Abstract
It is estimated that the rate of epigenetic changes may be orders of magnitude higher than that of genetic changes and that purely epigenetic mechanisms may explain why cancers arise with few or no recurrent mutations. However, supporting evidence remains limited, partly due to the cost of experimentally studying genome-wide epigenetic dysregulation. Since genome modification enzymes are recruited by long noncoding RNAs (lncRNAs) to specific genomic sites, analyzing differentially expressed genes and differentially methylated regions (DMRs) at the DNA binding sites of differentially expressed lncRNAs is important for uncovering epigenetic dysregulation. We performed RNA-seq and MeDIP-seq on a set of colorectal cancer (CRC) and normal colon samples and developed an analysis pipeline for combined analyses of gene expression, DNA methylation, and lncRNA/DNA binding. The genes identified in our data and important for CRC agree with widely reported findings. We found that aberrantly transcribed noncoding transcripts may epigenetically dysregulate genes, that correlated gene expression is significantly determined by epigenetic dysregulation, that differentially expressed noncoding transcripts and their epigenetic targets form distinct modules in different cancer cells, and that many hub lncRNAs in these modules are primate-specific. These results suggest that lncRNA-mediated epigenetic dysregulation greatly determines aberrant gene expression and that epigenetic dysregulation is highly species-specific. The analysis pipeline can effectively unveil cancer- and cell-specific modules of epigenetic dysregulation, and such modules may provide novel clues for identifying diagnostic, therapeutic, and prognostic targets for epigenetic dysregulation.
Keywords: Colorectal cancers, Differential gene expression, Epigenetic regulation, LncRNA/DNA binding, RNA-seq
1. Background
Mutations in genes and regulatory sequences in cancer cells accumulate as cancer grows. For years, researchers have hypothesized that mutations cause cancers and drive cancer evolution and have tried to identify them in different cancers [1]. This hypothesis, however, is not supported by many cancer genome sequencing studies. One study reported that only approximately 36% of mutations are expressed in primary triple-negative breast cancers [2]. Feinberg and colleagues recently estimated that 99.9% of mutational changes in cancers are not driver mutations [3]. Meanwhile, abundant long noncoding RNAs (lncRNAs) identified in mammalian genomes indicate that aberrant gene expression can be caused by lncRNA-mediated epigenetic dysregulation. Up to 40% of differentially expressed genes between humans and nonhuman primates may result from interspecies epigenetic differences [4], genomic regions enriched in noncoding RNAs and cis-regulatory elements host the majority of disease-related genetic variations [5], and growing evidence indicates that lncRNAs decisively regulate gene expression [6], [7]. These findings call for a systematic analysis of epigenetic dysregulation based on newly generated and publicly available cancer sequencing data.
Many lncRNAs can bind to DNA sequences and recruit DNA and histone modification enzymes to their binding sites [8], [9], [10]. Their genomic binding sites therefore determine their epigenetic target genes and transcripts [11], [12]. Since lncRNA sequences and DNA duplexes form DNA:RNA triplexes following noncanonical base-pairing rules [13], lncRNAs’ DNA binding motifs (also called triplex-forming oligonucleotides, TFOs) and genomic binding sites (also called triplex-targeting sites, TTSs) are predictable [14]. Thus, combining the analysis of gene expression, DNA methylation, and lncRNA/DNA binding is critical to uncovering the key players and extent of lncRNA-mediated epigenetic dysregulation in cancers.
Colorectal cancers (CRCs) occur widely in both developed and developing countries and cause many deaths annually [15]. Mutations and critical differentially expressed genes have been examined [16], [17], [18], but how and to what extent epigenetic dysregulation contributes to tumorigenesis and to cancer subtypes remains unclear. For example, a recent study by Merry and colleagues identified a lncRNA that regulated gene expression and DNA methylation in CRC but did not unveil how the lncRNA recognized its target genes [19]. In this study, we performed RNA sequencing (RNA-seq) and methylated DNA immunoprecipitation sequencing (MeDIP-seq) on 12 CRC samples and 3 normal colon tissue samples, analyzed aberrantly transcribed and differentially expressed coding and noncoding transcripts, analyzed differentially methylated regions (DMRs), and for the first time, explored the association between the aberrant transcription/differential expression of noncoding transcripts and the aberrant transcription/differential expression of protein-coding transcripts. We also examined several single-cell RNA-seq (scRNA-seq) datasets of CRC and found that the differential expression of lncRNAs and their potential epigenetic targets were highly correlated. Our analysis pipeline consists of widely used programs (Fig. 1). Our findings support each other, and well-known CRC-related genes were found to be dysregulated in our samples. The obtained results suggest that some aberrantly transcribed noncoding transcripts may have epigenetic regulatory functions, that epigenetic dysregulation greatly determines the correlated differential expression of genes, and that distinct epigenetic regulatory modules exist in different cancer cells. Since lncRNA expression and epigenetic regulation are highly tissue-specific, a combined genome-wide analysis of gene expression and lncRNA/DNA binding alone based on abundant publicly available RNA-seq data makes much sense.
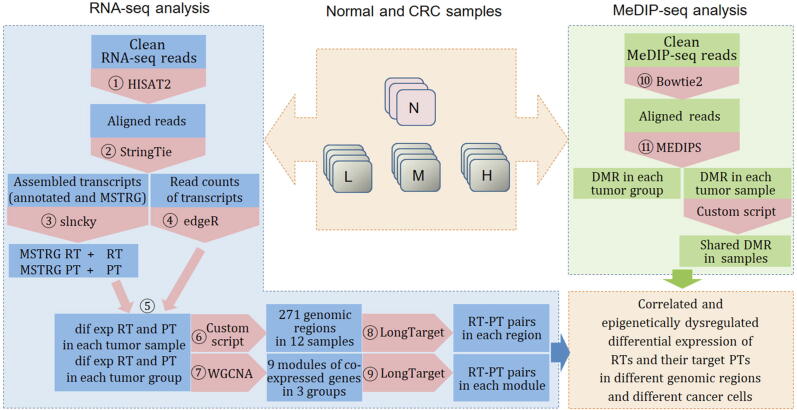
Fig. 1.
The data analysis pipeline. Orange boxes indicate programs; blue and green boxes indicate inputs and outputs. “N”, “L”, “M”, and “H” indicate the normal colon tissue group and low/medium/high differentiation CRC groups. For each sample, we used HISAT2 to align clean RNA-seq reads, used StringTie to assemble aligned reads into transcripts and genes (StringTie uses MSTRG to label unannotated transcripts and genes), used a script in the StringTie package to calculate the read count of each transcript, and used slncky to detect novel long noncoding transcripts from MSTRG transcripts. The combined use of StringTie and slncky identified four kinds of transcripts: MSTRG RT, RT, MSTRG PT, and PT (see abbreviations below). Then, the read counts of the four kinds of transcripts allowed edgeR to identify differentially expressed transcripts in each tumor sample and in each tumor group by performing a 1:3 comparison and a 4:3 comparison against the 3 normal samples. For each differentially expressed RT and MSTRG RT in each tumor sample, a simple script was used to identify the genomic region containing this RT and its nearby differentially expressed PTs/MSTRG PTs. With the differentially expressed and MSTRG-labeled RTs and PTs in each tumor group, WGCNA was used to identify the modules of coexpressed transcripts. A total of 271 genomic regions were identified in 12 tumor samples, and 9 modules were identified in 3 tumor groups. LongTarget was used to predict the TTSs of the differentially expressed and MSTRG-labeled RTs in each of the 271 genomic regions and the TTSs of the differentially expressed and MSTRG-labeled RTs in each module. Meanwhile, Bowtie2 was used to align the clean MeDIP-seq reads in each sample, and MEDIPS was used to identify DMRs in each tumor sample and in each tumor group by performing a 1:3 comparison and a 4:3 comparison against the 3 normal samples. The analyses following this pipeline thus unveil and verify correlated and epigenetically dysregulated gene expression in different genomic regions and in different cancer cells. For convenience, we sometimes use PT/MSTRG PT/RT/MSTRG RT indiscriminately to denote both genes and transcripts (and do not italicize gene names). Abbreviations: RT: lncRNA transcripts; PT: non-lncRNA transcripts (most are protein-coding transcripts); MSTRG RT: unannotated RT; MSTRG PT: unannotated PT; DMRs: differentially methylated regions; TFOs: triplex-forming oligonucleotides; TTSs: triplex-targeting sites; TF: transcription factor. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2. Materials and methods
2.1. Sample collection
Samples were provided by the Department of Pathology of Nanfang Hospital (The First Affiliated Hospital of Southern Medical University). Samples were collected from patients (regardless of age and sex) without any treatment during tumorectomy. Written informed consent to collect tissue samples during tumorectomy was obtained from all patients, and the study was performed in accordance with the Declaration of Helsinki and the Regulations of the Ethical Committee of Nanfang Hospital. Fifteen samples (3 normal and 12 tumor) with high library preparation quality were sequenced. The mean RNA integrity number (RIN) of the samples was 8.275, with a standard deviation (s.d.) of 0.5559, tested on an Agilent 2100 Bioanalyzer. According to the cell differentiation grades, 4 tumor samples (T19A, T50A, T95A, and T85A) were classified as grade 1 (high differentiation), 4 tumor samples (T111A, T132A, T160A, and T162A) as grade 2 (medium differentiation), and 4 tumor samples (T13A, T14A, T18A, and T2C) as grade 3 (low differentiation).
