SUMMARY:
Excessive levels of saturated fatty acids are toxic to cells, although the basis for this lipotoxicity remains incompletely understood. Here we analyzed the transcriptome, lipidome, and genetic interactions of human leukemia cells exposed to palmitate. Palmitate treatment increased saturated glycerolipids, accompanied by a transcriptional stress response, including upregulation of the ER stress response. A comprehensive genome-wide shRNA screen identified >350 genes modulating lipotoxicity. Among previously unknown genetic modifiers of lipotoxicity, depletion of RNF213, a putative ubiquitin ligase mutated in Moyamoya vascular disease, protected from lipotoxicity. On a broader level, integration of our comprehensive datasets revealed that changes of di-saturated glycerolipids, but not other lipid classes, are central to lipotoxicity in this model. Consistent with this, inhibition of ER-localized glycerol-phosphate acyltransferase activity protected from all aspects of lipotoxicity. Identification of genes modulating the response to saturated fatty acids may reveal novel therapeutic strategies for treating metabolic diseases linked to lipotoxicity.
Keywords: lipotoxicity, palmitate, saturated fatty acid, glycerolipid, Moyamoya disease
eTOC blurb:
Cellular lipotoxicity due to saturated fatty acids causes cell death and is thought to be a root cause of metabolic diseases. Piccolis, Bond et al. use unbiased analyses to reveal hundreds of genes modulating cellular palmitate lipotoxicity and implicate saturated glycerolipids as a causative factor.
INTRODUCTION
Lipid accumulation interferes with the normal functions of cells and tissues, a condition referred to as lipotoxicity (Brookheart et al., 2009; Shimabukuro et al., 1998). Lipotoxicity is thought to be an underlying cause for many metabolic diseases. For example, in lipodystrophy, lipids build up in tissues other than the adipose tissue, and in obesity, adipose cells are overwhelmed by lipid accumulation (Nagle et al., 2009; Shimabukuro et al., 1998). Toxic lipids in skeletal muscle, heart, liver, and pancreatic β-cells lead to obesity-associated diseases (Brookheart et al., 2009; Shimabukuro et al., 1998).
Saturated fatty acids, such as palmitate (C16:0), are particularly toxic to cells. Increased palmitate concentrations induce apoptosis (Paumen et al., 1997). Various factors have been implicated in palmitate-mediated cellular toxicity, including ceramides (Turpin et al., 2006), reactive oxygen species (Gao et al., 2010), endoplasmic reticulum (ER) stress, (Borradaile et al., 2006; Wei et al., 2006), and snoRNAs (Michel et al., 2011). Although genetic screens have been performed (Michel et al., 2011), comprehensive, global analyses of cellular toxicity due to saturated fatty acids are lacking. Saturated fatty acids have diverse fates, including incorporation into membrane lipids, storage in lipid droplets, serving as protein modifiers, or mitochondrial β-oxidation, and thus, multiple pathways might mediate or modulate palmitate toxicity in cells.
To unravel the contributors to palmitate-mediated toxicity, we combined complementary, unbiased approaches to examine the transcriptome, lipidome, and genetic modifiers of the response to saturated fatty acids in a cell-based model of lipotoxicity. Our results provide rich resources for investigating causative mechanisms for lipotoxicity and for identifying new drug targets. Highlighting this, we identify a putative E3 ligase and ER-localized glycerol-phosphate acyltransferase (GPAT) enzymes as central gatekeepers of cellular lipotoxicity.
RESULTS
Determining the Lipidome and Transcriptome of Palmitate-Induced Lipotoxicity
To probe the mechanisms of lipotoxicity in an unbiased manner, we first established a cellular model for palmitate lipotoxicity. Among several cell lines tested, we chose human leukemic K562 cells because they exhibit palmitate-induced lipotoxicity (Figure 1A) and are readily amenable to systematic genome perturbation (Bassik et al., 2013).
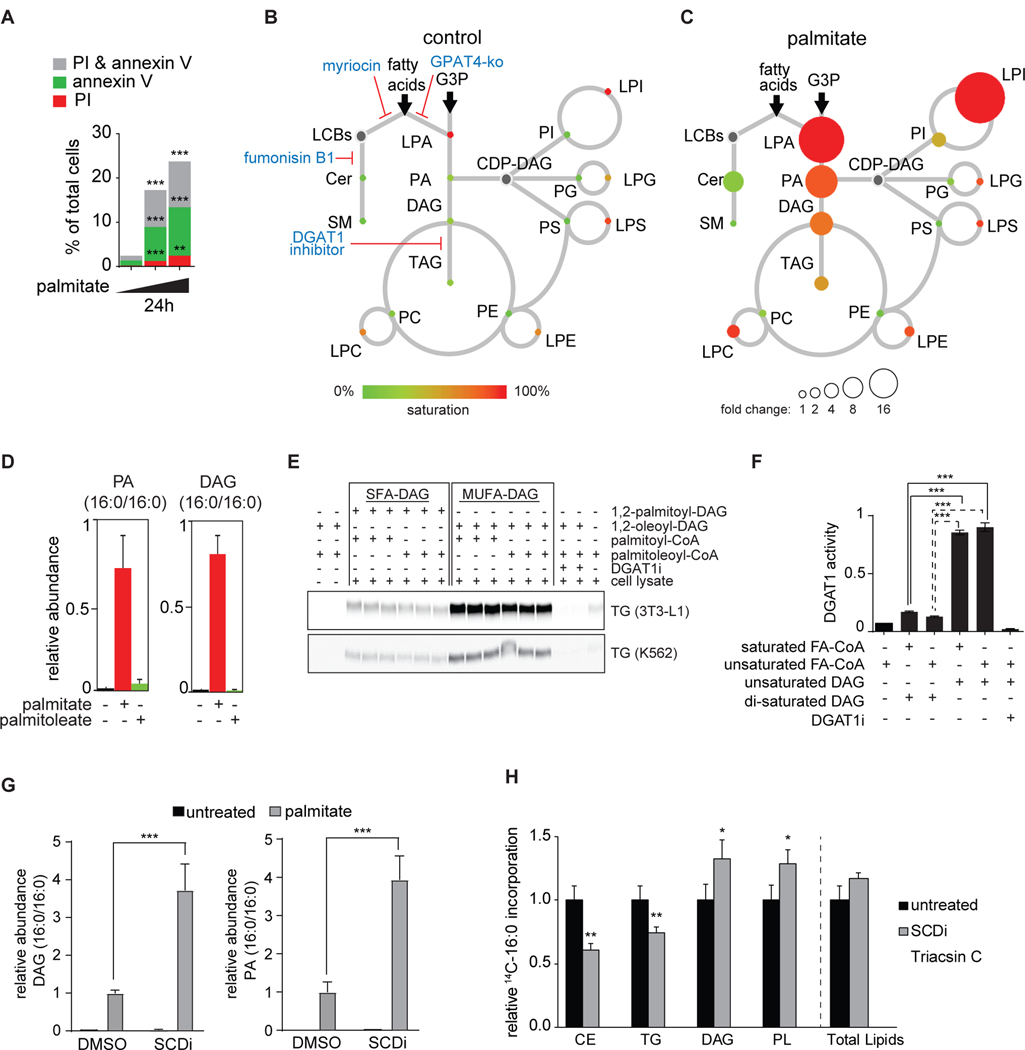
Figure 1. Lipidome of Palmitate-Induced Lipotoxicity in Human K562 Leukemia cells.
(A) Palmitate induces apoptotic cell death in K562 cells. Cell viability assay of K562 cells treated with increasing concentration of palmitate (0, 0.2 and 0.25 mM) for 24 h. Apoptotic cells were identified by propidium iodide (PI) and annexin V staining. n=3 for each treatment. **p < 0.01; ***p < 0.001.
(B) Lipidome of K562 cells in basal conditions. The scheme shows the relative levels of incorporation of exogenous fatty acids into sphingolipids and glycerophospholipids. Lipid classes identified by LC-MS2 analysis are presented as color-coded circles. The lipid species was designated as saturated if all of its fatty acid chains were saturated, or unsaturated if it had at least one unsaturated fatty acid chain. The percentage of saturated lipid species is shown for each class from green (low saturation) to red (high saturation). Lipid classes not identified are shown in grey. The size of the circles is set to the arbitrary unit of 1 for the control cells. G3P: glycerol-3-phosphate; LPA: lyso-phosphatidic acids; PA: phosphatidic acids; DAG: diacylglycerol; TAG: triacylglycerol; PC: phosphatidylcholine: PE: phosphatidylethanolamine; LPE: lyso-phosphatidylethanolamine; LPC: lyso-phosphatidylcholine; PS: phosphatidylserine; LPS: lyso-phosphatidylserine; PI: phosphatidylinositol; LPI: lyso-phosphatidylinositol; PG: phosphatidylglycerol; LPG: lyso-phosphatidylglycerol; Cer: ceramide; SM: sphingomyelin; LCB: long-chain base; CDP: cytidine diphosphate.
(C) As in Figure 1B, the lipidome of K562 cells treated with 0.2 mM palmitate for 20 h. The size of the circle is proportional to the change in abundance relative to the control sample. The complete dataset is provided in Table 1 “Lipidomics data”.
(D) Palmitate, but not palmitoleate, increases the number of di-saturated lipid species. Relative quantification for phosphatidic acid (PA, left panel) and diacylglycerol (DAG, right panel) identified by LC-MS2. K562 cells treated with control, 0.2 mM palmitate or 0.2 palmitoleate for 20 h. Lipid species changing the most upon palmitate treatment is shown for each class. n=3 for each treatment.
(E) Di-saturated diacylglycerol does not sustain DGAT1 activity in vitro. DGAT1 enzymatic activities were measured at Vmax in total cell lysate from differentiated 3T3-l1 adipocytes (upper panel) and K562 cells (lower panel). Each reaction contained 1,2-dioleoyl-glycerol or 1,2-dipalmitoyl-glycerol as diacyl-glycerol source. The DGAT1 inhibitor (15uM) was included as a control.
(F) Quantification of the TLC plates shown in E. n=3 for each treatment; ***p < 0.001.
(G) SCD inhibition (4uM) increases the accumulation of di-saturated PA and DAG compared to DMSO control cells. Relative quantification for phosphatidic acid (PA, left panel) and diacylglycerol (DAG, right panel) identified by LC-MS2. K562 cells treated with control or 0.2 mM palmitate. n=4 for each treatment. ***p < 0.001.
(H) Decreased partitioning of 14C-palmitate towards TG and CE. Cells were treated for 6 h with 0.2 mM nonradiolabeled palmitate and 0.15uCi 14C-palmitate. Triacsin C (10uM, an inhibitor of ACSL1, ACSL3 and ACSL4) was added to confirm ACSL-specific fatty acid uptake. Untreated and triascin C values are also shown in Figure 4F. n=5–6 for each treatment. *p < 0.05; **p < 0.01.
See also Figures S1.
To better understand how lipid metabolism is affected by palmitate toxicity, we measured the lipidome response of K562 cells to treatment with palmitate (complexed with albumin). In contrast to the lipidome of cells cultured under basal conditions, we found large increases in the relative amounts and saturation levels of many glycerolipids, including phosphatidic acid (PA), diacylglycerol (DAG), triacylglycerol (TG), and numerous lysophospholipids (Figure 1B, 1C and Table S1). In particular, many di-saturated glycerolipid species accumulated (Figure 1D). Typically, glycerolipids are saturated in the sn-1 position, owing to the substrate preference of GPAT enzymes (Haldar, 1994) and unsaturated in the sn-2 position, owing to the preferences of acylglycerol acyltransferase (AGPAT) enzymes (Agarwal et al., 2011; Prasad et al., 2011), as well as glycerophospholipid remodeling (Ellingson et al., 1970; Shindou et al., 2013). Accumulation of di-saturated glycerolipid species in response to excess palmitate is likely due to impaired flux of substrates for glycerolipid synthesis enzymes, as di-saturated PA is a poor substrate for lipin (a PA hydrolase) (Han and Carman, 2010). Additionally, we found that di-saturated diacylglycerol is a poor substrate for DGAT enzymes and TG synthesis in K562 cells and in 3T3-L1 fibroblasts (Figure 1E, 1F).
The hypothesis that di-saturated glycerolipids are toxic predicts that increasing the content of unsaturated fatty acids, particularly in the sn-2 position of the glycerol backbone, will prevent palmitate toxicity. Indeed, and in agreement with previous studies (Cimen et al., 2016; Eitel et al., 2002; Robblee et al., 2016), we found that adding palmitoleate (C16:1n7) with palmitate suppressed cellular toxicity, mitigated ER stress, increased the amounts of TG in cellular lipid droplets, and suppressed accumulation of di-saturated glycerolipids (Figure S1). Conversely, inhibition of SCD-1, the major Δ9 desaturase responsible for oxidizing palmitate (Strittmatter et al., 1974), worsened palmitate toxicity (Figure S1E). Highlighting the importance of fatty acid desaturation as a means of detoxification, SCD1 inhibition exacerbated the accumulation of di-saturated DAG and di-saturated PA (Figure 1G) and prevented the incorporation of radiolabeled palmitate into TG (Figure 1H), consistent with a previous report (Masuda et al., 2015).
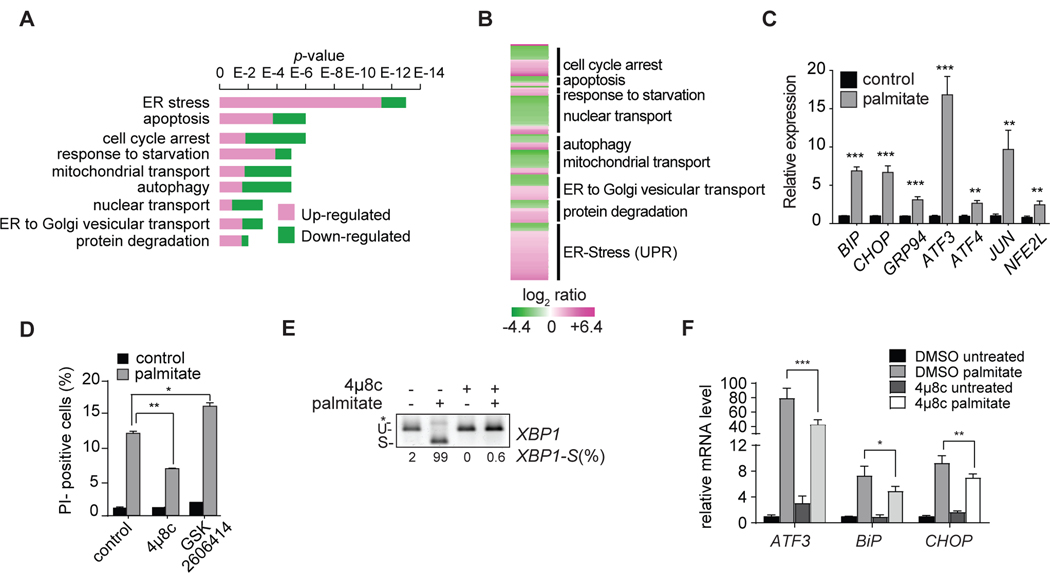
To determine the transcriptional response to saturated fatty acid exposure, we measured changes in gene expression with RNAseq methodology. Palmitate treatment resulted in widespread up-regulation of a host of cellular stress responses, including those related to membrane transport, autophagy, apoptosis, and the ER stress response (known as the unfolded protein response (UPR); Figures 2A, 2B). As suggested by previous studies (Borradaile et al., 2006; Kitai et al., 2013; Robblee et al., 2016), we found that numerous specific genes of ER stress were induced by palmitate lipotoxicity (Figure 2C). The complete transcriptome changes due to palmitate lipotoxicity are presented in Table S2. To test the hypothesis that cellular lipotoxicity is mediated by ER stress in K562 cells, we inhibited different branches of the UPR. Inhibiting the IRE1 signaling pathway (with the inhibitor 4μ8c) protected cells from palmitate-mediated lipotoxicity on K562 cells (Figure 2D and (Robblee et al., 2016), prevented splicing of XBP1 mRNA (Figure 2E), and attenuated induction of ER stress-response genes (Figure 2F). In contrast, inhibiting the PERK branch (with GSK2606414) appeared to aggravate the detrimental effects of palmitate (Figures S2 and 2D).
Figure 2. The Transcriptome of Palmitate-Induced Lipotoxicity in Human K562 Leukemia Cells.
(A) RNAseq analysis of genes expressed in K562 cells in response to palmitate versus control. The figure shows the p-values corresponding to the categories of genes shown in Figure 2B. Genes upregulated by palmitate are shown in magenta, and genes downregulated by palmitate are shown in green (p < 0.005). GO term enrichment has been performed using ClueGo in Cytoscape.
(B) RNAseq analysis of genes expressed in K562 cells in response to palmitate. Indicated are gene-expression categories as revealed by ClueGO analysis. Genes upregulated by palmitate are shown in magenta, and genes downregulated by palmitate are shown in green (p < 0.005). The complete dataset is provided in Table S2 “Gene expression profile data”.
(C) Increased activation of UPR target genes in K562 cells in response to palmitate. Relative mRNA quantification for main UPR target genes in basal (control) and treated (0.2 mM palmitate) cells. n=3 for each treatment. **p < 0.01; ***p < 0.001.
(D) Block of the IRE1 branch of the UPR protects K562 cells from palmitate-induced cell death. Propidium-iodide staining of K562 cells untreated or treated with 0.2 mM palmitate for 20 h. Cells were incubated with control vehicle or the IRE1 inhibitor 4μ8c (32 μM) or a PERK inhibitor GSK2606414 (15 nM) for 2 h before the incubation. n=3 for each treatment. *p < 0.05; **p < 0.01.
(E) Treatment with IRE1 inhibitor, 4μ8c, blocks palmitate-induced cleavage of XBP1 mRNA. U, unspliced; S, spliced; * unspecific band. XBP1 splicing is quantified as percentage of the spliced form over the total detected (XBP1-S(%)).
(F) IRE1 inhibition with 4μ8c mitigates induction of ER stress markers after palmitate treatment. n=3 for each treatment. *p < 0.05; **p < 0.01, ***p < 0.001;
See also Figures S2.
An shRNA Genome-Wide Screen Comprehensively Identifies Genes that Modify Palmitate-Induced Lipotoxicity
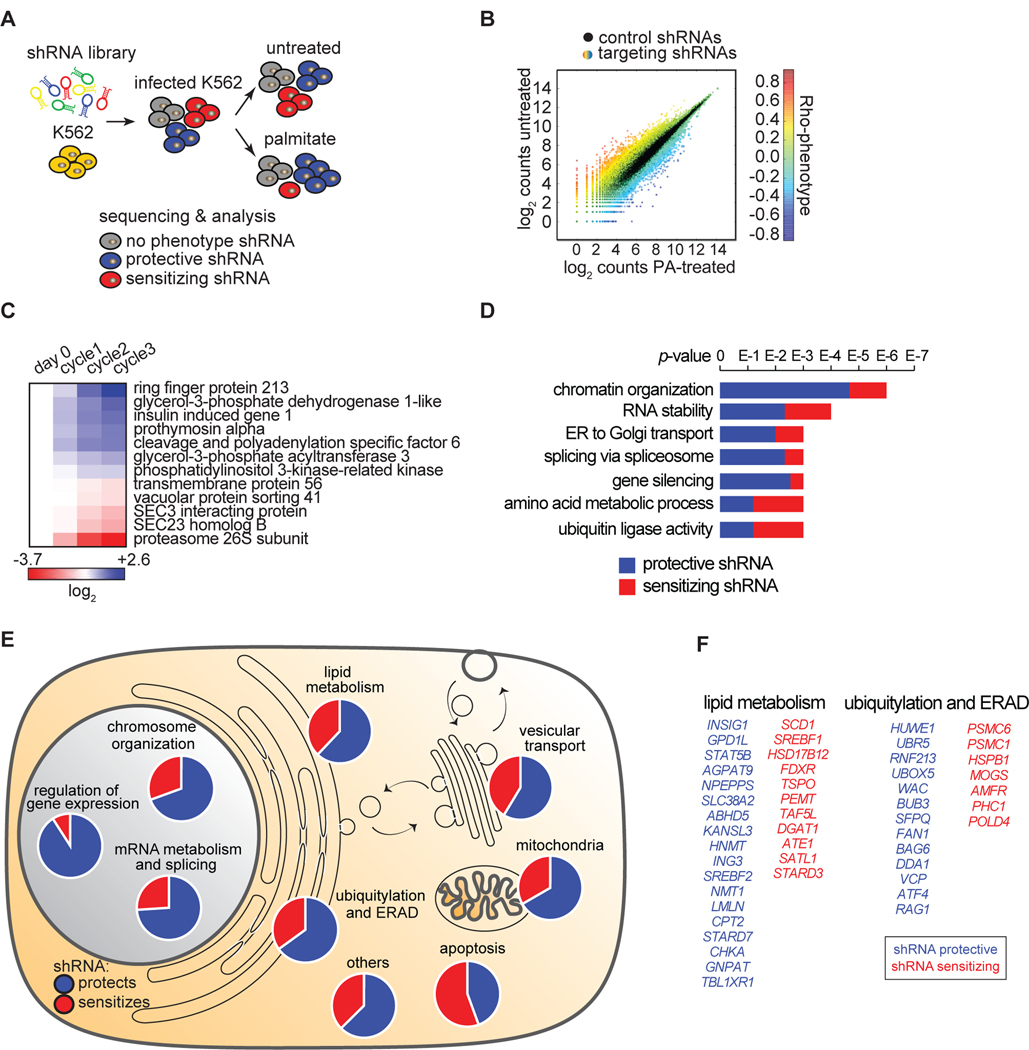
To identify the mechanisms mediating palmitate-induced cellular lipotoxicity, we used an unbiased, systematic genome perturbation screen with a high-coverage shRNA library (~25 shRNAs per gene) targeting more than 19,000 open reading frames (Bassik et al., 2013) (Figures 3A, S3A, S3B). Lipotoxic conditions were induced by treating K562 cells with 0.2 mM palmitate to achieve ~50% viability in 24 hours. This selection was repeated five times (see Materials and Methods), and treated and untreated (control) cells were compared. We determined the frequencies for each shRNA in the treated samples and controls by sequencing to obtain enrichment/de-enrichment ratios. For each gene, a phenotype (Rho) score was determined by comparing the frequency distributions of the targeting shRNAs (n ≥ 20) and control shRNAs (p <0.01) (Figure 3B and S3C). We independently performed the screen twice, and data from both biological replicates were highly correlated (Figure S3D). To validate the robustness of our analyses, we tested a subset of genes in an independent experimental design by challenging cells with fatty acids once and determining their growth by flow-cytometry analyses. The results showed a high degree of reproducibility with the original screen (Figure 3C, S3E).
Figure 3. Identification of Genes that Modify Lipotoxicity in K562 Human Leukemia Cells.
(A) Schematic of shRNA screen. K562 cells were infected with a complex lentiviral shRNA library (25 shRNAs/gene) as described in Methods. Cells were split into treated (0.2 mM palmitate) and untreated (control) samples. Upon five cycles of palmitate treatments, shRNAs were amplified by PCR and sequenced. Expected outcome: cells enriched in palmitate-treated sample (infected by protective shRNAs) are shown in blue, cells de-enriched in palmitate-treated sample (infected by sensitizing shRNAs) are shown in red, and cells equally present in treated and untreated samples (infected by shRNAs having no phenotype) are shown in grey.
(B) The screen output is shown. The absolute count for each shRNA from sequencing is shown as a dot. Each dot is the average among the biological replicate screens. Black dots are control shRNAs. Color-coded dots are targeting shRNA. The color varies according to the different phenotype.
(C) Screen candidates are confirmed by independent validation. The three shRNA-targeting sequences having the strongest phenotypes in the screen were cloned individually and then used to generate a stable knock-down for each of the 12 genes in the figure. To validate each candidate, we performed a growth competition assay between the knock-down cells (m-Cherry positive) and control cells (m-Cherry negative). The color code shows the enrichments in blue (for protective shRNAs) and de-enrichments in red (for sensitizing shRNAs) of knock-down cells compared to control cells upon three consecutive cycles of palmitate treatment. Flow cytometry was used to quantify each knockdown population. The variation, quantified after each individual palmitate treatment (cycle) compared to controls (day 0), is expressed as log2.
(D) Annotation of overrepresented terms identified among the screen candidates. Candidates are designated as genes that sensitize (red) or protect (blue) cells from palmitate-induced cell death. The analysis was performed using ClueGO in Cytoscape.
(E) Overview of screen results. The 355 genes targeted by the 234 protective shRNAs (in blue) and 121 sensitizing shRNAs (in red) are shown grouped by categories. Information on these genes is also presented in Table S3 “Screen result table”.
(F) Of the 355 genes identified, 29 genes were involved in lipid metabolism and 20 were involved in ubiquitylation and ERAD.
See also Figures S3.
Our screen yielded 121 hits in which mRNA depletion sensitized cells and 234 hits in which cells were protected from palmitate-induced cell death (Table S3). These hits spanned many cellular processes and compartments, including genes involved in mRNA metabolism, as reported (Caputa and Schaffer, 2016; Michel et al., 2011), transcription, membrane trafficking, protein degradation, and lipid metabolism (Figures 3D and 3E). Lists of the top protective and sensitizing genes for lipid metabolism and for ubiquitylation and ERAD are shown in Figure 3F.
SREBP1 Activation and Fatty Acid Desaturation Are Crucial for Protecting Cells from Palmitate Toxicity
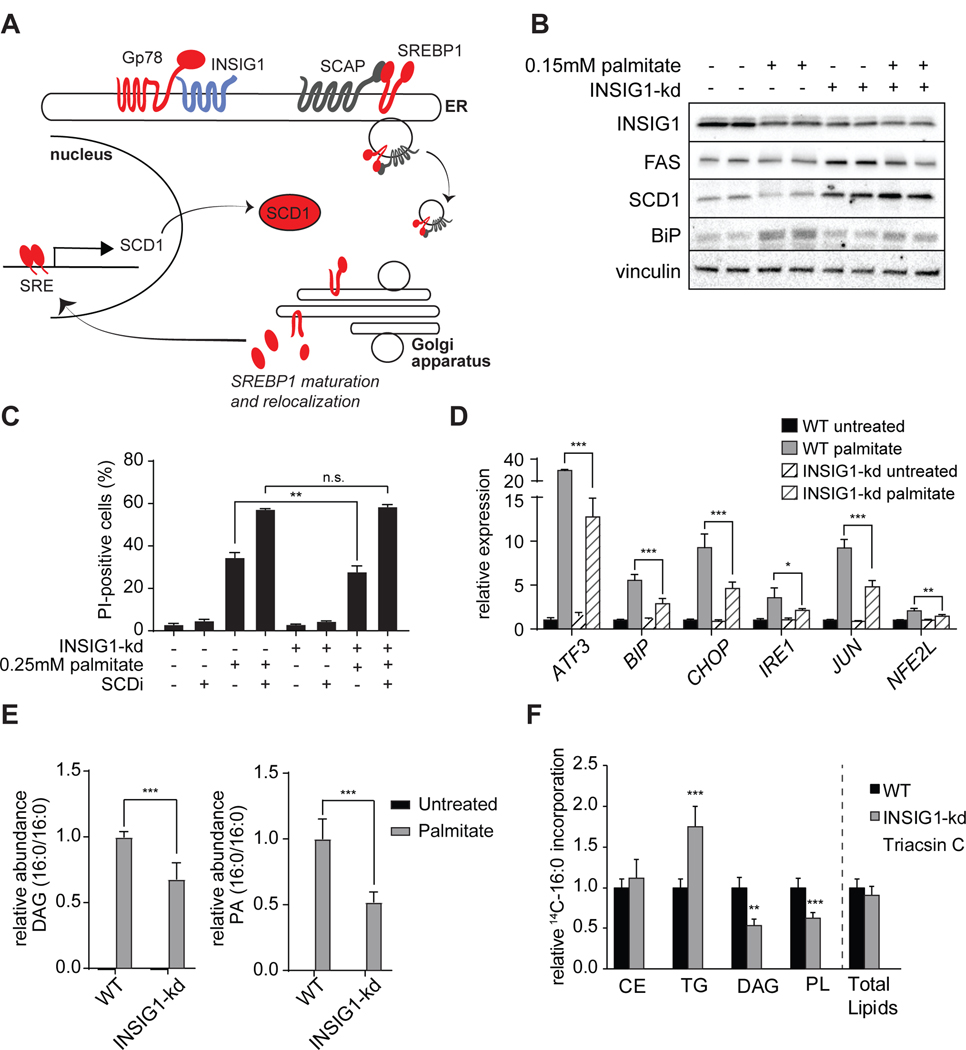
Mining our datasets, we analyzed the 29 genes of lipid metabolism (Figure 3E and 3F) that were identified by the shRNA screen. Among these were a number of genes expected as modifiers of palmitate lipotoxicity, including genes involved in the pathway of transcriptional control of lipid metabolism by the sterol-response element binding proteins (SREBP1, (Wang et al., 1993)). For instance, depletion of SREBP1, stearoyl-CoA desaturase 1 (SCD1), the primary Δ9 desaturase (oxidizing palmitate to palmitoleate) (Paton and Ntambi, 2009), and AMFR/Gp78 each sensitized the cells to palmitate (Figure 4A, S4A). Similarly, inhibition of SCD1 pharmacologically, as predicted (Green and Olson, 2011), led to sensitization to palmitate toxicity (Figure S1E). Depletion of each of these is predicted to decrease flux into the pathway of de novo fatty acid synthesis and desaturation, thus potentially crippling a homeostatic response of offsetting the increased levels of the saturated fatty acid palmitate with newly synthesized unsaturated fatty acids. On the other hand, our screen revealed that depletion of INSIG1, which should increase the flux through this pathway, protected cells (Figure 3C). In an independent experiment, INSIG1 deficiency (~45% knockdown) improved viability in response to palmitate treatment (Figure 4B and 4C). Further, INSIG1 knockdown blunted activation of ER stress markers (Figures 4B and 4D). Consistent with the role of INSIG1 in repressing transcriptional activation of SCD1, knockdown of INSIG1 elevated SCD1 protein levels (Figure 4B) and reduced the accumulation of di-saturated DAG and di-saturated PA (Figure 4E). To ensure that increased de novo lipogenesis did not cause reduced palmitate uptake as a contributing mechanism, we measured incorporation of radiolabeled palmitate into glycerolipids (Figure 4F). Total radiolabeled lipids were not altered between WT and INSIG1 knockdown cells. However, radiolabeled DAG was reduced and radiolabeled TG was elevated in INSIG1 knockdown cells. These results suggest that reduced toxic lipid accumulation in INSIG1 knockdown cells is due to improved detoxification rather than reductions in fatty acid uptake. Collectively, these results highlight the requirement for unsaturated fatty acids in relieving cellular toxicity, consistent with other reports (Flowers et al., 2008; Volmer et al., 2013). Because the generation of unsaturated fatty acids is important for the response to excess palmitate, likely other genes identified in the screen will ultimately prove to impact this pathway.
Figure 4. SREBP-Related Pathway Regulates Fatty Acid Desaturase and Palmitate-Induced Lipotoxicity.
(A) Schematic representation of the SREBP pathway in which GP78 degrades INSIG1 to free the SCAP-SREBP1 complex for trafficking from the ER to the Golgi apparatus, where cleavage liberates the SREBP1 transcription factor fragment from the membrane. This, in turn, leads to induction of target genes that have a sterol-response element (SRE) sequence in their promoter, such as SCD1. Aggravating hits are shown in red, protective hits are shown in blue.
(B) INSIG1 knockdown elevates FAS and SCD1 and reduces palmitate-induced upregulation of BIP. Western blot of WT and INSIG1-kd cells under basal condition or after palmitate treatment (0.15 mM, 20 h).
(C) INSIG1 knockdown modestly protects cells from palmitate-induced cell death, as measured by PI staining, and this protection is lost with SCD inhibition (4 μM). **p < 0.001.
(D) Unfolded protein response gene expression levels of WT and INSIG1-kd cells.
(E) Relative quantification for phosphatidic acid (PA, left panel) and diacylglycerol (DAG, right panel) in INISIG1-kd cells compared to WT cells before after after palmitate treatment, as identified by LC-MS2. K562 cells untreated or treated with 0.2 mM palmitate. n=3–4 for each treatment. ***p < 0.001.
(F) INSIG1-kd cells exhibit increased incorporation of radiolabeled palmitate into TG. Cells were treated for 6 h with 0.2 mM nonradiolabeled palmitate and 0.15μCi 14C-palmitate. Triacsin C (10uM, an inhibitor of ACSL1, ACSL3 and ACSL4) was added confirm ACSL-specific fatty acid uptake. Untreated and triacsin C values are also shown in Figure 1H. n=3 for each treatment. **p < 0.01; ***p < 0.001.
See also Figures S4.
Identification of ER-Localized GPAT Enzymes as Gate-Keepers of Cellular Lipotoxicity
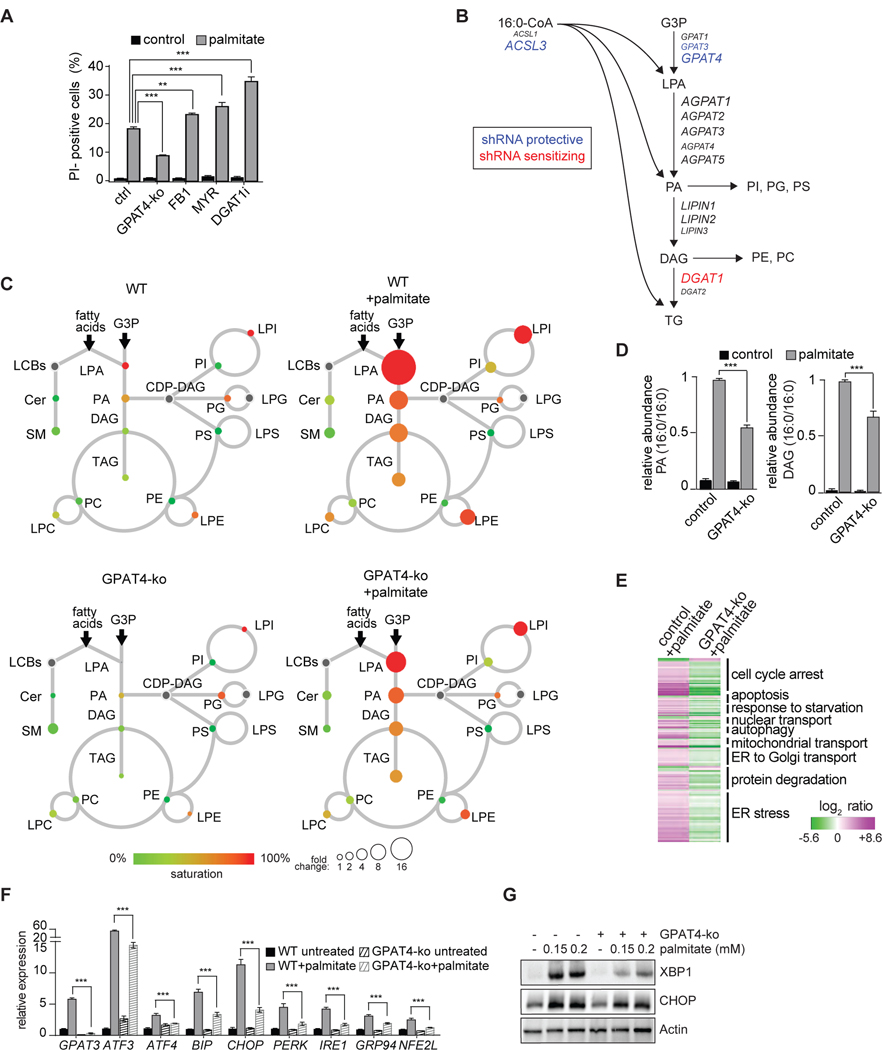
Because our lipidomics results suggested the hypothesis that di-saturated glycerolipid species are mediators of palmitate lipotoxicity, we analyzed screen hits from the glycerolipid (Kennedy) synthesis pathway (Figures 5A and 5B). The screen identified GPD1L, AGPAT9 (encoding GPAT3), and CHKA as genes whose depletion protected against palmitate lipotoxicity, and DGAT1 and PEMT as genes that sensitized to palmitate (Table S3). Inhibition of DGAT1 activity as an aggravator of cellular toxicity is consistent with a de-toxifying role of DGAT-mediated TG synthesis by reducing the burden of saturated glycerolipids through flux into neutral lipids (Chitraju et al., 2017; Listenberger et al., 2003). In contrast, inhibiting sphingolipid synthesis by myriocin or fumonisin B1 did not protect K562 cells from lipotoxicity (Figure 5A), suggesting intermediates of this pathway, such as sphingosines or ceramides, do not mediate palmitate-mediated lipotoxicity in the model system we employed.
Figure 5. Glycerolipid Synthesis-Related Pathway Is a Major Modifier of Palmitate-Induced Lipotoxicity.
(A) Blocking sphingolipid or triacylglycerol synthesis does not protect K562 cells against palmitate-induced cell death. Propidium-iodide staining of K562 cells were treated with fumonisin B (10 μM), myriocin (25 μM) or DGAT1 inhibitor (25 μM), and untreated or treated with 0.2 mM palmitate for 20 h. n=3 for each treatment. **p < 0.01; ***p < 0.001.
(B) Schematic representation of glycerolipid pathway. GPAT catalyzes the esterification of glycerol-3-phosphate and fatty acyl CoA to yield lysophosphatidic acid, which is further esterified by AGPAT to yield phosphatidic acid. In the next step the phosphate is hydrolyzed by lipin enzymes, yielding diacylglycerol (DAG). This can be used for the synthesis of glycerophospholipids, such as PC or PE, or for production of triacylglycerol by DGAT enzymes. Font size of proteins correlates to relative abundance (based on RNAseq data).
(C) Lipidome of control (top) and GPAT4-ko (bottom) K562 cells untreated (left) or treated (right) with palmitate (0.2 mM) for 20 h, as described in Figure 1B. The size of the circles is proportional to the fold-change, compared to untreated control cells. The complete dataset is provided in Table 1 “Lipidomics data”.
(D) Blocking glycerolipid synthesis by GPAT4 knockout reduces the palmitate-induced accumulation of di-saturated lipid species. Relative quantification for phosphatidic acid (PA, left panel) and DAG (right panel) identified by LC-MS2. Wild-type or GPAT4-ko K562 cells untreated or treated with 0.2 mM palmitate for 20 h. n=3 for each treatment. ***p < 0.001.
(E) Blocking glycerolipid synthesis protects against palmitate-induced activation of the UPR. Left lane, RNAseq data of wild-type K562 cells treated with 0.2 mM palmitate for 20 h. Genes (p < 0.005) are shown as log2 ratio of palmitate compared to the control and designated as upregulated (magenta) and downregulated (green). Right lane, RNAseq data of GPAT4-ko K562 cells treated with 0.2 mM palmitate for 20 h. Genes are shown as log2 ratio, compared to the wild-type treated with palmitate. The annotation was performed using ClueGO in Cytoscape. The complete dataset is provided in Table S2 “Gene expression profile data”.
(F) Blocking glycerophospholipid synthesis prevents palmitate-induced UPR target genes. Relative expression of main UPR target genes performed by qPCR. n=3 for each treatment; ***p < 0.001.
(G) Block of glycerophospholipid synthesis prevents transcription of XBP1. Western blot of total cell lysate from control and GPAT4-ko cells untreated or treated for 16 h with 0.2 mM palmitate.
See also Figures S5.
Since our initial screen identified the glycerolipid biosynthesis as a major pathway mediating lipotoxicity, we tested the role of lipid enzymes that decreased the levels of di-saturated glycerolipids. Specifically, we focused on the ER-localized GPAT enzymes, GPAT3 and GPAT4 (encoded by the AGPAT9 and AGPAT6 genes respectively), that catalyze the first step in glycerolipid biosynthesis (Takeuchi and Reue, 2009). Our screen identified GPAT3 as protective, but its close homologue GPAT4 is the more highly expressed ER-localized GPAT isoform in K562 cells. In follow up studies, combined knockdowns of GPAT3 and GPAT4 (protected against palmitate toxicity, despite the low efficiency of knockdowns (~25% knockdown efficiency; data not shown). Also, K562 cells lacking the more abundant GPAT4 (via Cas9-CRISPR knockout) were protected against palmitate toxicity (Figure 5A, Figure S5). We therefore focused our analyses in this study on GPAT4. In agreement with di-saturated glycerolipids as toxic metabolites during palmitate exposure, accumulation of di-saturated PA or DAG was reduced in cells lacking GPAT4 (Figure 5C and 5D). Gene expression changes induced by palmitate lipotoxicity were also prevented by GPAT4 deletion (Figure 5E), and the induction of ER stress by palmitate was largely prevented (Figure 5F and 5G). Thus, GPAT4 depletion emerges as a major protector of cellular lipotoxicity in this study.
Identification of the Disease Gene RNF213 as Mediator of Cellular Lipotoxicity
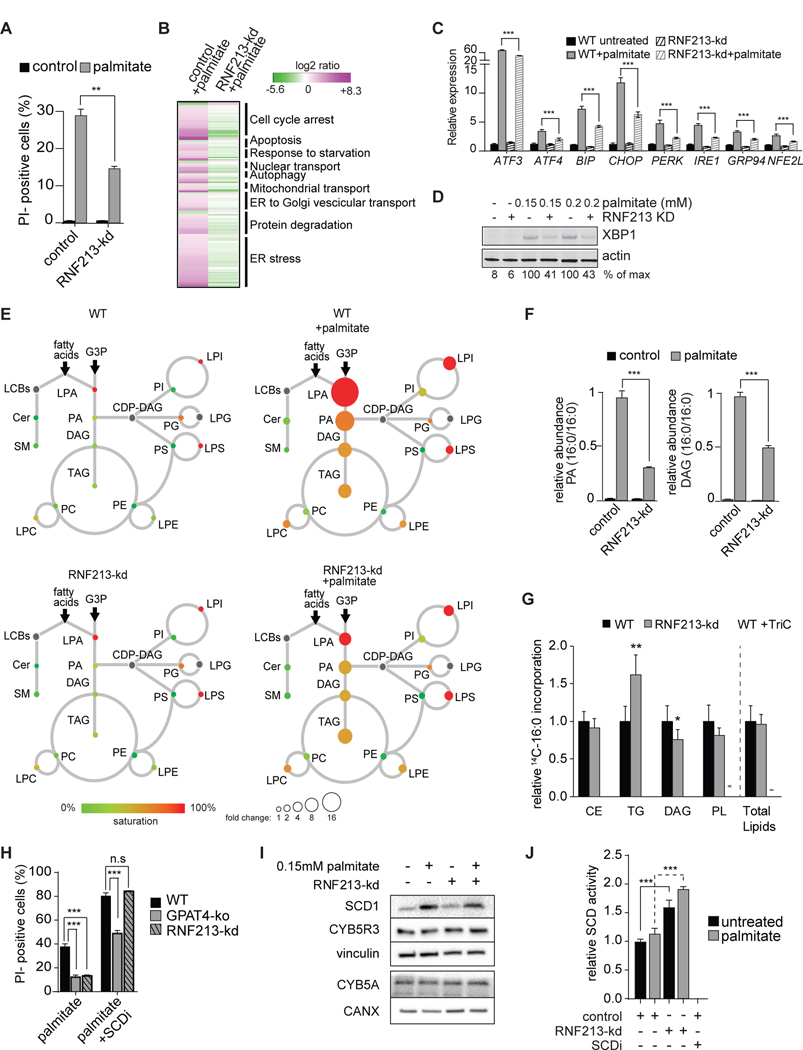
An intriguing screen hit whose knockdown protected against cellular lipotoxicity was RNF213 (Figure 3C), which encodes a protein of 5,256 amino acids with AAA-ATPase and RING-finger E3 ubiquitin-ligase domains (Kobayashi et al., 2015) The molecular function of the RNF213 protein is poorly understood. However, an R4810K mutation in RNF213, carried by as many as ~2.4% of the East and Southeast Asian populations (Liu et al., 2012), is associated with progressive, occlusive Moyamoya disease of the cerebrovascular arteries in humans (Liu et al., 2011). In addition, knockout of an RNF213 ortholog resulted in vascular abnormalities in zebrafish (Liu et al., 2011). Here we find that depletion of RNF213 in an independent experiment reduced palmitate-mediated cellular toxicity by ~50% (Figure 6A). RNF213 knockdown also lessened palmitate-induced cell death in other cell types, such as HepG2 and SUM159 cells (Figure S6A and S6B), although this effect was not found in HeLa cells, which required high levels of palmitate to effect cell death.
Figure 6. Depletion of the Putative E3 Ligase RNF213 Protects against Palmitate-Induced Lipotoxicity.
(A) Knockdown of RNF213 prevents palmitate-induced cell death. Propidium-iodide staining of control or RNF213-kd cells untreated or treated with 0.2 mM palmitate for 24 h. n=3 for each treatment. **p < 0.01.
(B) Knockdown of RNF213 protects against palmitate-induced activation of the UPR. Left lane, RNAseq data of wild-type K562 cells treated with 0.2 mM palmitate for 20 h. Genes (p<0.005) are shown as log2 ratio of palmitate compared to the control and designated as being upregulated (magenta) or downregulated (green). Right lane, RNAseq data of RNF213-ko K562 cells treated with 0.2 mM palmitate for 20 h. Genes are shown as log2 ratio compared to the control treated with palmitate. The complete dataset is provided in Table S2 “Gene expression profile data”.
(C) Knock-down of RNF213 prevents palmitate induction of UPR target genes. Relative expression of main UPR target genes performed by qPCR. n=3 for each treatment, ***p < 0.001.
(D) RNF213-kd protects cells against palmitate-induced UPR. Western blot of total cell lysate from control and RNF213-kd cells treated with vehicle or palmitate (0.15 or 0.2 mM) for 16 h. XBP1 protein is quantified as percentage over the max signal (100%) detected among all conditions.
(E) Lipidome of control K562 cells treated with palmitate (0.2 mM) for 20 h, as described in Figure 1B. The size of the circles is proportional to the fold-change, compared to untreated control cells. Lyso-phosphatidic acid (LPA) circle size was reduced 16-fold for visualization purposes. The complete dataset is provided in Table 1 “Lipidomics data”.
(F) Knockdown of RNF213 reduces accumulation of palmitate-induced di-saturated lipid species. Relative quantification for phosphatidic acid (PA, left panel) and diacylglycerol (DAG, right panel) identified by LC-MS2. Control or RNF213-kd K562 cells untreated or treated with 0.2 mM palmitate for 24 h. The lipid species changing the most upon palmitate treatment is shown for each class. n=3 for each treatment. ***p < 0.001.
(G) RNF213-kd cells exhibited unaltered total lipid synthesis but increased TG accumulation. Cells were treated for 6 h with 0.2 mM nonradiolabeled palmitate and 0.15uCi 14C-palmitate. Triacsin C (10uM, an inhibitor of ACSL1, ACSL3 and ACSL4) was added to confirm ACSL-specific fatty acid uptake. n=6 for each treatment. *p < 0.05, **p < 0.01.
(H) Inhibition of SCD1 exacerbates palmitate-induced cell death, an effect that is suppressed by GPAT inhibition but not by RNF213 knockdown. Propidium-iodide staining of GPAT4-ko or RNF213-kd cells treated with 0.2 mM palmitate for 20 h with or without SCD inhibitor (4 μM). n=3 for each treatment. ***p < 0.001; n.s., non-significant.
(I) SCD1 and cytochrome B5 reductase (CYB5R3) protein levels are not increased by RNF213 knockdown. Western blotting of total cell lysates from control and RNF213-kd cells untreated or treated for 16 h with 0.15 mM palmitate.
J) SCD activity is elevated in RNF213-kd cells compared to wildtype cells. In vitro Δ9 desaturase activity was determined in microsomes collected from WT and RNF213-kd cells untreated or treated with 0.15mM palmitate for 16 h and was measured as release of 3H from [9, 10-3H]-stearoyl-CoA. SCD inhibitor (4 μM) was included as a control. n=3 for each treatment. ***p < 0.001.
See also Figures S6.
Remarkably, depletion of RNF213 not only protected against lipotoxicity, but also normalized the palmitate-induced changes in gene expression, including the induction of ER stress genes (Figures 6B and 6C). Consistent with this, RNF213 deficiency reduced splicing of XBP1 mRNA during palmitate exposure (Figures 6D and S6C). However, the stress response per se was intact, inasmuch as perturbing ER calcium homeostasis (by blocking SERCA with thapsigargin) or inhibiting ER protein glycosylation (by tunicamycin) produced a comparable UPR in RNF213 depleted and wild-type cells (Figures S6D and S6E).
Maybe most surprisingly, depletion of RNF213 nearly completely normalized the cellular lipidome during palmitate exposure (Figures 6E and 6F), abrogating the accumulation of di-saturated glycerophospholipids. To confirm that RNF213kd protection does not stem from reduced palmitate uptake, we measured incorporation of radiolabeled palmitate into lipids. RNF213 knockdown did not alter total palmitate incorporation but did promote the partitioning of substrate towards triglycerides (Figure 6G). To unbiasedly identify proteins and pathways differentially regulated with RNF213 knockdown, we assessed total proteome and ubiquitylome changes. The results showed many changes due to RNF213 deficiency, but failed to identify any obvious direct targets based on criteria of increased levels in the total proteome and decreased levels of ubiquitylation (Table S4). Among proteins with decreased ubiquitylation in RNF213 knockdown after palmitate treatment were many members of the NF-κB pathway (Figure S6F). Consistent with this, we observed an RNF213-dependent increase in overall ubiquitylation and activation of NF-κB signaling and effectors (Figures S6G and S6H). This suggests palmitate triggers an NF-κB response, which is mitigated in the absence of RNF213. Taken together, our results highlight the protective effects of RNF213 depletion on palmitate-induced lipotoxicity via altered lipid metabolism, although they did not identify the direct target of the putative ubiquitin ligase.
RNF213 has been reported to modify non-mitochondrial oxygen consumption (Banh et al., 2016). In the context of lipotoxic stress, an important biochemical reaction that uses oxygen is the desaturation of palmitoyl-CoA to palmitoleoyl-CoA. Additionally, fatty acid desaturation promotes palmitate detoxification and storage into TGs (Figure 1H), an effect we also observed with RNF213 deficiency (Figure 6G). We thus tested functional interactions between RNF213, and the Δ9 desaturase SCD1. To address the alternative hypothesis that RNF213 limits incorporation of saturated fatty acids in cells, we assayed for genetic interactions of GPAT4 and RNF213. Whereas depletion of either GPAT4 or RNF213 protected against palmitate toxicity at baseline, the ability of RNF213 depletion to protect was selectively lost in the context of SCD1 inhibition (Figure 6H). This result suggests that RNF213 may act through effects on SCD1. Indeed, even though RNF213 did not directly affect SCD1 protein levels, or the levels of two proteins required for the desaturase reaction, cytochrome B5 reductase and cytochrome B5 (Figure 6I), RNF213-knockdown cells exhibited increased SCD activity in both basal conditions and after palmitate treatment, suggesting a possible regulatory effect of RNF213 on the SCD1 enzyme (Figure 6J).
Discussion
In this study, we used systematic, unbiased approaches to investigate the mechanisms of cellular lipotoxicity in cultured human cells. Examination of transcriptome changes implicated ER stress as a major mechanism of lipotoxic cell death in these cells, and this stress correlated with the accumulation of saturated fatty acids in glycerolipids (e.g., PA, DAG, lysophospholipids) as possible toxic mediators. Importantly, in addition to desaturation of fatty acids via upregulation of desaturation as a homeostatic mechanism confirmed in our shRNA screen, we also identified the flux of saturated fatty acids into glycerolipid synthesis via ER-localized GPAT enzymes, such as GPAT4, as a major pathway leading to ER stress and lipotoxic cell death. Importantly, another CRISPR-based gene inactivation screen in HeLa cells recently carried out by Zhu et al. ((Xiphias Ge Zhu) also identified GPAT4 as the major gatekeeper of lipotoxic lipids. Moreover, a recent study in vascular smooth muscle cells identified GPAT4 as a mediator of palmitate toxicity (Masuda et al., 2015). Thus, three studies, two using unbiased approaches, carried out independently in three cell types, identified ER-localized GPAT enzymes as mediators of lipotoxicity and suggest that inhibiting ER-localized GPAT (Cao et al., 2006; Yamashita et al., 2014) is a promising target for preventing lipotoxic cell injury and related disease.
Our findings and those of others suggest a model for cellular lipotoxicity from saturated fatty acids. Excess palmitate is incorporated in a wide variety of lipids, but specifically, the accumulation of di-saturated species of glycerolipids in the ER triggers sustained activation of the IRE1-branch of the UPR (Kitai et al., 2013). Downstream of persistent UPR activation, an apoptotic program is triggered by activation of JNK-kinases and caspases. Among the gene expression changes observed during palmitate treatment, one of the largest and most significant was the up-regulation of GPAT3 mRNA (Figure S4B), which adds saturated fatty acids to the glycerol backbone in the Kennedy pathway. This suggests a detrimental adaptive cellular response: cells attempt to cope with the increased availability of palmitate by up-regulating the enzyme that can incorporate this fatty acid into membrane lipids. However, because downstream enzymes cannot process them to TG, fully saturated membrane glycerolipids accumulate to toxic levels and trigger the UPR. Processes that increase fatty acid desaturation protect cells from saturated glycerolipid-mediated lipotoxicity. Our findings implicate the loss of homeostasis maintaining membrane lipid saturation as a major mechanism of lipotoxicity, at least some of which is mediated by UPR activation. Precisely how saturated lipids activate the UPR is unknown but likely involves IRE1 sensing changes in protein trafficking out of the ER or in membrane composition and is the subject of considerable investigation (Halbleib et al., 2017; Volmer et al., 2013). Consistent with our findings, knockout of GPAT4 in the ER largely prevents palmitate-induced lipotoxicity, suggesting GPAT4 inhibition as a potential therapeutic avenue.
Our screen also identified RNF213 depletion as protecting against palmitate-induced lipotoxicity. RNF213 encodes an interesting but poorly understood ~591-kDa protein that contains both AAA-ATPase and RING-finger E3 ubiquitin-ligase domains (Kamada et al., 2011). A missense mutation (R4810K) in the gene has been linked to Moyamoya vascular disease (Liu et al., 2011), and deletion of the gene in zebrafish results in abnormal vascular and muscle fiber development (Kotani et al., 2015). However, the function of the protein is poorly understood. A recent report linked RNF213 to non-mitochondrial oxygen consumption and suggested that RNF213 regulates one or more α-ketoglutarate-dependent dioxygenases, which are involved in hydroxylation and demethylation reactions (Banh et al., 2016). Further, a murine knockout for RNF213 led to protection against genetically induced diabetes (Kobayashi et al., 2013). If RNF213 is an E3 ligase, its direct target(s) are unknown. Our data suggest that RNF213 activity inhibits SCD1 specific activity. This could be through a direct effect, such as ubiquitin modification of SCD1 that reduces enzyme activity, or an indirect effect. We did not detect changes in the SCD1 protein size consistent with ubiquitylation, however. Although the mechanism remains to be determined, RNF213 clearly is a modulator of lipotoxicity from saturated fatty acids through a mechanism that has a major effect on the cell’s ability to store lipids in LDs. Downstream of lipotoxic stress, RNF213 deficiency blocks ER stress and the activation of the NF-κB pathway. It is possible that RNF213 has a more direct role in modulating these pathways, besides affecting SCD1. Of note, modulation of NF-κB in response to cellular stress could alter the expression of inflammatory cytokines, such as IL-6, which in turn have been linked to angiogenesis (Gopinathan et al., 2015; Huang et al., 2004; Karst et al., 2009), providing a possible link to Moyamoya disease. Future studies will be necessary to test this hypothesis in models of the diease.
Finally, we note that cellular lipotoxicity likely has multiple causes (Brookheart et al., 2009; Farese et al., 2012), and that our screen identifies those that are operative and contribute to lipotoxicity in the cell types we employed in this study. Nevertheless, we identified many novel genes that are potentially important for mediating palmitate-induced lipotoxicity, and our screen provides a high-confidence, comprehensive data set for genes that modulate lipotoxicity under chronic conditions. Further investigation of the identified hits may therefore provide novel targets for preventing or treating lipotoxicity-related diseases.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for reagent and resources should be directed to Robert V. Farese, Jr. (robert@hsph.harvard.edu) and Tobias C. Walther (twalther@hsph.harvard.edu).
METHOD DETAILS
Chemicals and Reagents
Palmitate was prepared by dissolving palmitic acid salt in 1–2 ml of water pre-warmed at ~80°C. The minimal amount of 1 N NaOH was added to achieve complete solubility. The mixture was then added drop-by-drop to BSA-containing RPMI pre-warmed at 37°C. After this procedure, a stock of 10 mM palmitate was generated. The ratio of palmitate to BSA was 6:1.
The SCD inhibitor (Abcam) was resuspended in DMSO and used at 4 μM for cell treatments. Fumonisin B1 (Sigma) was resuspended in DMSO and used at 50 μM. DGAT1 inhibitor ((Gluchowski et al., 2017; Liu et al., 2015); Merck) was resuspended in DMSO and used at 15uM for in vitro DGAT1 assays and 25uM for in vivo cell viability assays. Myriocin (Sigma) was resuspended in DMSO and used at 50 μM. The IRE1 inhibitor, 4μ8C (EMD Millipore), was resuspended in DMSO and used at 32 μM. The PERK inhibitor GSK2606414 (Tocris) was resuspended in DMSO and used at 15 nM. Thapsigargin (Sigma) was resuspended in DMSO and used at 15 or 3 μM. Tunicamycin (Sigma) was resuspended in DMSO and used at 1.5, 2 or 4 μg/ml.
Cell Culture and Treatments
K562 cells (ATCC CCL-243) were cultured in flasks using RPMI medium (Gibco) containing 25 mM Hepes and 2 mM glutamine. The medium was supplemented with 2 mM glutamine (Gibco), 10% FBS (Thermo) and Pen/Strep (Gibco). Cells were kept in active proliferation (max 100*104/ml) for the duration of the experiments. Cells reaching 100*104/ml were diluted 1/4 or 1/8 using fresh medium. Standard growing conditions were used (i.e., 37°C and 95% humidity). Cells were treated with palmitate or palmitoleate as indicated in figure legends. In general, high palmitate concentration (e.g., 0.2–0.25 mM for 24–28 h) were used for viability assays. Lower concentration (e.g., 0.15–0.2 mM for 16–20h) were used in all other experiments (including western blotting, RNAsq and lipidomics) to avoid carryover of more than 5–10% of death cells.
SUM159 cells (RRID: CVCL_5423, obtained from the lab of Tomas Kirchhausen) were maintained in DMEM/F-12 GlutaMAX (Life Technologies) with 5 ug/ml insulin (Cell Applications), 1 ug/ml 5% FBS (Life Technologies), 50 U/ml streptomycin and 50 U/ml penicillin. Hydrocortisone (Sigma), HepG2 cells (ATCC) cand HeLa cells (obtained from Thomas Kirchhausen) were cultured in DMEM supplemented with 10% FBS (Life Technologies) and 50 U/ml streptomycin and 50 U/ml penicillin.
Genome-Wide shRNA Screen
The genome-wide shRNA screen was performed as described (Kampmann et al., 2014) with modifications. Briefly, K562 cells were infected with nine libraries (labeled from A to L) containing 25 shRNA/gene. The nine libraries were then pooled together based on the percentage of infected cells for each library (evaluated by mCherry fluorescence) to maintain the same representation of each library. The representation of all shRNAs was efficiently maintained having 20 or more shRNAs for ~90% of all genes targeted (e.g., Figure S3A). Cells (1E9 cells/sample) were grown in spinner flasks in constant agitation and diluted ½ every day. At time 0 (T0), the initial population was divided in three parts: one T0 sample (frozen), one control, and one treated with palmitate (0.2 mM). We aimed to obtain 50% viability upon 24 h of palmitate treatment compared with control. After 24 h, viable cells were counted, and the sample was spun down, and the medium was replaced with medium lacking palmitate. Cells were allowed to recover for 2–3 days until the doubling time was 24 h again. After recovery, a new cycle of palmitate treatment was performed. A total of five cycles were performed. We calculated that palmitate-treated cells skipped ~six doublings, compared with control cells. Based on this calculation, we estimated that a gene having full protection would be enriched ~64 times in palmitate-treated sample compared with control. Such enrichment (including less effective phenotypes) was measured by sequencing.
We then proceeded to construct DNA libraries for the three samples obtained: T0, control, and palmitate treated (Bassik et al., 2013). Briefly, the genomic DNA was extracted, digested with PvuII (NEB) enzyme and run on agarose gel. The DNA was isolated around the 1.2-kb marker size and purified from the gel. PCR (100-μl reactions) was used to amplify the shRNAs from the genomic DNA. PCR products were separated on a gel, and the correct bands were excised, purified, and quantified by a Bioanalyzer (Agilent). The three samples were mixed at equimolar concentration and sequenced on an Illumina HiSeq 2000.
The screen was done in biological duplicates and analyzed using the GImap script suite available at http://gimap.ucsf.edu/ (Kampmann et al., 2013). We evaluated the correlation between replicate samples (e.g., Figure S3D). We evaluated the distribution of the non-targeting (control) shRNAs and the targeting shRNAs (e.g., Figure 3B). Because of the different performance of each library (Figure S3B), we decided to normalize all the libraries by all internal control shRNAs. Genes with no expression (RPKM<1) were excluded. The geometrical mean between replicate phenotypes was used to obtain the final Rho phenotype for each shRNA (e.g., Figure S4A). For each gene, we evaluated if the distribution of the targeting shRNA (N≥20) was significantly different from the control shRNA (p-value<0.01) using the Mann-Whitney U test (e.g., Figure S4A). GO term enrichment was performed using Cluego in Cytoscape. For each GO cluster identified, the GO term associated with the lowest p-value was picked as representative of the cluster.
Screen Validation and Generation of Stable Knockdown and Knockout Cells
Based on the GImap software (Kampmann et al., 2013), we identified the three strongest shRNAs for each of gene selected for validation. The shRNA oligonucleotide sequences (IDT) were annealed mixing 23 μl ddH2O +1 μl top oligo at 100 μM +1 μl bottom oligo at 100 μM + 25 μl of 2x Annealing Buffer (200 mM potassium acetate, 60 mM HEPES-KOH, pH 7.4, 4 mM Mg acetate). The mixture was incubated for 5 min at 95°C. The oligonucleotides were allowed to gradually anneal at room temperature (RT). This stock was diluted 1:200 prior to be used. Each annealed oligonucleotide was individually cloned into the pMK1222 using BSTXI as restriction sites. Each plasmid was sequenced to verify the sequence and then delivered to cell by lentivirus. To avoid off-target-effects of the shRNAs, the cells were infected with lentiviral particles carrying the three strongest shRNAs. The screen validation was performed on the infected cells (1–5% of infection) in 30 ml of T75 flasks. The number of infected cells was counted with flow cytometry monitoring of the m-Cherry reporter expressed by the pMK1222. The assay was done using the same set-up as the original screen, three cycles of 0.2 mM palmitate pulse for 24 hours, followed by 2 days of recovery. At the end of each cycle, the percentage of infected cells was counted and expressed as log2 ratio of infected over uninfected cells (Figure 3B). A pure cell line was obtained by FACS sorting for the stable Gpat3kd and RNF213kd. GPAT4KO cells were generated using inserting a guide targeting GPAT4 locus on the pSpCas9(BB)-2A-Puro (PX459) vector (Figure S5A). The genotype of the clone confirmed the presence of a premature stop codon (Figure S5B). Loss of GPAT4 was confirmed by LC-MS/MS (Figure S5C).
Lentivirus Production
Lentiviruses were generated in a small scale (one well of a six-well plate) using 293TF cells (Thermo). 250,000 cells were grown in 2 ml of complete growth medium. The expression vector and packaging vectors were transfected into 293TF by using Fugene 6 (Roche). The DNA mixture consisted of 0.75 μg of pMK1222 vector and 0.75 μg of lentiviral packaging mixture (equal parts pVSVG, pMDL, pRSV). Cells were then allowed to grow for 3 days. On the third day, the supernatant was collected and filtered through a 0.45-μm filter to remove cells/debris. K562 cells were infected mixing in equal parts of the viral filtered supernatant and growing K562 cells. At 24 hours after the infection, the medium was replaced, and the infection was verified by expression of mCherry.
Gene Expression Profiling and qPCR
Total RNA was isolated from K562 cells using an RNeasy Kit. The library construction and sequencing were performed by the TUFTS sequencing core (tucf-genomics.tufts.edu/). Briefly, the RNA was run on a Bioanlayzer (Agilent) to assess RNA quality and quantification. The biological triplicates were pooled at this point. The library preparation was performed using the TruSeq Stranded mRNA Library Prep Kit (Illumina). The samples were sequenced by HiSeq 2500, Rapid Paired-End 100 (RL-PE100) or HiSeq 2500, High Output v4 Paired-End 100 (HI-PE100, Illumina). All reads were mapped to the UCSC hg19 human reference genome using Tophat2 with multi-read correction, followed by normalization, quantification and differential expression analysis using Cuffdiff. The complete dataset is provided in Table S2 “Gene expression profile data”.
We validated selected targets with qPCR. RNA was reverse-transcribed to cDNA using iScript cDNA Synthesis Kit (Bio-Rad). qPCR was performed in triplicates using SYBR Green PCR Master Mix Kit (Applied Biosystems). The results were imported in Prism followed by statistical analysis (two-stage Benjamini, Krieger and Yekutieli t-test, FDR=1%). GO term enrichment was performed using the App Cluego of Cytoscape. For each GO cluster identified, the GO term associated with the lowest p-value was picked as representative of the cluster. Enrichments of terms containing low number of genes (n<15) were excluded from representation.
Microsome isolation
Cell pellets were resuspended in 1ml homogenizer buffer (250 mM sucrose, 20 mM Tris-HCl pH 7.4, 1 mM EDTA plus protease inhibitor), dounce homogenized and centrifuged for 10min at 8000xg. The supernatant was collected and centrifuged 100,000xg for 1 hour, The resulting pellet was resuspended in 0.1 M PK buffer (0.1 M K2HPO4 buffer adjusted to pH 7.2 with NaH2PO4).
Immunoblotting and XBP1 Splicing Measurements
Control cells and cells treated with palmitate were lysed using RIPA lysis buffer, complemented with both protease and phosphatase inhibitors (Roche). Cell lysates were briefly sonicated (5 sec at 30% output). Microsomes were used for measurement of cytochrome b5 protein abundance (Figure 6I). Samples were normalized by protein concentration (BCA) and incubated in Laemmli buffer for 5 min at 95°C.
Proteins were separated on 7–20% SDS-PAGE gel (Bio-rad) and transferred to PVDF membrane (i-Blot, Thermo). Western blot for INSIG1-kd and RNF213-kd experiments were conducted with wet transfer for 1 h at 115 V. The membranes were blocked in 5% BSA or 5% milk in TBS-T buffer for 1 h. Then, membranes were incubated with primary antibodies overnight at 4°C and with the appropriate secondary antibodies for 1 h at RT. WB were developed using Licor or femto-ECL. Quantification of XBP1 protein was done using FIJI (ImageJ) and expressed numerically from 0 to 100 (100 was set as max signal). Quantification of spliced XBP1 was also performed using FIJI and expressed as percentage of the XBP1 spliced form over the unspliced form. A rabbit polyclonal antibody against human INSIG1 was custom generated (GenScript, Piscataway, NJ), affinity purified and used at a 1:1,000 dilution. The peptide sequence used for the epitope was “CRQLAMGVPEKPHSD” and correlates to amino acids 264–277 of human INSIG1.
DGAT1 Assay
DGAT1 enzymatic activities were measured at Vmax in total cell lysates from K562 cells and from differentiated 3T3-L1 adipocytes. Samples were lysed in a buffer containing 250 mM sucrose and 50 mM Tris-HCl (pH 7.4), followed by sonication and normalization by protein concentration. The assay was performed in a 100 mM Tris-HCl (pH 7.4) buffer containing protease inhibitors (Roche). Each reaction contained: 10 μg sample, 100 μM of diacyl-glycerol and 25 μM of cold and hot (14C) acyl-CoA. 1,2-dioleoyl-sn-glycerol and 1,2-dipalmitoyl-glycerol (Avanti) were used as diacyl-glycerol source. Oleoyl-CoA and palmitoyl-CoA (hot form American Radiolabeled Chemicals and cold form Sigma) were used as source of activated fatty acid. Lipid mixture was sonicated prior to be added to samples to ensure appropriate solubilization. After the lipid mixture was added to the samples, the reaction occurred in a water bath at 37°C, as described (Cases et al., 2001; Yen et al., 2005). The DGAT1 inhibitor (Merck) was included as control and used at 15uM. The reaction was stopped by transferring the samples on ice and adding cold methanol/chloroform mixture. The extracted lipids were separated by TLC using hexane:diethyl ether:acetic acid (80:20:1) solvent system. A phosphorimager screen was used to develop and quantify the TG band of exposed TLC plates.
SCD Assay
The SCD assay was adapted from a protocol kindly provided by James Ntambi. Briefly, the assay premix consisted of 0.03 mM stearoyl-CoA (Sigma), 1 μCi/ml radioactive 3H stearoyl-CoA (American Radiolabeled Chemicals), 2 mM NADH (Sigma) added to 0.1 M PK buffer (0.1 M K2HPO4 buffer adjusted to pH 7.2 with NaH2PO4). Cold 1.5 mM stearoyl-CoA stock was prepared fresh in 10 mM NaC2H3O2/50% ethanol solution. Each assay point consisted in 180 μl of premix and ~100μg microsomes in 0.1 mM PK buffer. 4uM SCD inhibitor was added to the cell lysate before starting the reaction. The reaction was performed at 37°C for 30 min and then quenched with 200 μl of 6% perchloric acid. The organic material was absorbed by adding 700 μl of 10% (w/v) charcoal (Sigma) dissolved in PBS. The charcoal was spun down, and the aqueous phase was collected. Samples were counted in a scintillation conter and results were calculated as 3H counts per minute and normalized to untreated WT microsomes. All data points were measured in triplicates.
Incorporation of 14C-Palmitate into Lipids
20E4cells/mL cells were grown in RPMI complete media and supplemented with a mix containing 0.2mM nonradiolabeled palmitate (conjugated to BSA 6:1), additional 10uM BSA, 0.15uCi [1-14C] palmitic acid (mixed at 37° for 30 min). Cells were treated with 10uM Triacsin C (Santa Cruz, in DMSO) for 2 h prior to fatty acid treatment. After 6 h fatty acid treatment, cells were collected via centrifugation for 5min 1000xg and washed twice in cold PBS containing 0.5% BSA following by one wash in PBS without BSA. Lipids were extracted from cell pellets with addition of 3:2 hexane:isopropanol and 20min gentle shaking, followed by centrifugation 5min 5000xg. Lipid extracts were dried under air stream and neutral lipids were separated by TLC using hexane:diethyl ether:acetic acid (80:20:1). TLC plates were exposed to a phosphor imaging cassette and revealed by a phosphorimaging (Typhoon FLA 7000). Incorporation of radiolabeled palmitic acid into lipid classes (cholesterol esters, triglycerides, diacylglycerides and phospholipids) was measured as band intensity on TLC using Image J software. Remaining cell pellets were dried and proteins were solubilized using 0.3N NaOH in 0.1% SDS. Band intensity was normalized to protein concentration.
Flow Cytometry
K562 cells were cultured in 24-well plates using RPMI complete medium. Cells were diluted to 15*104/ml and treated as indicated in figure legends with the appropriate drugs or fatty acid for 24–28 h. At the time of collection, cells were washed with cold PBS and stained for Annexin V and/or propidium iodide, following the protocol in the Dead Cell Apoptosis Kit with Annexin V & Propidium Iodide (C.N. V13241, Thermo). Data were acquired on a FACSCalibur Analyzer (BD) and then analyzed off-line using FlowJo_V10.
RNF213 siRNA Knockdown in other Cell Lines
RNF213 knockdown was achieved through reverse siRNA transfection. SUM159, HepG2 and HeLa cells were seeded in 6-well plates at 150,000 cells/well and treated with 20nM RISC-free (control) or RNF213 siRNA (Dharmacon). Two days after siRNA treatment, cells were treated with the palmitate for 24 hours and processed for PI staining and flow cytometry.
Microscopy and BODIPY staining
K562 cells were imaged in RPMI complete media supplemented with palmitate (0.25 mM), palmitoleate (0.25 mM), or a mixture of palmitate (0.25 mM) and palmitoleate (0.25 mM) for 24 h. Live cells were stained with 0.5 ug/ml BODIPY 493/503 (Thermo Fisher) prior to imaging.
Lipidomics
For lipid analyses of WT, RNF213-kd and GPAT4-ko cells, the following was performed. K562 cells were cultured in complete RPMI medium and treated with the indicated drugs or fatty acids. Samples were collected by centrifugation and washed once with 155 mM ammonium acetate (AA) buffer, followed by centrifugation. The resulting pellet was resuspended in 800 μl of AA buffer and then sonicated for 5 sec at 30% output. Samples were normalized by protein concentration (BCA). The lipids were extracted using chloroform/methanol (2:1), as described in (Folch et al., 1957). UHPLC-ESI-MS/MS analyses were performed using a Dionex™ UltiMate 3000™ UHPLC system (Thermo) coupled to Q Exactive™ Hybrid Quadrupole-Orbitrap™ Mass Spectrometer (Thermo). 5 μl injections of samples were made onto a Accucore™ (Thermo) C18 LC column (2.6 μm solid-core particles, length of 30 mm). The eluent A consisted of acetonitrile/water (50:50 v/v) + 10 mM ammonium formate and 0.2% formic acid, and the eluent B consisted of methanol/isopropanol/water (10:88:2 v/v/v), 2 mM ammonium formate and 0.01 % formic acid. The following parameter was used for gradient elution of analytes (total run time of 46 min) at a constant flow rate of 500 μL per min. 35 % eluent B – 45 % eluent B over 5 min, 45 % eluent B – 85 % eluent B from min 5 – min 28, 85 % eluent B – 100% eluent B from min 28 – min 38, followed by an immediate drop back to 35% eluent B, which is held constant up to min 46 of each run. The column temperature was set to 35 °C, and the autosampler tray temperature was set to 10 °C. The ion source was a HESI II (Thermo) set in the following conditions: sheath gas flow rate set to 60; auxiliary flow rate set to 20; sweep gas flow rate set to 1; spray voltage (KV) set to 3.00; capillary temperature set to 285 °C, S-Lens RF level set to 45, and auxiliary temperature set to 370 °C. The mass spectrometer acquisition settings were as follows: Samples were run in either positive or in negative ion modes, under otherwise identical conditions. The instrument was set for Full Scan – top 15 data-dependent MS/MS. Full scan was set for a range of 250–1800 m/z. The mass resolution was set to 70,000; AGC target was set to 1e6, the C-trap ion accumulation time was set to 120 ms, Data-dependent MS/MS was set to a mass resolution of 30,000, AGC target was set to 5e5, the C-trap ion accumulation time was set to 120 ms, select ion exclusion was set to 8 s, and the HCD (higher-energy collisional dissociation) fragmentation ramp was set to 15, 25, and 35 NCE (normalized collision energy).
Mass spectrometry generated by the LC-MS/MS runs for extracted lipids were analyzed using LipidSearch™ software (Thermo) for the identification of lipid species, combining accurate-mass information from parent ion with MS2 information. The following parameters were used for batch analysis: Database – Q Exactive; Search Type – Product; Experiment Type (Exp Type) - LC-MS; Parent Tolerance (Parent Tol) - 0.1 Da; NL/Prec Tol – 0.5 Da; Precursor Tolerance (Prec Tol) - 10.0 ppm; Product Tolerance (15.0) ppm; Merge Range (Min) - 0.0; Minimal Peak Width (min) - 0.0; Threshold Type - Relative; Product Ion – 1.0%; m-score threshold – 2.0; Recalculate Isotope – ON; R.T. Interval (min) - 0.01; Execute Quantitation – ON; m/z Tolerance (m/z tol) - −10.0 / + 10.0; Tolerance Type – ppm; R.T. range (min) - −0.05 / +0.05; Toprank filter – ON; Main Node Filter – Main Isomer Peak; m-Score Threshold (Display) - 5.0; C-Score Threshold (Display) - 2.0; Fatty Acid Priority (FA Priority) - ON; ID Quality Filter (A; B; C). The following parameters were used for sample alignment: Search Type – Product; Experiment Type (ExpType) - LC – MS; Alignment Method – Max; R.T. Tolerance – 0.25; Calculate Unassigned Peak Area – ON; Filter Type – New Filter; Toprank Filter –ON; Main Node Filter – Main Isomer Peak; m-score threshold – 5.0; ID Quality – A,B,C. All lipid species identified using the LipidSearch™ software were manually curated after computational analysis. The final lipid quantification and representation was done using LiPa (Lipid Pathway) Viewer, a custom R-script.
Lipidomic analyses of INSIG1kd cells and SCD inhibition studies were conducted as follows. K562 cells were cultured in complete RPMI medium and treated with the indicated drugs or fatty acids. Samples were collected by centrifugation and washed once with PBS, followed by centrifugation. The resulting pellet was lysed by two sonication/snap freeze cycles in 0.1M HCl. Subsequently, lipids were extracted according to the Folch method (Folch et al., 1957). The organic phase of each sample werse then separated by an HPLC-MS method adopted from (Narvaez-Rivas and Zhang, 2016). HPLC analysis was performed employing a C30 reverse-phase column (Thermo Acclaim C30, 2.1 × 250 mm, 3 μm, operated at 55° C; Thermo Fisher Scientific) connected to a Dionex UltiMate 3000 HPLC system and a QExactive orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) probe. Dried lipid samples were dissolved in corresponding volumes of 2:1 methanol:chloroform (v/v) and 5 μl of each sample was injected, with separate injections for positive and negative ionization modes. Mobile phase A consisted of 60:40 water/acetonitrile, including 10 mM ammonium formate and 0.1% formic acid, and mobile phase B consisted of 90:10 2- propanol/acetonitrile, also including 10 mM ammonium formate and 0.1% formic acid. The elution was performed with a gradient of 90 minutes; during 0–7 minutes, elution starts with 40% B and increases to 55%; from 7 to 8 minutes, increase to 65% B; from 8 to 12 minutes, elution is maintained with 65% B; from 12 to 30 minutes, increase to 70% B; from 30 to 31 minutes, increase to 88% B; from 31 to 51 minutes, increase to 95% B; from 51 to 53 minutes, increase to 100% B; during 53 to 73 minutes, 100% B is maintained; from 73 to 73.1 minutes, solvent B was decreased to 40% and then maintained for another 16.9 minutes for column reequilibration. The flow-rate was set to 0.2 mL/min. The column oven temperature was set to 55° C, and the temperature of the autosampler tray was set to 4° C. The spray voltage was set to 4.2 kV, and the heated capillary and the HESI were held at 320° C and 300° C, respectively. The S-lens RF level was set to 50, and the sheath and auxiliary gas were set to 35 and 3 units, respectively. These conditions were held constant for both positive and negative ionization mode acquisitions. External mass calibration was performed using the standard calibration mixture every 7 days. MS spectra of lipids were acquired in full-scan/data-dependent MS2 mode. For the full-scan acquisition, the resolution was set to 70,000, the AGC target was 1e6, the maximum integration time was 50 msec, and the scan range was m/z = 133.4–2000. For data-dependent MS2, the top 10 ions in each full scan were isolated with a 1.0 Da window, fragmented at a stepped normalized collision energy of 15, 25, and 35 units, and analyzed at a resolution of 17,500 with an AGC target of 2e5 and a maximum integration time of 100 msec. The underfill ratio was set to 0. The selection of the top 10 ions was subject to isotopic exclusion with a dynamic exclusion window of 5.0 sec. Processing of raw data was performed using LipidSearch software (Thermo Fisher Scientific/Mitsui Knowledge Industries) (Taguchi and Ishikawa, 2010; Yamada et al., 2013). The assembled results were exported to R-Studio where all identified lipids were included for subsequent analyses if they fulfilled the following LipidSearch-based criteria: 1) reject equal to zero, 2) main grade A OR main grade B AND APValue<0.01 for at least three replicates, and 3) no missing values across all samples. Based on these filters, 656 out of 1901 lipids (SCD inhibition experiment) and 409 out of 1816 lipids (INSIG1-kd experiment) were present in all samples. Further quality controls were performed using pair-wise correlations between replicates and principal component analysis (PCA, FactomineR package (Lê et al., 2008), comparing sample groups. All replicates displayed high reproducibility (r-value>0.77 and p-value<0.01) and the PCA distinguished between sample groups on the first and second principal components with 95% confidence interval (results not shown). Manual curation was performed for DAG species, yielding 10 DAG species (SCD inhibition experiment) and 12 DAG species (INSIG1-kd experiment). Lipid abundance was normalized to protein concentration of cell lysates. Abundance of DAG species was normalized to extraction efficiency of internal standard TG(17:1/17:1/17:1) (Avanti).
To separate phosphatidates, HPLC-MS analysis was performed employing a Kinetex HILIC column (2.1 mm × 100 mm, 2.6 m, operated at 55˚ C; Phenomenex), connected to a Dionex UltiMate 3000 HPLC system and a QExactive orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) probe. Mobile phase A consisted of deionized water containing 10 mM ammonium formate and 0.5% formic acid. Mobile phase B consisted of 2-propanol/acetonitrile 5:2 (v/v) containing 10 mM ammonium formate and 0.5% formic acid as well. Gradient elution started at 5% A with a linear increase to 50% A over the course of 12 min followed by an isocratic elution with 50% A for 3 more min. Finally, the column was re-equilibrated for 15 min. The flow rate was set to 0.3 mL/min. Sample solvent was chloroform/methanol 1:1 (v/v) and 20 μL were injected. The temperature for the autosampler tray was set to 4° C. The spray voltage was set to 3.5 kV, and the heated capillary and the HESI were held at 350° C and 300° C, respec tively. The S-lens RF level was set to 50, and the sheath and auxiliary gas were set to 20 and 5 units, respectively. Data were acquired in negative ionization mode. The Orbitrap mass spectrometer was operated in data dependent analysis mode; a full scan spectrum was obtained in the Orbitrap mass analyzer at R = 70,000 while simultaneously tandem mass spectra of the ten most abundant full scan masses (selected from an inclusion list containing phosphatidic acid and lysophosphatidic acid molecular species) were obtained in the linear ion trap at a normalized collision energy of 35% and an exclusion duration of 10s. The assembled results were exported to R-Studio where all identified lipids were included for subsequent analyses if they fulfilled the following LipidSearch-based criteria: 1) reject equal to zero, 2) main grade A OR main grade B AND APValue<0.01 for at least three replicates, and 3) no missing values across all samples. Based on these filters, 62 out of 109 lipids (SCD inhibition experiment) and 46 out of 94 lipids (INSIG1-kd experiment) were present in all samples. Further quality controls were performed using pair-wise correlations between replicates and principal component analysis (PCA, FactomineR package (Lê et al., 2008), comparing sample groups. Manual curation was performed for PA species, yielding 10 PA species (SCD inhibition and INSIG1-kd experiment). Lipid abundance was normalized to protein concentration of cell lysates. Abundance of PA species was also normalized to extraction efficiency of internal standard PA(17:0/20:4) (Avanti).
Di-Gly Proteomics
Di-Gly sample preparation
The di-gly proteomics workflow was performed as described previously (Udeshi et al., 2013), with minor modification. K562 cells were labeled with SILAC RPMI 1640 media (Gibco). Control cells were labeled with heavy lysine (K8, Cambridge isotopes) and heavy arginine (R10, Cambridge isotopes); RNF213 knockdown cells were labeled with light lysine (K0, Cambridge isotopes) and light arginine (R0, Cambridge isotopes). Heavy and light cells were treated with 0.15 mM palmitate for 15 h, collected by centrifugation, washed with PBS, and an equal number of heavy and light labeled cells were mixed 1:1, pelleted, and snap-frozen. The cell pellet was lysed in Urea lysis buffer (8M Urea (Sigma), 75mM NaCl, 50mM Tris pH 8.2, 50mM chloroacetamide (Sigma), Complete protease inhibitor tablet (Roche)), incubated for 15 minutes on ice, then sonicated to ensure complete cell disruption. Disulfide bonds were reduced via treatment with TCEP at a final concentration of 5mM for 45 minutes, followed by alkylation of cysteine residues with a final concentration of 15mM chloroacetamide for 30 minutes at room temperature in the dark. 5mM final concentration of TCEP was added to quench the alkylation reaction for 15 minutes in the dark. Samples were then diluted with 50mM Tris pH 8.2 to reduce the concentration of urea to 4M, and digested with Lys-C (Wako, 10 ng/μL) for 2 hours. Samples were then diluted to 1.6M urea with 50mM Tris pH 8.2 and digested with trypsin (Promega) overnight at a ratio of 1:200 trypsin:substrate (37°C). ~30mg peptides were obtained following digestion (colorimetric peptide assay, Thermo) and acidified with Trifluoroacetic acid (TFA) to 0.4% (vol/vol) such that pH<2. Samples were centrifuged at 2500xg for 10 minutes at room temp to remove insoluble material; the supernatant containing digested peptides was collected. Peptides were desalted using a tC18 SepPak column (Waters WAT036790, 500mg bulk material, 6mL capacity). Samples were washed with 4×5mL 0.1% Formic acid (FA, Sigma) to remove residual TFA and eluted with 2×3mL 50% acetonitrile (ACN, Sigma)/0.1% FA. Eluates were snap frozen and lyophilized. Samples were reconstituted in 5mM ammonium formate (pH 10.0)/2% (vol/vol) ACN and fractionated via basic reversed phase (bRP) chromatography. We used an Agilent 1200 pump equipped with a degasser and a photodiode array (PDA) detector (set at 220 and 280 nm wavelength) from ThermoFisher Scientific. Peptides were subjected to a 60-min gradient in 10 mM ammonium bicarbonate pH 8 (5 to 90% acetonitrile) at a flow rate of 0.6 mL/min over an Agilent 300Extend C18 column (3.5 μm particles, 4.6 mm ID and 220 mm in length). The peptide mixture was fractionated into a total of 96 fractions, which were consolidated by pooling every 8th sample to obtain 8 samples total. Samples were subsequently acidified with 1% formic acid and vacuum centrifuged to near dryness. Di-gly antibody without conjugation to resin was a gift from Cell Signaling Technology. Antibody was crosslinked to protein A/G Ultralink resin (Thermo), and washed 3x with IAP buffer (50mM MOPS-NaOH pH 7.5, 10mM Na2HPO4, 50mM NaCl). 40μL of crosslinked bead slurry was added to Eppendorf tubes; peptides resuspended in IAP buffer were added to the slurry and incubated for 1.5 hours at 4°C with gentle end-over-end rotation (8 IPs in total for 8 fractions of peptides after bRP fractionation). Supernatant was transferred to a new tube of di-gly antibody-coupled resin for a second round of K-GG peptide depletion. Beads were washed 4x with cold IAP buffer, 1x with cold PBS, then eluted with 55μL 5% formic acid (10 min incubation, gently mixing tube). Samples were dried by vacuum centrifugation, resuspended in 5% FA/5% ACN, and desalted using STAGE tips.
QExactive parameters and SILAC quantification
All spectra were acquired on an Orbitrap QExactive mass spectrometer (Thermo Fisher) coupled to an with a Famos autosampler (LC Packings) and an Accela600 liquid chromatography (LC) pump (Thermo Fisher). Peptides were separated on an ~25cm, 100μm inner diameter microcapillary column packed with of Accucore C18 resin (2.6 μm, 150 Å, ThermoFisher); ~1μg peptides were loaded onto the column for each analysis. Peptides were separated with a 120 min gradient of 5–25% acetonitrile in 0.125% formic acid with a flow rate of ~300nL/min. The scan sequence began with an Orbitrap MS1 spectrum with the following parameters: resolution 70,000, scan range 300−1500 Th, automatic gain control (AGC) target 1 × 105, maximum injection time 250 ms, and centroid spectrum data type. We selected the top twenty precursors for MS2 analysis which consisted of HCD high-energy collision dissociation with the following parameters: resolution 17,500, AGC 1 × 105, maximum injection time 60 ms, isolation window 2 Th, normalized collision energy (NCE) 30, and centroid spectrum data type. The underfill ratio was set at 1%, which corresponds to a 1.1 × 104 intensity threshold. In addition, unassigned and singly charged species were excluded from MS2 analysis and dynamic exclusion was set to automatic. For data analysis, mass spectra were processed using a Sequest-based in-house software pipeline. MS spectra were converted to mzXML using a modified version of ReAdW.exe. MS2 spectra were searched with a database including all entries from the human UniProt database, concatenated with a reverse database composed of all protein sequences in reverse order. Searches were performed using a 50ppm precursor ion tolerance. Product ion tolerance was set to 0.03 Th. Oxidation of methionine residues (+15.995 Da), heavy lysine (K8) incorporation (+8.0142), and lysine ubiquitination (+ 114.04293 Da) were set as a variable modification; Carboxyamidomethylation of cysteine residues (+57.02146) was set as a static modification. Peptide spectral matches (PSMs) were altered to a 1% False discovery rate (FDR) (Elias and Gygi, 2007; 2010). PSM filtering was performed using a linear discriminant analysis, as described previously (Huttlin et al., 2010), while considering the following parameters: XCorr, ΔCn, missed cleavages, peptide length, charge state, and precursor mass accuracy. Peptide-spectral matches were identified, quantified, and collapsed to a 1% FDR and then further collapsed to a final protein-level FDR of 1%. Furthermore, protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides. To quantify the confidence of each ubiquitylation site, we used a modified version of Ascore (Beausoleil et al., 2006). To confidently localize ubiquitylation sites only the ones with Ascore values > 13 (p ≤ 0.05) were considered. SILAC quantified KGG sites were exported to excel and plotted according to the log2(RNF213kd/Control) values.
Whole Proteome Analysis
Samples were lysed in urea lysis buffer, reduced and alkylated, and digested as performed for the di-gly enrichment sample. The same parameters were also used for offline basic reversed phase separation (96 fraction); samples were combined in a serpentine pattern to obtain 24 samples. 12 non-consecutive samples were chosen for MS analysis. Mass spectrometry data were collected using an Orbitrap Fusion Lumos mass spectrometer (ThermoFisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC 1200 liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were separated on a 100 μm inner diameter microcapillary column packed with 35 cm of Accucore C18 resin (2.6 μm, 150 Å, ThermoFisher). For each analysis, we loaded ~2 μg onto the column. Separation was in-line with the mass spectrometer and was performed using a 2.5 hr gradient of 6 to 26% acetonitrile in 0.125% formic acid at a flow rate of ~450 nL/min. Each analysis used an MS2-based method. The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120,000; mass range 400−1400 m/z; automatic gain control (AGC) target 5 × 105; maximum injection time 100 ms). Precursors for MS2 analysis were selected using a Top10 method. MS2 analysis consisted of collision-induced dissociation (CID); AGC 2.0× 105; normalized collision energy (NCE) 35; maximum injection time 100 ms; normal scan rate, and isolation window of 1.6Da. For data analysis, mass spectra were processed using a Sequest-based in-house software pipeline. Searches were performed using a 50ppm precursor ion tolerance. Fragment ion tolerance was set to 1 Th. Oxidation of methionine residues (+15.995 Da), heavy lysine/arginine incorporation (K8, +8.0142/ R10, + 10.00826) were set as a variable modification; Carboxyamidomethylation of cysteine residues (+57.02146) was set as a static modification. Peptide spectral matches (PSMs) were altered to a 1% False discovery rate, and linear discriminant analysis was performed as detailed above. Protein assembly was guided by principles of parsimony to produce the smallest set of proteins necessary to account for all observed peptides. Data were exported to excel, where contaminant proteins (keratins) were filtered. A summed max intensity of the heavy plus light channel >10 was also applied as a filter, and log2(RNF213kd/Control) max median is shown.
Statistics
Results are expressed as mean ± SD. Unless otherwise noted, statistical significance was assessed via two-way ANOVA with Tukey’s post hoc test using Graph Pad Prism 7.
List of Plasmids and Primers
A list of all primers and plasmids used in this study is provided in Table S5.
Supplementary Material
Table S3. Screen result table, Related to Figure 3.
Table S4. Proteomics data, Related to Figure 6.
Table S5. List of primers, Related to STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| XBP1 | Biolegend | Cat # 619501 |
| Caspase-3 | Cell Signaling | Cat # 966 |
| Caspase-8 | Cell Signaling | Cat # 9746 |
| SAPK/JNK | Cell Signaling | Cat # 9252 |
| Phosphor-SAPK/JNK (Thr183/185) (G9) | Cell Signaling | Cat # 9255 |
| Actin | Cell Signaling | Cat # 3700 |
| Tubulin | Cell Signaling | Cat # 5346 |
| SCD1 | Cell Signaling | Cat # 2438 |
| Cytochrome b5 | Cell Signaling | Cat # 30311 |
| Cytochrome b5 reductase | Sigma | SAB4500655 |
| Vinculin | Cell Signaling | Cat # 4650 |
| BiP | Cell Signaling | Cat # 3177 |
| Calnexin | Cell Signaling | Cat # 2679 |
| FAS | Cell Signaling | Cat # 3180 |
| RNF213 | Sigma | HPA026790 |
| INSIG1 | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| BODIPY (493/503) | Thermo Fisher | Cat # D3922 |
| SCD inhibitor | Abcam | Cat #142089 |
| DGAT1 inhibitor | Merck & Co., Inc. | Liu et al., 2013 |
| Triacsin C | Santa Cruz | Cat # sc-200574A |
| 4u8c | EMD Millipore | Cat # 412512 |
| GSK2606414 | Tocris | Cat # 5107 |
| iScript cDNA Synthesis Kit | BioRad | Cat # 170-8891 |
| Power SYBR Green PCR Master Mix | Life Technologies | Cat # 4368706 |
| RNeasy RNA isolation kit | Qiagen | Cat # 79306 |
| 1,2-dioleoyl-sn-glycerol | Avanti | 800811O |
| 1,2-dipalmitoyl-sn-glycerol | Avanti | 800816 |
| Stearoyl [9,10-3H] Coenzyme A | American Radiolabeled Chemicals, Inc. | ART 0390 |
| Palmitic Acid [1-14C] | American Radiolabeled Chemicals, Inc. | ARC 0172A |
| Stearoyl coenzyme A | Sigma Aldrich | Cat # S0802 |
| NADH | Sigma Aldrich | Cat # N8129 |
| Protease inhibitor cocktail | Sigma-Aldrich | 118736170001 |
| PhosSTOP phosphatase inhibitor | Sigma-Aldrich | 4906837001 |
| Critical Commercial Assays | ||
| Dead Cell Apoptosis Kit | Thermo Fisher | V13242 |
| Experimental Models: Cell Lines | ||
| K562 | ATCC | CCL-243 |
| SUM159 | Laboratory of Tomas Kirchhausen | RRID:CVCL_5423 |
| HeLa | Laboratory of Wade Harper | N/A |
| HepG2 | ATCC | N/A |
| Oligonucleotides | ||
| RNF213 siRNA | Dharmacon | D-001220-01-05 |
| RISC-free (Control) siRNA | Dharmacon | L-023324-00-0005 |
| For primers for qPCR and cloning, see Table S5 | This paper | N/A |
| Deposited Data | ||
| Raw and processed RNAseq data | NCBI GEO | GSE125178 |
| Software and Algorithms | ||
| Lipid Search | Thermo Fisher | Taguchi et al., 2010 |
| ClueGo | Cytoscape | http://apps.cytoscape.org/apps/cluego |
Highlights.
Palmitate treatment of cells increased saturated glycerolipids and ER stress
Di-saturated glycerolipids are key to lipotoxicity in this model
Inhibiting ER-localized GPAT enzymes protected against lipotoxicity
Depletion of the putative E3 ligase RNF213 protected from lipotoxicity
ACKNOWLEDGEMENTS
We thank Abdullah Alfawaz for help with RT- qPCR analysis, Shirly Pinto and Merck & Co. Inc. for sharing DGAT inhibitors, James Ntambi for advice on SCD1, Gary Howard for editorial assistance, Nina Gluchowski for assistance with experiments, Niklas Mejhert for contribution towards lipidomics filtration method, Aditi Jatkar for design and optimization of INSIG1 antibody, Steve Gygi for the use of mass spectrometry facilities, and Cell Signaling Technology for the di-Gly antibody. This work was supported by the Swiss National Foundation fellowship (to M.P.), K99/R00 CA181494 (to M.K.), the National Science Foundation Graduate Research Fellowship under Grant No DGE1144152 and the Department of Defense (DoD) through the National Defense Science & Engineering Graduate Fellowship (NDSEG) (to C.B.K.J), K01 DK098285 (to J.A.P.), R01 DK101579 (to T.C.W and R.V.F), R37 NS083524 and R01NS110395 to J.W.H., the Mathers foundation (to T.C.W.), and funding from Pfizer (to R.V.F and T.C.W.). T.C.W is an investigator of the Howard Hughes Medical Institute.
Footnotes
DECLARATION OF INTERESTS
J.W.H. is a consultant and founder of Rheostat Therapeutics and a consultant for X-Chem Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agarwal AK, Sukumaran S, Cortes VA, Tunison K, Mizrachi D, Sankella S, Gerard RD, Horton JD, and Garg A. (2011). Human 1-acylglycerol-3-phosphate O-acyltransferase isoforms 1 and 2: biochemical characterization and inability to rescue hepatic steatosis in Agpat2(−/−) gene lipodystrophic mice. J Biol Chem 286, 37676–37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banh RS, Iorio C, Marcotte R, Xu Y, Cojocari D, Rahman AA, Pawling J, Zhang W, Sinha A, Rose CM, et al. (2016). PTP1B controls non-mitochondrial oxygen consumption by regulating RNF213 to promote tumour survival during hypoxia. Nature cell biology 18, 803–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassik MC, Kampmann M, Lebbink RJ, Wang S, Hein MY, Poser I, Weibezahn J, Horlbeck MA, Chen S, Mann M, et al. (2013). A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, and Gygi SP (2006). A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24, 1285–1292. [DOI] [PubMed] [Google Scholar]
- Borradaile NM, Han X, Harp JD, Gale SE, Ory DS, and Schaffer JE (2006). Disruption of endoplasmic reticulum structure and integrity in lipotoxic cell death. J Lipid Res 47, 2726–2737. [DOI] [PubMed] [Google Scholar]
- Brookheart RT, Michel CI, and Schaffer JE (2009). As a matter of fat. Cell Metab 10, 9–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Li JL, Li D, Tobin JF, and Gimeno RE (2006). Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc Natl Acad Sci U S A 103, 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputa G, and Schaffer JE (2016). RNA Regulation of Lipotoxicity and Metabolic Stress. Diabetes 65, 1816–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, and Farese RV Jr. (2001). Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem 276, 38870–38876. [DOI] [PubMed] [Google Scholar]
- Chitraju C, Mejhert N, Haas JT, Diaz-Ramirez LG, Grueter CA, Imbriglio JE, Pinto S, Koliwad SK, Walther TC, and Farese RV Jr. (2017). Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell metabolism 26, 407–418 e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimen I, Kocaturk B, Koyuncu S, Tufanli O, Onat UI, Yildirim AD, Apaydin O, Demirsoy S, Aykut ZG, Nguyen UT, et al. (2016). Prevention of atherosclerosis by bioactive palmitoleate through suppression of organelle stress and inflammasome activation. Sci Transl Med 8, 358ra126. [DOI] [PubMed] [Google Scholar]
- Eitel K, Staiger H, Brendel MD, Brandhorst D, Bretzel RG, Haring HU, and Kellerer M. (2002). Different role of saturated and unsaturated fatty acids in beta-cell apoptosis. Biochem Biophys Res Commun 299, 853–856. [DOI] [PubMed] [Google Scholar]
- Ellingson JS, Hill EE, and Lands WE (1970). The control of fatty acid composition in glycerolipids of the endoplasmic reticulum. Biochim Biophys Acta 196, 176–192. [DOI] [PubMed] [Google Scholar]
- Farese RV Jr., Zechner R, Newgard CB, and Walther TC (2012). The problem of establishing relationships between hepatic steatosis and hepatic insulin resistance. Cell metabolism 15, 570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers MT, Keller MP, Choi Y, Lan H, Kendziorski C, Ntambi JM, and Attie AD (2008). Liver gene expression analysis reveals endoplasmic reticulum stress and metabolic dysfunction in SCD1-deficient mice fed a very low-fat diet. Physiol Genomics 33, 361–372. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, and Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226, 497–509. [PubMed] [Google Scholar]
- Gao D, Nong S, Huang X, Lu Y, Zhao H, Lin Y, Man Y, Wang S, Yang J, and Li J. (2010). The effects of palmitate on hepatic insulin resistance are mediated by NADPH Oxidase 3-derived reactive oxygen species through JNK and p38MAPK pathways. J Biol Chem 285, 29965–29973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluchowski NL, Chitraju C, Picoraro JA, Mejhert N, Pinto S, Xin W, Kamin DS, Winter HS, Chung WK, Walther TC, et al. (2017). Identification and characterization of a novel DGAT1 missense mutation associated with congenital diarrhea. J Lipid Res 58, 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinathan G, Milagre C, Pearce OM, Reynolds LE, Hodivala-Dilke K, Leinster DA, Zhong H, Hollingsworth RE, Thompson R, Whiteford JR, et al. (2015). