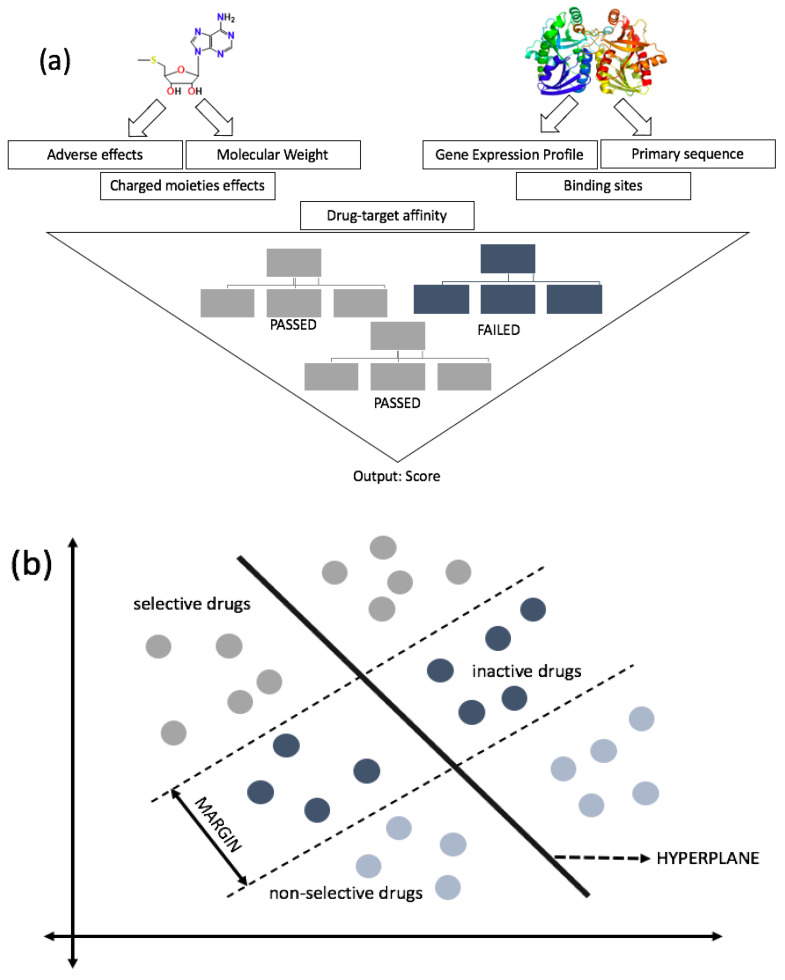
Figure 2.
Schematic view of drug development using random forest (RF) (a) and support vector machine (SVM) (b). (a) RF reaches the final decision of drugs by combining the results of randomly-created decision trees (three trees are shown for simplicity). There are multiple features that the computational queries look for in both target and drug. When there is a compatibility match, it proceeds to the next step to match additional features. A series of datasets is inputted into the query, and each tree is responsible for computing a prediction. The prediction picked by most trees is used for the next step. The system of using many decision trees is intended to minimize errors mathematically. (b) SVM utilizes similarities between the classes, called support vectors, to distinguish between the classes based on the trained features. It formulates hyperplanes that separate two classes (can be multiclass, if needed). SVM incorporates multiple training sets depending on the classifiers and formulates compounds’ status (active or inactive). During the process, compounds are separated into three sections: Non-selective compounds (active), selective compounds (active), and in the margin are inactive compounds. Although non-selective compounds are active, they are not selective towards the protein of interest. In contrast, selective compounds are active and selective towards the protein of interest.