2.2. cDNA library construction and RNA-seq
Total RNA was extracted from each of the samples, rRNAs were removed from the total RNA using the TruSeq PE Cluster Kit, and purified mRNAs were fragmented using fragmentation buffer. Short fragments were used to synthesize first-strand cDNA with the addition of random hexamer primers, and second-strand cDNA was synthesized using buffer, dNTPs, RNase H, and DNA polymerase I. Short double-stranded cDNA fragments were purified using the QIAquick PCR Extraction Kit and ligated with sequencing adapters. DNA fragments ranging from 100 to 500 bp were gel-purified and amplified by PCR. The amplified library was sequenced in paired-end reads on an Illumina HiSeq 2000 instrument. Library preparation and sequencing were performed by BGI Shenzhen (Shenzhen, China).
2.3. MeDIP library construction and MeDIP-seq
First, genomic DNA was extracted and sonicated to 100–500 bp. Second, DNA fragments were repaired to contain a 3′-dA overhang, and adapters were ligated at the ends using the Paired-End DNA Sample Prep Kit (Illumina). Third, DNA fragments were denatured and immunoprecipitated with 5-mC antibody using the Magnetic Methylated DNA Immunoprecipitation Kit (Diagenod), and q-PCR was performed to validate the enrichment efficiency of immunoprecipitation. Fourth, immunoprecipitated DNA fragments ranging from 100 to 500 bp were gel-purified and quantified using an Agilent 2100 Bioanalyzer. Finally, the qualified immunoprecipitated DNA library was sequenced in paired-end reads on an Illumina HiSeq 2000 instrument. Library preparation and sequencing were performed by BGI Shenzhen (Shenzhen, China).
2.4. RNA-seq read filtering, read mapping, and transcript assembly
Clean reads were obtained by using the SOAPnuke program (v1.5) to remove reads with adaptors and reads of low quality [20]. The SOAPnuke parameters for controlling read quality were Q ≤ 10 and 10% ‘N’s (reads with either more than 50% of bases with Q ≤ 10 or >10% 'N's were removed). The mean number of clean reads was 84,802,601 (s.d. = 13,391,825).
We used the HISAT2 package to map reads to the human genome build hg19 and used the hg19 GTF file (version GRCh37.75) from the Ensembl website to improve the mapping quality. The mean map rate and mean unique map rate was 96.67% and 71.05% (s.d. = 1.71% and 3.78%), respectively. Known splice sites and exons were extracted from the hg19 GTF file using hisat2_extract_exons.py and hisat2_extract_splice_sites.py. The hisat2-build program (with options ‘--ss’ and ‘--exon’) was used to index the reference genome hg19 using known splice sites and exons. The clean reads of each sample were mapped to hg19 using the HISAT2 program (v2.0.3, with default parameters) [21].
Upon transcript and gene annotation in hg19, we used StringTie (v1.2.2, with the ‘-G’ option to use the hg19 GTF file as the reference annotation file) [22] to assemble the aligned reads into transcripts in each sample. The assembled transcripts in all 15 samples were merged into a nonredundant set of transcripts using the ‘Transcript merge mode’ of StringTie (with the ‘--merge’ option and other parameters set to default values). To facilitate the use of edgeR to analyze differential expression, we used the prepDE.py program (with default parameters) in the StringTie package to calculate the read count of each transcript directly from the files generated by StringTie.
2.5. MeDIP-seq read filtering and read mapping
The SOAPnuke program (v1.5) was used to remove reads with adaptors and reads of low quality. The parameters for controlling read quality were Q ≤ 20 and 10% ‘N’s (reads with either more than 50% of bases with Q ≤ 20 or >10% ‘N’s were removed). Each sample has 102,040,816 clean reads. The clean MeDIP-seq reads were then mapped to hg19 using the Bowtie2 program (v2.2.5, with the options ‘-sensitive’ and ‘-end-to-end’) [23]. The mean map rate and mean unique map rate was 89.91% and 59.37% (s.d. = 1.31% and s.d. = 2.43%), respectively.
2.6. Identifying annotated and unannotated coding and noncoding transcripts
When assembling the aligned reads into transcripts and genes, StringTie uses MSTRG to label unannotated transcripts and assembles MSTRG transcripts into MSTRG genes. To investigate the epigenetic dysregulation mediated by both differentially expressed and aberrantly transcribed lncRNAs, we analyzed MSTRG RTs (both transcripts and genes). To do so, we used the slncky program (v1.0, with default parameters) [24] to identify novel (unannotated) long noncoding transcripts from other MSTRG transcripts upon gene annotation in the hg19 GTF file (in the study, other kinds of noncoding RNAs were not analyzed and here, annotated and unannotated noncoding transcripts mean those whose length is >200 bp). To examine whether MSTRG transcripts and genes were annotated in hg38, we used the liftOver function in the UCSC Genome Browser to transfer their genomic coordinates between hg19 and hg38.
The combined use of StringTie and slncky enabled us to identify four kinds of transcripts: annotated non-lncRNA transcripts (called PTs, because most of them are protein-coding transcripts), annotated long noncoding transcripts (called RTs, i.e., RNA transcripts), unannotated non-lncRNA transcripts (called MSTRG PTs), and unannotated long noncoding transcripts (called MSTRG RTs).
2.7. Detecting differentially expressed genes
We used prepDE.py (with default parameters) in the StringTie package to calculate the read count of each transcript in each sample (see Section 2.4), and the files of the read count matrix were used as the inputs for the edgeR program (v3.2.1) [25]. We used the exactTest function in edgeR to identify the differentially expressed genes in each tumor sample by comparing the read counts of transcripts in this sample against the read counts of transcripts in the 3 normal samples. We used the generalized linear model function in edgeR to identify differentially expressed genes in each tumor group by comparing the read counts of transcripts in this group against the read counts of transcripts in the 3 normal samples. In both situations, a transcript was assumed to be differentially expressed relative to the 3 normal samples if the criteria of FDR-adjusted p < 0.05 and absolute fold change > 2 (|FC|>2.0) were met.
2.8. Detecting DMRs
Compared with the DNA methylation signals in the 3 normal samples, the DMRs in each tumor group were identified using the MEDIPS program (window size = 300, p.adj = BH, diff.method = edgeR) with the criteria of FDR-adjusted p < 0.1 and |FC|>1.0 [26]. The DMRs in each tumor sample were identified using the same MEDIPS parameters and the same criteria. If some DMRs overlapped with each other in multiple tumor samples, they were merged into one region. A simple script was used to extract the DMR signals in a genomic region in each CRC group (and in each CRC sample) into a specific file (DMR track file, in bed format), which can be uploaded onto the UCSC Genome Browser as a custom track. To display the DMR tracks, display mode was set to dense.
