Abstract
Fusion of somatic cells to embryonic stem cells induces reprogramming of the somatic nucleus and can be used to study the effect of trans-acting factors from the pluripotent cell over the differentiated nucleus. However, fusion only occurs in a small fraction of the cells exposed to fusogenic conditions, hence the need for a protocol that produces high fusion rate with minimal cell damage, coupled with a method capable of identifying and selecting these rare events. Here, we describe a protocol to induce formation of bi-species mouse pluripotent/bovine somatic heterokaryons, as well as same-species homokaryons, using polyethylene glycol (PEG). To identify bi-species fusion products, heterokaryons were labeled using cell type-specific fluorescent antibodies and selected using imaging (Amnis ImageStream Mark II) and traditional (BD FACSAria I) flow cytometry. Heterokaryons selected with this method produced ES cell-like colonies in vitro. This procedure can be combined with downstream applications such as nucleic acid isolation for RT-PCR and RNA-Seq, and used as a tool to study somatic cell nuclear reprogramming.
Keywords: Cell fusion, Nuclear reprogramming, Polyethylene glycol, Homokaryon, Heterokaryon, Indirect immunofluorescence, Imaging, Flow cytometry, ImageStream
Introduction
Cell fusion is a process that involves combining the cytoplasmic membrane of two or more cells to form one single multinucleated cell (Yamanaka and Blau 2010). Nuclear reprogramming can be achieved by fusion of somatic cells to pluripotent cells such as embryonic stem (ES) cells (Tada et al. 2001; Silva et al. 2006; Sullivan et al. 2006), embryonal germ (EG) cells (Tada et al. 1997), or embryonal carcinoma (EC) cells (McBurney and Strutt 1979), suggesting the reprogramming activity of the pluripotent cell is predominant over the gene expression pattern of the somatic cell. The nuclei of somatic cells fused to ES cells reprogram faster (1 to 2 days) and with greater efficiency (up to 70%) than cells reprogrammed by transcription factor induction (Yamanaka and Blau 2010). Fused cells can become hybrids or heterokaryons depending if their nuclei fuse or remain intact, respectively. Gene expression changes in heterokaryons happen in the absence of cell division, without genomic mixing or transgenes, which makes cell fusion a powerful tool to study early modifications in gene expression. Moreover, the use of interspecies heterokaryons allows the discrimination of the transcriptome of each fusion partner, thus facilitating the screening of factors provided by the pluripotent cell, and the changes in gene expression induced in the somatic nucleus. The cell fusion approach has already provided valuable information about reprogramming (Brady et al. 2013) and differentiation (Wong et al. 2017) mechanisms in human and mouse. These processes are part of the foundations of regenerative medicine and developmental biology and are, to date, still not fully understood (Cyranoski 2014). Moreover, in other domestic and biomedically relevant species where pluripotent cell lines are not yet fully available (Koh and Piedrahita 2014; Ezashi et al. 2016), the interspecies heterokaryon approach could provide new insights as to which genes and mechanisms are essential in the establishment and maintenance of pluripotency.
Despite technological advances in cell sorting and transcriptome analysis, cell fusion has yet to be used to its full potential, and protocols designed specifically for the study of early heterokaryons in a nuclear reprogramming context are not easily available. Polyethylene glycol (PEG) is a polyether of repetitive ethylene oxide units that can be classified according to the number of ethylene oxide units or their approximate molecular weight (e.g. PEG-32 or PEG 1500, respectively), and is used as a drug delivery agent and in cosmetics (Leung 2014; D’souza Shegokar R and Shegokar 2016). PEG in the molecular weight range of 1000–3700 has been widely used to induce fusion in a laboratory setting, mainly due its low cytotoxicity, cost, and easiness to use, with a protocol that can be completed in less than 1 h (Davidson et al. 1976; Blau et al. 1983; Yang and Shen 2006; Yu et al. 2007; Zhang et al. 2007; Palermo et al. 2009; Pomerantz et al. 2009; Brady et al. 2013). Fusion can only be induced in a small fraction of the cells exposed to fusogenic conditions, and different cell types might require modifications of the method used in terms of PEG molecular weight, concentration, and/or time (Yang and Shen 2006). In a series of preliminary experiments, we determined that exposing mouse or bovine primary cells to PEG containing 10% DMSO produces the highest percentage of binucleated cells, with low cell death (Villafranca Locher 2018). Here, we produced interspecies heterokaryons formed by the fusion of mouse embryonic stem (mES) cells (C57BL/6 line) to bovine fetal fibroblasts (bFFs), and identified them by indirect immunostaining in live cells, using fluorescent antibodies targeting mES cell-specific marker SSEA-1 and bFF-specific marker CD44, as well as nuclear staining using Hoechst. The population of double-positive heterokaryons was identified first using the ImageStream Mark II imaging flow cytometer, and then selected using FACSAria I. Imaging flow cytometry was an essential step to identify the small heterokaryon cell population (< 1%). Because of its specificity, the heterokaryons selected with this method were suitable for downstream applications such as nucleic acid isolation for RT-PCR and RNA-Seq, or long-term culture to produce clonal hybrid colonies. Our selection protocol was also successful for production and selection of multinucleated homokaryons.
