Abstract
Firre encodes a lncRNA involved in nuclear organization. Here, we show that Firre RNA expressed from the active X chromosome maintains histone H3K27me3 enrichment on the inactive X chromosome (Xi) in somatic cells. This trans-acting effect involves SUZ12, reflecting interactions between Firre RNA and components of the Polycomb repressive complexes. Without Firre RNA, H3K27me3 decreases on the Xi and the Xi-perinucleolar location is disrupted, possibly due to decreased CTCF binding on the Xi. We also observe widespread gene dysregulation, but not on the Xi. These effects are measurably rescued by ectopic expression of mouse or human Firre/FIRRE transgenes, supporting conserved trans-acting roles. We also find that the compact 3D structure of the Xi partly depends on the Firre locus and its RNA. In common lymphoid progenitors and T-cells Firre exerts a cis-acting effect on maintenance of H3K27me3 in a 26 Mb region around the locus, demonstrating cell type-specific trans- and cis-acting roles of this lncRNA.
Subject terms: Nuclear organization, Epigenetics, Chromatin structure, Long non-coding RNAs, Nuclear organization
Firre encodes a lncRNA involved in nuclear organization in mammals. Here, the authors find that allelic deletion of Firre on the active X chromosome (Xa) results in dose-dependent loss of histone H3K27me3 on the inactive X chromosome (Xi), along with other trans-acting effects, including disruption of the perinuclear location and minor dysregulation of gene expression.
Introduction
X chromosome inactivation (XCI) is initiated by the long noncoding RNA (lncRNA) Xist, which becomes highly expressed on one allele, and coats the future inactive X chromosome (Xi) in cis1–5. Specific proteins that include components of the Polycomb repressive complexes PRC1 and PRC2 are recruited by Xist RNA to mediate serial layers of epigenetic modifications, resulting in gene silencing and heterochromatin formation2,6,7. Epigenetic hallmarks of the Xi include multiple repressive histone modifications such as ubiquitination of histone H2A at lysine 119 (H2AK119ubi), tri-methylation of histone H3 at lysine 27 (H3K27me3), and enrichment in the histone variant macroH2A18. Additional layers of control ensure stability of the silent state of the Xi, including DNA methylation of promoter-containing CpG islands, a shift to late replication, and spatial reorganization of the Xi within the nucleus9,10.
The Xi appears as the heteropycnotic Barr body usually located close to either the nuclear lamina or the periphery of the nucleolus11–15. These two locations are preferred sites of heterochromatin, not only for the Xi but also for other repressed regions of the genome, suggesting that their proximity helps maintain silent chromatin11,16. In particular, the perinucleolar space has a primary function in replication and maintenance of repressive chromatin state17,18. The factors and mechanisms that facilitate association of heterochromatic regions including the Xi to specific nuclear compartments such as the lamina or the nucleolus remain elusive. Xist RNA interaction with the lamin B receptor (LBR) has been proposed as a critical factor that recruits the Xi to the lamina and facilitates silencing19. Our previous studies suggest that perinucleolar positioning of the Xi may be facilitated by the lncRNA Firre20.
The Firre locus comprises conserved tandem repeats that bind CTCF specifically on the Xi but not on the Xa (active X chromosome)20–22. Despite sequence divergence between species, the conserved nature of the repeat locus suggests important roles in mammals21. Firre RNA is usually confined to the nucleus where it interacts with the nuclear matrix protein hnRNPU23,24. Multiple transcript isoforms including circular RNAs, further complicate an understanding of the roles of Firre in different cell types25. On the Xi the Firre locus contacts the Dxz4 locus that also binds CTCF only on the Xi26–28. Dxz4 is necessary for the formation of the bipartite structure of the Xi27,29–31. The Firre locus also interacts with several autosomal regions, consistent with a widespread role in nuclear architecture23,32. A Firre knockout (KO) mouse model is viable, but results in cell-type-specific defects in hematopoiesis that impact common lymphoid progenitors (CLPs)32,33. Importantly, these defects are rescued by ectopic expression of Firre from an autosomal location, thus defining a trans-acting role for Firre32,33. KO mice show organ-specific dysregulation of autosomal genes, consistent with physiological defects in distinct phases of hematopoiesis32.
Here, we investigate the role of Firre in maintenance of heterochromatin, gene expression, and 3D structure of the Xi by engineering allele-specific deletions of the Firre locus and by Firre knockdown (KD) in mouse cell lines and tissues. Depletion of Firre RNA reveals important roles in H3K27me3 enrichment on the Xi and in location of the Xi within the nucleus as shown by immunostaining, ChIP-seq, and CUT&RUN. Gene expression is disrupted, as is the 3D structure of the Xi as shown by RNA-seq, ATAC-seq, and Hi-C. Our results are supported by rescue experiments using cDNA transgenes. We demonstrate both trans- and cis-acting roles of Firre RNA and its locus, with evidence of cell-type-specific effects in cell lines and in vivo.
Results
Firre and CrossFirre are transcribed from the Xa
Allele-specific CRISPR/Cas9 editing of the Firre region was done in Patski cells, in which skewed XCI and species-specific SNPs allowed us to design guides to target the Xi from BL6 or the Xa from Mus spretus (spretus) (Supplementary Data 1). We isolated single-cell clones with either a ~160 kb deletion of Firre on the Xa (ΔFirreXa), a ~160 kb deletion of Firre on the Xi (ΔFirreXi), or a ~160 kb inversion of Firre on the Xi (InvFirreXi) (Fig. 1a). Deletion of the Firre locus on the Xa resulted in undetectable Firre expression by RT-PCR, while deletion on the Xi caused no change (Fig. 1a, b, Supplementary Data 2). Allele-specific RNA-seq analysis confirmed the absence of Firre transcripts from either Xa or Xi in ΔFirreXa, while control loci (Dxz4, Xist) showed no change (Supplementary Data 3).
Fig. 1. Firre and CrossFirre are expressed from the Xa.
a Genomic location of Firre and CrossFirre (Gm35612) on the mouse X chromosome (UCSC mm10 build 38 browser tracks). Note that Firre has multiple alternative transcripts. The locations of the CRISPR guide RNAs used to edit the locus (cut), and of the RT-PCR primer pairs to specifically detect Firre expression (F/R), strand-specific expression of CrossFirre (F1/R1), or strand-specific expression in a region of overlap between Firre and CrossFirre (F2/R2) are indicated. b RT-PCR analysis using the F/R primer pair detects Firre expression in WT, ΔFirreXi, and InvFirreXi, but not in ΔFirreXa. Firre expression was measured in n = 3 biologically independent samples per cell type. c Sanger sequencing analyses of a CrossFirre region (F1/R1) and of a region of overlap between Firre and CrossFirre (F2/R2) confirm heterozygosity of SNPs (BL6 on the Xi and spretus on the Xa) in each region assayed. Genomic DNA (gDNA) shows heterozygosity at the SNPs, while cDNA only shows expression from the spretus SNP (Xa). d Strand-specific analysis of CrossFirre and Firre was done using reverse transcription using either random primers, or F1, R1, F2, R2 primers, followed by PCR using F1/R1 or F2/R2 primer pairs. Firre and CrossFirre expression was measured in n = 3 biologically independent samples per cell type. Source data are provided as a Source Data file.
Our ΔFirreXa deletion includes the antisense transcript CrossFirre that partially overlaps Firre (Fig. 1a; Supplementary Fig. 1a)34. Note that, in contrast to ΔFirreXa, previously constructed Firre deletions in mouse ES cells and in a KO mouse do not include all of CrossFirre32,35. To test CrossFirre expression in edited Patski cells, we processed strand-specific RT-PCR using primers (F1/R1) that flank a 231 bp region in the middle of CrossFirre with no Firre overlap, and primers (F2/R2) that flank a 203 bp region overlapping the 3’ end of Firre (Fig. 1a). For the non-overlapped region, forward CrossFirre transcripts containing only SNPs from the Xa (spretus), were present in WT (wild-type), ΔFirreXi, and InvFirreXi, but absent in ΔFirreXa (Fig. 1c, d). For the overlapped region, transcripts from both directions (CrossFirre and Firre), again containing only SNPs from the Xa (spretus), were present in WT, ΔFirreXi, and InvFirreXi, but absent in ΔFirreXa (Fig. 1d). We found no evidence of miRNAs in the Firre/CrossFirre region using miRNA-seq in WT or ΔFirreXa, suggesting that the loci function independently of the small RNA pathway (Supplementary Data 4).
We conclude that Firre and CrossFirre are both transcribed from the Xa in Patski cells. Note that Firre was originally identified as a gene that escapes XCI in human and mouse20,23,28. However, our current results and those reported in Firre KO mouse ES cells and in KO mice clearly show that Firre is predominantly expressed from the Xa in fibroblasts32,35.