2.9. Detecting coexpressed modules of differentially expressed transcripts
Upon identifying the differentially expressed genes in each of the 3 tumor groups, we used the WGCNA R package (v1.49) to construct coexpression networks and identify modules of highly interconnected genes following the step-by-step network construction and module detection approach [27]. We used 1 and 0 to encode tumor and normal samples, and only one trait (the sample is CRC) was defined in the trait file. We used the pickSoftThreshold function to calculate the scale-free topology fit index R2 for multiple soft-thresholding powers, used the dynamic tree cut to detect modules, and performed module merging to merge modules with highly similar expression profiles. During the process, the parameters in the example of the R tutorial were used (e.g., deepSplit = 2, minClusterSize = minModuleSize = 30, and height cutoff = 0.25). GS (gene significance) and MM (module membership) were computed for every RT and PT in each module. GS and MM are the absolute values of the correlations between the transcript and CRC (the trait) and between the transcript and the module eigengene, respectively. With the settings GS > 0.7 (with p < 0.05) and MM > 0.7 (with p < 0.05), WGCNA identified 9 modules of coexpressed differentially expressed genes, including the H1, H2, and H3 modules in the high differentiation group, the M1, M2, and M3 modules in the medium differentiation group, and the L1, L2, and L3 modules in the low differentiation group. Connectivity measures the connection of a gene to all other genes in a module. We used the softConnectivity function to identify the most connected genes in each module.
2.10. Predicting the TTSs of RTs and MSTRG RTs
For each differentially expressed RT/MSTRG RT in each tumor sample, a simple script was used to identify the local genomic region in which this RT/MSTRG RT may play a regulatory role. The genomic region stretches from this RT/MSTRG RT upstream and downstream until either a normally expressed PT or another differentially expressed RT/MSTRG RT was met. If an RT/MSTRG RT was differentially expressed in multiple tumor samples, its local genomic regions in these samples were merged. In 12 tumor samples, 271 regions were identified, and we used LongTarget (v1.0, default parameters) to predict the TTSs of these RTs and MSTRG RTs in each of these regions [14]. This examines the epigenetic dysregulation in specific genomic regions of interest.
For each module of coexpressed genes identified by WGCNA in the 3 CRC groups, we used LongTarget (v1.0, default parameters) to predict the TTSs of RTs and MSTRG RTs in the genomic regions (including the promoter region +3500 bp upstream of the transcription start site) of PTs and MSTRG PTs. This examines the genome-wide epigenetic dysregulation in specific tumor cells.
LongTarget generated three files for each lncRNA/DNA binding prediction. Two class files (class1 and class2, in bedGraph format) describe the TTS distributions, and one sorted file contains the sequences of TFOs, coordinates of TTSs, and the rules of Hoogsteen and reverse Hoogsteen base pairing. We used the BLAT function in the UCSC Genome Browser to search the TFO sequence against the human genome hg19, and the hit perfectly matching the sequence pinpointed the TFO position. The class files and the DMR track files (see Section 2.8) were uploaded onto the UCSC Genome Browser as custom tracks. To display the TTS tracks, display mode was set to full, track height was set to 50, and other parameters were set to their default values.
2.11. Human transcription factors
To analyze the contribution of transcription factors to the dysregulation of gene expression, a list of 1978 human transcription factor genes (13849 transcripts) was obtained from a recent study [28]. We computed the ratio of differentially expressed TF transcripts to total TF transcripts and used LongTarget to predict how many differentially expressed TF transcripts were targets of differentially expressed RTs.
2.12. Analyzing scRNA-seq datasets
To verify our conclusions using scRNA-seq data, we downloaded multiple datasets of CRC tissue cells, CRC cell lines, and normal colon cells generated by a recent study [29]. We determined differentially expressed genes with |logFC|>1 and Q-value < 0.1, used the R program pheatmap to draw a heatmap of the lncRNA gene expression in different cells, and used LongTarget to predict the lncRNA TTSs in protein-coding genes in the two groups of data. The first group consisted of differentially expressed lncRNA genes and protein-coding genes in tumor cells, and the second group consisted of 90 highly expressed lncRNAs and all protein-coding genes in all cells. We also calculated the Spearman correlation between every lncRNA and every protein-coding gene in tumor cells to determine whether the correlation has any relationship with epigenetic regulation.
2.13. Detecting MSTRG RT expression in cell lines
We cultured cells of three cell lines, including FHC (ATCC® CRL-1831™, a normal human colon epithelial cell line), SW480 (ATCC® CCL-228™, a cell line derived from a grade 3–4 colon adenocarcinoma), and HCT116 ( ATCC® CCL-247™, a human colorectal carcinoma cell line ). Total RNA was extracted with Trizol and the concentration and purity were determined by OD260/280 using a Nanodrop (Agilent Technologies, Palo Alto, CA, USA). 1 μg RNA was reverse-transcribed into cDNA according to the manufacturer’s instructions (RR420A, Takara, Guangzhou, China). qRT-PCR was performed according to the manufacturer’s instructions (RR420A, Takara, Guangzhou, China) using a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Real-time quantitative PCR was performed to detect MSTRG.40340.1 and MSTRG.38311.2 mRNA expression in the FHC, SW480, and HCT116 cell lines. The primer sequences were as follows: MSTRG.40340.1, forward, 5′- TGACGTCCGATTAATCTCC-3′; MSTRG.40340.1, reverse, 5′-GTTGTTTGGACAAGCTAACA-3′; MSTRG.38311.2, forward, 5′-GAGGCATTGCTAATCTAGAAG-3′; MSTRG.38311.2, reverse, 5′-CGTAAATCCCCTCCATTATGTG-3′.
2.14. Availability of data and codes
The sequencing data can be downloaded from the NCBI GEO website (https://www.ncbi.nlm.nih.gov/geo) under the accession number GSE109204. LongTarget and MEDIPS results can be downloaded from the authors’ website (http://lncRNA.smu.edu.cn (in the OtherCodeData page)).
3. Results
3.1. Aberrantly transcribed long noncoding transcripts may dysregulate gene expression
To explore genome-wide lncRNA-mediated epigenetic dysregulation in CRC, we performed RNA-seq and MeDIP-seq on 12 CRC samples and 3 normal colon tissue samples. After obtaining RNA-seq data, we identified annotated and unannotated transcripts and differentially expressed transcripts. StringTie uses ‘MSTRG’ to label unannotated genes (e.g., MSTRG.38311) and transcripts (e.g., MSTRG.38311.2). MSTRG transcripts are assumed to be assembly artifacts or transcriptional noise [30], and in most studies, they are unexplored.
We evaluated the coding potential of all MSTRG transcripts using slncky [24] and classified all transcripts into four classes: annotated non-lncRNA transcripts (called PT, because most were protein-coding), annotated lncRNA transcripts (RT), unannotated non-lncRNA transcripts (MSTRG PT), and unannotated lncRNA transcripts (MSTRG RT) (for convenience, we sometimes also use PT/MSTRG PT/RT/MSTRG RT to denote genes) (Fig. 1). Many MSTRG genes (identified by StringTie upon gene annotation in the genome build hg19) overlapped with annotated genes in hg38 (Supplementary Table 1), indicating that some MSTRG transcripts are unannotated normal transcripts. However, some MSTRG transcripts had exons that were obviously different from the annotated exons, indicating that they were aberrantly transcribed. After the identification of differentially expressed transcripts by edgeR [25] and the four classes of transcripts, we examined four kinds of epigenetic dysregulation: RT → PT, RT → MSTRG PT, MSTRG RT → PT, and MSTRG RT → MSTRG PT, by predicting the TTSs of RTs/MSTRG RTs in specific genomic regions and in genes that form coexpression modules with these RTs/MSTRG RTs (see Section 3.3) [27] and by analyzing the DMRs in these genomic regions and genes.
