Abstract
Pulmonary arterial hypertension (PAH) describes a rare, progressive vascular disease caused by the obstruction of pulmonary arterioles, typically resulting in right heart failure. Whilst PAH most often manifests in adulthood, paediatric disease is considered to be a distinct entity with increased morbidity and often an unexplained resistance to current therapies. Recent genetic studies have substantially increased our understanding of PAH pathogenesis, providing opportunities for molecular diagnosis and presymptomatic genetic testing in families. However, the genetic architecture of childhood-onset PAH remains relatively poorly characterised. We sought to investigate a previously unsolved paediatric cohort (n = 18) using whole exome sequencing to improve the molecular diagnosis of childhood-onset PAH. Through a targeted investigation of 26 candidate genes, we applied a rigorous variant filtering methodology to enrich for rare, likely pathogenic variants. This analysis led to the detection of novel PAH risk alleles in five genes, including the first identification of a heterozygous ATP13A3 mutation in childhood-onset disease. In addition, we provide the first independent validation of BMP10 and PDGFD as genetic risk factors for PAH. These data provide a molecular diagnosis in 28% of paediatric cases, reflecting the increased genetic burden in childhood-onset disease and highlighting the importance of next-generation sequencing approaches to diagnostic surveillance.
Keywords: exome sequencing, molecular genetics, lung disease, paediatrics, pulmonary arterial hypertension
1. Introduction
Pulmonary arterial hypertension (PAH) is an uncommon vascular disorder that remains incurable despite significant advances in current treatment regimens. PAH is characterised by obstruction and occlusion of the pulmonary arterioles leading to progressive pulmonary arterial pressure overload, right ventricular hypertrophy and failure of the right side of the heart [1]. Typical histopathological features of PAH include marked vascular remodelling of the pulmonary arterioles as a consequence of exuberant proliferation of pulmonary artery endothelial (PAEC) and smooth muscle (PASMC) cells. In families, PAH predominantly segregates as an autosomal dominant trait displaying features of complex disease, namely variable expressivity, reduced penetrance and a gender bias favouring females [2]. Despite major advances in delineating PAH aetiology, the associated morbidity and early mortality burden of this disease remains perniciously high, with an average life expectancy of 3–5 years from diagnosis [3].
Paediatric PAH represents a clinically distinct form of disease. Although less prevalent, at an estimated 4.8–8.1 cases/million [4,5], than the adult form (15–50 cases/million) [6], childhood-onset PAH (cPAH) is marked by greater morbidity, depressed responses to therapy and poor survival metrics [7,8]. Taken together, these features suggest a specific genetic background. Putatively causal genetic variants have been described in at least 26 PAH risk genes [9,10,11,12,13,14], of which deleterious variation in BMPR2, encoding a type II receptor of the transforming growth factor beta (TGF-beta) superfamily, was firmly established as the major risk factor in adult PAH [13]. In paediatric cases, rare deleterious mutations of BMPR2 and other key components of the bone morphogenetic protein (BMP) signalling pathway have been determined to be rare causal factors. These include the receptor proteins activin A receptor-like type 1 (ACVRL1, encoding ALK1), BMPR1B and endoglin (ENG), as well as GDF2 (encoding the ligand BMP9) and the cytoplasmic signalling mediator SMAD9 [8,15,16,17,18,19,20].
Whole exome sequencing (WES) has been instrumental in the accelerated gene discovery in PAH, notably in genetic risk unrelated to the canonical BMP pathway. Moreover, these findings both expanded the mutation spectrum in cPAH and shed light on the epidemiology of this discrete disease entity. For example, mutations within the transcription factor genes T-box 4 (TBX4) and SRY-box 17 (SOX17) show enriched contribution to paediatric PAH compared to adult-onset, explaining up to 7.7% and 3.3% of paediatric cases, respectively [8,21,22]. Of note, SOX17 mutations show a strong correlation to PAH associated with congenital heart disease in both adult and paediatric disease [22,23]. More recent findings include loss-of-function variants in the ATP-binding cassette subfamily C member 8 (ABCC8) gene and in the endothelial cell-specific ligand BMP10 [24,25], whilst genome-wide gene burden approaches in predominantly adult cohorts identified the candidate risk genes ATPase 13A3 (ATP13A3), fibulin 2 (FBLN2) and platelet-derived growth factor D (PDGFD), among others [10,14]. These novel gene discoveries demonstrated the benefit of a granular approach to the genomic interrogation of discrete case cohorts.
While these data further developed the molecular architecture of PAH, the mutational landscape remains relatively poorly understood in paediatric cohorts, particularly for more recently reported genes [10,12]. Here, we sought to investigate a previously unsolved paediatric cohort, first, to better define the mutational background of cPAH, and second, to augment the existing body of data underlying this disease. Taken together, this study highlights the hitherto undefined role of ATP13A3 in the pathogenesis of paediatric PAH and, importantly, provides independent validation of putative gene-disease associations in BMP10 and PDGFD.
2. Materials and Methods
2.1. Patient Cohort and Clinical Assessment
Eighteen unrelated patients and their relatives were recruited from specialist PAH centres in the UK and the Netherlands. Ethical approval for the study was received from local ethics review boards and written informed consent was obtained from all participants or their legal guardian. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the St Thomas’ Hospital Research Ethics Committee (08/H0802/32).
