the market (Guo et al., 2020; Prompetchara et al., 2020; Roper and Rehm, 2009; Thanh Le et al., 2020). Various candidate vaccines in phase II and III clinical trials are shown in Table 3 .
Table 2.
Details of the vaccine platforms and candidate SARS-CoV-2 vaccines as of WHO data, 12 November 2020.
| Platform | Total no. of vaccine candidates | No. of vaccine candidates in clinical trials |
|---|---|---|
| RNA-based | 22 | 6 |
| DNA-based | 14 | 5 |
| Recombinant protein subunit-based | 56 | 15 |
| Inactivated virus | 15 | 7 |
| Virus-like particles | 16 | 2 |
| Replicating viral vector | 17 | 4 |
| Non-replicating viral vector | 19 | 9 |
| Live attenuated virus | 3 | 0 |
| Replicating bacteria vector | 1 | 0 |
| T-cell based | 1 | 0 |
| Total | 164 | 48 |
Table 3.
Major candidate vaccines in phase II and III trials as of WHO data, 12 November 2020.
| S. no. | Vaccine candidates, developers | Technology/platform | Current stage (participants) |
|---|---|---|---|
| 1 | BNT162 a1, b1, b2, c2, Pfizer/BioNTech | mRNA | Phase III cleared (38,000) |
| 2 | mRNA-1273, Moderna | Nanoparticle-based dispersion containing mRNA | Phase III cleared (30,000) |
| 3 | AZD1222, University of Oxford/AstraZeneca | Modified chimpanzee adenovirus (ChAdOx1) | Phase II-III (23,000) |
| 4 | COVAXIN, Bharat Biotech | Inactivated SARS-CoV-2 | Phase III (30,000) |
| 5 | Unnamed, Sinopharm | Inactivated SARS-CoV-2 (Vero cells) | Phase III (15,000) |
| 6 | CoronaVac, Sinovac | Inactivated SARS-CoV-2 | Phase III (10,490) |
In mid-November 2020, four vaccine developers reported data suggesting that their vaccines are highly effective. On 9 November, Pfizer and BioNTech released the first compelling evidence that their BNT162b2 vaccine is 90% effective in phase III trials (with more than 38,000 participants) (Callaway, 2020d). On 18 November, they reported that the vaccine's efficacy was more than 94% in high-risk groups and was consistent across racial groups with no serious side effects (Thomas, 2020). However, their vaccine presents major challenges in distribution, because it must be stored at –80 °C, and two doses are required for effective protection. On 11 November, the controversial Russian Gamaleya Research Institute's Sputnik V vaccine was reported to be 92% effective in preventing COVID-19 in an interim efficacy analysis report (with more than 16,000 participants) (Callaway, 2020c). On 16 November, Moderna reported 94.5% efficacy of the mRNA-1273 vaccine in phase III trials (with more than 30,000 participants) (Callaway, 2020a). Notably, Moderna's vaccine remains stable in conventional refrigerators for 1 month and in freezers for 6 months. On 18 November, the University of Oxford and Astrazeneca released the interim data from the AZD1222 clinical trials showing that the vaccine was highly effective in preventing COVID-19 with an average efficacy of 70% (with more than 23,000 participants) (Ramasamy et al., 2020). The data included people of all ages, raising hopes that it could protect age groups that are at high risk. However, their finding have raised several questions as the data were combined from two trials in two countries that used different protocols. On 20 November,Pfizer/BioNTech submitted a request to the U.S. Food and Drug Administration (FDA) for an emergency use authorization (EUA) of their vaccine. Once approved, this vaccine would be the first mRNA vaccine to get approval for public use. Other companies are also preparing the regulatory submission of the data for EUA to authoriteis around the world. However, recent reports on reinfections have prompted questions about the long-term immunity to SARS-CoV-2 and the prospects for a vaccine (Tillett et al., 2020; To et al., 2020). These four vaccine candidates show remarkable promise, with three exceeding 90% efficacy - an unexpectedly high rate. Notably, none reported significant safety issues, and one showed promise in older adults, a demographic that is particularly vulnerable to SARS-CoV-2 infection. However, it is vital to continue developing more vaccines as having multiple vaccines available for people worldwide will be essential to bring the pandemic under control. Besides, as ecouraging results emerge, the world must be prepared to address challenges linked to vaccine production, storage, distribution, and pricing. The details on the four vaccines are shown in Table 4 .
Table 4.
Details of the four vaccine candidates that are in the approval stage, as of 24 November 2020.
| S. no. | Vaccine Name | Developer | Type | Doses | Efficacy | Storage |
|---|---|---|---|---|---|---|
| 1 | mRNA-1273 | Moderna | mRNA | 2 | 95% | Normal refrigerator for 1 month and in −20 °C freezer for 6 months |
| 2 | BNT162b2 | Pfizer & BioNTech | mRNA | 2 | 95% | −80 °C |
| 3 | AZD1222 (also known as ChAdOx1 nCoV-19, Covishield) | University of Oxford & AstraZeneca | DNA (chimpanzee adenovirus vector) | 2 | 62-90% | Regular refrigerator temperature |
| 4 | Sputnik V | Gamaleya Research Institute | DNA (human adenovirus vector) | 2 | 92% | Regular refrigerator temperature |
8.2. Curative approach
In the absence of any clinically proven treatment strategy, the management and treatment of COVID-19 is mostly supportive, with the only aim being mortality reduction. Good hygiene, social distancing, and quarantine practices are recommended worldwide to reduce the transmission of the virus. Several repurposed drugs are being used, including antiviral and antimalarial drugs, as tactical treatments aiming to limit the adverse effects of viral pathogenesis. Antiviral drugs target specific proteins essential for the viral life cycle and disrupt various stages of viral growth. The antiviral drugs used against common RNA viruses include reverse transcriptase inhibitors (nucleoside reverse transcriptase inhibitor and nonnucleoside reverse transcriptase inhibitors) RdRp inhibitors, and protease inhibitors (target TMPRSS2 and other viral proteases that are involved in the processing of large polypeptide coded by ORF1) (Rajarshi et al., 2020). However, no evidence-based and clinically demonstrated strategy has been shown for the treatment of COVID-19. Most drugs currently in clinical trials are repurposed drugs designed for the treatment of other diseases (Dong et al., 2020; Gordon et al., 2020b; Guy et al., 2020; Tu et al., 2020). Several clinical trials are also underway to evaluate the suitability and efficacy of repurposed antiviral drugs, such as remdesivir, ribavirin, galidesivir, favipiravir, darunavir, oseltamivir, and arbidol, for treating COVID-19 (Calina et al., 2020; Dong et al., 2020; Tu et al., 2020). Several reports have indicated the development of resistance against antiviral therapies, owing to the rapid rate of mutation leading to the emergence of resistance in the viral strain. In such cases, a combination of these drugs, called the highly active antiviral therapy (HAAT) approach, has synergistic or additive effects of antiviral drugs, thus, delaying the progression of disease and increasing the survival rate of the patients. Various candidate drugs in phase II and III clinical trials are shown in Table 5 . The few drugs currently being used for the treatment and management of COVID-19 are discussed below and their structures are shown in Fig. 4 .