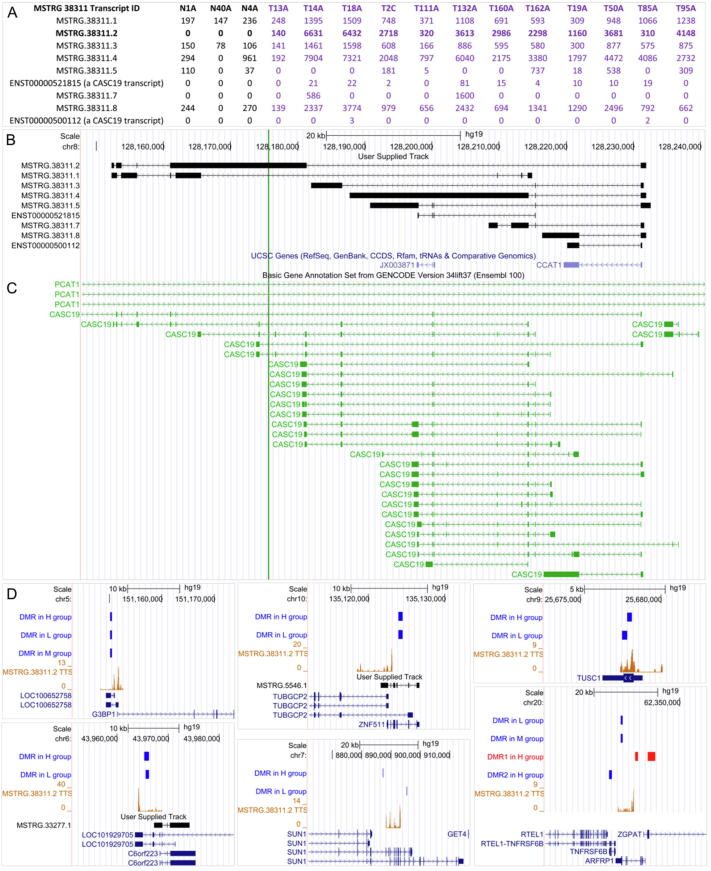
An impressive case of MSTRG genes is MSTRG.38311. This MSTRG gene was detected in 12 CRC samples, overlapped with the CRC-related lncRNA genes CCAT1 (colon cancer-associated transcript 1) and CASC19 (cancer susceptibility 19) [31], [32], [33], and comprised 9 transcripts (2 CASC19 transcripts and 7 MSTRG RTs) (Fig. 2). Specifically, MSTRG.38311.2 was not expressed in normal samples but was highly expressed in tumor samples, and some of its exons were much longer than CASC19 exons. These results indicate that some transcripts of MSTRG.38311 are aberrant transcripts instead of assembly artifacts or transitional noise. To make a further check, we used RT-PCR to detect the expression of MSTRG.38311.2 in one normal colon cell line and two CRC cell lines, and found that it was also highly expressed in the two CRC cell lines (Supplementary Fig. 1). We used LongTarget [14] to predict the TTSs of MSTRG.38311.2 in differentially expressed PTs and MSTRG PTs that were coexpressed with MSTRG.38311.2, and the results indicate that MSTRG.38311.2 has TTSs in many differentially expressed transcripts and MSTRG transcripts. Notably, the TFO of many TTSs was in an aberrant exon, and DMRs were detected in genomic regions of these differentially expressed transcripts and MSTRG transcripts (Fig. 2).
Fig. 2.
The expression levels, transcripts, exons, genomic position, and potential epigenetic targets of MSTRG.38311. (A) Multiple transcripts of MSTRG.38311 are highly expressed only in tumor samples. (B) MSTRG.38311 comprises 2 CASC19 transcripts and 7 MSTRG RTs. The TFO (indicated by the green bar) of many TTSs in differentially expressed transcripts and MSTRG transcripts is in an aberrantly transcribed exon. (C) MSTRG.38311 overlaps with the CRC-related lncRNA genes CASC19 and CCAT1. (D) The TTSs of MSTRG.38311.2 are predicted in differentially expressed PTs (including G3BP1, TUSC1, SUN1, and ARFRP1) and MSTRG PTs (including MSTRG.5546.1 and MSTRG.33277.1), and DMRs are detected in these PTs and MSTRG PTs. MSTRG.38311.2 and these PTs and MSTRG PTs are in the same coexpression module. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
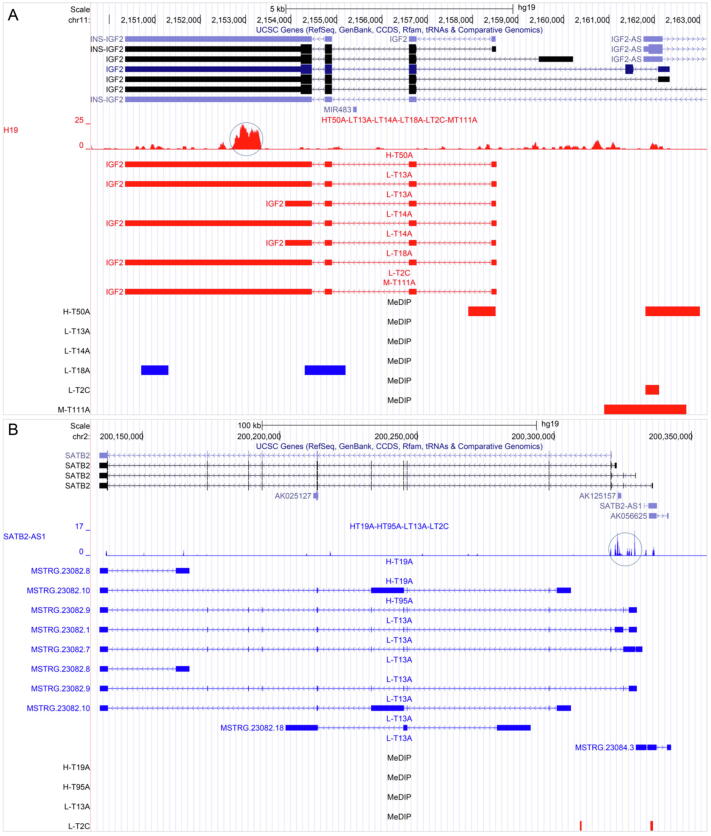
Next, we examined whether RTs and MSTRG RTs regulated nearby differentially expressed genes. In the 12 CRC samples, 271 differentially expressed noncoding transcripts (180 RTs and 91 MSTRG RTs) were identified, and centered on the genes of these transcripts, 271 local genomic regions were defined (see Section 2.10) (Supplementary Table 2). In many cases, regions of the same range were identified in multiple samples, and DMRs were identified in the genes in these regions. We examined whether these RTs and MSTRG RTs caused the aforementioned four kinds of epigenetic dysregulation in these regions. Many RTs and MSTRG RTs had clear TTSs in these regions (Supplementary Figs. 2–4). An impressive case is H19 and its target gene IGF2 (Fig. 3). The lncRNA H19 has been assumed to be a tumor suppressor because it regulates the imprinting of IGF2 [34]. However, H19 actually regulates many genes, and some recent findings indicate that H19 promotes oncogenesis [11], [35]. Our data indicate that both H19 and IGF2 were upregulated in multiple CRC samples, with DMRs in the local genomic region. These findings are consistent with reports that IGF2 is a driving factor in tumorigenesis and that the loss of imprinting on IGF2 occurs in CRC [36], [37], [38].
Fig. 3.
Some differentially expressed RTs have TTSs in their local genomic regions. From top to bottom in the two panels are tracks of the UCSC Genome Browser, including (a) the genome coordinates, (b) NCBI RefSeq genes, (c) TTSs (marked by the cycle) of the RT, (d) upregulated (in red) or downregulated (in blue) transcripts in specific tumor samples, and (e) DMRs (in red and blue, indicating increased and decreased methylation, respectively) in specific tumor samples. “H”, “M”, and “L” in sample names (e.g., H-T50A and L-T13A) indicate different tumor groups. (A) In the H19/IGF2 region in several tumor samples, H19 has a TTS in the last exon of IGF2, multiple H19 and IGF2 transcripts are upregulated, and six DMRs are detected in tumor samples. (B) In the SATB2-AS1/SATB2 region in several tumor samples, SATB2-AS1 has a TTS in the promoter region of SATB2, SATB2-AS1 expression is downregulated, multiple MSTRG.23082 transcripts are generated, and two DMRs are detected. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
If a dysregulated gene encodes a transcription factor (TF) or a lncRNA, it may intensify gene dysregulation by regulating its downstream targets. To evaluate this hypothesis, we computed the ratios of differentially expressed RTs to total RTs, differentially expressed PTs to total PTs, and differentially expressed TF transcripts to total TF transcripts, and further predicted how many differentially expressed RTs, PTs, and TF transcripts were targets of differentially expressed RTs. While only 0.548% RTs, 1.03% PTs, and 0.643% TF transcripts were differentially expressed, a large proportion of them had TTSs of the differentially expressed RTs (87.02%, 93.28%, and 91.01%, respectively) (Supplementary Fig. 5). These results indicate that the differential expression of many lncRNA genes and TF genes may be caused by lncRNA-mediated epigenetic dysregulation.