Materials and methods
Cell culture
Primary bovine fetal fibroblasts (bFFs) were derived from a male fetus of unknown genetic background obtained at an abattoir at gestation day 60. Cells were grown in a 5% CO2 in air incubator (Forma series II water jacketed incubator) at 38.5 °C on 10 cm tissue culture dishes (Falcon) in fibroblast medium: Dulbecco’s minimal essential medium (DMEM; Gibco) supplemented with 10% Fetal Bovine Serum (FBS; HyClone) and 50 µg/ml Gentamicin (Lonza). Medium was replaced every two to 3 days. Subculture was done with TrypLE Express (Gibco) before cells reached 80% confluence. Cells were passaged at least twice to ensure proper growth. Primary bFF lines used for all experiments were between passages 2 to 6. Mouse embryonic stem (mES) cells from the line C57BL/6 were purchased from ATCC (CRL-1002) and cultured in ES cell medium: DMEM with 15% ES cell-qualified fetal bovine serum (FBS; Gibco), 1X Non-Essential Amino Acids (HyClone), 1X Glutamax (Gibco), 1500 U/ml of ESGRO (Millipore), 0.55 mM beta-mercaptoethanol (Sigma), 1 μM PD0325901 (PD; Cayman chemical company), 3 μM CHIR99021 (CHIR; Cayman chemical company), and 50 μg/ml Gentamycin (Lonza); mES cells were cultured on a monolayer of gamma irradiated mouse embryonic fibroblasts (IRR-MEFs; StemGent). FACS-sorted heterokaryons and homokaryons were cultured on IRR-MEFs, ~ 200 sorted cells per 24-well plate well, in ES cell medium.
PEG fusion
We seeded 2 × 105 bFFs in one well of a 24-well tissue-culture treated Falcon polystyrene flat-bottom microplate (ThermoFisher) in ES cell medium, and 2 h later 6 × 105 mES cells were seeded in the same well, without replacing the culture medium. Cells were co-cultured in ES cell medium for 4 h before fusion. For fusion, monolayers were washed twice with 1 ml Dulbecco’s Phosphate-Buffered Saline (DPBS; HyClone) each time before addition of 200 µl PEG 1500 (Roche) plus 10% Dimethyl sulfoxide (DMSO; Sigma Aldrich) mixed and pre-warmed to 37 °C, for exactly 2 min at room temperature, followed by two successive washes with DPBS and one wash with ES cell medium (1 ml each one). Cells were cultured at 37 °C in ES cell medium until analysis. A co-culture control (bFF and mES cell plated as described but no fusion) was also included in one of the replicates.
Indirect immunostaining
Fused monolayers were dissociated to a monocellular suspension with TrypLE, centrifuged, resuspended in DNase solution (0.1 mg/ml deoxyribonuclease I in DMEM; Worthington biochemical corporation) and incubated for 15 min at room temperature. Following incubation, the solution was filtered through a 100 µM cell strainer (Thermo Fisher). Filtered cells were pelleted by gentle centrifugation and resuspended in DPBS plus 1% heat-inactivated FBS containing antibodies or Hoechst, incubated in the following order: anti-mouse/human CD44 (1:100 dilution, 30 min incubation; Biolegend cat#103001), rabbit anti-rat AlexaFluor® 488 (1:1000 dilution, 20 min incubation; Jackson Immunoresearch Inc., Cat#3125455003), anti-mouse SSEA-1 (1:100 dilution, 30 min incubation; Santa Cruz Biotechnology, sc-21702), goat anti-mouse Alexa Fluor® 647 (1:2000 dilution, 20 min incubation; Abcam ab150123), and Hoechst (15 min). In all cases, incubation was at 4 °C in the dark. Between staining steps, cells were pelleted by centrifugation and supernatant removed before resuspending in a new staining solution. After the last incubation step, cells were washed and resuspended in DPBS with 1% FBS. Controls (negative (unstained), single colors (CD44/A488, SSEA-1/A647, and Hoechst), secondary antibodies) were always prepared in parallel, following all steps described for samples.
Flow cytometry
Cells were stained as described and resuspended in DPBS with 1% FBS to a concentration of ~ 5 × 106 cells/ml for ImageStream Mark II (Amnis) and ~ 1 × 106 cells/ml for FACSAria I (BD) analysis. The ImageStream is an imaging flow cytometer that permits visualization (bright field and fluorescence) of cells directly in flow; the ImageStream does not perform sorting. The FACSAria is a sorting cytometer, and parameters identified on the ImageStream can be used to sort cells in the FACSAria. We first analyzed cells using ImageStream to identify location of bicolored, multinucleated heterokaryons. The preprocessing of the images and identification of the area with highest presence of heterokaryons was performed with IDEAS ImageStream Analysis Software. For all replicates, compensation was done with single color stained samples. The obtained parameters were used to collect heterokaryons with the FACSAria. Cells were sorted at low pressure, using a 100 μm nozzle aperture.
Results
Interspecies heterokaryons and homokaryons can be identified and selected using a combination of imaging and sorting flow cytometry
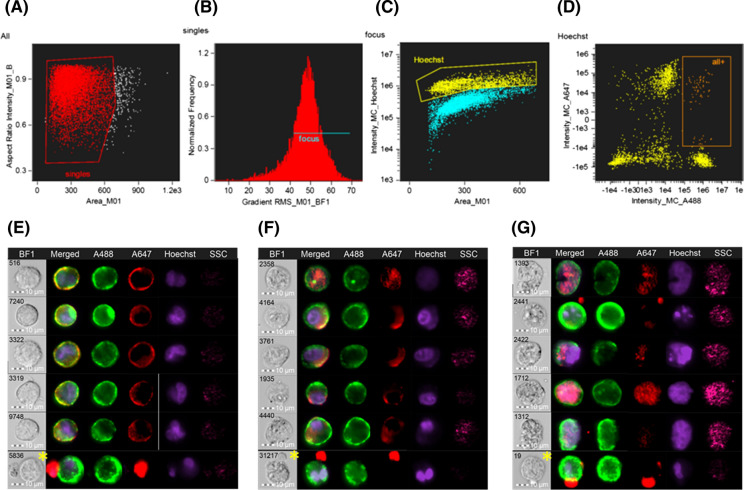
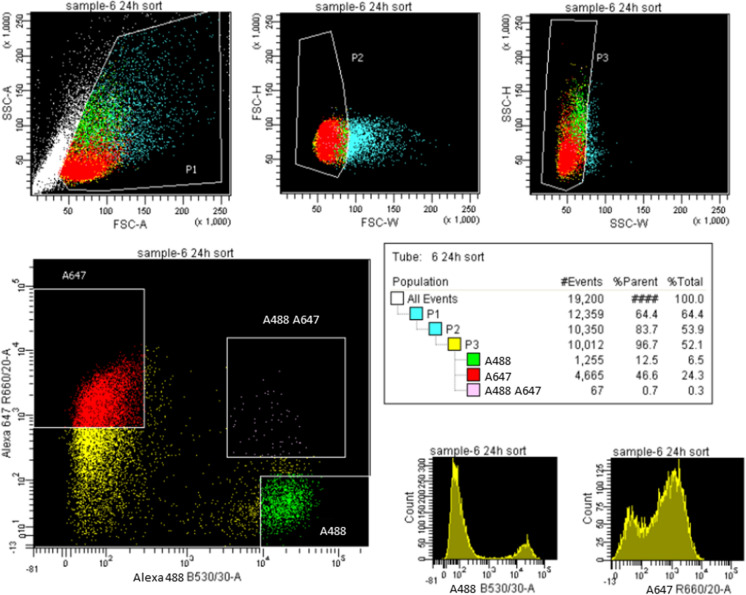
Fused monolayers of co-cultured bFFs and mES cells were stained and immediately analyzed using ImageStream, to identify the area in the histogram that contains double-positive heterokaryons (Fig. 1a–d). Distinct nuclei were observed in heterokaryons sampled at 24 and 48 h (Fig. 1e, f), whereas the double positive heterokaryons at 72 h presented enlarged and/or irregular nuclei (Fig. 1g). The parameters identified on the ImageStream were extrapolated to the FACSAria flow cytometer (Fig. 2) to sort an enriched population of heterokaryons. The expected yield of double positive heterokaryons is shown in Table 1.