Firre acts in trans to maintain PRC2 and H3K27me3 on the Xi
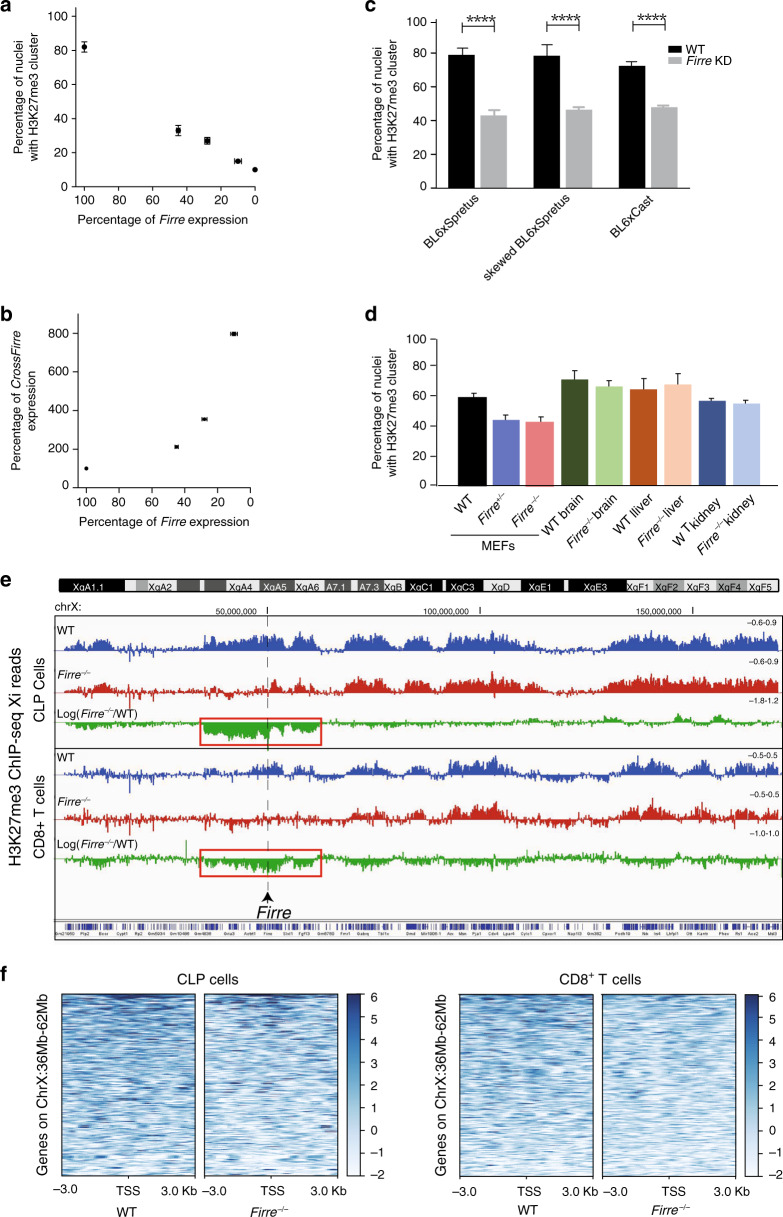
To determine whether any of the allelic alterations constructed, ΔFirreXa, ΔFirreXi, and InvFirreXi, influences epigenetic marks on the Xi, immunostainings for H3K27me3, H2AK119ubi, and macroH2A.1 were done in combination with Xist RNA-FISH to locate the Xi. The majority of nuclei (95 ± 3%) had one Xist cloud in all edited cell lines, indicating no disruption of Xist RNA coating (Fig. 2a). A strong H3K27me3 immunostaining cluster was observed on the Xi in 83 ± 2% of WT nuclei as expected. In contrast, only 9 ± 2% of nuclei with a H3K27me3 cluster were observed in ΔFirreXa, with most nuclei appearing uniformly mottled throughout (Fig. 2b; Table 1). There was no evidence of a complete loss of H3K27me3 over the Xi, which would have appeared as a “hole” with complete absence of immunostaining. H3K27me3 level throughout ΔFirreXa and WT nuclei was measured in regions outside the Xi using ImageJ to quantify fluorescence intensity by normalization to DNA or to histone panH436. No significant difference was detected in regions outside the Xi between ΔFirreXa and WT, but subtle or local changes undetectable by immunostaining cannot be excluded (Supplementary Fig. 2a, b). ΔFirreXi and InvFirreXi nuclei retained a strong H3K27me3 cluster on the Xi, consistent with retention of Firre RNA in these cells (Fig. 2b). Two other histone modifications known to be associated with XCI, H2AK119ubi and macroH2A.1, showed no changes in ΔFirreXa nuclei (Fig. 2c).
Fig. 2. Firre RNA acts in trans to maintain H3K27me3 on the Xi.
a–c A total of >300 Patski nuclei were scored per cell type over 3 independent experiments; significance was determined by one-sided Fisher exact test; bar plots are presented as mean values ± SEM; scale bars represent 10 µm. a Examples of nuclei after Xist RNA-FISH (green) and Hoechst 33342 staining (blue). The bar plot shows no significant differences among cell lines. b Examples of nuclei after H3K27me3 immunostaining (red) and Hoechst 33342 staining (blue). The bar plot shows significantly fewer nuclei with a H3K27me3 cluster in ΔFirreXa versus WT (p value = 4.63688e-94), but no change in ΔFirreXi nor InvFirreXi. c Examples of nuclei after macroH2A.1 or H2AK119ubi (red) immunostaining and Hoechst 33342 staining (blue). The bar plots show no significant differences between cell types. d Profiles of H3K27me3 ChIP-seq reads along the Xi in WT (blue), ΔFirreXa (red), and log2 ratio ΔFirreXa/WT (green). Box plots (log2) of H3K27me3 ChIP-seq reads in 100 bp Xi bins show a significantly lower median in ΔFirreXa (red) versus WT (blue) (Wilcoxon test: p value = 2.2e-16). The boxes demarcate the interquartile range (IQR) with median; whiskers ±1.5 times the IQR; outliers plotted as points. e Density histograms of the distribution of allelic proportions (Xa/(Xa+Xi)) of H3K27me3 peaks show a shift for the X chromosomes due to lower H3K27me3 on the Xi in ΔFirreXa (red) compared to WT (blue) (Wilcoxon test: −log10P = inf). f Heatmaps of H3K27me3 ChIP-seq reads 3 kb around transcription start sites (TSS) of genes on the Xi in ΔFirreXa versus WT. g. Metagene plots of average H3K27me3 occupancy at X-linked genes ((TSS to termination site (TES), not at scale)) in ΔFirreXa (Xi red Xa pink) versus WT (Xi blue, Xa purple). h Density histograms of the distribution of allelic proportions (Xa/(Xa+Xi)) of SUZ12 peaks show a shift for the X chromosomes due to lower SUZ12 on the Xi in ΔFirreXa (red) versus WT (blue) (Wilcoxon test: −log10P = 20.98). i Genome tracks demonstrating interactions between hnRNPK, EZH2, and SUZ12 with Firre RNA based on RIP-seq data in trophoblast and embryo stem cells39,40.
Table 1.
Percentages of nuclei with a H3K27me3 cluster on the Xi.
| Cell lines | Genotype | Percent nuclei |
|---|---|---|
| Patski cells derived from kidney from an F1 embryo (BL6 HprtBM3 × spretus) | WT | 83% |
| ∆FirreXa | 9% | |
| ∆FirreXa+mtransgene | 38% | |
| ∆FirreXa+htransgene | 21% | |
| ∆FirreXi | 77% | |
| InvFirreXi | 79% | |
| Firre shRNA KD | 33% | |
| Firre siRNA KD | 27% | |
| Firre si/shRNA KD | 15% | |
| Primary MEFs derived from an F1 embryo (BL6 × spretus) | Control | 79% |
| Firre KD | 43% | |
| Primary MEFs derived from an F1 embryo with skewed XCI (BL6 XistΔ × spretus) | Control | 80% |
| Firre KD | 45% | |
| Primary MEFs derived from an F1 embryo (BL6 × castaneus) | Control | 78% |
| Firre KD | 47% | |
| Primary MEFs derived from a Firre KO mouse model, with or without induction of Firre by doxycycline | Firre+/+ control | 58% |
| Firre+/− heterozygote | 43% | |
| Firre−/− homozygote | 41% | |
| Firre+/− tg;rtTA; -Dox | 52% | |
| Firre+/− tg;rtTA; +Dox | 91% | |
| Firre−/− tg;rtTA; -Dox | 49% | |
| Firre−/− tg;rtTA; +Dox | 83% | |
| Tissues derived from a Firre KO mouse model | Firre+/+ brain | 71% |
| Firre−/− brain | 70% | |
| Firre+/+ liver | 67% | |
| Firre−/− liver | 69% | |
| Firre+/+ kidney | 56% | |
| Firre−/− kidney | 58% |
The table lists each cell line, including its origin, its genotype, and the percentage of nuclei with a H3K27me3 cluster on the Xi.
Next, allele-specific profiles of H3K27me3 were generated by ChIP-seq, which demonstrated a chromosome-wide decrease on the Xi in ΔFirreXa (Fig. 2d). The loss of H3K27me3 in mutant cells was quantified by counting unique reads mapped to the Xa, Xi, and autosomes. In WT the Xi/Xa read ratio was 2.45, reflecting the characteristic H3K27me3 enrichment on the Xi. In contrast, this ratio was only 1.60 in ΔFirreXa, representing a significant decrease on the Xi (Fig. 2d). The BL6/spretus read ratios for autosomes in WT and ΔFirreXa remained similar, 1.04 and 1.14, respectively. The 34% percent decrease in the level of H3K27me3 on the Xi as measured by ChIP-seq is lower than that measured by immunostaining (89%), which is expected based on known differences between methodologies. While immunostaining captures condensation of the Xi and thus results in many nuclei with a visible H3K27me3 cluster, ChIP-seq reveals uneven distribution of H3K27me3 along the Xi and thus an apparently lesser enrichment37. In fact, the 34% change in H3K27me3 we measured by ChIP-seq in ΔFirreXa versus WT is similar to that reported between undifferentiated and differentiated mouse ES cells, in which the onset of XCI causes only a 40% increase in H3K27me3 level as measured by ChIP-seq, while over 90% of nuclei acquire a visible H3K27me3 cluster, as determined by immunostaining37.
Next, we calculated allelic proportions of SNP read coverage for each H3K27me3 peak covered by at least five SNP reads. In both WT and ΔFirreXa the distribution of allelic proportions (spretus/(spretus + BL6)) for the autosomes centered close to the anticipated 0.5, reflecting a similar enrichment between alleles (Fig. 2e). In contrast, the distribution of allelic proportions for the X chromosomes (Xa/(Xa + Xi)) centered at ~0.35 in WT, consistent with H3K27me3 enrichment on the Xi, but was markedly shifted to higher values (~0.5) in ΔFirreXa, supporting the 34% decrease in H3K27me3 on the Xi (Fig. 2e). Heatmaps and metagene plots of allelic ChIP-seq data further demonstrate a dramatic loss of H3K27me3 around the transcription start site and throughout the body of X-linked genes in mutant cells (Fig. 2f, g). LINE and SINE repeats also show lower H3K27me3 on the Xi in ΔFirreXa (Supplementary Fig. 2c). Finally, we confirmed a decrease in H3K27me3 in ΔFirreXa using CUT&RUN in a separate study38.
Next, we investigated SUZ12, a subunit of the PRC2 complex, using CUT&RUN, which showed a decrease on the Xi in ΔFirreXa. Again, the distribution of allelic proportions (Xa/(Xa+Xi)) for each SUZ12 peak showed a pronounced shift toward higher values (~0.75) for the X chromosomes in ΔFirreXa versus WT, while allelic proportions for the autosomes (0.5) did not change (Fig. 2h). Reanalysis of published datasets of RNA/protein interactions in mouse cells confirmed Firre RNA interactions with PRC components. Analysis of RIP-seq data showed Firre RNA interactions with two components of PRC2, EZH2 and SUZ12, and with hnRNPK, a protein that recruits the noncanonical PCGF3/5-PRC1 (Fig. 2i; Supplementary Data 5)39,40. Analysis of data obtained by CLIP-seq and PAR-CLIP, two UV-crosslinking-based methods for highly accurate mapping of RNA-protein interactions, confirmed Firre RNA interactions with EZH2 and detected interactions with JARID2 and RBFOX2, two proteins known to help recruit PRC2 to chromatin, and with CBX7, a component of the canonical PRC1 complex implicated in H3K27me3 deposition41–45 (Supplementary Data 5).