3.2. Epigenetic dysregulation is polymorphic
Compared with the patterns of mutations in cancers [38], to what extent epigenetic dysregulation varies in patients has been less explored [39]. To address this question, we examined differentially expressed transcripts and DMRs in the 12 tumor samples. Few differentially expressed transcripts and DMRs were found in many samples, and many differentially expressed transcripts and DMRs were found in few samples. The upregulation of RTs and MSTRG RTs was more prominent than the upregulation of PTs and MSTRG PTs, and the loss of DNA methylation was more prominent than the gain of DNA methylation (Supplementary Fig. 6–7; Supplementary Table 3–8). These findings indicate the highly polymorphic nature of epigenetic dysregulation and help explain the high intratumor heterogeneity of CRCs [18], [40].
3.3. Epigenetic dysregulation significantly determines correlated gene expression
Many studies have examined the correlation between differentially expressed genes, but few have explored to what extent the correlation is determined by epigenetic regulation and dysregulation. To address this question, we first used edgeR to identify differentially expressed transcripts and then used WGCNA to identify modules of coexpressed differentially expressed transcripts [27]. WGCNA identified 9 modules of coexpressed transcripts, including the H1, H2, and H3 modules in the high differentiation group, the M1, M2, and M3 modules in the medium differentiation group, and the L1, L2, and L3 modules in the low differentiation group. These 9 modules consist of 49,849 RT-PT (including MSTRG RTs and MSTRG PTs) pairs. To examine how many RT-PT pairs have a potential epigenetic regulation relationship, we predicted the TTSs of RTs in the genomic region of PTs. In 11,806 pairs (23.7%), the RT had a TTS in the PT (Supplementary Table 9), indicating that a great portion of correlated differential expression was determined by epigenetic dysregulation.
Specifically, we identified the foremost epigenetic regulators (the RTs that have TTSs in most PTs) and the foremost epigenetic targets (the PTs that contain TTSs of most RTs), which may reflect key features of epigenetic dysregulation in CRC. Four H19 transcripts (H19-002, H19-003, H19-004, and H19-015) had 1499 RT-PT pairs, in line with previous findings that lncRNA H19 is a master epigenetic regulator [11], [35]. Other RTs with TTSs in many PTs included AC010127.3, RP11-698N11.4, MSTRG.38311 (see Fig. 1), MSTRG.32134 (which overlaps the cancer-related microRNA gene MIR146A [41]), MSTRG.11306, and MSTRG.23870. PTs containing TTSs of most RTs included GNB2L1 [42], MUC4 [43], [44], VEGFA [45], CA1 [46], PDE9A, SLC35F2 [47], SLC6A19 [48], MDFI [49], and OAZ1 [50]. All of these genes are reported to be involved in CRC or other cancers. MSTRG.11306 and MSTRG.38311 are two impressive RT genes because nearly all of their targets have high WGCNA gene significance and module membership values (GS > 0.7 with p < 0.05 and MM > 0.7 with p < 0.05). Gene significance and module membership measure the correlation of the expression of a gene with CRC and with other genes in the module, respectively. MSTRG.11306.1 and MSTRG.38311.2 have TTSs at CDK4-005 and CLN3-001 that encode two cell cycle proteins, and MSTRG.11306.1 has TTSs at VEGFA-009, VEGFA-012, and VEGFA-023 that are widely involved in cancers [51], [52]. These results support that there is a strong link between coexpressed and epigenetically regulated genes in RT-PT pairs.
Furthermore, we examined whether epigenetic dysregulation has distinct features in CRCs of different differentiation grades. Of the 11,806 RT-PT pairs, 105 had the same expression change in all three tumor groups (Supplementary Table 10). MSTRG.11306.1 was co-upregulated with 36 target transcripts, and MSTRG.38311.2 was co-upregulated with 37 target transcripts. We examined RT-PT pairs that are specific to different cell differentiation grades and identified the most featured ones. An interesting case is the upregulated MYC transcript ENST00000520751. The TTSs of 9 RTs (including the four H19 transcripts, MSTRG.11306.1, MSTRG.38311.2, and three other RTs) were predicted in this MYC transcript. In the L2 module, the MYC transcript ENST00000520751 was coexpressed with 8 of the 9 RTs, but in none of the H modules was it coexpressed with these 9 RTs. Another case is the two SPEF2 transcripts ENST00000282469 and ENST00000440995. The TTSs of 31 RTs were predicted in the genomic regions of the two transcripts. Of note, all of the RT-ENST00000282469 pairs were in the L1 module, while all of the RT-ENST00000440995 pairs were in the H3 module. These results indicate that distinct RT-PT pairs are present in different cancer cells. To identify the key RT-PT pairs in the 9 modules, we used the WGCNA parameter Connectivity > 0.7 (together with GS > 0.7 and MM > 0.7) to obtain the top 30% most connected RTs and PTs. The results indicate that specific highly connected RTs are enriched in the H, M, and L groups (Table 1).
Table 1.
Numbers of RT-PT pairs of highly connected RTs and PTs in specific groups (Supplementary Table 9).
| Specific RTs |
Specific PTs |
||||||
|---|---|---|---|---|---|---|---|
| Ensembl ID (Gene name) | PT number |
Ensembl ID (Gene name) | RT number |
||||
| H | M | L | H | M | L | ||
| ENSG00000260495 (RP11-55K13.1) | 86 | 0 | 0 | ENSG00000065328 (MCM10) | 4 | 1 | 1 |
| ENSG00000261589 (CTC-462L7.1) | 40 | 0 | 0 | ENSG00000008300 (CELSR3) | 5 | 0 | 0 |
| ENSG00000188206 (HNRNPU-AS1) | 0 | 0 | 40 | ENSG00000141002 (TCF25) | 3 | 0 | 1 |
| MSTRG.11306 (MSTRG.11306) | 0 | 38 | 0 | ENSG00000004866 (ST7) | 4 | 0 | 0 |
| ENSG00000259959 (RP11-121C2.2) | 0 | 35 | 0 | ENSG00000135451 (TROAP) | 4 | 0 | 0 |
| ENSG00000251363 (RP11-129M6.1) | 0 | 0 | 27 | ENSG00000173467 (AGR3) | 4 | 0 | 0 |
| ENSG00000272841 (RP3-428L16.2) | 25 | 0 | 0 | ENSG00000147140 (NONO) | 4 | 0 | 0 |
| ENSG00000259969 (RP11-999E24.3) | 21 | 0 | 0 | ENSG00000177606 (JUN) | 4 | 0 | 0 |
| ENSG00000259886 (U82695.10) | 14 | 0 | 0 | ENSG00000050426 (LETMD1) | 0 | 0 | 2 |
| ENSG00000185186 (LINC00313) | 0 | 0 | 6 | ENSG00000169710 (FASN) | 0 | 0 | 2 |
3.4. Distinct epigenetic regulatory networks in cells of different differentiation grades
After identifying 9 modules of coexpressed differentially expressed genes in the 3 tumor groups, we used different conditions to filter the RTs, PTs, MSTRG RTs, and MSTRG PTs in each module to identify critical RT-PT pairs. First, we used the WGCNA parameters GS > 0.7 and MM > 0.7 to identify transcripts highly related to CRC and highly connected to other genes in specific modules. This condition filtered out considerable RTs and PTs in the H2, H3, M1, and L1 modules but few RTs and PTs in the other modules (Table 2). Second, we used the condition that an RT has a TTS in at least one PT. Notably, this condition, which identifies RTs and PTs with potential epigenetic regulation, did not influence any modules, indicating that differentially expressed transcripts in coexpression modules were greatly determined by epigenetic regulation (Table 2). Furthermore, we combined the two conditions to filter RT-PT pairs in each module. No or few pairs were removed from the H1, M2, L2, and L3 modules under the combined conditions (Table 2). For example, the H1 and L2 modules comprised 29 RTs + 489 PTs and 28 RTs + 346 PTs before filtering and comprised 25 RTs + 486 PTs and 25 RTs + 334 PTs after filtering. These results indicate that the link between correlated transcripts and epigenetically regulated transcripts is especially strong in these modules.
Table 2.
Number of correlated and epigenetically regulated RTs and PTs in modules under different filtering conditions.
| H1 | H2 | H3 | M1 | M2 | M3 | L1 | L2 | L3 | |
|---|---|---|---|---|---|---|---|---|---|
| RTs/PTs | 29/489 | 19/146 | 17/288 | 10/173 | 7/185 | 7/54 | 37/348 | 28/346 | 26/247 |
| RTs/PTs #1 | 29/489 | 9/67 | 2/51 | 1/11 | 7/182 | 7/51 | 0/23 | 27/340 | 21/224 |
| RTs/PTs #2 Pairs |
25/486 3274 |
14/83 238 |
15/266 1535 |
8/171 727 |
6/170 385 |
4/21 34 |
36/346 3471 |
25/340 2784 |
23/223 653 |
| RTs/PTs #1#2 Pairs |
25/486 3274 |
6/40 72 |
2/18 22 |
0/0 0 |
6/167 380 |
4/20 33 |
0/0 0 |
25/334 2748 |
17/202 452 |
#1: The condition is GS > 0.7 and MM > 0.7.
#2: The condition is that an RT has a TTS in at least one PT.
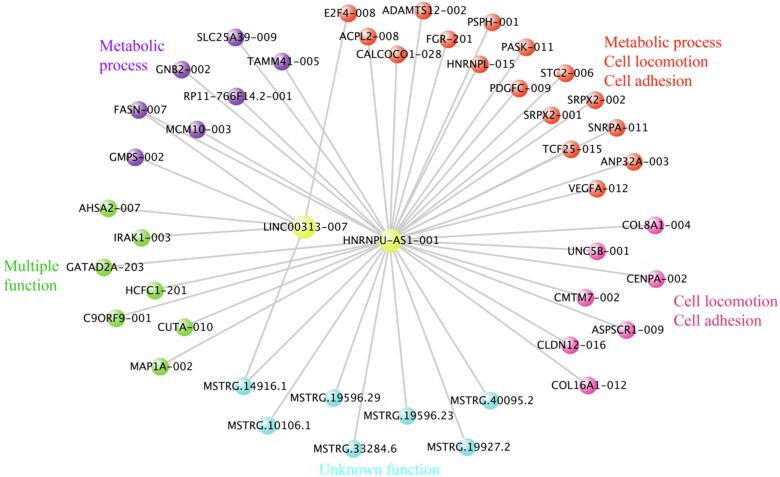
Motivated by the above results, we tried to identify the hub RTs in each module by further filtering RTs and PTs using the WGCNA parameter Connectivity > 0.7. Six networks consisting of hub RTs and their potential epigenetic targets were obtained. Using the DAVID database [53], we found that these epigenetic target transcripts are enriched in specific biological functions, such as cell locomotion and cell adhesion (Fig. 4; Supplementary Fig. 8). Interestingly, some hub nodes were MSTRG RTs, supporting that these MSTRG RTs are unlikely transcriptional noise or assembly artifacts, and most hub RTs may be primate-specific lncRNAs (Supplementary Fig. 9), suggesting that a great portion of differentially expressed genes between humans and other species may result from interspecies epigenetic differences [4]. These findings suggest that epigenetic dysregulation is also, to a great extent, species-specific.
Fig. 4.
A network consisting of differentially expressed hub RTs and differentially expressed PTs, which have potential epigenetic regulation, exists in each module. Shown is the network in the L2 module. This network contains the top 30% most connected RTs and their target PTs. Orthologous sequences of LINC00313 exons are identified only in simians, and HNRNPU-AS1 has a region conserved in mammals and a new primate-specific region (Supplementary Fig. 9). Gene enrichment analysis revealed that these PTs are involved in cell locomotion, cell adhesion, the cell cycle, and metabolic processes.
3.5. Analysis of differentially expressed genes in single-cell RNA-sequencing datasets
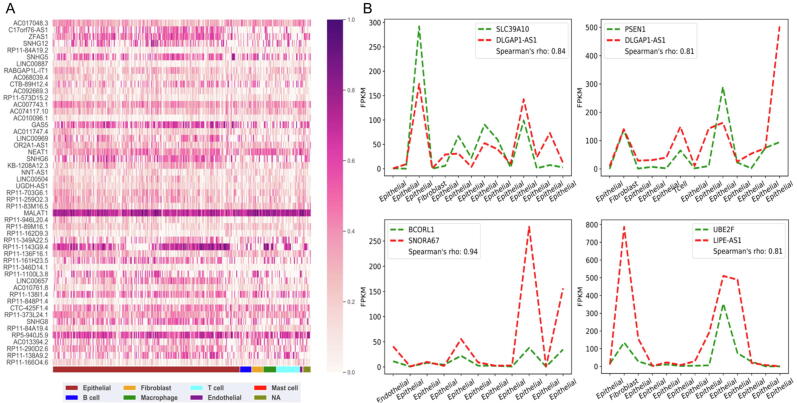
To examine and verify whether lncRNAs significantly dysregulate gene expression in CRC, we analyzed the datasets from a recent single-cell RNA-seq (scRNA-seq) study [29]. The expression levels of lncRNAs varied greatly across cells (Fig. 5A). We detected differentially expressed genes in tumor cells under the conditions of |logFC|>1 and Q-value < 0.1, predicted the TTSs of differentially expressed lncRNAs in differentially expressed protein-coding genes, and found that the lncRNA RP11-254F7.2 in myeloid cells has TTSs in nearly all of the differentially expressed protein-coding genes. RP11-254F7.2 is also a simian-specific lncRNA (Supplementary Fig. 9). Furthermore, we predicted the TTSs of lncRNAs that have the highest expression in some tumor cells in protein-coding genes, calculated the Spearman correlation between the expression of these lncRNAs and the expression of the protein-coding genes that have TTSs and found that the correlation is very strong in many cells (Fig. 5B; Supplementary Table 11). This correlation, together with the cell-specific expression of lncRNAs, supports the conclusion that lncRNA-mediated epigenetic dysregulation occurs distinctly in cancer cells.
Fig. 5.
LncRNA-mediated epigenetic regulation contributes to dysregulated gene expression in single cells. (A) Heatmap of lncRNA expression (the FPKM data are log-transformed) in different kinds of cells, indicating lncRNA cell-specific expression profiles. (B) Several examples showing that the expression levels (Y-axis) of lncRNAs and of their target genes in different cells (X-axis) are highly correlated (indicated by red and green dashed lines). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
How gene expression is epigenetically dysregulated in cancers has drawn increasing attention [39], [54], [55], [56]. As multiomics sequencing data are being generated at an unprecedented pace and many lncRNAs epigenetically regulate gene expression by recruiting enzymes such as DNA methyltransferases (DNMTs) and polycomb repressive complex 2 (PRC2) to specific genomic sites, it is important to combine the analysis of sequencing data with the analysis of lncRNA/DNA binding. In this study, we performed RNA-seq and MeDIP-seq on a set of CRC samples and normal colon tissues and developed an analysis pipeline to perform the combined analysis. In the pipeline, MeDIP-seq data can be replaced by RRBS-seq (reduced-representation bisulfite sequencing) or WGBS-seq (whole-genome bisulfite sequencing) data. Focusing on which lncRNAs possibly regulate which target genes in colorectal cancer cells, we obtained four novel findings. First, some aberrantly transcribed noncoding transcripts may epigenetically dysregulate gene expression. Second, lncRNA-mediated epigenetic dysregulation greatly determines the correlated differential expression of genes. Third, distinct epigenetic regulatory modules exist in different CRC cells. Fourth, many hub lncRNAs in these modules are primate-specific. These findings indicate important yet unexplored roles of MSTRG transcripts in CRC and other cancers and indicate that epigenetic dysregulation is highly species-specific. The identified critical RT-PT pairs, especially hub RTs and their targets, should be promising diagnostic, therapeutic, and prognostic targets of cancers. An important question to be further examined is to what extent critical RT-PT pairs and epigenetic regulatory modules are cancer-specific.
Our results agree with and are supported by reported experimental findings. First, aberrantly transcribed genes were detected in CRC in a previous study (which were called “differentially spliced genes” and “tumor RNA-seq reads”) (see Supplementary Fig. 6, 7 in [17]) but were not analyzed. Second, many MSTRG RTs highly expressed in CRC samples overlapped with annotated genes; an example is MSTRG.38311, which overlapped with the two important CRC-related lncRNA genes CCAT1 and CASC19 [31], [32], [33]. Third, many epigenetic targets of the differentially expressed RTs and MSTRG RTs were CRC-related or tumorigenesis-related. Specifically, the epigenetic targets of MSTRG.11306.1 and MSTRG.38311.2 include the transcripts of CDK4, CLN3, and VEGFA, which are widely involved in cancers. In addition, some MSTRG RTs were highly connected to other transcripts in coexpression modules; this can unlikely occur by chance.
When a cell divides, downstream proofreading is also required for faithful mitotic transmission of DNA methylation and histone modification. This proofreading causes a genome-wide lag between the copying of genetic and epigenetic information, and this lag not only causes inconsistency between the genome and epigenome but also amplifies and propagates epigenetic errors [57]. In addition to this lag of epigenome proofreading, our finding that aberrantly transcribed noncoding transcripts may epigenetically dysregulate gene expression further supports the estimate that the rate of epigenetic changes may be orders of magnitude higher than that of genetic changes [58] and the viewpoint that purely epigenetic mechanisms may explain tumors that arise with few or no recurrent mutations [39].
Experimentally revealing the lag of epigenome proofreading and the function of MSTRG transcripts is difficult; thus, computationally unveiling in what cells and to what extent epigenetic regulation goes wrong with publicly available sequencing data is valuable. Two notes on using the pipeline to perform more analyses are as follows. First, since many MSTRG genes and transcripts overlap genes and transcripts annotated in the genome build hg38, fewer MSTRG genes and transcripts would be reported if hg38 is used to assemble transcripts. Second, since lncRNA expression and epigenetic regulation are highly tissue-specific, the combined RNA-seq data analysis and lncRNA/DNA binding analysis alone (without DNA methylation data) may be satisfactorilly reliable.
5. Author’s contributions
H.Z. and S.H. designed the study and drafted the manuscript. J.C. and Y.D. collected and processed the samples. J.C. and F.Z. performed RT-PCR. BGI-Shenzhen (China) performed RNA-seq and MeDIP-seq. S.H., H.G. and G.Y. performed the data analysis. All authors have read the manuscript and consent to its publication.
6. Availability of data and code
The RNA-seq and MeDIP-seq data were deposited in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo) under the accession number GSE109204. LongTarget and MEDIPS results are available at the authors’ website (http://lncRNA.smu.edu.cn).
CRediT authorship contribution statement
Sha He: Conceptualization, Methodology, Writing - original draft. Juanzhi Chen: Validation. Huan Gao: Methodology. Guixian Yang: Methodology. Feixiang Zhang: Validation. Yanqing Ding: Supervision, Resources. Hao Zhu: Supervision, Conceptualization, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the Guangzhou Science and Technology Innovation Committee (201607010067). We thank the National Supercomputing Shenzhen Center for technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.11.004.
Contributor Information
Yanqing Ding, Email: dyqgz@126.com.
Hao Zhu, Email: zhuhao@smu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Mwenifumbo J.C., Marra M.A. Cancer genome-sequencing study design. Nat Rev Genet. 2013;14(5):321–332. doi: 10.1038/nrg3445. [DOI] [PubMed] [Google Scholar]
- 2.Shah S.P., Roth A., Goya R., Oloumi A., Ha G., Zhao Y., Turashvili G., Ding J., Tse K., Haffari G., Bashashati A., Prentice L.M., Khattra J., Burleigh A., Yap D., Bernard V., McPherson A., Shumansky K., Crisan A., Giuliany R., Heravi-Moussavi A., Rosner J., Lai D., Birol I., Varhol R., Tam A., Dhalla N., Zeng T., Ma K., Chan S.K., Griffith M., Moradian A., Cheng S.-W., Morin G.B., Watson P., Gelmon K., Chia S., Chin S.-F., Curtis C., Rueda O.M., Pharoah P.D., Damaraju S., Mackey J., Hoon K., Harkins T., Tadigotla V., Sigaroudinia M., Gascard P., Tlsty T., Costello J.F., Meyer I.M., Eaves C.J., Wasserman W.W., Jones S., Huntsman D., Hirst M., Caldas C., Marra M.A., Aparicio S. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486(7403):395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg A.P., Koldobskiy M.A., Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17(5):284–299. doi: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernando-Herraez, I., et al., DNA Methylation: Insights into Human Evolution. PLoS Genet, 2015. 11(12): p. e1005661. [DOI] [PMC free article] [PubMed]
- 5.Hrdlickova B., de Almeida R.C., Borek Z., Withoff S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta (BBA) – Mol Basis Dis. 2014;1842(10):1910–1922. doi: 10.1016/j.bbadis.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Yang L., Lin C., Jin C., Yang J.C., Tanasa B., Li W., Merkurjev D., Ohgi K.A., Meng D.a., Zhang J., Evans C.P., Rosenfeld M.G. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013;500(7464):598–602. doi: 10.1038/nature12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalwa M., Hänzelmann S., Otto S., Kuo C.-C., Franzen J., Joussen S., Fernandez-Rebollo E., Rath B., Koch C., Hofmann A., Lee S.-H., Teschendorff A.E., Denecke B., Lin Q., Widschwendter M., Weinhold E., Costa I.G., Wagner W. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016;44(22):10631–10643. doi: 10.1093/nar/gkw802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J.T. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev. 2009;23(16):1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai M.-C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotake Y., Nakagawa T., Kitagawa K., Suzuki S., Liu N., Kitagawa M., Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene. 2011;30(16):1956–1962. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabory A., Ripoche M.-A., Le Digarcher A., Watrin F., Ziyyat A., Forne T., Jammes H., Ainscough J.F.X., Surani M.A., Journot L., Dandolo L. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136(20):3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- 12.Chu C., Qu K., Zhong F., Artandi S., Chang H. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abu Almakarem, A.S., et al., Comprehensive survey and geometric classification of base triples in RNA structures. Nucleic Acids Res, 2012. 40(4): p. 1407–23. [DOI] [PMC free article] [PubMed]
- 14.Lin J., Wen Y., He S., Yang X., Zhang H., Zhu H. Pipelines for cross-species and genome-wide prediction of long noncoding RNA binding. Nat Protoc. 2019;14(3):795–818. doi: 10.1038/s41596-018-0115-5. [DOI] [PubMed] [Google Scholar]
- 15.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012: Global Cancer Statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 16.Calon A. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ongen H., Andersen C.L., Bramsen J.B., Oster B., Rasmussen M.H., Ferreira P.G., Sandoval J., Vidal E., Whiffin N., Planchon A., Padioleau I., Bielser D., Romano L., Tomlinson I., Houlston R.S., Esteller M., Orntoft T.F., Dermitzakis E.T. Putative cis-regulatory drivers in colorectal cancer. Nature. 2014;512(7512):87–90. doi: 10.1038/nature13602. [DOI] [PubMed] [Google Scholar]
- 18.Sottoriva A., Kang H., Ma Z., Graham T.A., Salomon M.P., Zhao J., Marjoram P., Siegmund K., Press M.F., Shibata D., Curtis C. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merry C.R., Forrest M.E., Sabers J.N., Beard L., Gao X.-H., Hatzoglou M., Jackson M.W., Wang Z., Markowitz S.D., Khalil A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum Mol Genet. 2015;24(21):6240–6253. doi: 10.1093/hmg/ddv343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, Y., et al., SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. Gigascience, 2018. 7(1): p. 1–6. [DOI] [PMC free article] [PubMed]
- 21.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 2016;17:19. doi: 10.1186/s13059-016-0880-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lun A.T., Chen Y., Smyth G.K. It's DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol Biol. 2016;1418:391–416. doi: 10.1007/978-1-4939-3578-9_19. [DOI] [PubMed] [Google Scholar]
- 26.Lienhard, M., et al., MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics, 2014. 30(2): p. 284–6. [DOI] [PMC free article] [PubMed]
- 27.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahrami S., Ehsani R., Drablos F. A property-based analysis of human transcription factors. BMC Res Notes. 2015;8:82. doi: 10.1186/s13104-015-1039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Courtois E.T., Sengupta D., Tan Y., Chen K.H., Goh J.J.L., Kong S.L., Chua C., Hon L.K., Tan W.S., Wong M., Choi P.J., Wee L.J.K., Hillmer A.M., Tan I.B., Robson P., Prabhakar S. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet. 2017;49(5):708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 30.Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghafouri‐Fard S., Taheri M. Colon cancer‐associated transcripts 1 and 2 : roles and functions in human cancers. J Cell Physiol. 2019;234(9):14581–14600. doi: 10.1002/jcp.28176. [DOI] [PubMed] [Google Scholar]
- 32.Kalmár A., Nagy Z.B., Galamb O., Csabai I., Bodor A., Wichmann B., Valcz G., Barták B.K., Tulassay Z., Igaz P., Molnár B. Genome-wide expression profiling in colorectal cancer focusing on lncRNAs in the adenoma-carcinoma transition. BMC Cancer. 2019;19(1) doi: 10.1186/s12885-019-6180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozawa T., Matsuyama T., Toiyama Y., Takahashi N., Ishikawa T., Uetake H., Yamada Y., Kusunoki M., Calin G., Goel A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28(8):1882–1888. doi: 10.1093/annonc/mdx248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 35.Anastasiadou E., Jacob L.S., Slack F.J. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui H. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299(5613):1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 37.Sakatani T. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307(5717):1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas, N., Comprehensive molecular characterization of human colon and rectal cancer. Nature, 2012. 487(7407): p. 330–7. [DOI] [PMC free article] [PubMed]
- 39.Flavahan W.A., Gaskell E., Bernstein B.E. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348) doi: 10.1126/science.aal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landan G., Cohen N.M., Mukamel Z., Bar A., Molchadsky A., Brosh R., Horn-Saban S., Zalcenstein D.A., Goldfinger N., Zundelevich A., Gal-Yam E.N., Rotter V., Tanay A. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44(11):1207–1214. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- 41.Iacona J.R., Lutz C.S. miR-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip Rev RNA. 2019;10(4) doi: 10.1002/wrna.1533. [DOI] [PubMed] [Google Scholar]
- 42.Cheng S., Ren J., Su L., Liu J., Liu Q., Zhou J., Ye X., Zhu N. O-GlcNAcylation of the signaling scaffold protein, GNB2L1 promotes its degradation and increases metastasis of gastric tumours. Biochem Biophys Res Commun. 2016;478(4):1497–1502. doi: 10.1016/j.bbrc.2016.08.074. [DOI] [PubMed] [Google Scholar]
- 43.Niv Y., Rokkas T. Mucin expression in colorectal cancer (CRC): systematic review and meta-analysis. J Clin Gastroenterol. 2019;53(6):434–440. doi: 10.1097/MCG.0000000000001050. [DOI] [PubMed] [Google Scholar]
- 44.Krishn S.R., Kaur S., Smith L.M., Johansson S.L., Jain M., Patel A., Gautam S.K., Hollingsworth M.A., Mandel U., Clausen H., Lo W.-C., Fan W.-T., Manne U., Batra S.K. Mucins and associated glycan signatures in colon adenoma–carcinoma sequence: prospective pathological implication(s) for early diagnosis of colon cancer. Cancer Lett. 2016;374(2):304–314. doi: 10.1016/j.canlet.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Terme M., Pernot S., Marcheteau E., Sandoval F., Benhamouda N., Colussi O., Dubreuil O., Carpentier A.F., Tartour E., Taieb J. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res. 2013;73(2):539–549. doi: 10.1158/0008-5472.CAN-12-2325. [DOI] [PubMed] [Google Scholar]
- 46.Kummola L. Expression of a novel carbonic anhydrase, CA XIII, in normal and neoplastic colorectal mucosa. BMC Cancer. 2005;5:41. doi: 10.1186/1471-2407-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winter G.E., Radic B., Mayor-Ruiz C., Blomen V.A., Trefzer C., Kandasamy R.K., Huber K.V.M., Gridling M., Chen D., Klampfl T., Kralovics R., Kubicek S., Fernandez-Capetillo O., Brummelkamp T.R., Superti-Furga G. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat Chem Biol. 2014;10(9):768–773. doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kessler M.D., Bateman N.W., Conrads T.P., Maxwell G.L., Dunning Hotopp J.C., O’Connor T.D. Ancestral characterization of 1018 cancer cell lines highlights disparities and reveals gene expression and mutational differences: ancestral analysis of cancer cell lines. Cancer. 2019;125(12):2076–2088. doi: 10.1002/cncr.32020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J., Chen C., Bi X., Zhou C., Huang T., Ni C., Yang P., Chen S., Ye M., Duan S. DNA methylation of CMTM3, SSTR2, and MDFI genes in colorectal cancer. Gene. 2017;630:1–7. doi: 10.1016/j.gene.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 50.Patil S., Arakeri G., Alamir A.W.H., Awan K.H., Baeshen H., Ferrari M., Patil S., Fonseca F.P., Brennan P.A. Role of salivary transcriptomics as potential biomarkers in oral cancer: a systematic review. J Oral Pathol Med. 2019;48(10):871–879. doi: 10.1111/jop.12895. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi N., Iwasa S., Taniguchi H., Sasaki Y., Shoji H., Honma Y., Takashima A., Okita N., Kato K., Hamaguchi T., Shimada Y., Yamada Y. Prognostic role of ERBB2, MET and VEGFA expression in metastatic colorectal cancer patients treated with anti-EGFR antibodies. Br J Cancer. 2016;114(9):1003–1011. doi: 10.1038/bjc.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jang K. VEGFA activates an epigenetic pathway upregulating ovarian cancer-initiating cells. EMBO Mol Med. 2017;9(3):304–318. doi: 10.15252/emmm.201606840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Ju H.X., An B., Okamoto Y., Shinjo K., Kanemitsu Y., Komori K., Hirai T., Shimizu Y., Sano T., Sawaki A., Tajika M., Yamao K., Fujii M., Murakami H., Osada H., Ito H., Takeuchi I., Sekido Y., Kondo Y. Distinct profiles of epigenetic evolution between colorectal cancers with and without metastasis. Am J Pathol. 2011;178(4):1835–1846. doi: 10.1016/j.ajpath.2010.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee D.S. An epigenomic roadmap to induced pluripotency reveals DNA methylation as a reprogramming modulator. Nat Commun. 2014;5:5619. doi: 10.1038/ncomms6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen H., Laird P.W. Interplay between the cancer genome and epigenome. Cell. 2013;153(1):38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charlton J., Downing T.L., Smith Z.D., Gu H., Clement K., Pop R., Akopian V., Klages S., Santos D.P., Tsankov A.M., Timmermann B., Ziller M.J., Kiskinis E., Gnirke A., Meissner A. Global delay in nascent strand DNA methylation. Nat Struct Mol Biol. 2018;25(4):327–332. doi: 10.1038/s41594-018-0046-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siegmund K.D., Marjoram P., Woo Y.-J., Tavare S., Shibata D. Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci. 2009;106(12):4828–4833. doi: 10.1073/pnas.0810276106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.