Table 5.
Candidate drugs in phase II and III trials.
| S. no. | Target | Drug candidates | Current stage |
|---|---|---|---|
| 1 | RdRP | Remdesivir | Phase II/III |
| 2 | Favipiravir | Phase III | |
| 3 | Ribavirin | Phase II | |
| 4 | Oseltamivir | Phase III | |
| 5 | Galidesivir | Phase II | |
| 6 | Sofosbuvir | Phase II/III | |
| 7 | EIDD-2801 | Phase II | |
| 8 | 3CL protease | Lopinavir/ritonavir | Phase II |
| 9 | Ivermectin | Phase III | |
| 10 | S-protein/ACE2 fusion | Arbidol | Phase III |
| 11 | HIV protease | Darunavir | Phase III |
| 12 | Nucleic acid synthesis | Clevudine | Phase II |
| 13 | DPP4 | Brensocatib | Phase III |
| 14 | Reverse transcriptase | Truvada | Phase III |
| 15 | Acidification of endosomes | Hydroxychloroquine/chloroquine | Phase III |
Fig. 4.
2D structures of the drugs currently used for the treatment of COVID-19.
In an international coalition, the WHO recently conducted a multi-center, open-label global trial, called the SOLIDARITY trial, to investigate the potential of four repurposed drugs—remdesivir, hydroxychloroquine, lopinavir/ritonavir, and interferon-β1a—for the treatment of COVID-19. The trial enrolled approximately 12,000 patients in 500 hospital sites in more than 30 countries. The interim results of the SOLIDARITY trial showed that the drugs had little or no effect on the mortality of hospitalized patients with COVID-19 (Pan et al., 2020). However, because the data were not scrutinized and reviewed, several questions on the report remain. The trial design also prioritized broad access, thereby resulting in significant heterogeneity in trial adoption, implementation, controls, and patient populations. Thus, scientists believe that whether any conclusive findings can be drawn from the trial results is unclear. The data also suggested that antiviral monotherapy for moderately to severely ill patients with COVID-19 might not be sufficient (Cao and Hayden, 2020).
8.2.1. Remdesivir
Remedesivir, the most promising antiviral drug being used for COVID-19 treatment, is a broad-spectrum antiviral drug, repurposed for single-stranded RNA viruses (such as SARS, MERS, and Ebola) (Mulangu et al., 2019). It is an adenosine nucleoside analog prodrug that inhibits the activity of the RdRp enzyme by integrating into newly synthesized viral RNA strands, thus, delaying the chain termination of viral RNA and inhibiting the replication and synthesis of RNA (Saha et al., 2020; Wang et al., 2020a). Recent data from a randomized, open-label, phase III clinical trial, including 584 patients with moderate COVID-19 have indicated that a 5-day course of remdesivir has statistically significantly better results than standard care at 11 days of treatment. The clinical importance of this finding is uncertain (Spinner et al., 2020). Recent data from a randomized, double-blind, placebo-controlled trial of intravenous remdesivir have indicated that this treatment is statistically superior to placebo in shortening the time to recovery in adults (Beigel et al., 2020). Another nucleoside analog inhibitor, EIDD-2801, has been found to be effective against remdesivir-resistant SARS-CoV; EIDD-2801 decreases the replication and pathogenesis of CoVs similarly to remdesivir (Sheahan et al., 2020).
8.2.2. Favipiravir
Favipiravir is a prodrug with a similar mechanism of action to that of remdesivir. In cells, it is converted into an active phosphoribosylated form (favipiravir-RTP), which binds and inhibits viral RdRP. However, being a guanine analog, it structurally differs from remdesivir (Furuta et al., 2017). It induces mutations in the viral RdRP complex, thus resulting in a large proportion of nonviable viruses within the total virus population. The drug was approved as the first anti-COVID 19 drug in China in March 2020, after a clinical trial showed promising data with very few adverse effects. The drug causes a rapid decrease in viral load within 4 days and demonstrates up to 88% clinical improvement in patients with mild to moderate COVID-19.
8.2.3. Ivermectin
Ivermectin is an anti-parasitic and antiviral drug approved by the FDA for the treatment of parasitic worms, HIV, and dengue (Wagstaff et al., 2012). It suppresses host cellular processes by inhibiting nuclear transport by importin α/β1, thus, leading to a decrease in viral replication (Caly et al., 2012). Ivermectin also inhibits the replication of SARS-CoV-2 in vitro, and a single dose can result in a ∼5000-fold reduction in viral RNA at 48 h (Caly et al., 2020). The drug is also in clinical trials for treating COVID-19. A recent observational retrospective study of patients with COVID-19 has reported significantly lower mortality with ivermectin treatment (Rajter et al., 2020).
8.2.4. Lopinavir/ritonavir
Lopinavir and ritonavir is a combination drug used for the treatment of HIV. These compounds are aspartate protease inhibitors, and therefore, are presumed to inhibit the SARS-CoV-2 3CLpro enzyme (Furuta et al., 2017). The drugs are effective against SARS-CoV and MERS-CoV in vitro and animal models; however, they have not found to be useful for treating severe COVID-19 in a randomized, controlled, open-label trial in hospitalized adults patients (Cao et al., 2020).
8.2.5. Hydroxychloroquine/chloroquine
Hydroxychloroquine/chloroquine are widely known anti-malarial and anti-autoimmune drugs (Savarino et al., 2003). Some data suggest that these drugs also inhibit the replication of CoVs (Vincent et al., 2005) and cause a rapid decrease in SARS-CoV-2 viral load. In the initial stages of the pandemic, the drugs were widely used worldwide, because clinical studies reported that the administration of hydroxychloroquine improves the conditions of patients with COVID-19. A further study has confirmed that the combination of hydroxychloroquine with azithromycin is more effective than hydroxychloroquine alone (Arshad et al., 2020). Emerging preclinical and clinical data indicate that neither chloroquine nor hydroxychloroquine provides clinical benefit against COVID-19 (Fatima et al., 2020; Funnell et al., 2020). The (Randomised Evaluation of COVid-19 thERapY) trial on hydroxychloroquine, which enrolled more than 15,000 patients from 175 NHS hospitals in the UK, has concluded that there is no beneficial effect of hydroxychloroquine in patients hospitalized with COVID-19 (Horby and Landray, 2020). In June of 2020, the FDA confirmed that hydroxychloroquine/chloroquine alone is not effective in treating COVID-19, and their risks may outweigh their potential benefits. Therefore, the FDA revoked the emergency use of hydroxychloroquine/chloroquine to treat COVID-19.