We conclude that Firre RNA transcribed from the Xa specifically helps target PRC1 and PRC2 complexes to the Xi for maintenance of H3K27me3, consistent with a trans-acting effect in Patski cells. In support we find that several proteins implicated in the PRC complexes are Firre RNA interactors.
Dose-dependent and cell-type-specific effects of Firre
To quantify the effects of Firre, we knocked it down in a dose-dependent manner using shRNA and siRNA treatments of an independent Patski isolate to achieve KD levels ranging from 43, 28, and 10% of WT. This caused a parallel reduction in percentages of nuclei with a H3K27me3 cluster to 33%, 27%, and 15%, respectively, versus 83% in WT (p < 0.0001) (Fig. 3a; Table 1). Importantly, this reveals a dose-dependent effect of Firre RNA on H3K27me3 enrichment on the Xi (Fig. 3a). Concomitantly, a dose-dependent increase in CrossFirre RNA inversely correlates to the amount of Firre RNA, consistent with Firre repressing the antisense transcript (Fig. 3b).
Fig. 3. Dose-dependent effects of Firre RNA on H3K27me3 on the Xi in cell lines and in vivo.
a Percentage of nuclei with a H3K27me3 cluster after KD in Patski cells relative to Firre expression level measured by qRT-PCR in n = 3 biologically independent samples. Data presented as mean values ± SEM. b The level of CrossFirre expression is inversely related to that of Firre after Firre KD. Expression measured by qRT-PCR in n = 3 biologically independent samples. Data presented as mean values ± SEM. c, d A total of >300 Patski nuclei were scored per cell type over three independent experiments; significance was determined by one-sided Fisher exact test; bar plots are presented as mean values ± SEM; scale bars represent 10 µm. c Bar plots show significantly fewer H3K27me3 clusters after Firre KD in nuclei from three primary MEFs compared to mock treatment (p value = 2.89075e-12, 1.41519e-32, and 8.95838e-10, respectively). Primary MEFs were derived from a (BL6 × spretus) embryo with a spretus Xi, and from embryos either (BL6 × spretus) or (BL6 × castaneus) with random XCI (Table1). d Bar plots show a lower (but not significantly) percentage of H3K27me3 cluster in nuclei from MEFs derived from Firre+/− and Firre−/− KO embryos than in controls (p value = 0.0774 and 0.0567, respectively). No significant differences were found in brain, liver, and kidney from Firre KO mice compared to WT (p value = 0.9127, 0.8796, and 0.762, respectively). e Profiles of H3K27me3 ChIP-seq reads along the X chromosomes from CLPs and CD8 + T cells from WT (blue), Firre−/− (red), and log2 ratio Firre−/−/WT (green) show a significant decrease covering ~26 Mb around the Firre locus in mutant CLPs, and to a lesser extent CD8 + T cells (Supplementary Data 6). f Heatmaps of H3K27me3 ChIP-seq reads located 3 kb around the transcription start sites (TSS) of X-linked genes that map within the 26 Mb region around Firre in WT and Firre−/− CLPs and CD8 + T cells.
A separate KD to deplete Firre RNA to 33% of WT in primary mouse embryonic fibroblasts (MEFs) independently derived from a BL6 × spretus F1 embryo with random XCI significantly decreased the percentage of nuclei with a H3K27me3 cluster to 43%, versus 79% in controls (p < 10−4) (Fig. 3c; Table 1). To exclude species-specific effects, we tested MEFs from a BL6 × spretus F1 embryo with an XCI pattern opposite to that in Patski cells (i.e., the Xi is from spretus), and MEFs from a BL6 × castaneus F1 embryo. After KD in these lines Firre RNA level was lowered to 21 and 15% of WT, causing a significant reduction to 45 and 47% of nuclei with a H3K27me3 cluster, compared to 80 and 78% in controls, respectively (p < 10−4) (Fig. 3c; Supplementary Fig. 2d; Table 1). No significant change in CrossFirre was found in those lines (Supplementary Fig. 2e). We then examined cells and tissues from a Firre KO mouse model32,33. MEFs from heterozygous (Firre+/−) and homozygous (Firre−/−) KO female embryos showed a modest reduction to 43 and 41% of nuclei with a H3K27me3 cluster, respectively, compared to 58% in a control littermate (Firre+/+) (Fig. 3d; Table 1). H3K27me3 immunostaining in liver, kidney, and brain sections derived from Firre−/− female mice showed no significant decrease in nuclei with a cluster (Fig. 3d; Table 1).
Next, we performed H3K27me3 ChIP-seq analysis of sorted CLPs and CD8 + T cells derived from Firre−/− KO female mice. Surprisingly, we found a striking loss of H3K27me3 in a ~26 Mb (ChrX: 36–62 Mb) region centered around the Firre locus, together with a slight decrease across the entire X chromosome, but not the autosomes (Fig. 3e, Supplementary Fig. 3a, b). Heatmaps of control and Firre−/− KO CLPs and CD8 + T cells showed a loss of H3K27me3 around the transcription start site of genes located within this 26 Mb region (Fig. 3f, Supplementary Data 6). Although we assume that this local loss of H3K27me3 affects the Xi and not the Xa, allelic analysis was not possible since KO mice are homozygous BL6. The 26 Mb region around Firre contains 266 curated genes, 15 involved in immunity (e.g., Nkap, Sash3, Atp11c, Sh2d1a, Elf446–48), and 15 potentially exhibiting haploinsufficiency effects in human (e.g., GPC349) (Supplementary Data 6). However, no specific expression changes have been reported for any of these genes in CLPs from KO mice33.
In summary, we found a dose-dependent effect of Firre RNA in maintenance of H3K27me3 on the Xi in several types of fibroblasts. Lack of detectable changes in three tissues from Firre KO mice suggests that the role of Firre may be cell-type- and tissue-specific, as proposed for other lncRNAs50. Consistent with the role of Firre in hematopoiesis, local changes were observed in CLPs and CD8 + T cells around the Firre locus, revealing a cell-type-specific role in cis-maintenance of H3K27me333.
Rescue of H3K27me3 on the Xi by Firre/FIRRE cDNA transgenes
To confirm a causative role of Firre RNA on H3K27me3 enrichment on the Xi, ΔFirreXa cells were transfected with a mouse Firre cDNA transgene that lacks the first five 5’ exons of Firre and is expressed from a CMV promoter (Supplementary Fig. 1b). By RNA-seq Firre expression was restored to a near-normal level after transfection. Ectopic expression of the mouse cDNA in ΔFirreXa+mtransgene cells rescued the presence of a H3K27me3 cluster from 9 to 38% of nuclei, supporting a trans-acting role (Fig. 4a, b, Table 1). This partial rescue is not readily explained by heterogeneous transgene levels among cells, since a cloned transgenic line with stable high Firre expression rescued to a similar level (34%). Firre RNA-FISH in this clone showed association of the lncRNA to the Xist cloud in 15% of cells, consistent with partial rescue (Fig. 4c). Incomplete rescue is likely due to the partial cDNA transgene that may lack functional RNA motifs, isoforms, and regulatory elements (Supplementary Fig. 1b). Interestingly, we also observed partial rescue after ectopic expression of a human FIRRE cDNA in ΔFirreXa+htransgene cells (Fig. 4a, b; Table 1). Next, we examined Firre+/− and Firre−/− MEFs from KO mice with an ectopic doxycycline (DOX)-inducible plasmid copy of a complete Firre cDNA integrated in the genome. After Firre expression induction (10–20 fold) a strong increase to 91 and 83% of nuclei with a H3K27me3 cluster was found compared to 43 and 41% in noninduced KO cells, respectively (Fig. 4d; Table 1).
Fig. 4. Ectopic expression of Firre/FIRRE RNA partially restore H3K27me3 on the Xi.
a Examples of nuclei after H3K27me3 immunostaining (red) and Hoechst 33342 staining (blue) in WT, ΔFirreXa, and ΔFirreXa transfected with a mouse Firre transgene (ΔFirreXa+mtransgene) or a human FIRRE transgene (ΔFirreXa+htransgene). b The bar plot shows a significantly higher percentage of nuclei with a H3K27me3 cluster in cells with a mouse or human Firre/FIRRE transgene, compared to ΔFirreXa (p value = 2.8075e-16 and 0.0000134599, respectively). c Examples of nuclei after RNA-FISH for Xist (green) and Firre (red) in a ΔFirreXa+mtransgene cell clone with high expression of the mouse transgene. The upper two nuclei show association between Firre and Xist signals (seen in 15% of nuclei), and the lower nuclei, lack of association. d The bar plot shows the percentage of nuclei with a H3K27me3 cluster in MEFs derived from Firre+/− and Firre−/− females that harbor a doxycycline (Dox) inducible transgene (Firre+/− tg;rtTA; -Dox; Firre+/− tg;rtTA; +Dox; Firre−/− tg;rtTA; -Dox; Firre−/− tg;rtTA; +Dox), compared to WT and mutants. The percentage of nuclei with a H3K27me3 cluster increases significantly in Firre+/− tg;rtTA and Firre−/− tg;rtTA MEFs after addition of doxycycline (Dox+) (p value=4.31567e-30 and 3.51193e-20, respectively). A total of >300 nuclei scored for the presence of a H3K27me3 cluster (a, b, d), or for Xist and Firre RNA-FISH signals (c) per cell type over three independent experiments; significance was determined by one-sided Fisher exact test; bar plots are presented as mean values ± SEM; scale bars represent 10 µm.
We conclude that loss of H3K27me3 on the Xi in mutant cells is rescuable by a cDNA transgene, supporting Firre trans-acting role. Surprisingly, a human FIRRE transgene can also rescue to some extent, suggesting functional compatibility between species despite sequence divergence. Results of induction of a transgene in the mouse KO model further support a dose-dependent trans-effect of Firre RNA on H3K27me3 enrichment on the Xi.
Firre acts in trans to maintain nuclear location of the Xi
We next examined the effects of allelic Firre mutations on the Xi location relative to the nucleolus and lamina. In WT, ΔFirreXi, and InvFirreXi H3K27me3 and nucleophosmin (NPM1) immunostaining was used to locate the Xi and the nucleoli, respectively (Fig. 5a). Since the H3K27me3 cluster is compromised in ΔFirreXa, Xist RNA-FISH was applied in combination with NPM1 immunostaining. The Xi location was scored in WT nuclei as adjacent to the nuclear periphery (70%), the nucleolus surface (50%), or neither (8%) (Fig. 5b). Note that in 28% of nuclei the Xi was close to both the periphery and the nucleolus. Loss of Firre RNA in ΔFirreXa caused significant reductions in Xi-nuclear periphery and Xi-nucleolus associations to 20 and 22% of nuclei, respectively (p < 0.0001) (Fig. 5b). Importantly, ectopic expression of a cDNA transgene in ΔFirreXa+mtransgene significantly (but not completely) rescued these associations (Fig. 5b). Deletion or inversion of Firre on the Xi did not alter its location (Fig. 5a, b). In primary MEFs independently derived from a BL6 × spretus F1 mouse with random XCI Firre KD also caused a decrease in Xi-nuclear periphery and Xi-nucleolus associations (Fig. 5c).
Fig. 5. Firre RNA acts in trans to maintain Xi location.
a–c A total of >300 nuclei per cell type over 3 independent experiments were scored for the location of the Xi marked by a H3K27me3 cluster or an Xist cloud relative to the nuclear periphery or the nucleolus; significance was determined by one-sided Fisher exact test; bar plots are presented as mean values ± SEM; scale bars represent 10 µm. a Examples of nuclei after H3K27me3 immunostaining (red) to locate the Xi in WT, ΔFirreXi, and InvFirreXi, or Xist RNA-FISH (red) to locate the Xi in ΔFirreXa since there is no H3K27me3 cluster in these nuclei. Nuclei were also immunostained for NPM1 (green) to locate the nucleolus. b Bar plots show a significant decrease in periphery- and nucleolus association of the Xi in ΔFirreXa compared to WT (p value = 3.95632e-40 and 3.75497e-14, respectively), but no significant changes in ΔFirreXi and InvFirreXi (p value = 0.6968). Ectopic expression of a mouse transgene in ΔFirreXa+mtransgene partly rescues the Xi location to 41% at the periphery and 34% at the nucleolus (p value = 6.62701e-10 and 0.000265481, respectively). c Bar plots show a significant decrease in periphery- and nucleolus association of the Xi after Firre KD in primary MEFs derived from an F1 embryo (BL6 × spretus) (p value = 1.13757e-09 and 0.0000634923, respectively). d Density histograms of the distribution of allelic proportions (Xa/(Xa + Xi)) of CTCF peaks show a shift in the distribution for the X chromosomes due to a decrease in CTCF on the Xi in ΔFirreXa (red) compared to WT (blue). In ΔFirreXa+mtransgene (brown) this distribution becomes binomial due to partial restoration of CTCF on the Xi. e Plots of Xi-associated (common +Xi-specific) CTCF peak density (counts binned within 100 kb windows) along the Xi for WT (blue) and ΔFirreXa (red). To account for differences in the number of SNP-covered peaks due to sequencing depth, the binned counts are scaled by a factor obtained from the between-sample ratios of autosomal diploid SNP-covered peaks.
CTCF has been implicated in nucleolus association of genomic regions, and we have previously shown that Ctcf KD reduces Xi-nucleolus associations in Patski cells20,51. Here, we profiled allelic CTCF binding by CUT&RUN, which demonstrated a loss of CTCF binding on the Xi in ΔFirreXa (Fig. 5d). The distribution of allelic proportions (Xa/(Xa + Xi)) for CTCF peaks on the X chromosomes showed a pronounced shift toward higher values (~0.85) in ΔFirreXa compared to WT (~0.65), while allelic proportions (spretus/(spretus + BL6)) for the autosomes were close to the anticipated 0.5 (Fig. 5d). Thus, while there is less CTCF binding on the Xi versus Xa in WT as expected, CTCF binding is even lower in ΔFirreXa (Fig. 5e). In ΔFirreXa+mtransgene the distribution of allelic proportions for the X chromosomes becomes binomial, indicating partial restoration of CTCF on the Xi (Fig. 5d).
Taken together, our results show that Firre RNA transcribed from the Xa or from an ectopic cDNA transgene can influence in trans the Xi location within the nucleus. Our results further suggest a potential cooperation between Firre RNA and CTCF in maintenance of Xi location.
Loss of Firre affects gene expression in Patski cells
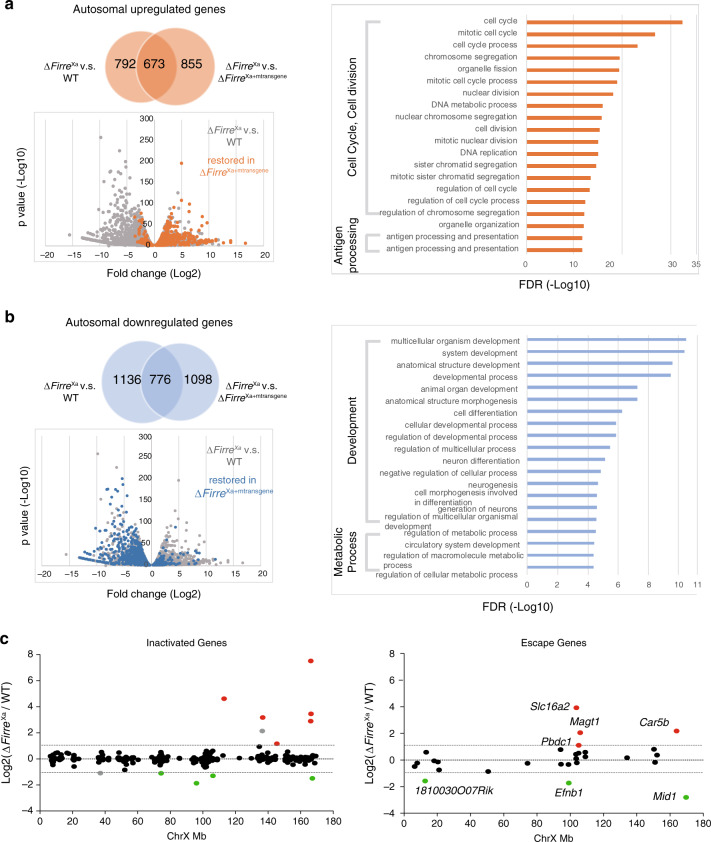
Next, we examined changes in total gene expression (autosomal and X-linked genes without allele discrimination) in ΔFirreXa. About 11 and 14% of genes with expression ≥1TPM in at least one condition were upregulated and downregulated in ΔFirreXa versus WT, respectively, (Supplementary Data 7 and 8). A large proportion (46 and 40%, respectively) of dysregulated genes were rescued in ΔFirreXa+mtransgene (Fig. 6a, b, Supplementary Fig. 4a, Supplementary Data 7). To rule out effects of aneuploidy between cell lines results were confirmed for genes located on diploid chromosomes in WT and ΔFirreXa (Supplementary Fig. 4b, Supplementary Data 9). GO analysis show that the top 20 GO terms for genes upregulated in ΔFirreXa and rescued in ΔFirreXa+mtransgene are related to cell cycle, DNA replication, chromosome segregation, and immune cell function, while downregulated genes are implicated in development, differentiation, and metabolism (Fig. 6a, b; Supplementary Data 10). In human HEK293 cells FIRRE RNA KD resulted in dose-dependent cell death, consistent with a role in cell growth and survival (Supplementary Fig. 4c).
Fig. 6. Loss of Firre RNA affects gene expression.
a Upregulated genes in ΔFirreXa and Gene Ontology (GO) term enrichment. The Venn diagram shows the number of upregulated genes in ΔFirreXa versus WT, and in ΔFirreXa versus ΔFirreXa+mtransgene, with the overlapping gene set representing upregulated genes in ΔFirreXa that are rescued by transgene expression. The scatter plot shows dysregulated genes in ΔFirreXa versus WT (grey), with genes rescued by reduced expression in ΔFirreXa+mtransgene versus ΔFirreXa (more than 2-fold; p value < 0.05 by the Wald test) highlighted in orange. The top 20 GO terms of overlapping upregulated genes in ΔFirreXa versus WT, which are rescued in ΔFirreXa+mtransgene are listed. The X-axis indicates the FDR (−log10). b Downregulated genes in ΔFirreXa and Gene Ontology (GO) term enrichment. The Venn diagram shows the number of downregulated genes in ΔFirreXa versus WT, and in ΔFirreXa versus ΔFirreXa+mtransgene, the overlapping gene set representing downregulated genes in ΔFirreXa that are rescued by transgene expression. The scatter plot shows dysregulated genes in ΔFirreXa versus WT (gray), with genes rescued by increased expression in ΔFirreXa+mtransgene versus ΔFirreXa (more than 2-fold; p value <0.05 by the Wald test) highlighted in blue. The top 20 GO terms of overlapping downregulated genes in ΔFirreXa versus WT, which are rescued in ΔFirreXa+mtransgene are listed. The X-axis indicates the FDR (−log10). c Xi-expression fold changes for genes that are subject to or escape XCI between ΔFirreXa and WT. Upregulated genes are in red and downregulated genes in green. To note, one upregulated gene and one gene subject to XCI are in gray, as they showed more than 2-fold change in expression but with a p value >0.05 by the Wald test in ΔFirreXa versus WT. Genes are ordered from centromere to telomere along the Xi.
To determine whether gene expression was disrupted on the Xi upon loss of Firre RNA we evaluated allelic gene expression. Only 6/352 genes known to be subject to XCI showed >2-fold upregulation in ΔFirreXa, suggesting minor Xi reactivation (Fig. 6c, Supplementary Data 3 and 7). Four of the reactivated genes were located at the telomeric end of the Xi where higher accessibility and decreased contact density were observed by ATAC-seq and Hi-C (see below). Although their number is small and we cannot rule out a chance occurrence, more genes known to escape XCI were dysregulated (10–14%) from the Xi than genes subject to XCI (2–3%) (Fig. 6c, Supplementary Fig. 4d, Supplementary Data 7). A majority of dysregulated X-linked genes (67%) were rescued in ΔFirreXa+mtransgene (Supplementary Fig. 4e, Supplementary Data 3 and 7).
Metagene plots of H3K27me3 along autosomal and X-linked genes grouped based on their expression changes in ΔFirreXa, together with profiles of individual genes showed a small increase in H3K27me3 for downregulated genes, and very little or no change for upregulated or unchanged genes (Supplementary Fig. 5a, b). Note that 90% of dysregulated genes have no H3K27me3 enrichment in either WT or ΔFirreXa. Additional CUT&RUN analyses of two active epigenetic marks, H3K36me3 and H3K4me3, showed no significant changes on the Xi in ΔFirreXa, consistent with little Xi reactivation (Supplementary Fig. 5c, d)
Thus, the loss of Firre RNA causes widespread changes in gene expression in large part rescued by a cDNA transgene. Very little reactivation and few changes in escape genes occur on the Xi despite the observed loss of H3K27me3, consistent with this epigenetic mark representing only one layer of XCI control20.
Allelic alterations of the Firre locus change Xi structure
We evaluated chromatin accessibility in Firre mutants by ATAC-seq. As expected, in WT the distribution of allelic proportions centered at 0.5 for autosomes, but was skewed towards the Xa (0.95) for the X chromosomes, indicative of lower chromatin accessibility on the Xi. This pattern remained similar in ΔFirreXa, consistent with near absence of gene reactivation. Plots of ATAC peak density further captured the low accessibility profiles of the Xi in both WT and ΔFirreXa (Fig. 7a, b). However, a higher peak density was observed in the telomeric region of the Xi in ΔFirreXa where reactivated genes are located (Fig. 6c; 7c). ATAC-seq patterns were unchanged in ΔFirreXi and InvFirreXi (Supplementary Fig. 6a, b). To determine whether Dxz4 and Firre may have synergistic cis-effects on chromatin accessibility on the Xi, ATAC-seq was done on a double-mutant line ΔFirreXi/ΔDxz4Xi. Interestingly, a pronounced shift to lower values in the distribution of allelic proportions for the X chromosomes (peak at ~0.55) was observed in this double-mutant compared to either ΔDxz4Xi (peak at ~0.85) or ΔFirreXi (peak at ~1), indicating increased chromatin accessibility on the Xi (Fig. 7d, e).
Fig. 7. Chromatin accessibility after allelic Firre deletions and a Firre/Dxz4 deletion.
a Density histograms of the distribution of allelic proportions (spretus/(spretus + BL6)) of ATAC peaks along the autosomes and the X chromosomes for WT (blue) and ΔFirreXa (red). No shift is observed (Wilcoxon test: −log10P = 32). b. Percentages of ATAC peaks along the autosomes and the X chromosomes classified as spretus-specific, BL6-specific, or both show no differences between WT (blue) and ΔFirreXa (red). c. Plots of Xi-associated (common +Xi-specific) ATAC peak density (counts binned within 500 kb windows) along the Xi show increased accessibility at the telomeric end of the Xi in ΔFirreXa (red) versus WT (blue). To account for differences in the number of SNP-covered peaks between samples due to sequencing depth, the binned counts are scaled by a factor obtained from the between-sample ratios of autosomal diploid SNP-covered peaks. d Density histograms of the distribution of allelic proportions (spretus/(spretus + BL6)) of ATAC peaks show a shift to a lower Xa/(Xa + Xi) ratio in the double-mutant ΔFirreXi/ΔDxz4Xi (purple), compared to ΔDxz4Xi (black) and ΔFirreXi (green), consistent with increased accessibility on the Xi (Wilcoxon test: −log10P = 35). e Percentages of ATAC peaks in ΔDxz4Xi (black), ΔFirreXi (green), and ΔFirreXi/ΔDxz4Xi (purple) along the autosomes and the X chromosomes classified as spretus-specific, BL6-specific, or both show an increase on the BL6 Xi in the double mutant.
Based on high-resolution allele-specific contact maps generated by DNase Hi-C the Xi bipartite structure was retained in ΔFirreXi, but contacts increased between the superdomains and decreased within each superdomain, suggesting that the Firre locus acts in cis to help shape the 3D structure (Fig. 8a). Contacts within regions flanked by Dxz4 and Xist (ChrX:75–100 Mb) and to a lesser extent, flanked by Firre and Dxz4 (ChrX:50–75 Mb) were increased, suggesting disruption of contacts emanating from Dxz4 (Fig. 8a, Supplementary Fig. 7a). Similarly, InvFirreXi nuclei showed persistence of the Xi bipartite structure, but also a redistribution of contacts around Firre, including loss of proximal contacts (ChrX:5–50 Mb) and gain of distal contacts (ChrX:50–75 Mb), which was confirmed by virtual 4 C (Fig. 8a, b; Supplementary Fig. 7a). The boundary at or near the Firre locus on the WT Xi was maintained upon deletion or inversion of the locus (Supplementary Fig. 7b). While the loss of Firre RNA in ΔFirreXa did not perturb the bipartite structure of the Xi, there were some changes in contact distribution, suggesting trans-effects (Fig. 8c, Supplementary Fig. 7c). Specifically, contacts increased in the region flanked by Dxz4 and Xist (ChrX:75–100 Mb), and diminished in the very distal telomeric region (ChrX:165–170 Mb), consistent with an increase in chromatin accessibility and gene expression (Figs. 6c, 7c, 8c). Deletion of Firre on the Xa also resulted in contact changes on the Xa, including loss of the boundary at or close to the locus, as confirmed by insulation score analysis (Fig. 8d; Supplementary Fig. 7d, e).
Fig. 8. Changes in the X 3D structure after Firre deletion or inversion.
a Pearson correlated-transformed differential contact maps of the Xi at 500 kb resolution highlight differences between ΔFirreXi and WT, and between InvFirreXi and WT. The color scale shows differential Pearson correlation values, with loss and gain of contacts in the mutants versus WT appearing blue and red, respectively. See text for description of changes. b Virtual 4 C plots derived from Hi-C data at 500 kb resolution using Firre as the viewpoint on the Xi in WT (blue), ΔFirreXi (green) and InvFirreXi (gray) show an increase in contacts between Firre and Dxz4 in InvFirreXi. c Pearson correlated-transformed differential contact maps of the Xa and Xi at 500 kb resolution to highlight differences between ΔFirreXa and WT. The color scale shows differential Pearson correlation values, with loss and gain of contacts in ΔFirreXa versus WT appearing blue and red, respectively. See text for description of changes. d Pearson correlated-transformed contact maps (40 kb resolution) of the Xa for 4 Mb around the Firre locus highlight the loss of the strong boundary between TADs on the Xa in ΔFirreXa versus WT (see Supplementary Fig. 7c for corresponding maps of the Xi).
Taken together, our results indicate cooperation between Firre and Dxz4 in repression of chromatin accessibility on the Xi, with each locus contributing to the two superdomains separation. Firre contacts with other regions on the Xi appear orientation-dependent, reminiscent to the orientation-dependent contacts made by Dxz429. Firre RNA exerts trans-effects on the Xi 3D structure, potentially secondary to losses of H3K27me3 and CTCF binding.
Discussion
Studies of lncRNAs support the notion that these molecules can either spread in cis from their genomic locus or localize to cellular compartments away from their own locus of transcription to perform essential functions in regulating gene expression52–55. Here, we report that the lncRNA Firre transcribed from the Xa acts in trans and in cis on the Xi to maintain its epigenetic features, nuclear location, and 3D structure. LncRNAs have important roles in the structure of nuclei where they fold into higher-order structures and act in cooperation with proteins including chromatin-modifying complexes56. Xist represents the quintessential example of a lncRNA that spreads along the Xi in cis to recruit a series of proteins including components of the PRC complexes that implement chromatin modifications such as H3K27me32,3,6,19,56. We find that maintenance of H3K27me3 on the Xi mediated by Firre RNA involve the PRC complexes, which is supported by our reanalyses of RNA/protein interaction datasets39–45. Among Firre interactors EZH2 and SUZ12 represent core subunits of PRC2, while JARID2 and RBFOX2 are cofactors that directly interact with RNA57–59. PRC2 recruitment of the noncanonical PCGF3/5-PRC1 complex is facilitated by the Firre interactor hnRNPK60, while CBX7 is a subunit of the canonical PRC1 complex61. A new method to detect protein-RNA interactions (incPRINT) also identifies JARID2, EPC1, CTCF, and hnRNPU as Firre interactors62. Most Firre interactors have previously been implicated in XCI, and more specifically in H3K27me3 enrichment on the Xi (Supplementary Data 5)2,6,63–66. Colocalization of Firre RNA to the Xi was seen in only 15% of nuclei, suggesting dynamic binding of the lncRNA. Interestingly, single particle tracking of endogenous EZH2 and SUZ12 in human cells reveals rapid diffusion of PRC2 through the nucleus, with only ~20% chromatin-bound67.
Trans-effects have been reported for several lncRNAs. For example, Fendrr and Pint recruit PRC2 for H3K27 tri-methylation of loci located on other chromosomes68,69. Meg3 also recruits PRC2 components, JARID2 and EZH2, to facilitate H3K27me3 deposition and repression of genes in trans42,70. Meg3 has an additional cis-acting role by sequestration of PRC2 to prevent DNA methylation-induced repression of genes within the Meg3-Mirg imprinting cluster71,72. Interestingly, we observe a local loss of H3K27me3 centering around the Firre locus and extending to 26 Mb in CLPs and CD8 + T cells from Firre KO female mice, suggesting that Firre may also act in cis to maintain H3K27me3 in certain cell types. Importantly, defects in hematopoiesis that impact CLPs have been observed in Firre KO mice32,33. Local cis-effects have also been demonstrated for the imprinted lncRNAs Airn and Kcnq1ot1, which recruit the PRCs via hnRNPK to silence Mb-sized regions on one allele in cis39. Our results based on KD and transgenic rescue reveal dose-dependent effects of Firre RNA on H3K27me3 enrichment on the Xi, reminiscent of the dose-dependent silencing effects of Airn and Kcnq1ot139.
The nucleolus has emerged as a platform for the organization of chromatin enriched in repressive histone modifications16–18,73. For example, loss of NPM1 results in deformed nucleoli and redistribution of H3K27me374. Depletion of Firre RNA limits association of the Xi to the nucleolus and nuclear periphery, supporting coordinated roles in Xi positioning and maintenance of H3K27me320. Two studies corroborate our findings: indeed, relocation of the Xi to the nucleolus during the cell cycle is required to maintain H3K27me3, and sparser H3K27me3 on the Xi occurs when it is kept away from the nucleolus or nuclear periphery75,76. Deletion of Xist also decreases both H3K27me3 enrichment and nucleolar association of the Xi77. However, we did not observe disruption of Xist RNA expression nor coating of the Xi in ΔFirreXa nuclei, suggesting that Firre acts independently of Xist. On the other hand, CTCF, a protein that facilitates interactions between chromatin and the nucleolus may act in concert with Firre to help Xi positioning51. In addition to CTCF-DNA interactions CTCF-RNA interactions via specific zinc fingers represent important structural components of genome organization78. Such dual interactions characterize Firre since CTCF not only binds to the locus on the Xi, but also interacts with the RNA20,21,62. Thus, in the absence of Firre RNA disrupted Xi anchoring to the nucleolar periphery could result from the significant loss of CTCF binding observed along the entire Xi, including at the Firre locus. Supporting this notion, RNA depletion using RNase A disrupts the local chromatin environment around CTCF binding sites and the structural integrity of heterochromatin38. We cannot exclude other heterochromatin factors, for example, EZH1 or histone H1, both downregulated in ΔFirreXa and rescued in transgenic lines79,80. Considering the 3D structure of the Xi, the Firre locus helps insulate the two superdomains of the Xi perhaps via CTCF binding and acts in synergy with Dxz4 to compact the Xi perhaps via a superloop with Dxz427.
Our rescue experiments imply that a subset of Firre exons are sufficient for partial maintenance of H3K27me3 on the Xi and its location in the nucleus. It will be interesting to map additional functional units within the Firre locus, which may provide a more complete rescue. The Firre locus harbors several conserved local repeats and many of these do not overlap exons, except for the RRD repeats that display 65% identity between human and mouse, which makes them good candidates for the rescue function observed21,23. Notably, these repeats bind the nuclear matrix protein hnRNPU known to associate with Xist, the Xi, and other genomic regions20,21,24,81–84.
Our findings of dysregulated genes implicated in cell cycle and development upon loss of Firre RNA support a role in cell growth, a common finding for other lncRNAs85. Both Firre KO mice and mice with Firre overexpression have abnormalities specifically in hematopoiesis, implying the importance of Firre dosage32,33. In human loss of FIRRE RNA causes dysregulation of inflammatory gene expression, while amplification of FIRRE causes congenital abnormalities and is associated with decreased survival rates in cancer, further supporting FIRRE dosage effects24,85,86.
Despite ample evidence supporting their biological relevance in cell systems lncRNAs are often dispensable for survival of vertebrate organisms. For example, mice homozygous for deletions of regions harboring 1243 noncoding sequences had no distinguishable phenotypes87. Similarly, mice with deletions of nine conserved lncRNAs including Malat were viable with no obvious abnormalities50,88. Firre KO mice have few abnormal phenotypes except for abnormalities in B- and T-cell physiology33. This dearth of phenotypes could be explained by alternative pathways, possibly other lncRNAs that may compensate during development, providing redundancy in the system89. A recent publication reports that following 10 days of differentiation, mouse ES cells with a Firre deletion on the Xa, the Xi or on both alleles show no effects on H3K27me3 on the Xi, which differs from our results based on mutations induced in differentiated fibroblasts35. Thus, phenotypic effects apparently depend on the developmental timing of Firre mutations, and mutations at early stages may allow compensatory pathways to effectively help survival. Indeed, Firre KO ES cells display a marked decrease in growth rate, supporting the existence of critical mechanisms for survival of subsets of cells and subsequent differentiation/development23. In addition, differences in the effects of Firre mutations may reflect developmental stage-specific composition of PRC complexes90.
Another important consideration is cell type specificity. There was no apparent loss of H3K27me3 on the Xi in brain, kidney, and liver from Firre KO mice. However, a local disruption of H3K27me3 around Firre was found in CLPs and CD8 + T cells, suggesting that Firre RNA may act in cis in certain cell types. In a CRISPRi phenotypic screen a majority of lncRNAs only displayed phenotypes in a single cell type50. A new approach, PIRCh-seq, designed to find association between RNA and specific chromatin modifications reports Firre interactions with H3K27me3 chromatin in MEFs, but not in ES cells, highlighting differences between cell types91. Furthermore, mouse and human Firre/FIRRE exhibit diverse expression patterns across tissues, e.g., enrichment in neural crest but depletion in lung fibroblasts23. Firre KO mice show organ-dependent gene dysregulation, with a larger number of dysregulated genes in spleen, which can be rescued by ectopic Firre expression supporting a trans-acting role in vivo32,33. The gene expression changes we observed in kidney-derived Patski cells differ from those reported in Firre KO mouse spleen, again suggesting tissue-specificity. In immune cells where Firre RNA exerts a local cis-effect on the Xi, it remains to be determined whether Firre might be expressed from the Xi. Interestingly, female lymphocytes lack Xist clouds and H3K27me3 foci, and show reactivation of the Xi92. Further analyses to distinguish alleles is needed to better understand Firre local effects in immune cells.
Methods
Cell lines
The Patski cell line, originally derived from embryonic kidney (18.5 dpc) from a cross between a female C57BL/6 J (BL6) with an HprtBM3 mutation and a male Mus spretus (spretus), was previously selected in HAT (hypoxanthine-aminopterin-thymidine) medium so that the Xi is always from BL693,94. Primary MEF cultures were derived from a 13.5 dpc female embryo resulting from a BL6 × Mus spretus cross, which results in skewed inactivation of the spretus X chromosome due to an Xist mutation on the BL6 X chromosome95. Primary MEFs were also derived from BL6 × Mus spretus and BL6 × Mus castaneus 13.5 dpc female F1 embryos with random XCI. Cells were cultured as described and the presence of normal X chromosomes verified by karyotyping29.
For ectopic Firre expression assays in Patski cells and in MEFs, a mouse Firre cDNA plasmid (mtransgene; Dharmacon BC055934) or a human FIRRE cDNA plasmid (htransgene; Dharmacon BC038558) were each transfected together with the selectable marker pPGK-Puro plasmid (gift from R. Jaenisch; Addgene 11349) into ΔFirreXa cells using lipofectamine 3000 (Invitrogen). Blast searches were performed to map homology of the cDNAs to the reference genomes (Supplementary Fig. 1b). After transfection, ΔFirreXa+mtransgene and ΔFirreXa+htransgene were selected in Eagle’s medium with 2 µg/ml puromycin for 72 h, followed by recovery in Eagle’s medium with 1 µg/ml puromycin for 10 days. A ΔFirreXa+mtransgene clone with high Firre expression was also isolated.
KO mouse tissues and isolation of CLPs and CD8 + T cells
Mice used in this study were maintained in pathogen-specific-free facilities under the supervision of either the University of Washington Institutional Animal Care and Use Committee (Protocol number 2254) or Harvard University’s Institutional Animal Care Committee. We complied with the ethical regulations for animal testing and research in accordance with these committees. Tissues collected from a Firre KO mouse model included liver, kidney, and brain from heterozygous (Firre+/−) and homozygous (Firre−/–) mutants, and from control female mice verified by genotyping33. Ectopic expression of Firre induced by doxycycline (DOX) injection in mice was done as described33. MEFs were derived from mutant (Firre+/−, Firre−/−) and control female 13.5 dpc embryos.
We isolated CLPs live [Lin–Sca-1locKitloIL7Rα+] by fluorescence-activated cell sorting (FACS) from WT (age: 35–43 days) and Firre−/− (age: 35 days) female mice to generate two replicate samples per genotype, consisting of two mice per replicate. Bone marrow was isolated from both femurs and tibias of each mouse by removing the end caps and flushing the bone marrow with a 27 G syringe containing staining media (DMEM, Gibco, 11995–073) with 5% fetal bovine serum (FBS, Gibco, 26140079) and 10 mM EDTA into a 50 mL conical tube. Cells pelleted by centrifugation at 1200 rpm were subjected to lineage depletion according to the manufacturer’s protocol (Miltenyi Biotec, 130–090–858). Lineage-depleted bone marrow was resuspended with the following antibodies (1:100): PE/Cy7 anti-mouse CD127 (IL-7Rα) clone A7R34 (Biolegend, 135014), Alexa Fluor 488 anti-mouse CD117 (c-Kit) clone 2B8 (Biolegend, 105816), PE/Dazzle-594 anti-mouse Ly-6A/E (Sca-1) clone D7 (Biolegend, 108138), APC anti-mouse CD34 clone HM34 (Biolegend, 128612), PE anti-mouse CD135 clone A2F10 (Biolegend, 135306), and Pacific Blue anti-mouse Lineage Cocktail (20 μL per 1 × 106 cells) clones 17A2/RB6-8C5/RA3-6B2/Ter-119/M1/70 (Biolegend, 133310). Zombie Aqua Fixable Viability Kit (Biolegend, 423101) was used as a viability stain according to the manufacturer’s protocol. Samples were incubated on ice in the dark for 60 min, washed twice with staining media, and resuspended before sorting by FACS (BD Aria).
To isolate CD8 + T-cells peripheral blood was collected from mice by cardiac puncture into a tube containing 4% citrate. Red blood cells were lysed for 15 min at room temperature using BD Pharm Lyse (BD, 555899). Cells were washed twice with staining media and the following antibodies were added (1:100) to each sample prior to incubation for 30 min at room temperature: PE anti-mouse CD3 clone 17A2 (Biolegend, 100205), Alexa Fluor 700 anti-mouse CD8a clone 53–6.7 (Biolegend, 100730), APC anti-mouse CD19 clone 6D5 (Biolegend, 115512), Alexa Fluor 488 anti-mouse NK-1.1 clone PK136 (Biolegend, 108718), PE/Dazzle-594 anti-mouse CD4 clone GK1.5 (Biolegend, 100456), TruStain FcX (anti-mouse CD16/32) clone 93 (1:50) (Biolegend, 101319). Zombie Aqua Fixable Viability Kit (Biolegend, 423101) was used as a viability stain. Cells were washed twice with staining media and sorted by FACS (BD Aria).
Allele-specific CRISPR/Cas9 editing and RNAi KD
For allele-specific CRISPR/Cas9 editing of the endogenous Firre locus, three highly specific sgRNAs with BL6 or spretus SNPs at the PAM site and with low off-target scores were chosen and aligned back to the reference genome using BLAT (UCSC) to verify specificity (Supplementary Data 1). The sgRNAs cloned into p × 330 plasmids (Addgene) were transfected into WT Patski cells using Ultracruz reagents (Santa Cruz). A Patski line with a deletion of Dxz4 was also transfected to generate a double-mutant ΔFirreXi/ΔDxz4Xi 29. Single-cell derived colonies were selected and deletions or inversions of the targeted Firre locus verified using PCR and Sanger sequencing to confirm allele-specific editing (Supplementary Data 2). Firre RNAi KD was performed as described20. Cells were harvested after double shRNA/siRNA treatment and qRT-PCR performed to verify KD efficiency.
Immunofluorescence, RNA-FISH, and DNA-FISH
Immunofluorescence was done on cells grown on chamber slides, fixed with paraformaldehyde, permeabilized, and blocked as described previously20. Mouse liver, kidney, and brain were embedded in a cassette and sectioned by the University of Washington histopathology service center. Tissue sections (5 µm) were permeabilized using 0.5% Triton X-100 for 10 min and fixed in 4% paraformaldehyde for 10 min. After incubation with a primary antibody specific for H3K27me3 (Upstate/Millipore, #07–449), H2AK119ubi (Cell Signaling, #8240 S), macroH2A (Abcam, #ab37264), or NPM1 (Abcam, #ab10530) overnight at 4 °C in a humidified chamber cells/tissue sections were washed in 1× PBS (phosphate-buffered saline) buffer and incubated with a secondary antibody conjugated to Texas Red (anti-rabbit, Vector, # TI-1000) or fluorescein (anti-mouse, Vector, # FI-2100). RNA-FISH for Xist was done using a labeled 10 kb cDNA plasmid (pXho, which contains most of Xist exon 1) as described20. RNA-FISH for Firre was done in a clonal ΔFirreXa+mtransgene line that overexpresses Firre RNA using a labeled plasmid probe containing Firre cDNA. DNA-FISH was done using labeled BAC probes containing Firre (RP24-322N20) or Dxz4 (RP23-299L1)29. Images were acquired with a Zeiss fluorescence microscope equipped with an image capture system ZEN 2.3.
Slides were examined by fluorescence microscopy to score the number of nuclei with enrichment in each histone modification on the Xi, using Xist RNA-FISH for Xi identification. A minimum of 300 nuclei were scored per cell type by at least two observers. Measurements of overall H3K27me3 staining intensity outside the Xi cluster were done in a minimum of 300 nuclei using ImageJ36. To minimize experimental variance, a mixture of 80% ΔFirreXa and 20% WT cells were grown in the same chamber prior to immunostaining with H3K27me3 and a control histone panH4 (Abcam ab10158), together with DNA counterstaining using Hoechst 33342. Selected nuclear areas away from the Xi were used for measurement of H3K27me3 median intensity, with same-area normalization to either Hoechst 33342 or panH4 staining. Comparisons between WT and ΔFirreXa were done by calculating either the H3K27me3 staining intensity versus Hoechst 33342 and panH4 for WT and ΔFirreXa nuclei separately, or the relative H3K27me3 staining intensity in ΔFirreXa versus WT nuclei present in the same microscope field. The location of the Xi with respect to the nuclear periphery and the edge of the nucleolus (labeled with NPM1) was recorded in at least 300 nuclei.
Allelic in situ Hi-C, CUT&RUN, ATAC-, RNA-, and ChIP-seq
In situ DNase Hi-C was done on intact nuclei from ΔFirreXa, ΔFirreXi, InvFirreXi, and WT as described29,30. Hi-C libraries were sequenced using 150 bp paired-end reads (Supplementary Data 11). ATAC-seq on ΔFirreXa, ΔFirreXi, InvFirreXi, ΔFirreXi/ΔDxz4Xi, and WT, and RNA-seq on ΔFirreXa, ΔFirreXa+mtransgene, and WT were done as previously described29. ChIP-seq was done on ΔFirreXa and WT using an antibody for H3K27me3 and an established protocol20. CUT&RUN was done on ΔFirreXa, ΔFirreXa+mtransgene, and WT using an antibody for CTCF (Upstate/Millipore, 07-729-25UL), and on ΔFirreXa and WT using an antibody for SUZ12 (Abcam ab12073) using a published protocol96. H3K27me3 ChIP analyses of CLPs and CD8 + T cells were done using the TrueMicroChIP kit (Diagenode, C01010130) according to the manufacturer’s protocol. Briefly, we sheared fixed chromatin from ~32,000 CLPs and ~50,000 CD8 + T cells in separate 1.5 mL TPX plastic tubes (Diagenode, C3001001050) using the following cycles on a Bioruptor (Diagenode): eight cycles of 5 min (30 ON, 30 OFF). Between each cycle samples were placed on ice. Sheared chromatin was then immunoprecipitated using 1 µL of H3K27me3 antibody (Diagenode, C15410069), retaining 10% of the chromatin as input. ChIP products were eluted in 12 µL and libraries prepared using the MicroPlex Library Preparation kit v2 (Diagenode, C05010012). Ten microleter of ChIP products were used as input, iPCRtagT1-T12 were used for 12 amplification cycles. After library clean up, the libraries were analyzed on a BioAnalyzer DNA-HS chip.
ATAC-seq, ChIP-seq, and CUT&RUN libraries for Patski cells were sequenced as 75 bp pair-end reads (Supplementary Data 11). RNA-seq libraries for Patski cells were sequenced as 75 bp single-end reads (Supplementary Data 11 ). Sequencing datasets were analyzed to assign reads to the spretus or BL6 genomes using a previously developed allele-specific data analysis pipeline29. RNA-seq reads were mapped to the UCSC mm10 (NCBI build v38) refSeq BL6 mouse transcriptome. Tophat2 (v 2.0.12) (calling bowtie2 (v2.2.3)) was used to perform single-end mapping allowing six mismatches. Mapped reads were assigned to refSeq genes using HT-seq(v0.11.0) and counts were converted into TPMs using custom R scripts. DE analysis was performed using DESeq2. ATAC-seq, ChIP-seq, and CUT&RUN reads were mapped to the BL6 mouse genome using the NCBI build v38/mm10 reference genome assembly obtained from the UCSC Genome Browser using BWA-MEM (v0.7.3) in paired-end mode using default parameters. Peaks were called using MACS2. DNase Hi-C reads were mapped to the BL6 genome using the NCBI build v38/mm10 reference genome assembly obtained from the UCSC Genome Browser and a pseudo-spretus genome using BWA-MEM (v0.7.3) in single-end mode using default parameters29. Micro-RNA-seq (less than 200nt) was done in ΔFirreXa and WT Patski cells by BGI Genomics (https://www.bgi.com/us/). ChIP-seq libraries for CLPs and CD8 + T cells were sequenced as 150 bp pair-end reads. Sequencing datasets were analyzed to assign reads to the BL6 genome using the NCBI build v38/mm10 reference genome assembly obtained from the UCSC Genome Browser using Bowtie2. Differential tracks and heatmaps were generated by Deeptools. GO analysis was done using http://geneontology.org/.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by grants GM131745 (CMD) and GM127327 (XD) from the National Institute of General Medical Sciences, by grant DK107979 (JS and WSN) from the National Institutes of Health Common Fund 4D Nucleome, and by a HHMI Faculty Scholars grant (J.L.R.). We thank the HSCRB Flow Cytometry Core for assistance with cell sorting, the Bauer Sequencing Core at Harvard University for sequencing, and Nydia Chang for assistance with mouse husbandry.
Source data
Author contributions
H.F., G.B., X.D., W.S.N., and C.M.D. conceived the study; H.F. and X.D. constructed the mutant cell lines; J.P.L. and J.L.R. constructed the mouse model; J.T. and S.H. performed CUT&RUN; H.F., G.N.F., Z.D., and J.S. performed RNA-seq, ATAC-seq, ChIP-seq, and Hi-C assays; G.B. and H.F. performed data analyses; H.F., G.B., X.D., W.S.N., and C.M.D. wrote the manuscript, with input from all authors.
Data availability
All sequencing data that support the findings of this study have been deposited in the National Centre for Biotechnology Information GEO and are accessible through the GEO SuperSeries “GSE59779”. Publicly available RNA Immunoprecipitation data for Fig. 2i were obtained from NCBI GEO “GSE118402” (RIP-seq_TSC HNRNPK) and “GSE137491” (WT_1_ESC_SUZ12_RIPseq and WT_1_ESC_EZH2_RIPseq). All other data and the scripts used for the analyses that support the findings of this study are available within the article and its Supplementary Information files or from the corresponding authors upon reasonable request. Source data are provided with this paper. A reporting summary for this Article is available as a Supplementary Information file. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: He Fang, Giancarlo Bonora.
Contributor Information
Xinxian Deng, Email: dengx2@uw.edu.
William S. Noble, Email: wnoble@uw.edu
Christine M. Disteche, Email: cdistech@uw.edu
Supplementary information
Supplementary information is available for this paper at 10.1038/s41467-020-19879-3.
References
- 1.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat. Rev. Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 2.Galupa R, Heard E. X-chromosome inactivation: a crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 2018;52:535–566. doi: 10.1146/annurev-genet-120116-024611. [DOI] [PubMed] [Google Scholar]
- 3.Engreitz JM, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simon MD, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–469. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 6.Mira-Bontenbal H, Gribnau J. New Xist-interacting proteins in X-chromosome inactivation. Curr. Biol. 2016;26:R338–R342. doi: 10.1016/j.cub.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 7.Pinheiro, I. & Heard, E. X chromosome inactivation: new players in the initiation of gene silencing. F1000Res 6 Faculty Rev-344. (2017). [DOI] [PMC free article] [PubMed]
- 8.Żylicz JJ, et al. The implication of early chromatin changes in X chromosome inactivation. Cell. 2019;176:182–197.e23. doi: 10.1016/j.cell.2018.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonora G, Disteche CM. Structural aspects of the inactive X chromosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:1733. doi: 10.1098/rstb.2016.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jégu T, Aeby E, Lee JT. The X chromosome in space. Nat. Rev. Genet. 2017;18:377–389. doi: 10.1038/nrg.2017.17. [DOI] [PubMed] [Google Scholar]
- 11.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 12.Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 13.Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am. J. Hum. Genet. 1962;14:135–148. [PMC free article] [PubMed] [Google Scholar]
- 14.Rego A, Sinclair PB, Tao W, Kireev I, Belmont AS. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J. Cell Sci. 2008;121:1119–1127. doi: 10.1242/jcs.026104. [DOI] [PubMed] [Google Scholar]
- 15.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 16.Padeken J, Heun P. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 2014;28:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Belagal P, et al. Decoding the principles underlying the frequency of association with nucleoli for RNA polymerase III-transcribed genes in budding yeast. Mol. Biol. Cell. 2016;27:3164–3177. doi: 10.1091/mbc.e16-03-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S, Deerinck TJ, Ellisman MH, Spector DL. The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CK, et al. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354:468–472. doi: 10.1126/science.aae0047. [DOI] [PubMed] [Google Scholar]
- 20.Yang F, et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hacisuleyman E, Shukla CJ, Weiner CL, Rinn JL. Function and evolution of local repeats in the Firre locus. Nat. Commun. 2016;7:11021. doi: 10.1038/ncomms11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barutcu AR, Maass PG, Lewandowski JP, Weiner CL, Rinn JL. A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus. Nat. Commun. 2018;9:1444. doi: 10.1038/s41467-018-03614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hacisuleyman E, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu Y, et al. The NF-κB-responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J. Immunol. 2017;199:3571–3582. doi: 10.4049/jimmunol.1700091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izuogu OG, et al. Analysis of human ES cell differentiation establishes that the dominant isoforms of the lncRNAs RMST and FIRRE are circular. BMC Genomics. 2018;19:276. doi: 10.1186/s12864-018-4660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Darrow EM, et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl Acad. Sci. USA. 2016;113:E4504–E4512. doi: 10.1073/pnas.1609643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horakova AH, Moseley SC, McLaughlin CR, Tremblay DC, Chadwick BP. The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum. Mol. Genet. 2012;21:4367–4377. doi: 10.1093/hmg/dds270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonora G, et al. Orientation-dependent Dxz4 contacts shape the 3D structure of the inactive X chromosome. Nat. Commun. 2018;9:1445. doi: 10.1038/s41467-018-03694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng X, et al. Bipartite structure of the inactive mouse X chromosome. Genome Biol. 2015;16:152. doi: 10.1186/s13059-015-0728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giorgetti L, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535:575–579. doi: 10.1038/nature18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andergassen D, et al. In vivo Firre and Dxz4 deletion elucidates roles for autosomal gene regulation. elife. 2019;8:e47214. doi: 10.7554/eLife.47214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewandowski JP, et al. The Firre locus produces a trans-acting RNA molecule that functions in hematopoiesis. Nat. Commun. 2019;10:5137. doi: 10.1038/s41467-019-12970-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andergassen D, et al. Mapping the mouse Allelome reveals tissue-specific regulation of allelic expression. elife. 2017;6:e25125. doi: 10.7554/eLife.25125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Froberg JE, Pinter SF, Kriz AJ, Jégu T, Lee JT. Megadomains and superloops form dynamically but are dispensable for X-chromosome inactivation and gene escape. Nat. Commun. 2018;9:5004. doi: 10.1038/s41467-018-07446-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luense S, et al. Quantification of histone H3 Lys27 trimethylation (H3K27me3) by high-throughput microscopy enables cellular large-scale screening for small-molecule EZH2 inhibitors. J. Biomol. Screen. 2015;20:190–201. doi: 10.1177/1087057114559668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marks H, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–1373. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakur, J., He, F., Trizia, L., Christine, D. & Steven, H. Architectural RNA is required for heterochromatin organization. biorvix10.1101/784835 (2019).
- 39.Schertzer MD, et al. lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG Island DNA. Mol. Cell. 2019;75:523–537.e10. doi: 10.1016/j.molcel.2019.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garland W, et al. A functional link between nuclear RNA decay and transcriptional control mediated by the polycomb repressive complex 2. Cell Rep. 2019;29:1800–1811.e6. doi: 10.1016/j.celrep.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei C, et al. RBFox2 binds nascent RNA to globally regulate polycomb complex 2 targeting in mammalian genomes. Mol. Cell. 2016;62:875–889. doi: 10.1016/j.molcel.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaneko S, et al. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell. 2014;53:290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 2013;20:1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavares L, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenberg M, et al. Denaturing CLIP, dCLIP, pipeline identifies discrete RNA footprints on chromatin-associated proteins and reveals that CBX7 targets 3’ UTRs to regulate mRNA expression. Cell Syst. 2017;5:368–385.e15. doi: 10.1016/j.cels.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang YH, et al. 2B4-SAP signaling is required for the priming of naive CD8. Oncoimmunology. 2017;6:e1267094. doi: 10.1080/2162402X.2016.1267094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nat. Immunol. 2009;10:618–626. doi: 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao F, Cai Y, Kapranov P, Xu D. Reverse-genetics studies of lncRNAs-what we have learnt and paths forward. Genome Biol. 2020;21:93. doi: 10.1186/s13059-020-01994-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/S1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 52.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem. Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 54.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engreitz JM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engreitz JM, Ollikainen N, Guttman M. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 57.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol. Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.da Rocha ST, et al. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol. Cell. 2014;53:301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 60.Pintacuda G, et al. hnRNPK recruits PCGF3/5-PRC1 to the Xist RNA B-repeat to establish polycomb-mediated chromosomal silencing. Mol. Cell. 2017;68:955–969.e10. doi: 10.1016/j.molcel.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 62.Graindorge A, et al. In-cell identification and measurement of RNA-protein interactions. Nat. Commun. 2019;10:5317. doi: 10.1038/s41467-019-13235-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brockdorff N. Polycomb complexes in X chromosome inactivation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20170021. doi: 10.1098/rstb.2017.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu Z, Carter AC, Chang HY. Mechanistic insights in X-chromosome inactivation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20160356. doi: 10.1098/rstb.2016.0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brockdorff N, Bowness JS, Wei G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 2020;34:733–744. doi: 10.1101/gad.337196.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monfort, A. & Wutz, A. The B-side of Xist. F1000Res 9 F1000 Faculty Rev-55. (2020).
- 67.Youmans DT, Schmidt JC, Cech TR. Live-cell imaging reveals the dynamics of PRC2 and recruitment to chromatin by SUZ12-associated subunits. Genes Dev. 2018;32:794–805. doi: 10.1101/gad.311936.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marín-Béjar O, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14:R104. doi: 10.1186/gb-2013-14-9-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yen YP, et al. locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. elife. 2018;7:e38080. doi: 10.7554/eLife.38080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Das PP, et al. PRC2 is required to maintain expression of the maternal Gtl2-Rian-Mirg locus by preventing de novo DNA methylation in mouse embryonic stem cells. Cell Rep. 2015;12:1456–1470. doi: 10.1016/j.celrep.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Padeken J, et al. The nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol. Cell. 2013;50:236–249. doi: 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 74.Holmberg Olausson K, Nistér M, Lindström MS. Loss of nucleolar histone chaperone NPM1 triggers rearrangement of heterochromatin and synergizes with a deficiency in DNA methyltransferase DNMT3A to drive ribosomal DNA transcription. J. Biol. Chem. 2014;289:34601–34619. doi: 10.1074/jbc.M114.569244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lyu G, et al. Changes in the position and volume of inactive X chromosomes during the G0/G1 transition. Chromosome Res. 2018;26:179–189. doi: 10.1007/s10577-018-9577-0. [DOI] [PubMed] [Google Scholar]
- 76.Stewart ER, et al. Maintenance of epigenetic landscape requires CIZ1 and is corrupted in differentiated fibroblasts in long-term culture. Nat. Commun. 2019;10:460. doi: 10.1038/s41467-018-08072-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Colognori D, Sunwoo H, Kriz AJ, Wang CY, Lee JT. Xist deletional analysis reveals an interdependency between Xist RNA and polycomb complexes for spreading along the inactive X. Mol. Cell. 2019;74:101–117.e10. doi: 10.1016/j.molcel.2019.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saldaña-Meyer R, et al. RNA interactions are essential for CTCF-mediated genome organization. Mol. Cell. 2019;76:412–422.e5. doi: 10.1016/j.molcel.2019.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ezhkova E, et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JM, et al. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci. Rep. 2015;5:16714. doi: 10.1038/srep16714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev. Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 82.Pullirsch D, et al. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development. 2010;137:935–943. doi: 10.1242/dev.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521:232–236. doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan H, et al. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res. 2018;28:192–202. doi: 10.1101/gr.224576.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goff LA, Rinn JL. Linking RNA biology to lncRNAs. Genome Res. 2015;25:1456–1465. doi: 10.1101/gr.191122.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Beltrán-Anaya FO, Cedro-Tanda A, Hidalgo-Miranda A, Romero-Cordoba SL. Insights into the regulatory role of non-coding RNAs in cancer metabolism. Front. Physiol. 2016;7:342. doi: 10.3389/fphys.2016.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nóbrega MA, Zhu Y, Plajzer-Frick I, Afzal V, Rubin EM. Megabase deletions of gene deserts result in viable mice. Nature. 2004;431:988–993. doi: 10.1038/nature03022. [DOI] [PubMed] [Google Scholar]
- 88.Han X, et al. Mouse knockout models reveal largely dispensable but context-dependent functions of lncRNAs during development. J. Mol. Cell Biol. 2018;10:175–178. doi: 10.1093/jmcb/mjy003. [DOI] [PubMed] [Google Scholar]
- 89.Rossi A, et al. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- 90.Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27:2663–2677. doi: 10.1101/gad.225888.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fang J, et al. PIRCh-seq: functional classification of non-coding RNAs associated with distinct histone modifications. Genome Biol. 2019;20:292. doi: 10.1186/s13059-019-1880-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang J, et al. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl Acad. Sci. USA. 2016;113:E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lingenfelter PA, et al. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat. Genet. 1998;18:212–213. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 94.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Berletch JB, et al. Escape from X inactivation varies in mouse tissues. PLoS Genet. 2015;11:e1005079. doi: 10.1371/journal.pgen.1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Skene PJ, Henikoff JG, Henikoff S. Targeted in situ genome-wide profiling with high efficiency for low cell numbers. Nat. Protoc. 2018;13:1006–1019. doi: 10.1038/nprot.2018.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All sequencing data that support the findings of this study have been deposited in the National Centre for Biotechnology Information GEO and are accessible through the GEO SuperSeries “GSE59779”. Publicly available RNA Immunoprecipitation data for Fig. 2i were obtained from NCBI GEO “GSE118402” (RIP-seq_TSC HNRNPK) and “GSE137491” (WT_1_ESC_SUZ12_RIPseq and WT_1_ESC_EZH2_RIPseq). All other data and the scripts used for the analyses that support the findings of this study are available within the article and its Supplementary Information files or from the corresponding authors upon reasonable request. Source data are provided with this paper. A reporting summary for this Article is available as a Supplementary Information file. Source data are provided with this paper.