Clinical diagnoses were established by PAH specialists in accordance with the classification of the World Symposium on Pulmonary Hypertension [26]. All patients previously underwent conventional genetic screening for BMPR2, ACVRL1 and ENG, and were found to be mutation-negative [16,27]. Following pedigree analysis and clinical examination, patients were classified as having either idiopathic PAH (IPAH; n = 8), PAH associated with congenital heart disease (APAH-CHD; n = 7) or heritable PAH with one or more affected relatives (HPAH; n = 3).
2.2. Exome Sequencing
Genomic DNA was extracted from peripheral blood using standard methodologies. Whole exome sequencing for all probands was performed by Novogene Ltd. Briefly, exome libraries were captured using the SureSelect Human All Exon kit (Agilent Technologies, Santa Clara, CA, USA), and enriched fragments underwent paired-end sequencing on an Illumina platform. Quality control filtration of raw FASTQ files discarded read pairs containing adapter contamination and those with <80% of base calls achieving a Phred score of Q30. Sequence alignment to human reference genome assembly GRCh37 was undertaken using Burrows–Wheeler Aligner [28] and Picard MarkDuplicates (Genome Analysis Tool Kit (GATK), Broad Institute, Cambridge, MA, USA) was used to flag PCR duplicate reads for removal. GATK (v3.8) software was used for single nucleotide variant (SNV) and insertion and deletion (indel) detection [29]. Variant annotation was performed using ANNOtate VARiation (ANNOVAR) [30].
2.3. Candidate Gene Analysis
Annotated WES profiles for each patient were screened for rare, deleterious variants in 26 established and emerging PAH risk genes: ABCC8, ACVRL1, AQP1, ATP13A3, BMP10, BMPR1B, BMPR2, CAV1, EIF2AK4, ENG, EVI5, FBLN2, GDF2, GGCX, KCNA5, KCNK3, KDR, KLF2, KLK1, NOTCH3, PDGFD, SMAD1, SMAD4, SMAD9, SOX17 and TBX4. Rare variants were defined as those with a minor allele frequency of ≤1 in 10,000 (MAF ≤ 0.0001), as reported in the Genome Aggregation Database control population (gnomAD v2.1.1; https://gnomad.broadinstitute.org). Splice region and exonic variants with likely functional consequences (nonsense, frameshift, nonframeshift indels and missense SNVs) were retained. Copy number variants were not considered in the analysis presented herein. The functional impact of missense variants was informed by in silico pathogenicity predictions. Specifically, variants with a Combined Annotation Dependent Depletion (CADD) score of ≥20 (https://cadd.gs.washington.edu) were considered potentially causal [31]. Additional evidence for pathogenicity was obtained from observing at least one deleterious score across Sorting Intolerant from Tolerant (SIFT; ≤0.05), PolyPhen-2 (≥0.909) and MutationTaster2 (annotation “disease causing”) algorithms [32,33,34]. The location of each variant was inspected for correlation with conserved protein domains, providing further confirmation of likely pathogenicity.
Candidate variants were visualised and inspected for artefacts using Integrative Genomics Viewer (IGV) to exclude false positives [35,36]. All identified variants were validated in an independent sample by Sanger sequencing (Eurofins Genomics, Ebersberg, Germany) and tested for familial co-segregation where DNA samples were available.
3. Results
3.1. WES Analysis of the cPAH Cohort
Overall, 92% of base calls achieved a mean quality score of Q30. A mean depth of coverage of 65x (range: 54x–78x) was achieved for all samples, with an average of 91% (range: 86–94%) of bases achieving >20x coverage. Manual inspection of all PAH risk genes using IGV confirmed sufficient coverage of ≥10x across all exonic regions and intron–exon boundaries.
A targeted variant analysis of previously reported risk genes in our panel of childhood-onset IPAH, APAH and HPAH patients revealed putatively pathogenic missense variants in ABCC8, ATP13A3, BMP10, PDGFD and SMAD9 (Table 1). All variants were validated by Sanger sequencing. Based on our exclusion criteria, no other likely causal variants in the reported PAH risk genes were identified in these patients.
Table 1.
Variants Identified in Childhood-Onset Pulmonary Arterial Hypertension.
| Patient | Sex | Age of Onset | Diagnosis | Variant Identified | Mutation Category | GnomAD MAF | Variant Identifier(s) | CADD Score | SIFT Score | PolyPhen2 (HumVar) | Mutation Taster |
|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | M | 9 y | APAH–CHD (ASD) |
ATP13A3 (exon 11) NM_024524.4 c.1148C>A (p.Thr383Lys) |
Missense * | - | Novel | 32 | 0.001, D | 0.991, D | 1.0, D |
| P2 | F | 3 y | IPAH |
BMP10 (exon 1) NM_014482.3 c.247G>A (p.Glu83Lys) |
Missense | 0.000009 | dbSNP: rs1192957334; Not in ClinVar |
28 | 0.047, D | 0.999, D | 1.0, D |
| P3 | F | <5 y | IPAH |
PDGFD (exon 4) NM_025208.5 c.550G>A (p.Glu184Lys) |
Missense | - | dbSNP: rs769639743; Not in ClinVar |
28.2 | 0.007, D | 0.980, D | 0.9998, D |
| P4 | M | 19 m | IPAH |
ABCC8 (exon 7) NM_000352.6 c.1069G>A (p.Val357Ile) |
Missense | 0.000018 | dbSNP: rs771392416; Not in ClinVar |
23 | 0.235, T | 0.042, B | 1.0, D |
| P5 | F | 4.5 y | HPAH (mother also affected) |
SMAD9 (exon 6) NM_001127217.3 c.1117G>A (p.Val373Ile) |
Missense | 0.00013 | dbSNP: rs140504903; ClinVar: VCV000311894 |
27 | 0.001, D | 0.936, D | 1.0, D |
APAH–CHD: PAH associated with congenital heart disease; ASD: atrial septal defect; B: benign; CADD: Combined Annotation Dependent Depletion; D: damaging (SIFT), probably damaging (PolyPhen2) or disease causing (Mutation Taster); IPAH: idiopathic pulmonary arterial hypertension; HPAH: heritable pulmonary arterial hypertension; MAF: minor allele frequency in gnomAD v2.1.1 (controls); m: months; SIFT: Sorting Independent from Tolerant; T: tolerated; y: years. * The ATP13A3 missense variant in Patient 1 is located within the consensus splice region.
3.2. Novel Association of ATP13A3 with Paediatric APAH-CHD
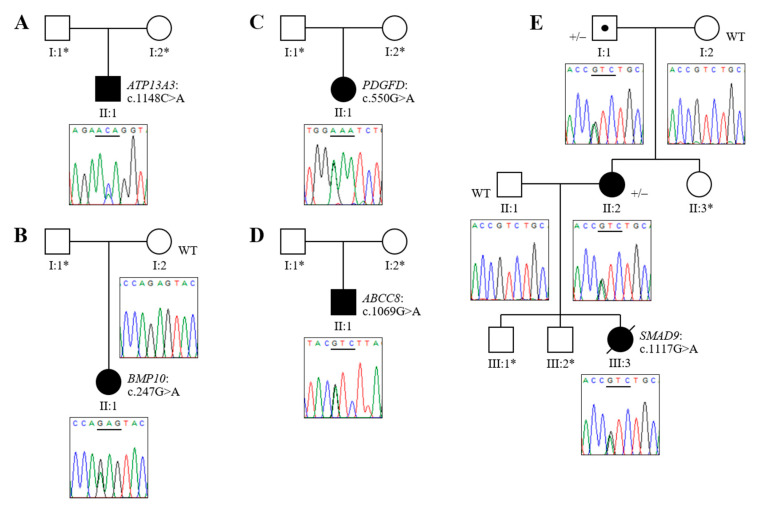
We identified a novel missense variant in ATP13A3 (c.1148C>A, p.Thr383Lys) in a male child with APAH–CHD. To the best of our knowledge, this represents the first finding of a molecular defect in this risk gene in a paediatric case. The patient was diagnosed with PAH at nine years of age, associated with a secundum atrial septal defect (Figure 1A). All in silico predictions were consistent with a high probability of pathogenicity (Table 1). Parental samples were not available to conduct familial segregation analysis.
Figure 1.
Sequence chromatograms and familial segregation of identified variants in childhood-onset pulmonary arterial hypertension (cPAH). (A) Patient 1 has a c.1148C>A missense mutation in ATP13A3; (B) Patient 2 has a heterozygous BMP10 c.247G>A mutation; (C) Patient 3 is heterozygous for a c.550G>A mutation in PDGFD; (D) Patient 4 has a c.1069G>A missense variant in the ABCC8 gene; (E) Patient 5 carries a heterozygous SMAD9 c.1117G>A missense variant, which co-segregates with the disease (+/−) in his affected mother and maternal grandfather, who is an obligate carrier. Unaffected family members available for testing are all wild-type (WT). The horizontal line underlines the mutated codon. * DNA sample not available.
While annotated as a missense variant, further analysis of the ATP13A3 c.1148C>A variant revealed that the mutation is located 3 bp from the intron–exon boundary and one nucleotide upstream of the highly conserved donor splicing motif of exon 12 (CAG|gtagga). An independent interrogation of in silico data using the NNSplice tool in MutationTaster revealed a potential disruption of the donor splice site, likely resulting in the formation of an aberrant gene transcript or premature stop codon leading to nonsense-mediated decay of the messenger RNA. However, further work is required on the cDNA level to experimentally validate this predicted deleterious effect.
3.3. Novel Mutations of BMP10, PDGFD and ABCC8 in Childhood IPAH
In the IPAH cohort we detected a BMP10 variant (c.247G>A, p.Glu83Lys) in a female Dutch patient diagnosed with severe disease at 36 months. The patient was stably treated on oral dual therapy (bosentan and sildenafil) but died at 18 years of age. This mutation, which is observed in 1/109,376 control individuals in gnomAD and was not previously associated with disease in a PAH clinical setting, provides the first independent confirmation of BMP10 as a PAH risk gene [25]. All bioinformatic predictions were consistent with a high likelihood of pathogenicity (Table 1). Familial co-segregation analysis confirmed the unaffected mother was wild-type but, as no DNA was available from the father, it was not possible to determine whether this variant had arisen de novo in the proband (Figure 1B).
PDGFD is a newly identified risk gene in PAH [14]; this analysis offers the first independent validation of its causal association with PAH, as well as confirming a specific role in the development of paediatric disease through the identification of a rare missense variant (c.550G>A, p.Glu184Lys) in a female cPAH patient (Figure 1C). The variant, which is not present in population control databases, gave a CADD score of 28.2 and was predicted to be deleterious by all three missense prediction algorithms (Table 1).
A heterozygous missense variant in ABCC8 (c.1069G>A, p.Val357Ile), novel in the context of PAH, was observed in a male patient diagnosed at 19 months of age (Figure 1D). While the CADD score and MutationTaster prediction were strongly indicative of a deleterious impact, the conclusions of pathogenicity based on other in silico markers were ambiguous, likely due to the interchangeability of valine and isoleucine residues at this position across related proteins in lower organisms (Table 1). Unavailability of parental DNA samples precluded familial segregation analysis of the PDGFD and ABCC8 variants.
3.4. Analysis of HPAH Samples Identifies a Potential Molecular Defect of SMAD9
An examination of WES data from the three HPAH probands revealed a missense variant in SMAD9 (c.1117G>A, p.Val373Ile) in a single family. Following a diagnosis of PAH, the proband died at five years of age. The affected amino acid residue displays high conservation across species, reflected in high deleterious scores across all in silico prediction algorithms employed (Table 1). Co-segregation was confirmed in two affected family members and an obligate carrier (Figure 1E). In addition, the candidacy of this variant as a causal defect in PAH was underpinned by the same finding in an HPAH patient reported in the ClinVar database (SCV000384082). However, as an observed allele frequency of >0.0001 in European controls exceeded our threshold for inclusion, this was resultantly considered to be a variant of unknown significance (VUS) in the absence of compelling functional data.
4. Discussion
This study applied a targeted 26 gene analysis to WES data in a European paediatric cohort, elucidating candidate molecular mechanisms in five patients to further illuminate the genetic architecture of early-onset PAH. It must be noted that all candidate variants presented in this analysis were categorised as variants of unknown significance in accordance with American College of Medical Genetics and Genomics (ACMG) guidelines [37], including those that met our criteria as being highly deleterious. Evidence of variant pathogenicity derived from population frequency, bioinformatic predictions and the biological relevance of these genes to PAH pathology is considered herein.
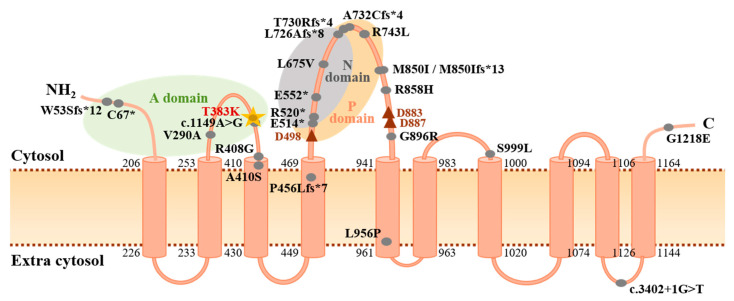
While ATP13A3 was recently identified and functionally characterised as being causative in adult PAH [10], this study is the first to implicate heterozygous variation of ATP13A3 in paediatric-onset cases. The gene encodes a P-type ATPase for which the substrate specificity and biological role remain poorly defined [38], but is a known transmembrane protein that localises to endosomal compartments and is likely involved in polyamine transport, key molecules for cell growth and proliferation [39,40]. Recent functional work demonstrated that ATP13A3 mRNA is expressed in PASMCs, whilst loss of ATP13A3 inhibits endothelial cell proliferation and increases apoptosis, consistent with disease initiation models in PAH [10]. The novel c.1148C>A (p.Thr383Lys) variant identified in this study alters a highly conserved amino acid residue located within the actuator domain, which functions to dephosphorylate the catalytic phosphorylation domain (Figure 2). The variant is predicted to disrupt splicing and result in nonsense-mediated decay. Although molecular defects in the ATP13A3 gene clearly underlie susceptibility to PAH [10,12,20,41], the mechanism of pathogenicity remains to be defined. Further functional analyses are therefore warranted to elucidate whether ATP13A3 loss-of-function represents a molecular mechanism independent to the BMP signalling pathway, and thus a new therapeutic target.
Figure 2.
Protein structure of ATP13A3 highlighting mutations identified in PAH. Topological analysis of ATP13A3 according to UniProtKB protein component and site position data (ID: Q9H7F0) and published reports [38,40]. Likely pathogenic PAH mutations (CADD ≥ 15) reported in the literature are indicated by the filled grey circles [10,12,20,41]. The c.1148C>A (p.T383K) mutation identified in this study is highlighted by the gold star and is located adjacent to a previously reported splice-region variant [10]. Numbers indicate amino acid positions at each end of the 10 transmembrane domains. The red triangles denote essential asparagine residues (D498: active catalytic site; D883, D887: Mg2+ binding sites). A domain: actuator domain; C: carboxyl terminus; N domain: nucleotide binding domain; NH2: amino terminus; P domain: phosphorylation domain.
The WES analysis workflow in this study identified a novel BMP10 c.247G>A (p.Glu83Lys) missense variant with consistent in silico markers of pathogenicity. The residue is highly conserved across species and is located within the conserved TGF-beta propeptide domain, where a BMP10 nonsense mutation was previously identified in cPAH [25]. This observation supports accumulating evidence implicating BMP10 loss-of-function in PAH pathogenesis [25,42]. BMP10 encodes a high affinity ligand that activates the ALK1 and BMPR2 endothelial cell surface receptor within the BMP signalling pathway [43]. The gene product shares 65% amino acid sequence identity with its BMP9 paralogue, encoded by the GDF2 gene. GDF2 variant enrichment was recently reported in three large PAH cohorts, strongly implicating the BMP9/BMP10/ALK1/BMPR2 signalling pathway in PAH pathobiology [10,12,20]. Both in vitro and in vivo studies propose a model where BMP9 and BMP10 act interchangeably and with compensatory functionality via ALK1 to regulate endothelial cell migration and proliferation, maintaining endothelial quiescence [44,45]. Based on evidence of sequence homology, mature peptide structural similarities and reported functional redundancy of these gene products, it is proposed that the BMP10 missense variant reported here may have a similar pathogenic mechanism to those observed in GDF2; further exploration on both the genetic and functional level is now warranted.
PDGFD missense variants were recently associated with PAH pathogenesis in a case-control analysis of a large, combined cohort [14]. The identification of a novel c.550G>A (p.Glu184Lys) variant of this gene in the cPAH panel analysed here serves to establish PDGFD as a contributing factor to childhood disease and provides independent validation of the discovery study. PDGF family signalling pathways play important roles in vascular pathologies, including mediation of remodelling processes that are known to occur in PAH [46]. The plausibility of this gene contributing to PAH risk is further enhanced by cell-type-specific gene expression studies using single cell RNA-seq data, demonstrating high expression in endothelial cells within heart and lung tissue, a key site of disease potentiation in PAH [14]. Of interest, PDGFD presents an exciting drug-targeting candidate, with studies of the tyrosine kinase inhibitor imatinib, a known pharmacological inhibitor of PDGF signalling, demonstrating 15-fold reductions in PDGFD expression in cardiac tissue [47] and suppression of medial thickening via receptor inhibition in patient-derived in vitro PASMC models [48].
The novel c.1069G>A (p.Val357Ile) missense variant identified in ABCC8 describes an amino acid substitution in the highly conserved transmembrane domain 1 of the mature and functionally important SUR1 product. This finding supports previous investigations that indicate pathogenic variation in ABCC8 underlies 2–5% of PAH cases, including 10 previous reports of paediatric-onset cases [8,12,24]. The gene encodes the SUR1 subunit of KATP potassium channels located in cardiac smooth muscle cells. Of note, the variant identified in this study was previously reported to cause hyperinsulinism in compound heterozygosity with a second ABCC8 allele, although without any clinical signs of PAH [49]. Whole cell electrophysiology and radium efflux assays of reported missense variants have demonstrated loss of KATP channel activity [24]. Taken together, these data suggest that loss of SUR1-dependent channel function represents a pathogenic mechanism in PAH, although the primary physiological role remains unknown. These findings are of clinical pertinence, as ABCC8 is considered a possible therapeutic target wherein impaired KATP activity could be pharmacologically rescued in vitro using diazoxide, a selective potassium-channel opening drug [24]. This may have further implications for the development of diazoxide-induced pulmonary hypertension in children with hyperinsulinaemic hypoglycaemia [50,51].
The SMAD9 c.1117G>A (p.Val373Ile) variant detected in an HPAH case in this study is predicted to be deleterious by in silico analysis, further supported by familial co-segregation and its location with the C-terminal Mad Homology 2 (MH2) domain, a region of extensive evolutionary conservation. SMAD1, SMAD4 and SMAD9 encode signalling intermediaries that act as downstream mediators of the BMPR2 pathway. Variants in SMAD9 were previously identified in three patients [17,52,53], and were recently validated in both adult-onset and paediatric PAH [12,20,22]. Functional analyses of the impact of SMAD9 variants on SMAD-mediated signalling demonstrated impaired responses to ligand stimulation and reduced transcriptional activation of the downstream BMP target gene, ID2 [17,52]. Although this variant was deemed a VUS based on our minor allele frequency threshold boundary, it is noteworthy that it was previously detected in an independent patient with PAH. This speaks to the challenges of setting an appropriate MAF cut-off when investigating rare diseases, especially in the context of notable reduced penetrance. Whereas most studies consider an approximation of disease allele frequency equivalent to population disease prevalence, inheritance-based calculations derived from Hardy–Weinberg equilibrium with adjustment for nonpenetrant alleles may be a more appropriate method to avoid false negative findings.
5. Conclusions
Taken together, these findings provide data supporting wider locus heterogeneity of paediatric PAH than previously reported and expand the allelic series of variation in known cPAH genetic risk factors. This study supports the inclusion of these genes in screening panels in future work for diagnostic surveillance of paediatric cases. In particular, we provide independent validation of the recently identified risk factors BMP10 and PDGFD, and the first report of an ATP13A3 variant in a paediatric case, warranting further analysis of understudied cellular pathways key to PAH pathogenesis. Notably, although novel gene detection was beyond the scope of this study in the context of extensive locus heterogeneity, this report emphasises the importance of the rigorous analysis of well-defined case cohorts to delineate the molecular basis of cPAH as a framework for future gene discovery initiatives.
Author Contributions
Conceptualization, R.D.M. and L.S.; formal analysis, S.M.G. and M.A.K.; funding acquisition, R.C.T., R.D.M. and L.S.; investigation, S.M.G., C.E.B., R.M.F.B., R.C.T., R.D.M. and L.S.; resources, R.M.F.B., R.C.T. and L.S.; visualization, S.M.G. and L.S.; writing—original draft preparation, S.M.G.; writing—review and editing, R.D.M. and L.S. All authors have read and agreed to the published version of the manuscript.
Funding
The study was funded by a British Heart Foundation programme grant (RG/08/006/25302) to R.C.T. and a pilot research grant from the Molecular and Clinical Sciences Research Institute at St George’s, University of London (L.S. and R.D.M.). L.S. is supported by the Wellcome Trust Institutional Strategic Support Fund (204809/Z/16/Z) awarded to St. George’s, University of London.
Conflicts of Interest
The University Medical Center Groningen contracts with Actelion, GSK and Lilly for steering committee and advisory board activities of Berger. All other authors declare no conflicts of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Machado R.D., Eickelberg O., Elliott C.G., Geraci M.W., Hanaoka M., Loyd J.E., Newman J.H., Phillips J.A., Soubrier F., Trembath R.C., et al. Genetics and genomics of pulmonary arterial hypertension. J. Am. Coll. Cardiol. 2009;54:S32–S42. doi: 10.1016/j.jacc.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machado R.D., Aldred M.A., James V., Harrison R.E., Patel B., Schwalbe E.C., Gruenig E., Janssen B., Koehler R., Seeger W., et al. Mutations of the TGF-beta type II receptor BMPR2 in pulmonary arterial hypertension. Hum. Mutat. 2006;27:121–132. doi: 10.1002/humu.20285. [DOI] [PubMed] [Google Scholar]
- 3.O’Callaghan D.S., Humbert M. A critical analysis of survival in pulmonary arterial hypertension. Eur. Respir. Rev. 2012;21:218–222. doi: 10.1183/09059180.00003512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Loon R.L.E., Roofthooft M.T.R., Hillege H.L., ten Harkel A.D.J., van Osch-Gevers M., Delhaas T., Kapusta L., Strengers J.L.M., Rammeloo L., Clur S.-A.B., et al. Pediatric pulmonary hypertension in the Netherlands: Epidemiology and characterization during the period 1991 to 2005. Circulation. 2011;124:1755–1764. doi: 10.1161/CIRCULATIONAHA.110.969584. [DOI] [PubMed] [Google Scholar]
- 5.Li L., Jick S., Breitenstein S., Hernandez G., Michel A., Vizcaya D. Pulmonary arterial hypertension in the USA: An epidemiological study in a large insured pediatric population. Pulm. Circ. 2017;7:126–136. doi: 10.1086/690007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humbert M., Sitbon O., Chaouat A., Bertocchi M., Habib G., Gressin V., Yaici A., Weitzenblum E., Cordier J.-F., Chabot F., et al. Pulmonary arterial hypertension in France: Results from a national registry. Am. J. Respir. Crit. Care Med. 2006;173:1023–1030. doi: 10.1164/rccm.200510-1668OC. [DOI] [PubMed] [Google Scholar]
- 7.Ivy D. Pulmonary hypertension in children. Cardiol. Clin. 2016;34:451–472. doi: 10.1016/j.ccl.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu N., Gonzaga-Jauregui C., Welch C.L., Ma L., Qi H., King A.K., Krishnan U., Rosenzweig E.B., Ivy D.D., Austin E.D., et al. Exome sequencing in children with pulmonary arterial hypertension demonstrates differences compared with adults. Circ. Genom. Precis. Med. 2018;11:e001887. doi: 10.1161/CIRCGEN.117.001887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chida A., Shintani M., Matsushita Y., Sato H., Eitoku T., Nakayama T., Furutani Y., Hayama E., Kawamura Y., Inai K., et al. Mutations of NOTCH3 in childhood pulmonary arterial hypertension. Mol. Genet. Genom. Med. 2014;2:229–239. doi: 10.1002/mgg3.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gräf S., Haimel M., Bleda M., Hadinnapola C., Southgate L., Li W., Hodgson J., Liu B., Salmon R.M., Southwood M., et al. Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat. Commun. 2018;9:1416. doi: 10.1038/s41467-018-03672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrell N.W., Aldred M.A., Chung W.K., Elliott C.G., Nichols W.C., Soubrier F., Trembath R.C., Loyd J.E. Genetics and genomics of pulmonary arterial hypertension. Eur. Respir. J. 2019;53:1801899. doi: 10.1183/13993003.01899-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu N., Pauciulo M.W., Welch C.L., Lutz K.A., Coleman A.W., Gonzaga-Jauregui C., Wang J., Grimes J.M., Martin L.J., He H., et al. Novel risk genes and mechanisms implicated by exome sequencing of 2572 individuals with pulmonary arterial hypertension. Genome Med. 2019;11:69. doi: 10.1186/s13073-019-0685-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Southgate L., Machado R.D., Gräf S., Morrell N.W. Molecular genetic framework underlying pulmonary arterial hypertension. Nat. Rev. Cardiol. 2020;17:85–95. doi: 10.1038/s41569-019-0242-x. [DOI] [PubMed] [Google Scholar]
- 14.Zhu N., Swietlik E.M., Welch C.L., Pauciulo M.W., Hagen J.J., Zhou X., Guo Y., Karten J., Pandya D., Tilly T., et al. Rare variant analysis of 4241 pulmonary arterial hypertension cases from an international consortium implicate FBLN2, PDGFD and rare de novo variants in PAH. bioRxiv. 2020:550327. doi: 10.1101/2020.05.29.124255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trembath R.C., Thomson J.R., Machado R.D., Morgan N.V., Atkinson C., Winship I., Simonneau G., Galie N., Loyd J.E., Humbert M., et al. Clinical and molecular genetic features of pulmonary hypertension in patients with hereditary hemorrhagic telangiectasia. N. Engl. J. Med. 2001;345:325–334. doi: 10.1056/NEJM200108023450503. [DOI] [PubMed] [Google Scholar]
- 16.Harrison R.E., Berger R., Haworth S.G., Tulloh R., Mache C.J., Morrell N.W., Aldred M.A., Trembath R.C. Transforming growth factor-beta receptor mutations and pulmonary arterial hypertension in childhood. Circulation. 2005;111:435–441. doi: 10.1161/01.CIR.0000153798.78540.87. [DOI] [PubMed] [Google Scholar]
- 17.Shintani M., Yagi H., Nakayama T., Saji T., Matsuoka R. A new nonsense mutation of SMAD8 associated with pulmonary arterial hypertension. J. Med. Genet. 2009;46:331–337. doi: 10.1136/jmg.2008.062703. [DOI] [PubMed] [Google Scholar]
- 18.Chida A., Shintani M., Nakayama T., Furutani Y., Hayama E., Inai K., Saji T., Nonoyama S., Nakanishi T. Missense mutations of the BMPR1B (ALK6) gene in childhood idiopathic pulmonary arterial hypertension. Circ. J. 2012;76:1501–1508. doi: 10.1253/circj.CJ-11-1281. [DOI] [PubMed] [Google Scholar]
- 19.Levy M., Eyries M., Szezepanski I., Ladouceur M., Nadaud S., Bonnet D., Soubrier F. Genetic analyses in a cohort of children with pulmonary hypertension. Eur. Respir. J. 2016;48:1118–1126. doi: 10.1183/13993003.00211-2016. [DOI] [PubMed] [Google Scholar]
- 20.Wang X.-J., Lian T.-Y., Jiang X., Liu S.-F., Li S.-Q., Jiang R., Wu W.-H., Ye J., Cheng C.-Y., Du Y., et al. Germline BMP9 mutation causes idiopathic pulmonary arterial hypertension. Eur. Respir. J. 2019;53:1801609. doi: 10.1183/13993003.01609-2018. [DOI] [PubMed] [Google Scholar]
- 21.Kerstjens-Frederikse W.S., Bongers E.M.H.F., Roofthooft M.T.R., Leter E.M., Douwes J.M., Van Dijk A., Vonk-Noordegraaf A., Dijk-Bos K.K., Hoefsloot L.H., Hoendermis E.S., et al. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J. Med. Genet. 2013;50:500–506. doi: 10.1136/jmedgenet-2012-101152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu N., Welch C.L., Wang J., Allen P.M., Gonzaga-Jauregui C., Ma L., King A.K., Krishnan U., Rosenzweig E.B., Ivy D.D., et al. Rare variants in SOX17 are associated with pulmonary arterial hypertension with congenital heart disease. Genome Med. 2018;10:56. doi: 10.1186/s13073-018-0566-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hiraide T., Kataoka M., Suzuki H., Aimi Y., Chiba T., Kanekura K., Satoh T., Fukuda K., Gamou S., Kosaki K. SOX17 mutations in Japanese patients with pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2018;198:1231–1233. doi: 10.1164/rccm.201804-0766LE. [DOI] [PubMed] [Google Scholar]
- 24.Bohnen M.S., Ma L., Zhu N., Qi H., McClenaghan C., Gonzaga-Jauregui C., Dewey F.E., Overton J.D., Reid J.G., Shuldiner A.R., et al. Loss-of-function ABCC8 mutations in pulmonary arterial hypertension. Circ. Genom. Precis. Med. 2018;11:e002087. doi: 10.1161/CIRCGEN.118.002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eyries M., Montani D., Nadaud S., Girerd B., Levy M., Bourdin A., Trésorier R., Chaouat A., Cottin V., Sanfiorenzo C., et al. Widening the landscape of heritable pulmonary hypertension mutations in paediatric and adult cases. Eur. Respir. J. 2019;53:1801371. doi: 10.1183/13993003.01371-2018. [DOI] [PubMed] [Google Scholar]
- 26.Simonneau G., Montani D., Celermajer D.S., Denton C.P., Gatzoulis M.A., Krowka M., Williams P.G., Souza R. Haemodynamic definitions and updated clinical classification of pulmonary hypertension. Eur. Respir. J. 2019;53:1801913. doi: 10.1183/13993003.01913-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haarman M.G., Kerstjens-Frederikse W.S., Vissia-Kazemier T.R., Breeman K.T.N., Timens W., Vos Y.J., Roofthooft M.T.R., Hillege H.L., Berger R.M.F. The genetic epidemiology of pediatric pulmonary arterial hypertension. J. Pediatr. 2020;225:65–73.e5. doi: 10.1016/j.jpeds.2020.05.051. [DOI] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013;76:7.20.1–7.20.41. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 35.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson J.T., Thorvaldsdóttir H., Wenger A.M., Zehir A., Mesirov J.P. Variant Review with the Integrative Genomics Viewer. Cancer Res. 2017;77:e31–e34. doi: 10.1158/0008-5472.CAN-17-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kühlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 39.Madan M., Patel A., Skruber K., Geerts D., Altomare D.A., Iv O.P. ATP13A3 and caveolin-1 as potential biomarkers for difluoromethylornithine-based therapies in pancreatic cancers. Am. J. Cancer. Res. 2016;6:1231–1252. [PMC free article] [PubMed] [Google Scholar]
- 40.Sørensen D.M., Holemans T., van Veen S., Martin S., Arslan T., Haagendahl I.W., Holen H.W., Hamouda N.N., Eggermont J., Palmgren M., et al. Parkinson disease related ATP13A2 evolved early in animal evolution. PLoS ONE. 2018;13:e0193228. doi: 10.1371/journal.pone.0193228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerche M., Eichstaedt C.A., Hinderhofer K., Grünig E., Tausche K., Ziemssen T., Halank M., Wirtz H., Seyfarth H.-J. Mutually reinforcing effects of genetic variants and interferon-β 1a therapy for pulmonary arterial hypertension development in multiple sclerosis patients. Pulm. Circ. 2019;9:2045894019872192. doi: 10.1177/2045894019872192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hodgson J., Swietlik E.M., Salmon R.M., Hadinnapola C., Nikolic I., Wharton J., Guo J., Liley J., Haimel M., Bleda M., et al. Characterization of GDF2 Mutations and Levels of BMP9 and BMP10 in Pulmonary Arterial Hypertension. Am. J. Respir. Crit. Care Med. 2020;201:575–585. doi: 10.1164/rccm.201906-1141OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David L., Mallet C., Mazerbourg S., Feige J.-J., Bailly S. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood. 2007;109:1953–1961. doi: 10.1182/blood-2006-07-034124. [DOI] [PubMed] [Google Scholar]
- 44.Ricard N., Ciais D., Levet S., Subileau M., Mallet C., Zimmers T.A., Lee S.-J., Bidart M., Feige J.-J., Bailly S. BMP9 and BMP10 are critical for postnatal retinal vascular remodeling. Blood. 2012;119:6162–6171. doi: 10.1182/blood-2012-01-407593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tillet E., Bailly S. Emerging roles of BMP9 and BMP10 in hereditary hemorrhagic telangiectasia. Front. Genet. 2014;5:456. doi: 10.3389/fgene.2014.00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Folestad E., Kunath A., Wågsäter D. PDGF-C and PDGF-D signaling in vascular diseases and animal models. Mol. Asp. Med. 2018;62:1–11. doi: 10.1016/j.mam.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Burke M.J., Walmsley R., Munsey T.S., Smith A.J. Receptor tyrosine kinase inhibitors cause dysfunction in adult rat cardiac fibroblasts in vitro. Toxicol. In Vitro. 2019;58:178–186. doi: 10.1016/j.tiv.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 48.Morii C., Tanaka H.Y., Izushi Y., Nakao N., Yamamoto M., Matsubara H., Kano M.R., Ogawa A. 3D in vitro model of vascular medial thickening in pulmonary arterial hypertension. Front. Bioeng. Biotechnol. 2020;8:482. doi: 10.3389/fbioe.2020.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melikyan M., Kareva M., Petraykina E., Averyanova J., Christesen H. Hypoglycaemia in children [abstract]; Proceedings of the 51st Annual Meeting of the European Society for Paediatric Endocrinology (ESPE); Leipzig, Germany. 20–23 September 2012. [Google Scholar]
- 50.Timlin M.R., Black A.B., Delaney H.M., Matos R.I., Percival C.S. Development of pulmonary hypertension during treatment with diazoxide: A case series and literature review. Pediatr. Cardiol. 2017;38:1247–1250. doi: 10.1007/s00246-017-1652-3. [DOI] [PubMed] [Google Scholar]
- 51.Chen S.C., Dastamani A., Pintus D., Yau D., Aftab S., Bath L., Swinburne C., Hunter L., Giardini A., Christov G., et al. Diazoxide-induced pulmonary hypertension in hyperinsulinaemic hypoglycaemia: Recommendations from a multicentre study in the United Kingdom. Clin. Endocrinol. 2019;91:770–775. doi: 10.1111/cen.14096. [DOI] [PubMed] [Google Scholar]
- 52.Nasim M.T., Ogo T., Ahmed M., Randall R., Chowdhury H.M., Snape K.M., Bradshaw T.Y., Southgate L., Lee G.J., Jackson I., et al. Molecular genetic characterization of SMAD signaling molecules in pulmonary arterial hypertension. Hum. Mutat. 2011;32:1385–1389. doi: 10.1002/humu.21605. [DOI] [PubMed] [Google Scholar]
- 53.Drake K.M., Zygmunt D., Mavrakis L., Harbor P., Wang L., Comhair S.A., Erzurum S.C., Aldred M.A. Altered MicroRNA processing in heritable pulmonary arterial hypertension: An important role for Smad-8. Am. J. Respir. Crit. Care Med. 2011;184:1400–1408. doi: 10.1164/rccm.201106-1130OC. [DOI] [PMC free article] [PubMed] [Google Scholar]