Abstract
Cell-free protein synthesis has been developed as a critical platform in synthetic biology. Unlike the cell-based synthesis system, cell-free system activates transcriptional and translational mechanisms in vitro, and can control protein synthesis by artificially adding components or chemicals. However, the control method puts forward higher requirements in terms of accurate and non-toxic control, which cannot be achieved by chemical substances. For cell-free system, physical signal is a kind of ideal spatiotemporal control approach to replace chemical substances, realizing high accuracy with little side effect. Here we review the methods of using physical signals to control gene expression in cell-free systems, including studies based on light, temperature, electric field, and magnetic force. The transfer of these switches into cell-free system further expands the flexibility and controllability of the system, thus further expanding the application capability of cell-free systems. Finally, existing problems such as signal source and signal transmission are discussed, and future applications in pharmaceutical production, delivery and industrial production are further looked into.
Keywords: Cell-free system, Physical stimuli, Spatiotemporal control, Synthetic biology
Abbreviations: CFPS, Cell-free Protein Synthesis; TCSs, Two-component Systems; S-DM-Azo, 2,6-dimethyl-4-(methylthio) zobenzene-4′-carobxylic acid; RNATs, RNA thermometers; DEP, Dielectrophoresis; DMF, Digital Microfluidic
1. Introduction
Cell-free protein synthesis (CFPS) has developed rapidly in recent years [1]. CFPS activates biological machinery without the use of living cells, allowing direct control of transcription, translation and metabolism in an open environment [2]. It eliminates membrane closure [3,4], and is not restricted by most intracellular metabolic pathways, providing simpler and faster engineering solutions to design with extreme freedom than cell system. In CFPS, by adding specific substances, the biosynthetic reaction is initiated and completes within a few hours [5], offering rapid iterative “design-build-test” cycles [6]. Given the above superiorities, CFPS has been widely used in protein synthesis, including biodrug [7], membrane protein [8], unnatural protein [9,10], biological diagnosis [[11], [12], [13]], genetic circuit design [14,15], and high-throughput analysis [16]. However, different applications have specific requirements in time and space, and the transcription and translation process of the system needs precise initiation and control to better realize these applications.
Therefore, one of the major objectives and challenges for cell-free systems is appropriate spatial and temporal control of the transcription or translation. Openness is a unique characteristic of cell-free systems. The addition of inorganic ions and organics at a specific time can flexibly control the transcriptional and translation process in a cell-free system. In recent years, to address these challenges, researchers have proposed a spatiotemporal control method based on physical elements as stimulus signals, including light [4,5,17], temperature [18], electric field [19] and magnetic force [20] in CFPS. Physical signals with low toxicity and side effects are better than chemical substances at switching genes on and off [21]. More importantly, the physical signal can be flexibly adjusted in CFPS, so as to achieve immediate and local signal transmission and activation, and the rate will not be limited by other metabolic paths in vitro. The introduction of the physical signal switch into CFPS increases the potential of the system in health care, industrial production, synthetic biology education, and eventually achieving creating intelligent synthetic life.
Here, we mainly review the progress on the control of gene transcription and translation in CFPS by using physical signals with two types of classification, including direct response and indirect response (Fig. 1). Direct response involves directly changing the structure of the regulatory elements, while indirect response involves altering the position of the elements. In addition to summarizing the control principles (Table 1) and technical characteristics (Table 2) of each control method, the application fields of the system also prospect, the possible defects are discussed, and the future research direction is further proposed.
Fig. 1.
Schematic illustration of the gene expression control systems in CFPS. Strategies shown in the top half part are based on the principle of direct response regulated by light and temperature. Strategies shown in the bottom half part are based on the principle of indirect response regulated by electric field and magnetic force.
Table 1.
Summary of the gene expression control systems in CFPS.
| Sorting | Physical signal | Key elements | Mechanism | Induction Multiple | Ref. |
|---|---|---|---|---|---|
| Direct response | Light | Two-component system: YF1/FixJ | 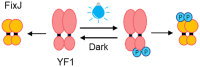 |
6 | [5] |
| One-component system: EL222 | 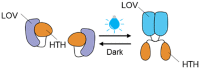 |
>10 | [17] | ||
| Azobenzene-tethered photoresponsive T7 promoter |  |
7.1 | [30] | ||
| Temperature | RNA thermometer |  |
11 | [18] | |
| Indirect response | Electric field | DNA compartment on a chip |  |
– | [19] |
| DMF board |  |
– | [42] | ||
| Magnetic force | Magnetic beads coated with DNA |  |
– | [20] |
Table 2.
Summary of technical details in different CFPS system. Citations indicate where more information about the systems in the context of cell-free can be found.
| Physical signal | Key elements | Cell-free system | Reaction format | Volume | Pros | Cons | Ref. |
|---|---|---|---|---|---|---|---|
| Light | YF1/FixJ | E. coli extract | Batch; One compartment |
20 μL | Abundant components responding to different wavelengths | Slow signal relay and reversal rates; Poor portability |
[5] |
| EL222 | E. coli extract | Batch; One compartment |
25 μL | Good portability; Fast response |
Limited response wavelength | [17] | |
| Azobenzene-tethered promoter | PURE | Batch; One compartment |
– | Avoiding complex protein interactions | Tedious preparation process | [30] | |
| Temperature | RNAT | PURE | Batch; One compartment |
– | Flexible and varied means of temperature control | Low expression | [18] |
| Electric field | DNA brush on a chip | PURE | Batch; Fixed in compartment on a chip |
Compartment: R = 35 μm; H = 3 μm |
Novel manipulation of cell-free expression | Reduced component performance by electrothermal effect | [19] |
| DMF board | E. coli extract | Batch; Multiple droplet compartments |
20 μL | Remoted control of cell-free expression | Limited control scale | [42] | |
| Magnetic force | Magnetic beads coated with DNAs | E. coli extract | Batch; One compartment |
24 μL | Simple control method and spatiotemporal principle | The attachment effectiveness of DNA on magnetic beads | [20] |
2. Direct response - control of elements
Direct method is to change the state of the corresponding regulatory element by physical stimuli, which may be allosteric, phosphorylated, or specifically altered in the structure of the DNA. Specific elements that respond to physical stimuli can bind to gene promoters to determine whether genes are expressed or not. At present, two physical stimuli, light and temperature, have been well developed to respond directly to genetic elements, on the basis of which the applications of these systems in cell-free systems have been achieved (Table 1).
2.1. Control by light
Light possesses many advantages as an ideal genetic control switch, including fast input speed, good spatiotemporal conversion, low toxicity, and low cost of light source device. Light-controlled gene synthesis belongs to the field of optogenetics technology, which uses light to remotely control modified genes. At present, light control systems are mainly based on two-component systems (TCSs) [22], one-component systems [23] and photoreceptors [24]. Their introduction into CFPS could enrich the optogenetics research means, provide better guidance for the design of light control elements, and provide guidance for studying the interaction between proteins.
The two-component system (TCSs) depends on the phosphorylation between bicomponent proteins under different light conditions, ranging from ultraviolet light to near-infrared light, and light signal controls gene expression under the matched promoter [25]. Zhang et al. constructed two plasmids named pDark and pLight using TCSs YF1/FixJ that responds to blue light, to realize cell-free light-controlling gene expression with Escherichia coli extract under darkness and blue light in the batch, respectively [5]. This work successfully transplanted the TCSs into CFPS and achieved light control, further demonstrating its potential in education, healthcare and artificial cell construction. However, the control signal studied so far is limited to blue light, whose phototoxicity might destroy the expressed proteins, and the subsequent in vivo application of artificial cells constructed by CFPS is limited. To address these challenges, an alternative solution is to use TCSs with longer wavelengths, such as CcaS/CcaR responding to green light [26], Cph8/OmpR responding to red light [27], and BphP1/PpsR2 responding to near-infrared light [28]. It also could achieve flexible cooperative regulation of CFPS using different wavelengths of light, to enrich the applications of cell-free light-controlled system. However, the TCSs are intrinsically complex, relatively slow in signal relay and reversal, and less portable. All of these are the directions for further efforts and attempts to construct cell-free optogenetic systems in the future.
To avoid the possible drawbacks brought by TCSs, the system that only requires a single element to achieve control was developed, the so-called one-component system. Jayaraman et al. used EL222 protein-based blue light-inducible promoter to construct the light-controlled system in CFPS with E. coli extract [17]. EL222 protein forms homologous dimers when irradiated by blue light, and the dimers would bind to the PBind promoter, recruiting relevant RNA polymerase to the promoter region to initiate transcription [29]. This work proved the possibility of dynamic control of target gene expression, but this light control system is not currently being developed for engineering applications. However, in the one-component system, the components that can be selected to respond to long wavelengths are limited compared with TCSs, which may limit its development for CFPS.
In addition to the above light-controlled methods by directly interacting macromolecular photosensitive proteins with promoters, organic small molecular structures that respond to specific wavelengths of light are also used to control the cell-free expression. Kamiya et al. tethered 2,6-dimethyl-4-(methylthio) azobenzene-4′-carobxylic acid (S-DM-Azo) units to the T7 promoter region and achieved photoswitching of gene expression using visible light in CFPS with PURE system [30]. In this system, azobenzene can induce reversible formation and dissociation of the DNA duplex upon light irradiation [31]. The system avoids complex protein-promoter interactions, but the sample preparation procedure was laborious, and heating to above the Tm of the template was required to induce photoisomerization, limiting its practical applications. In addition to azobenzene, the photoswitches commonly used also include stilbenes [32], hemithioindigos [33], spiropyrans [34], diarylethenes [35], and fulgides [36]. By modularizing these conversion units into DNA strands, it is also possible to reversibly control cell-free gene transcription [37].
At present, cell-free optogenetic system is the main means of direct response, but there are still some challenges to be solved for its development. The establishment of the system requires that the optically responsive elements originally existing in vivo should be transplanted into the cell-free system solution. To overcome the differences between in vivo and in vitro and improve the efficiency, the prototype design should be carried out, and the key influencing factors of the system should be explored. One important problem of cell-free optogenetic system is that light can be refracted in the solution. The effective transmission of light in solution is also the direction of future research.
2.2. Control by temperature
Temperature is another physical signal with high flexibility and stability. Compared with the light control, there are more methods to control and change the temperature of the system, including external triggers such as focused ultrasound, infrared light, and magnetic particle hyperthermia [38]. In general, there are heat-shock protein promoters [39], thermo-sensitive repressor [40], and RNA thermometers (RNATs) that could induce the transcription process [41]. Changes in temperature alter the structure of these elements, thereby altering the process of gene transcription. It is of great significance for the intelligent creation of artificial life systems to give cell-free systems with the ability to respond to temperature.
Though there have been various ways to achieve temperature control in cells, only RNATs have been transplanted into cell-free systems in previous studies. RNAT loop unfolds when the temperature exceeds a defined threshold, to release the RBS for subsequent gene expression. Jia et al. designed three different threshold temperature RNAT switches that can be activated at 35 °C, 37 °C, or 40 °C in cell-free PURE system [18]. This study further used this system to create a temperature-sensitive protocell model for potential drug delivery applications. However, the expression of a high threshold switch under thermal activation is relatively low, which may be related to the stability of the secondary structure in the system.
From a technical point of view, heat-shock promoters and heat repressor proteins are also feasible means of abundant cell-free temperature control in the future. Compared with RNA thermometers, the interactions between promoters and related proteins are more complex and may be associated with metabolic pathways in the body, making transplantation more difficult. Another idea is about miniaturization, which has a great prospect in the application fields such as therapy and medicine. Currently, magnetic nanoparticles are used to be coated with microcapsules to generate heat under the action of alternating magnetic field, so as to achieve the purpose of heat induction.
3. Indirect response - control of contact
Indirect methods mainly use external physical signals to control the location of elements involved in transcription and translation, thus controlling the process of gene expression. This control mode does not change the activity and state of biological components, but is more like a button that controls the contact between elements and DNA. The indirect methods are mainly achieved via electric field and magnetic force (Table 1), while it is difficult for the two signals to directly control gene expression mainly due to the insufficient research on the related elements.
3.1. Control by electric field
Compared with the precise and complex control of genetic elements by light and temperature, electric field control is simpler and more flexible. The biomolecules under a nonuniform E-field could be trapped and polarized using dielectrophoresis (DEP). The other way is to turn on gene expression by manipulating the droplet's motion directly with the electric field. It is also worth noting that, for the cell-free system, the manipulation using electric field can be carried out without considering the survival and growth of cells.
One way that the electric field controls cell-free gene expression is to use DEP technology. Efrat et al. used DEP to demonstrate the electric field on/off switch response of gene expression in the PURE CFPS system on a chip [19]. Various forms of DEP can be applied to trap, manipulate and separate biomolecules to controlling the contact between the DNA brush attached to the surface in the trap and the elements, such as ribosomes, RNA polymerases, newborn RNAs and proteins, so that controlling protein synthesis. Based on the electronic control system, the biochip was constructed and demonstrated protein synthesis oscillations successfully, which further improved the potential of electric control. However, the electrothermal effect may occur when the electric field interacts with the solution system, which affects the performance of the components in the system. In the future, by combining the design principles of integrated circuits with the powerful information processing capability of biological systems, the capability of biochips to process biological information can be expanded. At the same time, further development of genetic elements that can accurately respond to electric fields would enrich the means of electric field control.
An alternative approach to controlling gene expression using electric fields takes a more macroscopic approach. There are also studies achieved controlling gene expression into proteins using digital microfluidic (DMF), which can directly manipulate single droplets. Liu et al. used real-time remote-controlled DMF technology to start cell-free synthetic reaction successfully [42]. By controlling the movement and mixing of the drops containing different components through real-time DMF, cell-free protein expression reaction started on the board. This work enables the automation of the preparation of cell-free system solutions, which can save time and manpower costs. However, limited by the number of plate electrode arrays, only the motion of small droplets can be controlled. Moreover, this control plate can only initiate the gene expression by electric fields, while it cannot be stopped without the addition of an exogenous transcriptional inhibitor, because the components in the droplet are sufficiently mixed.
Electric-field regulation is usually used to regulate complex neural signaling pathways in vivo, mainly to control the expression of some factors and enzymes [43], which is difficult to be transplanted to in vitro system. At present, the method of electric-field control in CFPS is relatively simple. In addition to controlling the location of key components, the combination of electric field and materials science improved the accuracy of spatiotemporal control, such as hydrogels that release substances in response to electric field stimuli [44].
3.2. Control by magnetic force
Magnetic force is a noteworthy control signal. Magnetic force has a stronger ability to penetrate tissues than light signals, and it does not cause global effects or local thermal damage like temperature signals, nor does it need to consider the potential harm of electric current to life activities. Although the regulation of magnetic field on life activities has been studied, the regulatory elements for gene expression have not been developed, so magnetic field control is still indirect.
The magnetic force controlling cell-free gene expression mainly relies on nanomagnetic beads. Finkler et al. developed a cell-free magnetic gene expression control system based on E. coli extract using the magnetic beads connected to target DNA [20]. Specifically, the DNA templates bound to magnetic beads can accumulate under magnetic control, thus stopping elements in the solution system from participating in DNA transcription and translation. This method can precisely regulate gene expression only through the spatiotemporal separation of nucleic acids. The magnetic control system is simple in principle and can be applied to microfluidics or non-regular amino acid embedded proteins. However, this control method requires attaching the target DNA to the nanoparticles, which cannot guarantee that each particle will adhere effectively.
4. Conclusion and perspective
At present, the research on cell-free gene expression by the direct response realized in CFPS is mainly based on the light and temperature signals as switches. The signal response circuit originally existed in the cell is transplanted to the cell-free system, and the protein expression was successfully induced and controlled in vitro by using two kinds of physical signals, light and temperature. The direct response method has been preliminarily applied to simple dynamic demonstration, control of pharmaceutical protein synthesis and artificial cells, showing certain application potential. The indirect response is a simple control method that can bypass the complex biological metabolic interaction network to turn on and off the biological response of gene expression. The ability of electric and magnetic forces to control the elements allows them to respond indirectly, and the researchers have built systems for controlling gene expression that is sensitive and fast. Such systems have shown potential for mass control of synthesis and non-natural amino acids embedded in proteins, but have not been further explored for application.
The physical signal-controlling system has been developed with certain control ability, but it still needs to be further improved. For a system that responds directly, overcoming differences between in vivo and in vitro can improve the control effect of components, so it is necessary to screen out key influencing factors to reduce such differences. And rapid prototyping and high-throughput screening are also advantaging cell-free systems. Besides, direct methods require several hours to respond, and the development of systems with shorter response time will greatly improve its application ability. For systems that respond indirectly, the flexibility of the control system can be broadened by combining the control system with new materials. For example, the electro-responsive hydrogels are currently being carried out in drug delivery, and the combination of material and elements is worth considering.
In addition, this system has great application potential in both engineering manufacturing and medical health. CFPS controlled by physical signals has not been put into engineering manufacturing practice. Engineering proteins with complex structures put forward higher requirements on the synthesis ability, so it is necessary to further improve the ability to modify the high-level structure of proteins in cell-free systems. In recent years, the construction of artificial cells with cell-free system has increased. Studies have used light and temperature signals to control protein expression in artificial cells, preliminarily proving that protein synthesis can be controlled at the cell scale. Since artificial cells may be responsible for the production and delivery of drugs in vivo, the transmission of physical signals in vivo and the activity of protein production require further study.
In general, the development of controlled protein synthesis in cell-free systems will greatly promote the development of targeted drug synthesis and delivery technologies, and the flexible control of protein production in vitro will further promote the progress in the field of industrial protein manufacturing.
Ethics approval
This article does not contain any studies with human participants or experimental animals performed by any of the authors.
CRediT authorship contribution statement
Junzhu Yang: Conceptualization, Validation, Writing - original draft. Yuan Lu: Writing - review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Key R&D Program of China (Grant No. 2018YFA0901700), the National Natural Science Foundation of China (Grant No. 21878173 and 21706144) and the Beijing Natural Science Foundation (Grant No. 2192023).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Silverman A.D., Karim A.S., Jewett M.C. Cell-free gene expression: an expanded repertoire of applications. Nat Rev Genet. 2020;21:151–170. doi: 10.1038/s41576-019-0186-3. [DOI] [PubMed] [Google Scholar]
- 2.Lu Y. Cell-free synthetic biology: engineering in an open world. Synth Syst Biotechnol. 2017;2:23–27. doi: 10.1016/j.synbio.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yanagisawa T., Takahashi M., Mukai T., Sato S., Wakamori M., Shirouzu M. Multiple site-specific installations of Nε-monomethyl-L- lysine into histone proteins by cell-based and cell-free protein synthesis. Chembiochem. 2014;15:1830–1838. doi: 10.1002/cbic.201402291. [DOI] [PubMed] [Google Scholar]
- 4.Karig D.K., Iyer S., Simpson M.L., Doktycz M.J. Expression optimization and synthetic gene networks in cell-free systems. Nucleic Acids Res. 2012;40:3763–3774. doi: 10.1093/nar/gkr1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang P., Yang J., Cho E., Lu Y. Bringing light into cell-free expression. ACS Synth Biol. 2020;9:2144–2153. doi: 10.1021/acssynbio.0c00211. [DOI] [PubMed] [Google Scholar]
- 6.Moore S.J., MacDonald J.T., Freemont P.S. Cell-free synthetic biology for in vitro prototype engineering. Biochem Soc Trans. 2017;45:785–791. doi: 10.1042/BST20170011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pardee K., Slomovic S., Nguyen P.Q., Lee J.W., Donghia N., Burrill D. Portable, on-demand biomolecular manufacturing. Cell. 2016;167:248–259. doi: 10.1016/j.cell.2016.09.013. e12. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs M.L., Boyd M.A., Kamat N.P. Diblock copolymers enhance folding of a mechanosensitive membrane protein during cell-free expression. Proc Natl Acad Sci U S A. 2019;116:4031–4036. doi: 10.1073/pnas.1814775116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin R.W., Des Soye B.J., Kwon Y.C., Kay J., Davis R.G., Thomas P.M. Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat Commun. 2018;9:1–9. doi: 10.1038/s41467-018-03469-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao W., Bu N., Lu Y. Efficient incorporation of unnatural amino acids into proteins with a robust cell-free system. Methods Protoc. 2019;2:16. doi: 10.3390/mps2010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen K.Y., Cameron L., Chappell J., Jensen K., Bell D.J., Kelwick R. A cell-free biosensor for detecting quorum sensing molecules in P. Aeruginosa-infected respiratory samples. ACS Synth Biol. 2017;6:2293–2301. doi: 10.1021/acssynbio.7b00219. [DOI] [PubMed] [Google Scholar]
- 12.Salehi A.S.M., Shakalli Tang M.J., Smith M.T., Hunt J.M., Law R.A., Wood D.W. Cell-free protein synthesis approach to biosensing hTRβ-specific endocrine disruptors. Anal Chem. 2017;89:3395–3401. doi: 10.1021/acs.analchem.6b04034. [DOI] [PubMed] [Google Scholar]
- 13.Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 14.Pardee K., Green A.A., Ferrante T., Cameron D.E., Daleykeyser A., Yin P. Paper-based synthetic gene networks. Cell. 2014;159:940–954. doi: 10.1016/j.cell.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi M.K., Chappell J., Hayes C.A., Sun Z.Z., Kim J., Singhal V. Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription-translation (TX-TL) systems. ACS Synth Biol. 2015;4:503–515. doi: 10.1021/sb400206c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha A., Acharya B.N., Priya R., Tripathi N.K., Shrivastava A., Rao M.K. Development of nsP2 protease based cell free high throughput screening assay for evaluation of inhibitors against emerging Chikungunya virus. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-018-29024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaraman P., Yeoh J.W., Jayaraman S., Teh A.Y., Zhang J., Poh C.L. Cell-free optogenetic gene expression system. ACS Synth Biol. 2018;7:986–994. doi: 10.1021/acssynbio.7b00422. [DOI] [PubMed] [Google Scholar]
- 18.Jia H., Heymann M., Härtel T., Kai L., Schwille P. Temperature-sensitive protein expression in protocells. Chem Commun. 2019;55:6421–6424. doi: 10.1039/c9cc02734c. [DOI] [PubMed] [Google Scholar]
- 19.Efrat Y., Tayar A.M., Daube S.S., Levy M., Bar-Ziv R.H. Electric-field manipulation of a compartmentalized cell-free gene expression reaction. ACS Synth Biol. 2018;7:1829–1833. doi: 10.1021/acssynbio.8b00160. [DOI] [PubMed] [Google Scholar]
- 20.Finkler M., Ott A. Bead-based assay for spatiotemporal gene expression control in cell-free transcription–translation systems. Biotechniques. 2019;66:29–33. doi: 10.2144/btn-2018-0097. [DOI] [PubMed] [Google Scholar]
- 21.Lu Y., Sun W., Gu Z. Stimuli-responsive nanomaterials for therapeutic protein delivery. J Contr Release. 2014;194:1–19. doi: 10.1016/j.jconrel.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.West A.H., Stock A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/S0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z., Zhang J., Jin J., Geng Z., Qi Q., Liang Q. Programming bacteria with light-sensors and applications in synthetic biology. Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.02692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kong S.G., Okajima K. Diverse photoreceptors and light responses in plants. J Plant Res. 2016;129:111–114. doi: 10.1007/s10265-016-0792-5. [DOI] [PubMed] [Google Scholar]
- 25.Möglich A., Ayers R.A., Moffat K. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol. 2009;385:1433–1444. doi: 10.1016/j.jmb.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose Y., Shimada T., Narikawa R., Katayama M., Ikeuchi M. Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci U S A. 2008;105:9528–9533. doi: 10.1073/pnas.0801826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tabor J.J., Levskaya A., Voigt C.A. Multichromatic control of gene expression in escherichia coli. J Mol Biol. 2011;405:315–324. doi: 10.1016/j.jmb.2010.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong N.T., Olson E.J., Tabor J.J. Engineering an E. coli near-infrared light sensor. ACS Synth Biol. 2018;7:240–248. doi: 10.1021/acssynbio.7b00289. [DOI] [PubMed] [Google Scholar]
- 29.Motta-Mena L.B., Reade A., Mallory M.J., Glantz S., Weiner O.D., Lynch K.W. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamiya Y., Takagi T., Ooi H., Ito H., Liang X., Asanuma H. Synthetic gene involving azobenzene-tethered t7 promoter for the photocontrol of gene expression by visible light. ACS Synth Biol. 2015;4:365–370. doi: 10.1021/sb5001092. [DOI] [PubMed] [Google Scholar]
- 31.Liang X., Mochizuki T., Asanuma H. A supra-photoswitch involving sandwiched DNA base Pairs and azobenzenes for light-driven nanostructures and nanodevices. Small. 2009;5:1761–1768. doi: 10.1002/smll.200900223. [DOI] [PubMed] [Google Scholar]
- 32.Waldeck D.H. Photoisomerization dynamics of stilbenes in polar solvents. J Mol Liq. 1993;57:127–148. doi: 10.1016/0167-7322(93)80051-V. [DOI] [Google Scholar]
- 33.Lougheed T., Borisenko V., Hennig T., Rück-Braun K., Woolley G.A. Photomodulation of ionic current through hemithioindigo-modified gramicidin channels. Org Biomol Chem. 2004;2:2798–2801. doi: 10.1039/B408485C. [DOI] [PubMed] [Google Scholar]
- 34.Minkin V.I. Photoswitchable molecular systems based on spiropyrans and spirooxazines. In: Feringa B.L., Browne W.R., editors. Molecular switches. second ed. Wiley-VCH; Weinheim: 2011. pp. 37–80. [DOI] [Google Scholar]
- 35.Irie M. Diarylethenes for memories and switches. Chem Rev. 2000;100:1685–1716. doi: 10.1021/cr980069d. [DOI] [PubMed] [Google Scholar]
- 36.Yokoyama Y. Fulgides for memories and switches. Chem Rev. 2000;100:1717–1740. doi: 10.1021/cr980070c. [DOI] [PubMed] [Google Scholar]
- 37.Szymański W., Beierle J.M., Kistemaker H.A.V., Velema W.A., Feringa B.L. Reversible photocontrol of biological systems by the incorporation of molecular photoswitches. Chem Rev. 2013;113:6114–6178. doi: 10.1021/cr300179f. [DOI] [PubMed] [Google Scholar]
- 38.Piraner D.I., Abedi M.H., Moser B.A., Lee-Gosselin A., Shapiro M.G. Tunable thermal bioswitches for in vivo control of microbial therapeutics. Food, Pharm Bioeng Div. 2017;2:695–702. doi: 10.1038/nchembio.2233. 2017 Core Program Area 2017 AIChE Annu Meet. [DOI] [PubMed] [Google Scholar]
- 39.Rome C., Couillaud F., Moonen C.T.W. Spatial and temporal control of expression of therapeutic genes using heat shock protein promoters. Methods. 2005;35:188–198. doi: 10.1016/j.ymeth.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Valdez-Cruz N.A., Caspeta L., Pérez N.O., Ramírez O.T., Trujillo-Roldán M.A. Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters. Microb Cell Factories. 2010;9:1–16. doi: 10.1186/1475-2859-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sen S., Apurva D., Satija R., Siegal D., Murray R.M. Design of a toolbox of RNA thermometers. ACS Synth Biol. 2017;6:1461–1470. doi: 10.1021/acssynbio.6b00301. [DOI] [PubMed] [Google Scholar]
- 42.Liu D., Yang Z., Zhang L., Wei M., Lu Y. Cell-free biology using remote-controlled digital microfluidics for individual droplet control. RSC Adv. 2020;10:26972–26981. doi: 10.1039/D0RA04588H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen E.B., Wishner J., Slowinska K. The effect of pulsed electric field on expression of ECM proteins: collagen, elastin, and MMP1 in human dermal fibroblasts. J Electroanal Chem. 2018;812:265–272. doi: 10.1016/j.jelechem.2018.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomer R., Dimitrijevic D., Florence A.T. Electrically controlled release of macromolecules from cross-linked hyaluronic acid hydrogels. J Contr Release. 1995;33:405–413. doi: 10.1016/0168-3659(94)00115-B. [DOI] [Google Scholar]