8.2.6. Arbidol
Arbidol (umifenovir), a broad-spectrum drug, has been found to be effective against several viral diseases, such as influenza, adenovirus, respiratory syncytial virus, and Ebola, and has been used for decades in China and Russia for treating respiratory viral infections (Blaising et al., 2014; Boriskin et al., 2008). It is currently being repurposed for the treatment of COVID-19. Arbidol is approved in China for the clinical treatment of COVID-19. In the influenza virus, it binds the cell surface glycoprotein hemagglutinin and prevents the fusion of the viral membrane with the receptor (Kadam and Wilson, 2017). Arbidol targets the S-protein/ACE2 interaction, thereby inhibiting viral entry by interfering with viral binding to host cells (Padhi et al., 2020a; Wang et al., 2020d). It also inhibits postentry stages by blocking intracellular vesicle trafficking (Wang et al., 2020d). Clinical trials comparing the efficacy of arbidol with favipiravir are also being conducted (Chen et al., 2020a). Some patients have recovered successfully from COVID-19 after receiving a combination arbidol and lopinavir/ritonavir treatment (Lim et al., 2020; Wang et al., 2020e). Arbidol monotherapy has been found to be superior to lopinavir/ritonavir (Zhu et al., 2020) as well as favipiravir (Chen et al., 2020a).
8.2.7. Corticosteroids
Corticosteroids, such as dexamethasone, are glucocorticoid receptor agonists and immunosuppressive agents. On the basis of data from the RECOVERY trial, the WHO REACT (Rapid Evidence Appraisal for COVID-19 Therapies) working group has reported that critically ill patients with COVID-19 are less likely to die when administered corticosteroids than people who do not receive this treatment (Horby et al., 2020; The, 2020). A meta-analysis that pooled data from seven randomized clinical trials conducted in 12 countries has indicated that the systemic administration of corticosteroids, such as dexamethasone, to critically ill patients with COVID-19 is associated with lower mortality, thus suggesting that steroids may be used as a part of the standard treatment for people with severe COVID-19 (The, 2020). However, the safety and efficacy of corticosteroids for the treatment of COVID-19 have not been rigorously studied.
8.2.8. Other drugs
Beyond the above-discussed drugs, several others are being tested for their ability to treat COVID-19, such as ribavirin, fedratinib, fluvoxamine, oseltamivir, darunavir, sofosbuvir, clevudine, brensocatib, truvada, galedesivir, nafamostat, lianhuaqingwen, and baricitinib (De Clercq, 2004; Elfiky, 2020; Koren et al., 2003; Li et al., 2020a; Runfeng et al., 2020; Sarzi-Puttini et al., 2020; Velavan and Meyer, 2020; Wang et al., 2020b; Wu and Yang, 2020).
8.3. Strategic management approach
A potential alternative, but a reliable approach to ameliorate the adverse effects of COVID-19 aims to boost and strengthen the immune system, thus, allowing the adaptive immune component to be neutralized and combatting SARS-CoV-2. In this approach, several natural product-based formulations are used in the management of the disease, given that herbal medicines have the potential to treat COVID-19. Several herbal formulations have been recommended for the management of COVID-19, such as Indian giloy (Tinospora cordifolia) and ashwagandha; these have shown to be effective in the management COVID-19 in India. Giloy/amrita in the Indian system of medicine has substantial therapeutic properties and is recognized for its immunomodulatory properties and ability to boost the nonspecific immune system by acting on macrophages (Kapil and Sharma, 1997; Nair et al., 2006; Sharma et al., 2012). Withania somnifera, commonly known as ashwagandha/Indian ginseng, has been recognized in Ayurveda for its excellent antioxidative, anti-inflammatory, and immunomodulatory activities (Bharti et al., 2016). Withanone, the active ingredient of ashwagandha, has been shown to be a potent phytochemical for controlling SARS-CoV-2 entry into the host cell; withanone can dock effectively at the interface of the ACE2-RBD complex, thus, decreasing the RBD-ACE2 interaction (Balkrishna et al., 2020). Withanone obtained from Withania somnifera and caffeic acid phenethyl ester obtained from bee propolis have the potential to effectively interact with the substrate-binding pocket of viral main protease and block the RNA replication machinery of SARS-CoV-2. Moreover, these natural compounds also modulate the host surface-bound protease TMPRSS2, thereby inhibiting viral entry into host cells (Kumar et al., 2020).
9. Conclusions
The COVID-19 pandemic has sparked a research revolution, as scientists are working at breakneck speed to understand the disease and find a cure. Scientists have demonstrated how rapidly they can adapt to re-emerging and emerging threats. Non-COVID-19 research has taken a back seat during the pandemic. The past year has seen rapid advances in understanding of the biology, pathogenesis, and clinical characteristics of the disease. There are still several unknowns about the mechanisms of SARS-CoV-2 biology and pathogenesis. Epidemiologists are predicting short- and long-term projections to prepare for and potentially mitigate the spread and impact of the disease. However, the pandemic has exposed our susceptibly to microbial pathogens and gaps in our therapeutic arsenal. COVID-19 is not the first pandemic that humanity has faced, and it will not be the last. However, it has taught us how unprepared the world was for such an outbreak, and provided us with lessons in preparing and responding to virus warnings in the future. We hope that the continued research will lead to breakthroughs and help us be better prepared for future outbreaks.
Authors' Contributions
SKM and TT conceived the idea and wrote the manuscript.
Declaration of Competing Interest
The authors declare no competing financial interests.
Funding
The authors declare that there are no funding sources to be acknowledged.
References
- Al Bari M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5:e00293. doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Krammer F. SARS-CoV-2 vaccines: status report. Immunity. 2020;52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arshad S., Kilgore P., Chaudhry Z.S., Jacobsen G., Wang D.D., Huitsing K., Brar I., Alangaden G.J., Ramesh M.S., McKinnon J.E., O'Neill W., Zervos M. Treatment with hydroxychloroquine, azithromycin, and combination in patients hospitalized with COVID-19. Int. J. Infect. Dis. 2020;97:396–403. doi: 10.1016/j.ijid.2020.06.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrani P., Hassan M.I. SARS-CoV-2 mediated lung inflammatory responses in host: targeting the cytokine storm for therapeutic interventions. Mol. Cell Biochem. 2020:1–13. doi: 10.1007/s11010-020-03935-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G.J., Esshaki D.J., Thomas W.D., Jr., Ambrosino D.M. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkrishna A., Pokhrel S., Singh J., Varshney A. Withanone from Withania somnifera may inhibit novel Coronavirus (COVID-19) entry by disrupting interactions between viral S-protein receptor binding domain and host ACE2 receptor. Res. Sq. 2020 (PrePrint) [Google Scholar]
- Banerjee, A., Kulcsar, K., Misra, V., Frieman, M., Mossman, K., 2019. Bats and coronaviruses. viruses 11. [DOI] [PMC free article] [PubMed]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fätkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C. Remdesivir for the treatment of Covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benvenuto D., Giovanetti M., Ciccozzi A., Spoto S., Angeletti S., Ciccozzi M. The 2019-new coronavirus epidemic: evidence for virus evolution. J. Med. Virol. 2020;92:455–459. doi: 10.1002/jmv.25688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Muller M.A., Lavender H., Gnirss K., Nehlmeier I., Niemeyer D., He Y., Simmons G., Drosten C., Soilleux E.J., Jahn O., Steffen I., Pohlmann S. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 2011;85:13363–13372. doi: 10.1128/JVI.05300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava, H.D., 2020. Corona virus History, WebMD medical reference.
- Bharti V.K., Malik J.K., Gupta R.C. Elsevier; 2016. Ashwagandha: multiple health benefits, Nutraceuticals; pp. 717–733. [Google Scholar]
- Bi, Q., Lessler, J., Eckerle, I., Lauer, S.A., Kaiser, L., Vuilleumier, N., Cummings, D.A.T., Flahault, A., Petrovic, D., Guessous, I., Stringhini, S., Azman, A.S., 2020. Household transmission of SARS-COV-2: insights from a population-based serological survey. medRxiv, 2020.2011.2004.20225573.
- Bianchi M., Benvenuto D., Giovanetti M., Angeletti S., Ciccozzi M., Pascarella S. Sars-CoV-2 envelope and membrane proteins: structural differences linked to virus characteristics? Biomed. Res. Int. 2020 doi: 10.1155/2020/4389089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black S. The costs and effectiveness of large Phase III pre-licensure vaccine clinical trials. Expert Rev. Vaccines. 2015;14:1543–1548. doi: 10.1586/14760584.2015.1091733. [DOI] [PubMed] [Google Scholar]
- Blaising J., Polyak S.J., Pecheur E.I. Arbidol as a broad-spectrum antiviral: an update. Antivir. Res. 2014;107:84–94. doi: 10.1016/j.antiviral.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat. Microbiol. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Boriskin Y.S., Leneva I.A., Pecheur E.I., Polyak S.J. Arbidol: a broad-spectrum antiviral compound that blocks viral fusion. Curr. Med. Chem. 2008;15:997–1005. doi: 10.2174/092986708784049658. [DOI] [PubMed] [Google Scholar]
- Braciale T.J., Hahn Y.S. Immunity to viruses. Immunol. Rev. 2013;255:5–12. doi: 10.1111/imr.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calina D., Docea A.O., Petrakis D., Egorov A.M., Ishmukhametov A.A., Gabibov A.G., Shtilman M.I., Kostoff R., Carvalho F., Vinceti M., Spandidos D.A., Tsatsakis A. Towards effective COVID-19 vaccines: Updates, perspectives and challenges (Review) Int. J. Mol. Med. 2020;46:3–16. doi: 10.3892/ijmm.2020.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. COVID vaccine excitement builds as Moderna reports third positive result. Nature. 2020;587:337–338. doi: 10.1038/d41586-020-03248-7. [DOI] [PubMed] [Google Scholar]
- Callaway E. Making sense of coronavirus mutations. Nature. 2020;585:174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- Callaway E. Russia announces positive COVID-vaccine results from controversial trial. Nature. 2020 doi: 10.1038/d41586-020-03209-0. [DOI] [PubMed] [Google Scholar]
- Callaway E. What Pfizer's landmark COVID vaccine results mean for the pandemic. Nature. 2020 doi: 10.1038/d41586-020-03166-8. [DOI] [PubMed] [Google Scholar]
- Caly L., Druce J.D., Catton M.G., Jans D.A., Wagstaff K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., Wagstaff K.M., Jans D.A. Nuclear trafficking of proteins from RNA viruses: potential target for antivirals? Antiviral Res. 2012;95:202–206. doi: 10.1016/j.antiviral.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Cao B., Hayden F.G. Antiviral monotherapy for hospitalised patients with COVID-19 is not enough. Lancet. 2020;396:1310–1311. doi: 10.1016/S0140-6736(20)32078-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. New Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Zhang, Y., Huang, J., Yin, P., Cheng, Z., Wu, J., Chen, S., Zhang, Y., Chen, B., Lu, M., Luo, Y., Ju, L., Zhang, J., Wang, X., 2020a. Favipiravir versus Arbidol for COVID-19: a randomized clinical trial. medRxiv, 2020.2003.2017.20037432.
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., Xia J., Yu T., Zhang X., Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Antivirals and antiviral strategies. Nature Rev. Microbiol. 2004;2:704–720. doi: 10.1038/nrmicro975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., Snijder E.J., Kikkert M., van Hemert M.J. Host factors in coronavirus replication. Curr. Top Microbiol. Immunol. 2018;419:1–42. doi: 10.1007/82_2017_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit E., Van Doremalen N., Falzarano, Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Micobiol. 2016;14:523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degtyarev M., De Maziere A., Orr C., Lin J., Lee B.B., Tien J.Y., Prior W.W., van Dijk S., Wu H., Gray D.C., Davis D.P., Stern H.M., Murray L.J., Hoeflich K.P., Klumperman J., Friedman L.S., Lin K. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J. Cell Biol. 2008;183:101–116. doi: 10.1083/jcb.200801099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- EA J.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima U., Rizvi S.S.A., Fatima S., Hassan M.I. Impact of Hydroxychloroquine/Chloroquine in COVID-19 Therapy: Two Sides of the Coin. J. Interferon Cytokine Res. 2020;40:469–471. doi: 10.1089/jir.2020.0105. [DOI] [PubMed] [Google Scholar]
- Follis K.E., York J., Nunberg J.H. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350:358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2016;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier R.A., Kuiken T., Schutten M., van Amerongen G., van Doornum G.J., van den Hoogen B.G., Peiris M., Lim W., Stohr K., Osterhaus A.D. Aetiology: Koch's postulates fulfilled for SARS virus. Nature. 2003;423:240. doi: 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell S.G.P., Dowling W.E., Muñoz-Fontela C., Gsell P.S., Ingber D.E., Hamilton G.A., Delang L., Rocha-Pereira J., Kaptein S., Dallmeier K.H., Neyts J., Rosenke K., de Wit E., Feldmann H., Maisonnasse P., Le Grand R., Frieman M.B., Coleman C.M. Emerging preclinical evidence does not support broad use of hydroxychloroquine in COVID-19 patients. Nature Commun. 2020;11:4253. doi: 10.1038/s41467-020-17907-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y., Komeno T., Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proceed. Jpn. Acad., Ser. B. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Muller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., Niemeyer D., Schneider H., Drosten C., Pohlmann S. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., Jureka A.S., Obernier K., Guo J.Z., Batra J., Kaake R.M., Weckstein A.R., Owens T.W., Gupta M., Pourmal S., Titus E.W., Cakir M., Soucheray M., McGregor M., Cakir Z., Jang G., O'Meara M.J., Tummino T.A., Zhang Z., Foussard H., Rojc A., Zhou Y., Kuchenov D., Hüttenhain R., Xu J., Eckhardt M., Swaney D.L., Fabius J.M., Ummadi M., Tutuncuoglu B., Rathore U., Modak M., Haas P., Haas K.M., Naing Z.Z.C., Pulido E.H., Shi Y., Barrio-Hernandez I., Memon D., Petsalaki E., Dunham A., Marrero M.C., Burke D., Koh C., Vallet T., Silvas J.A., Azumaya C.M., Billesbølle C., Brilot A.F., Campbell M.G., Diallo A., Dickinson M.S., Diwanji D., Herrera N., Hoppe N., Kratochvil H.T., Liu Y., Merz G.E., Moritz M., Nguyen H.C., Nowotny C., Puchades C., Rizo A.N., Schulze-Gahmen U., Smith A.M., Sun M., Young I.D., Zhao J., Asarnow D., Biel J., Bowen A., Braxton J.R., Chen J., Chio C.M., Chio U.S., Deshpande I., Doan L., Faust B., Flores S., Jin M., Kim K., Lam V.L., Li F., Li J., Li Y.L., Li Y., Liu X., Lo M., Lopez K.E., Melo A.A., Moss F.R., 3rd, Nguyen P., Paulino J., Pawar K.I., Peters J.K., Pospiech T.H., Jr, Safari M., Sangwan S., Schaefer K., Thomas P.V., Thwin A.C., Trenker R., Tse E., Tsui T.K.M., Wang F., Whitis N., Yu Z., Zhang K., Zhang Y., Zhou F., Saltzberg D., Hodder A.J., Shun-Shion A.S., Williams D.M., White K.M., Rosales R., Kehrer T., Miorin L., Moreno E., Patel A.H., Rihn S., Khalid M.M., Vallejo-Gracia A., Fozouni P., Simoneau C.R., Roth T.L., Wu D., Karim M.A., Ghoussaini M., Dunham I., Berardi F., Weigang S., Chazal M., Park J., Logue J., McGrath M., Weston S., Haupt R., Hastie C.J., Elliott M., Brown F., Burness K.A., Reid E., Dorward M., Johnson C., Wilkinson S.G., Geyer A., Giesel D.M., Baillie C., Raggett S., Leech H., Toth R., Goodman N., Keough K.C., Lind A.L., Klesh R.J., Hemphill K.R., Carlson-Stevermer J., Oki J., Holden K., Maures T., Pollard K.S., Sali A., Agard D.A., Cheng Y., Fraser J.S., Frost A., Jura N., Kortemme T., Manglik A., Southworth D.R., Stroud R.M., Alessi D.R., Davies P., Frieman M.B., Ideker T., Abate C., Jouvenet N., Kochs G., Shoichet B., Ott M., Palmarini M., Shokat K.M., García-Sastre A., Rassen J.A., Grosse R., Rosenberg O.S., Verba K.A., Basler C.F., Vignuzzi M., Peden A.A., Beltrao P., Krogan N.J. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020 doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., Tummino T.A., Hüttenhain R., Kaake R.M., Richards A.L., Tutuncuoglu B., Foussard H., Batra J., Haas K., Modak M., Kim M., Haas P., Polacco B.J., Braberg H., Fabius J.M., Eckhardt M., Soucheray M., Bennett M.J., Cakir M., McGregor M.J., Li Q., Meyer B., Roesch F., Vallet T., Mac Kain A., Miorin L., Moreno E., Naing Z.Z.C., Zhou Y., Peng S., Shi Y., Zhang Z., Shen W., Kirby I.T., Melnyk J.E., Chorba J.S., Lou K., Dai S.A., Barrio-Hernandez I., Memon D., Hernandez-Armenta C., Lyu J., Mathy C.J.P., Perica T., Pilla K.B., Ganesan S.J., Saltzberg D.J., Rakesh R., Liu X., Rosenthal S.B., Calviello L., Venkataramanan S., Liboy-Lugo J., Lin Y., Huang X.P., Liu Y., Wankowicz S.A., Bohn M., Safari M., Ugur F.S., Koh C., Savar N.S., Tran Q.D., Shengjuler D., Fletcher S.J., O'Neal M.C., Cai Y., Chang J.C.J., Broadhurst D.J., Klippsten S., Sharp P.P., Wenzell N.A., Kuzuoglu-Ozturk D., Wang H.Y., Trenker R., Young J.M., Cavero D.A., Hiatt J., Roth T.L., Rathore U., Subramanian A., Noack J., Hubert M., Stroud R.M., Frankel A.D., Rosenberg O.S., Verba K.A., Agard D.A., Ott M., Emerman M., Jura N., von Zastrow M., Verdin E., Ashworth A., Schwartz O., d'Enfert C., Mukherjee S., Jacobson M., Malik H.S., Fujimori D.G., Ideker T., Craik C.S., Floor S.N., Fraser J.S., Gross J.D., Sali A., Roth B.L., Ruggero D., Taunton J., Kortemme T., Beltrao P., Vignuzzi M., García-Sastre A., Shokat K.M., Shoichet B.K., Krogan N.J. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He Z.X., Liu L., Shan H., Lei C.L., Hui D.S.C. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.R., Cao Q.D., Hong Z.S., Tan Y.Y., Chen S.D., Jin H.J., Tan K.S., Wang D.Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil. Med. Res. 2020;7:11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D. Angiotensin receptor blockers as tentative SARS-CoV-2 therapeutics. Drug. Dev. Res. 2020;81:537–540. doi: 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. a first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton T. Bats may be SARS reservoir. Jama. 2005;294:2291. doi: 10.1001/jama.294.18.2291. [DOI] [PubMed] [Google Scholar]
- Han S. Clinical vaccine development. Clin. EXP Vaccine Res. 2015;4:46–53. doi: 10.7774/cevr.2015.4.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon C.C., Lam T.Y., Shi Z.L., Drummond A.J., Yip C.W., Zeng F., Lam P.Y., Leung F.C. Evidence of the recombinant origin of a bat severe acute respiratory syndrome (SARS)-like coronavirus and its implications on the direct ancestor of SARS coronavirus. J. Virol. 2008;82:1819–1826. doi: 10.1128/JVI.01926-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Landray M. University of Oxford; Oxford, UK: 2020. No clinical benefit from use of hydroxychloroquine in hospitalised patients with COVID-19. [Google Scholar]
- Horby P., Lim W.S., Emberson J.R., Mafham M., Bell J.L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., Prudon B., Green C., Felton T., Chadwick D., Rege K., Fegan C., Chappell L.C., Faust S.N., Jaki T., Jeffery K., Montgomery A., Rowan K., Juszczak E., Baillie J.K., Haynes R., Landray M.J. Dexamethasone in Hospitalized patients with Covid-19 - preliminary report. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S., Pan J., Chen Y., Yang Y., Xu J., Peng Y., Wu Y., Li Z., Zhu Y., Tien P., Guo D. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:5288–5295. doi: 10.1128/JVI.79.9.5288-5295.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J., Li S., Yount B., Smith A., Sturges L., Olsen J.C., Nagel J., Johnson J.B., Agnihothram S., Gates J.E., Frieman M.B., Baric R.S., Donaldson E.F. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J. Virol. 2012;86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y., Tanaka N., Tanaka Y., Inoue S., Morita K., Zhuang M., Hattori T., Sugamura K. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 2007;81:8722–8729. doi: 10.1128/JVI.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79:14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.H., Cai L., Cheng Z.S., Cheng H., Deng T., Fan Y.P., Fang C., Huang D., Huang L.Q., Huang Q., Han Y., Hu B., Hu F., Li B.H., Li Y.R., Liang K., Lin L.K., Luo L.S., Ma J., Ma L.L., Peng Z.Y., Pan Y.B., Pan Z.Y., Ren X.Q., Sun H.M., Wang Y., Wang Y.Y., Weng H., Wei C.J., Wu D.F., Xia J., Xiong Y., Xu H.B., Yao X.M., Yuan Y.F., Ye T.S., Zhang X.C., Zhang Y.W., Zhang Y.G., Zhang H.M., Zhao Y., Zhao M.J., Zi H., Zeng X.T., Wang Y.Y., Wang X.H. A rapid advice guideline for the diagnosis and treatment of 2019 novel coronavirus (2019-nCoV) infected pneumonia (standard version) Mil. Med. Res. 2020;7:4. doi: 10.1186/s40779-020-0233-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam R.U., Wilson I.A. Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol. Proceed. Natl. Acad. Sci. 2017;114:206–214. doi: 10.1073/pnas.1617020114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapil A., Sharma S. Immunopotentiating compounds from Tinospora cordifolia. J. Ethnopharmacol. 1997;58:89–95. doi: 10.1016/s0378-8741(97)00086-x. [DOI] [PubMed] [Google Scholar]
- Kikkert M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate. Immun. 2020;12:4–20. doi: 10.1159/000503030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking Changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 Virus. Cell. 2020;182 doi: 10.1016/j.cell.2020.06.043. 812-827.e819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren G., King S., Knowles S., Phillips E. Ribavirin in the treatment of SARS: a new trick for an old drug? CMAJ. 2003;168:1289–1292. [PMC free article] [PubMed] [Google Scholar]
- Kramps T., Probst J. Messenger RNA-based vaccines: progress, challenges, applications. Wiley Interdiscip. Rev. RNA. 2013;4:737–749. doi: 10.1002/wrna.1189. [DOI] [PubMed] [Google Scholar]
- Kumar V., Dhanjal J.K., Kaul S.C., Wadhwa R., Sundar D. Withanone and caffeic acid phenethyl ester are predicted to interact with main protease (M(pro)) of SARS-CoV-2 and inhibit its activity. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Zhou Y., Zhang M., Wang H., Zhao Q., Liu J. Updated approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 2020;64 doi: 10.1128/AAC.00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early Transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B.T., Zhang S., Wang L.F. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N., Murakami A., He Y., Marasco W.A., Guan Y., Choe H., Farzan M. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. Embo. J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J., Jeon S., Shin H.Y., Kim M.J., Seong Y.M., Lee W.J., Choe K.W., Kang Y.M., Lee B., Park S.J. Case of the index patient who caused tertiary transmission of COVID-19 infection in korea: the application of lopinavir/ritonavir for the treatment of COVID-19 infected pneumonia monitored by quantitative RT-PCR. J. Korean. Med. Sci. 2020;35:e79. doi: 10.3346/jkms.2020.35.e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. How COVID-19 can damage the brain. Nature. 2020;585:342–343. doi: 10.1038/d41586-020-02599-5. [DOI] [PubMed] [Google Scholar]
- Masters P.S., Perlman S. Wolters Kluwer Health/Lippincott Williams & Wilkins; Philadelphia: 2013. Fields virology. [Google Scholar]
- Mulangu S., Dodd L.E., Davey R.T., Jr, Tshiani Mbaya O., Proschan M., Mukadi D., Lusakibanza Manzo M., Nzolo D., Tshomba Oloma A, Ibanda A. A randomized, controlled trial of Ebola virus disease therapeutics. New Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P.D., Simon A.E. New insights into the mechanisms of RNA recombination. Virology. 1997;235:1–9. doi: 10.1006/viro.1997.8681. [DOI] [PubMed] [Google Scholar]
- Nair P.K., Melnick S.J., Ramachandran R., Escalon E., Ramachandran C. Mechanism of macrophage activation by (1,4)-alpha-D-glucan isolated from Tinospora cordifolia. Int. Immunopharmacol. 2006;6:1815–1824. doi: 10.1016/j.intimp.2006.07.028. [DOI] [PubMed] [Google Scholar]
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogrady B. What the data say about asymptomatic COVID infections. Nature. 2020 doi: 10.1038/d41586-020-03141-3. [DOI] [PubMed] [Google Scholar]
- Padhi, A., Seal, A., Tripathi, T., 2020a. How does arbidol inhibit the novel coronavirus SARS-CoV-2? atomistic insights from molecular dynamics simulations. ChemRxiv.
- Padhi A.K., Kalita P., Zhang K.Y.J., Tripathi T. High throughput designing and mutational mapping of RBD-ACE2 interface guide non-conventional therapeutic strategies for COVID-19. . 2020 bioRxiv. [Google Scholar]
- Padhi, A.K., Shukla, R., Tripathi, T., 2020c. Rational design of the remdesivir binding site in the RNA-dependent RNA Polymerase of SARS-CoV-2: implications for potential resistance. bioRxiv. [DOI] [PMC free article] [PubMed]
- Padhi A.K., Tripathi T. Can SARS-CoV‐2 accumulate mutations in the S‐Protein to increase pathogenicity? ACS Pharmacol. Transl. Sci. 2020;3:1023–1026. doi: 10.1021/acsptsci.0c00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palayew A., Norgaard O., Safreed-Harmon K., Andersen T.H., Rasmussen L.N., Lazarus J.V. Pandemic publishing poses a new COVID-19 challenge. Nat. Hum. Behav. 2020;4:666–669. doi: 10.1038/s41562-020-0911-0. [DOI] [PubMed] [Google Scholar]
- Pan, H., Peto, R., Karim, Q.A., Alejandria, M., Henao-Restrepo, A.M., García, C.H., Kieny, M.-P., Malekzadeh, R., Murthy, S., Preziosi, M.-P., Reddy, S., Periago, M.R., Sathiyamoorthy, V., Røttingen, J.-A., Swaminathan, S., 2020. Repurposed antiviral drugs for COVID-19 –interim WHO SOLIDARITY trial results. medRxiv, 2020.2010.2015.20209817.
- Paterson R.W., Brown R.L., Benjamin L., Nortley R., Wiethoff S., Bharucha T., Jayaseelan D.L., Kumar G., Raftopoulos R.E., Zambreanu L., Vivekanandam V., Khoo A., Geraldes R., Chinthapalli K., Boyd E., Tuzlali H., Price G., Christofi G., Morrow J., McNamara P., McLoughlin B., Lim S.T., Mehta P.R., Levee V., Keddie S., Yong W., Trip S.A., Foulkes A.J.M., Hotton G., Miller T.D., Everitt A.D., Carswell C., Davies N.W.S., Yoong M., Attwell D., Sreedharan J., Silber E., Schott J.M., Chandratheva A., Perry R.J., Simister R., Checkley A., Longley N., Farmer S.F., Carletti F., Houlihan C., Thom M., Lunn M.P., Spillane J., Howard R., Vincent A., Werring D.J., Hoskote C., Jäger H.R., Manji H., Zandi M.S. The emerging spectrum of COVID-19 neurology: clinical, radiological and laboratory findings. Brain. 2020 doi: 10.1093/brain/awaa240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, G.L., Antiviral agents, An Introduction to Medicinal Chemistry 4ed. Oxford University Press, pp. 475–518.
- Pelkmans L., Helenius A. Insider information: what viruses tell us about endocytosis. Curr. Opin. Cell Biol. 2003;15:414–422. doi: 10.1016/s0955-0674(03)00081-4. [DOI] [PubMed] [Google Scholar]
- Perrier A., Bonnin A., Desmarets L., Danneels A., Goffard A., Rouille Y., Dubuisson J., Belouzard S. The C-terminal domain of the MERS coronavirus M protein contains a trans-Golgi network localization signal. J Biol Chem. 2019;294:14406–14421. doi: 10.1074/jbc.RA119.008964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020 doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Qu X.X., Hao P., Song X.J., Jiang S.M., Liu Y.X., Wang P.G., Rao X., Song H.D., Wang S.Y., Zuo Y., Zheng A.H., Luo M., Wang H.L., Deng F., Wang H.Z., Hu Z.H., Ding M.X., Zhao G.P., Deng H.K. Identification of two critical amino acid residues of the severe acute respiratory syndrome coronavirus spike protein for its variation in zoonotic tropism transition via a double substitution strategy. J. Biol. Chem. 2005;280:29588–29595. doi: 10.1074/jbc.M500662200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarshi K., Khan R., Singh M.K., Ranjan T., Ray S., Ray S. Essential functional molecules associated with SARS-CoV-2 infection: Potential therapeutic targets for COVID-19. Gene. 2020 doi: 10.1016/j.gene.2020.145313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajter J.C., Sherman M.S., Fatteh N., Vogel F., Sacks J., Rajter J.J. Use of Ivermectin is associated with lower mortality in hospitalized patients with coronavirus disease 2019: the ICON study. Chest. 2020 doi: 10.1016/j.chest.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., Voysey M., Aley P.K., Angus B., Babbage G., Belij-Rammerstorfer S., Berry L., Bibi S., Bittaye M., Cathie K., Chappell H., Charlton S., Cicconi P., Clutterbuck E.A., Colin-Jones R., Dold C., Emary K.R.W., Fedosyuk S., Fuskova M., Gbesemete D., Green C., Hallis B., Hou M.M., Jenkin D., Joe C.C.D., Kelly E.J., Kerridge S., Lawrie A.M., Lelliott A., Lwin M.N., Makinson R., Marchevsky N.G., Mujadidi Y., Munro A.P.S., Pacurar M., Plested E., Rand J., Rawlinson T., Rhead S., Robinson H., Ritchie A.J., Ross-Russell A.L., Saich S., Singh N., Smith C.C., Snape M.D., Song R., Tarrant R., Themistocleous Y., Thomas K.M., Villafana T.L., Warren S.C., Watson M.E.E., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Faust S.N., Pollard A.J. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2020 doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper R.L., Rehm K.E. SARS vaccines: where are we? Expert. Rev. Vaccines. 2009;8:887–898. doi: 10.1586/erv.09.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe C.L., Fleming J.O., Nathan M.J., Sgro J.Y., Palmenberg A.C., Baker S.C. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J. Virol. 1997;71:6183–6190. doi: 10.1128/jvi.71.8.6183-6190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runfeng L., Yunlong H., Jicheng H., Weiqi P., Qinhai M., Yongxia S., Chufang L., Jin Z., Zhenhua J., Haiming J. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Sharma A.R., Bhattacharya M., Sharma G., Lee S.S., Chakraborty C. Probable molecular mechanism of remdesivir for the treatment of COVID-19: need to know more. Arch Med Res. 2020:30699. doi: 10.1016/j.arcmed.2020.05.001. S 0188-4409–30698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarzi-Puttini P., Giorgi V., Sirotti S., Marotto D., Ardizzone S., Rizzardini G., Antinori S., Galli M. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin. Exp. Rheumatol. 2020;38:337–342. [PubMed] [Google Scholar]
- Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases. The Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki S.G., Sawicki D.L. Coronavirus transcription: a perspective. Curr. Top Microbiol. Immunol. 2005;287:31–55. doi: 10.1007/3-540-26765-4_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:1–22. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrezenmeier E., Dorner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- Sharma U., Bala M., Kumar N., Singh B., Munshi R.K., Bhalerao S. Immunomodulatory active compounds from Tinospora cordifolia. J. Ethnopharmacol. 2012;141:918–926. doi: 10.1016/j.jep.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schäfer A., Dinnon K.H., 3rd, Stevens L.J., Chappell J.D., Lu X., Hughes T.M., George A.S., Hill C.S., Montgomery S.A., Brown A.J., Bluemling G.R., Natchus M.G., Saindane M., Kolykhalov A.A., Painter G., Harcourt J., Tamin A., Thornburg N.J., Swanstrom R., Denison M.R., Baric R.S. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z., Hu Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008;133:74–87. doi: 10.1016/j.virusres.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu M. In: Cytokine Storm Syndrome. Cron R, B.E., editor. Springer; 2019. Clinical features of Cytokine Storm Syndrome; pp. 31–41. [Google Scholar]
- Sieczkarski S.B., Whittaker G.R. Dissecting virus entry via endocytosis. J. Gen. Virol. 2002;83:1535–1545. doi: 10.1099/0022-1317-83-7-1535. [DOI] [PubMed] [Google Scholar]
- Singer, J., Gifford, R., Cotten, M., Robertson, D., 2020. CoV-GLUE: a web application for tracking SARS-CoV-2 genomic variation. Preprints.
- Singhal T. A review of Coronavirus Disease-2019 (COVID-19) Indian J. Pediatr. 2020:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinner C.D., Gottlieb R.L., Criner G.J., Arribas López J.R., Cattelan A.M., Soriano Viladomiu A., Ogbuagu O., Malhotra P., Mullane K.M., Castagna A., Chai L.Y.A., Roestenberg M., Tsang O.T.Y., Bernasconi E., Le Turnier P., Chang S.C., SenGupta D., Hyland R.H., Osinusi A.O., Cao H., Blair C., Wang H., Gaggar A., Brainard D.M., McPhail M.J., Bhagani S., Ahn M.Y., Sanyal A.J., Huhn G., Marty F.M. Effect of Remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA. 2020;324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh Le T., Andreadakis Z., Kumar A., Gomez Roman R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020 doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- The, W.H.O.R.E.A.f.C.-T.W.G. Association between administration of systemic corticosteroids and mortality among critically Ill patients with COVID-19: a meta-analysis. JAMA. 2020;324:1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. The New York Times; New York: 2020. Pfizer Says New Results Show Vaccine Is Safe and 95% Effective; p. 7. ed. Sulzberger, A.G., New York. [Google Scholar]
- Thompson M.R., Kaminski J.J., Kurt-Jones E.A., Fitzgerald K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 2011;3:920–940. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillett R.L., Sevinsky J.R., Hartley P.D., Kerwin H., Crawford N., Gorzalski A., Laverdure C., Verma S.C., Rossetto C.C., Jackson D., Farrell M.J., Van Hooser S., Pandori M. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Hung I.F., Ip J.D., Chu A.W., Chan W.M., Tam A.R., Fong C.H., Yuan S., Tsoi H.W., Ng A.C., Lee L.L., Wan P., Tso E., To W.K., Tsang D., Chan K.H., Huang J.D., Kok K.H., Cheng V.C., Yuen K.Y. COVID-19 re-infection by a phylogenetically distinct SARS-coronavirus-2 strain confirmed by whole genome sequencing. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler P., Staker B., Prasad S.G., Menon S., Tang J., Parsons T., Ryan D., Fisher M., Williams D., Dales N.A., Patane M.A., Pantoliano M.W. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., Lai W.Y., Yang D.M., Chou S.J., Yang Y.P., Wang M.L., Chiou S.H. A review of SARS-CoV-2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varatharaj A., Thomas N., Ellul M.A., Davies N.W.S., Pollak T.A., Tenorio E.L., Sultan M., Easton A., Breen G., Zandi M., Coles J.P., Manji H., Al-Shahi Salman R., Menon D.K., Nicholson T.R., Benjamin L.A., Carson A., Smith C., Turner M.R., Solomon T., Kneen R., Pett S.L., Galea I., Thomas R.H., Michael B.D. Neurological and neuropsychiatric complications of COVID-19 in 153 patients: a UK-wide surveillance study. Lancet Psychiatry. 2020 doi: 10.1016/S2215-0366(20)30287-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan T.P., Meyer C.G. The COVID‐19 epidemic. Trop. Med. Int. Health. 2020;25:278. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:1–10. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell. 2020 doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff K.M., Sivakumaran H., Heaton S.M., Harrich D., Jans D.A. Ivermectin is a specific inhibitor of importin α/β-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281-292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020:94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., Arledge K.C., Chen Y.H., Zhang L., Wang X. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894-904.e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Cao R., Zhang H., Liu J., Xu M., Hu H., Li Y., Zhao L., Li W., Sun X., Yang X., Shi Z., Deng F., Hu Z.A.-O., Zhong W., Wang M.A.-O. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6:28. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chen X., Lu Y., Chen F., Zhang W. Clinical characteristics and therapeutic procedure for four cases with 2019 novel coronavirus pneumonia receiving combined Chinese and Western medicine treatment. Biosci. Trends. 2020;14:64–68. doi: 10.5582/bst.2020.01030. [DOI] [PubMed] [Google Scholar]
- Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H., Bai R., Teng J.L., Tsang C.C., Wang M., Zheng B.J., Chan K.H., Yuen K.Y. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang z., wang j., sheng j., quan l., xia z., tan w., cheng g., jiang t. genome composition and divergence of the novel Coronavirus (2019-nCoV) originating in China. Cell Host. Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Yang X.O. TH17 responses in cytokine storm of COVID-19: an emerging target of JAK2 inhibitor Fedratinib. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Peng G., Wilken M., Geraghty R.J., Li F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012;287:8904–8911. doi: 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z., Yang L., Ren X., Zhang J., Yang F., Zhang S., Jin Q. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J. Infect. Dis. 2016;213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhu Y., Liu M., Lan Q., Xu W., Wu Y., Ying T., Liu S., Shi Z., Jiang S., Lu L. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol. Immunol. 2020;17:765–767. doi: 10.1038/s41423-020-0374-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Du L., Ojcius D.M., Pan C., Jiang S. Measures for diagnosing and treating infections by a novel coronavirus responsible for a pneumonia outbreak originating in Wuhan, China. Microbes Infect. 2020;22:74–79. doi: 10.1016/j.micinf.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Ledezma C.,C, D.F.C., Medina E., Sinche F., Santiago Vispo N., Dahoumane S.A., Alexis F. Biomedical science to tackle the COVID-19 pandemic: current status and future perspectives. Molecules. 2020;25 doi: 10.3390/molecules25204620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehra Z., Luthra M., Siddiqui S.M., Shamsi A., Gaur N.A., Islam A. Corona virus versus existence of human on the earth: a computational and biophysical approach. Int. J. Biol. Macromol. 2020;161:271–281. doi: 10.1016/j.ijbiomac.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.H., Li B., Zheng X.S., Yang X.L., Hu B., Wang Y.Y., Xiao G.F., Yan B., Shi Z.L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes. Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B.J., Wong K.H., Zhou J., Wong K.L., Young B.W., Lu L.W., Lee S.S. SARS-related virus predating SARS outbreak. Hong Kong. Emerg. Infect. Dis. 2004;10:176–178. doi: 10.3201/eid1002.030533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong N.S., Zheng B.J., Li Y.M., Poon, Xie Z.H., Chan K.H., Li P.H., Tan S.Y., Chang Q., Xie J.P., Liu X.Q., Xu J., Li D.X., Yuen K.Y., Peiris, Guan Y. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Lu Z., Xu T., Chen C., Yang G., Zha T., Lu J., Xue Y. Arbidol monotherapy is superior to lopinavir/ritonavir in treating COVID-19. J. Infect. 2020;81:e21–e23. doi: 10.1016/j.jinf.2020.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]