Fig. 1.
Identification of heterokaryons using the ImageStream Mark II imaging flow cytometer. Gating strategy: only single cells (a) in focus (b) were considered and plotted as intensity of Hoechst vs. cell area in the bright field image (c); a population of high intensity of Hoechst was further selected and plotted as intensity of Alexa 488 vs. Alexa 647 (d). In this scatterplot, a population of high A488 and high A647 was found to contain a majority of heterokaryons (orange rectangle). Representative images of heterokaryons found in this area at 24, 48, and 72 h are shown in e–g, respectively. It was still possible to find some false positive cells in this subpopulation (yellow asterisks) but it was not possible to gate these events out. Parameters identified on the ImageStream were used to sort heterokaryons in the FACSAria
Fig. 2.
Representative image of gating strategy used to select heterokaryons in the FACSAria I. Heterokaryons are present in the top right quadrant of the A647 vs A488 scatterplot
Table 1.
Approximate expected yield of heterokaryons at different time points
| Yield | 24 h | 48 h | 72 h |
|---|---|---|---|
| Number of events | 150,000 | 150,000 | 150,000 |
| Percentage (% parent (A488 + A647 +)) | 0.7–0.1% | 0.2–0.1% | 0.2–0.1% |
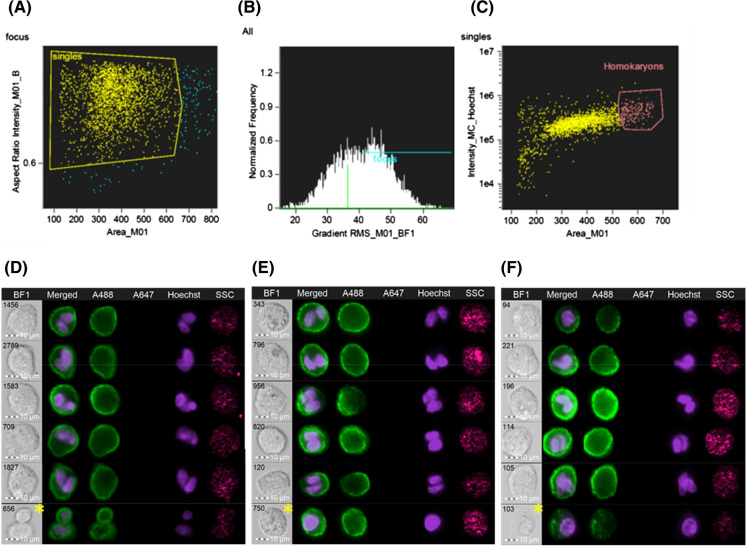
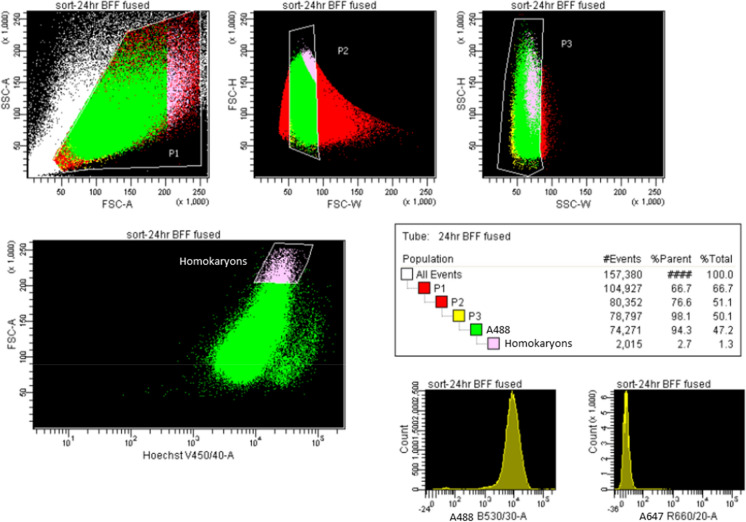
Somatic bFF/bFF homokaryons were also identified based on cell size and intensity of cell type-specific dye (Fig. 3a–c). Distinct nuclei were observed at all time points (Fig. 3d–f). Gating strategy for FACSAria is presented in Fig. 4. Expected yield of double positive bFF homokaryons is shown in Table 2.
Fig. 3.
Identification of homokaryons using ImageStream Mark II imaging flow cytometer. Similar to heterokaryons, only single cells (a) in focus (b) were were plotted as intensity of Hoechst versus cell area in the bright field image (c); in that plot, a section containing a majority of homokaryons was found. Representative images of homokaryons found (d) 24 h, (e) 48 h, and (f) 72 h after fusion are shown. It was still possible to find some false positive cells in this subpopulation (yellow asterisks) but it was not possible to gate these events out. Parameters identified on the ImageStream were used to sort homokaryons in the FACSAria
Fig. 4.
Representative image of gating strategy used to select bFF homokaryons using FACSAria I
Table 2.
Approximate expected yield of homokaryons at different time points
| Yield | 24 h | 48 h | 72 h |
|---|---|---|---|
| Number of events | 150,000 | 150,000 | 150,000 |
| Percentage (% parent (A488 +)) | 2.7% | 2.1% | 4.9% |
Hybrids can be produced from sorted heterokaryons
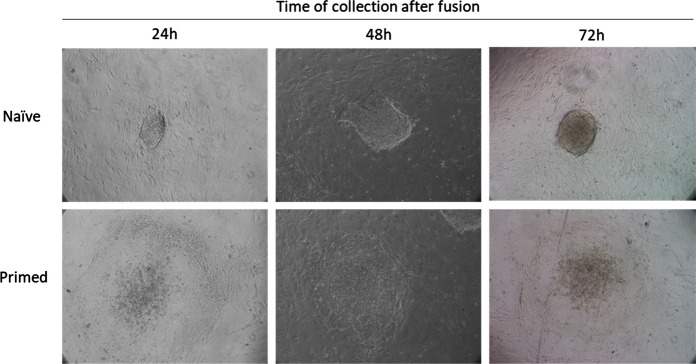
We selected 600 double-positive heterokaryons and 600 bFF homokaryons both at 24, 48, and 72 h after fusion using FACS, and cultured on IRR-MEFs in ES cell culture media. Colonies were visible in heterokaryon cultures as early as 24 h, and it was possible to observe two distinct types of colony morphologies at all time points: tightly packed colonies with a defined border (“naïve” morphology), and colonies with an irregular undefined border (“primed”), as shown in Fig. 5. Colonies were counted 6 days after plating (Table 3).
Fig. 5.
Representative images of colonies observed after six-day culture of FACS sorted mouse/bovine heterokaryons, selected at different time points after fusion with PEG plus 10% DMSO. Colonies with naïve or primed morphology were observed at all time points. Images are 10X
Table 3.
Number of colonies observed after 6-day culture of heterokaryons and homokaryons selected at different time points after fusion
| Morphology | 24 h | 48 h | 72 h | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heterok. | Homok. | Heterok. | Homok. | Heterok. | Homok. | |||||||
| # | % | # | % | # | % | # | % | # | % | # | % | |
| Naïve | 14 | 2.3 | 0 | 0.0 | 26 | 4.3 | 0 | 0.0 | 45 | 7.5 | 2 | 0.3 |
| Primed | 14 | 2.3 | 1 | 0.2 | 33 | 5.5 | 0 | 0.0 | 63 | 10.5 | 0 | 0.0 |
| Total colonies | 28 | 4.7 | 1 | 0.2 | 59 | 9.8 | 0 | 0.0 | 108 | 18.0 | 2 | 0.3 |
Two distinct colony morphologies were observed at each time point. Percentage indicated is based on the 600 cells initially sorted
Discussion
The interspecies cell fusion approach holds great potential for the study of early events during nuclear reprogramming (Yamanaka and Blau 2010). Although cell fusion has been used for decades, most reports focus on the study of established hybrid colonies, which form days after the initial fusion event (Miller et al. 1976; Cowan et al. 2005; Yu et al. 2006; Imai et al. 2020). It is known that not all fusion products form hybrids, making it difficult to estimate how many cells initially fused. Efficiency of cell fusion also varies depending on the cell type(s), the fusogen, and the method used to determine fusion. For example, Blau et al. (1983) fused human amniocytes and mouse myotubes with PEG, and observed an average of 73% of heterokaryon formation by visual inspection of multinucleated cells (Blau et al. 1983). In contrast, Brady et al. (2013) used PEG to fuse GFP+ mouse ESCs and dsRed+ human primary fibroblasts, and used FACS to select for double positive heterokaryons, obtaining 1.16% of heterokaryons on a first sort, which was later increased to 51.6% after a second sort and 77.8% after enrichment (Brady et al. 2013). Here, using FACS and cell type-specific antibodies SSEA-1 and CD44, respectively, we found that less than 1% of the cells initially fused form double positive heterokaryons on a first sort.
Upon fusion, heterokaryons can originate proliferative hybrids which form ES cell-like colonies when placed in stem cell culture conditions (Yamanaka and Blau 2010). We observed ~ 5% of sorted heterokaryons form colonies when cultured. Interestingly, hybrid colonies obtained at different time points after fusion presented different proportions of naïve and primed morphologies, with the number of primed colonies increasing over 48 and 72 h. While heterokaryons collected at 24 and 48 h after fusion presented distict binucleated morphology, cells obtained at 72 h after fusion presented irregular nuclei. Abnormal karyotypes and chromosomal loss has been described in hybrid cell lines of interspecies origin (Nowak-Imialek et al. 2010; Serov et al. 2011), which may explain the low percentage of heterokaryons forming colonies, the higher proportion of primed colonies, and raises concerns over the use of hybrids for the study of pluripotency. In addition, we observed colony-like structures in cultured BFF homokaryons, which is in line with our previous observation about bFFs and other rapidly growing bovine primary cells forming aggregates morphologically similar to ES cell colonies when cultured in confluent monolayers. In that same study, we also showed that bovine fibroblasts growing in clusters express Alkaline phosphatase, a commonly used marker for mouse ES and iPS cells (Villafranca Locher 2018); we therefore did not use this staining to characterize hybrid colonies.
It has been described that reprogramming happens in distinct phases early after expression of pluripotency factors in the somatic nucleus (Polo et al. 2012; Sancho-Martinez and Izpisua Belmonte 2013; Knaupp et al. 2017). In this context, if heterokaryons are to be used to study nuclear reprogramming, this should happen before nuclear fusion and cell division occurs. In this study, nuclei remained intact up to 48 h after fusion. Our method is therefore relevant for the study of early reprogramming events. The bi-species approach permits the differentiation of transcripts from the individual fusion partners, and is thus ideal for the study of nuclear reprogramming in species in which pluripotent cell lines are not yet available. We are currently using this method to analyze the bovine transcriptome upon fusion with mES cells.
Conclusions
We present a method to consistently produce bi-species somatic/pluripotent (bFF/mES cell) heterokaryons, using 50% PEG 1500 plus 10% DMSO. Multinucleated cells were detected by indirect immunofluorescence in live cells, using cell-type specific markers. We used ImageStream imaging flow cytometry to identify the cell population of interest and selected these events using FACS. The selected heterokaryons can be used directly for RNA isolation for RT-PCR or RNA-Seq, or cultured in stem cell conditions where they are capable of producing hybrid colonies. The method described here could be adapted to other cell types and species, and aid in the understanding of early nuclear reprogramming mechanisms. Our method is also suitable for production of same species homokaryons.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Blau HM, Chiu C-P, Webster C. Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell. 1983;32:1171–1180. doi: 10.1016/0092-8674(83)90300-8. [DOI] [PubMed] [Google Scholar]
- Brady JJ, Li M, Suthram S, et al. Early role for IL-6 signalling during generation of induced pluripotent stem cells revealed by heterokaryon RNA-Seq. Nat Cell Biol. 2013;15:1244–1252. doi: 10.1038/ncb2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1374. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- Cyranoski D. Stem cells: the black box of reprogramming. Nature. 2014;516:162–164. doi: 10.1038/516162a. [DOI] [PubMed] [Google Scholar]
- D’souza Shegokar R, AA, Shegokar R. Polyethylene glycol (PEG): a versatile polymer for pharmaceutical applications. Expert Opin Drug Deliv. 2016;13:1257–1275. doi: 10.1080/17425247.2016.1182485. [DOI] [PubMed] [Google Scholar]
- Davidson RL, O’Malley KA, Wheeler TB. Polyethylene glycol-induced mammalian cell hybridization: effect of polyethylene glycol molecular weight and concentration. Somatic Cell Genet. 1976;2:271–280. doi: 10.1007/BF01538965. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Yuan Y, Roberts RM. Pluripotent stem cells from domesticated mammals. Annu Rev Anim Biosci. 2016;4:223–253. doi: 10.1146/annurev-animal-021815-111202. [DOI] [PubMed] [Google Scholar]
- Imai H, Kusakabe KT, Kiso Y, et al. Induction of pluripotency in mammalian fibroblasts by cell fusion with mouse embryonic stem cells. Biochem Biophys Res Commun. 2020;521:24–30. doi: 10.1016/j.bbrc.2019.10.026. [DOI] [PubMed] [Google Scholar]
- Knaupp AS, Buckberry S, Pflueger J, et al. Transient and permanent reconfiguration of chromatin and transcription factor occupancy drive reprogramming. Cell Stem Cell. 2017;21:834–845.e6. doi: 10.1016/j.stem.2017.11.007. [DOI] [PubMed] [Google Scholar]
- Koh S, Piedrahita JA. From “ES-like” cells to induced pluripotent stem cells: a historical perspective in domestic animals. Theriogenology. 2014;81:103–111. doi: 10.1016/j.theriogenology.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H-W. Polyethylene glycol. Third Edit: Elsevier; 2014. [Google Scholar]
- McBurney MW, Strutt B. Fusion of embryonal carcinoma cells to fibroblast cells, cytoplasts, and karyoplasts. Developmental properties of viable fusion products. Exp Cell Res. 1979;124:171–180. doi: 10.1016/0014-4827(79)90267-2. [DOI] [PubMed] [Google Scholar]
- Miller OJ, Miller Da, Dev VG, et al. Expression of human and suppression of mouse nucleolus organizer activity in mouse-human somatic cell hybrids. Proc Natl Acad Sci USA. 1976;73:4531–4535. doi: 10.1073/pnas.73.12.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak-Imialek M, Kues WA, Rudolph C, et al. Preferential loss of porcine chromosomes in reprogrammed interspecies cell hybrids. Cell Reprogram. 2010;12:55–65. doi: 10.1089/cell.2009.0045. [DOI] [PubMed] [Google Scholar]
- Palermo A, Doyonnas R, Bhutani N, et al. Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional. FASEB J. 2009;23:1431–1440. doi: 10.1096/fj.08-122903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz JH, Mukherjee S, Palermo AT, Blau HM. Reprogramming to a muscle fate by fusion recapitulates differentiation. J Cell Sci. 2009;122:1045–1053. doi: 10.1242/jcs.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho-Martinez I, Izpisua Belmonte JC. Stem cells: surf the waves of reprogramming. Nature. 2013;493:310–311. doi: 10.1038/493310b. [DOI] [PubMed] [Google Scholar]
- Serov OL, Matveeva NM, Khabarova AA (2011) Reprogramming mediated by cell fusion technology. In: International review of cell and molecular biology. pp 155–190 [DOI] [PubMed]
- Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- Sullivan S, Pells S, Hooper M, et al. Nuclear reprogramming of somatic cells by embryonic stem cells is affected by cell cycle stage. Cloning Stem Cells. 2006;8:174–188. doi: 10.1089/clo.2006.8.174. [DOI] [PubMed] [Google Scholar]
- Tada S, Tada T, Lefebvre L, et al. Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, et al. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/S0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Villafranca Locher MC (2018) Fusion of bovine fibroblasts to mouse embryonic stem cells: a model to study nuclear reprogramming. Virginia Polytechnic Institute and State University
- Wong WT, Matrone G, Tian X, et al. Discovery of novel determinants of endothelial lineage using chimeric heterokaryons. Elife. 2017;6:1–20. doi: 10.7554/eLife.23588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Shen MH. Polyethylene glycol-mediated cell fusion. In: Pells S, editor. Nuclear Reprogramming. Methods in Molecular Biology. Totowa: Humana Press; 2006. pp. 59–66. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Slukvin II, Thomson JA. Human Embryonic Stem Cells Reprogram Myeloid Precursors Following Cell-Cell Fusion. Stem Cells. 2006;24:168–176. doi: 10.1634/stemcells.2005-0292. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik M, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang F, Pomerantz JH, Sen G, et al. Active tissue-specific DNA demethylation conferred by somatic cell nuclei in stable heterokaryons. Proc Natl Acad Sci. 2007;104:4395–4400. doi: 10.1073/pnas.0700181104. [DOI] [PMC free article] [PubMed] [Google Scholar]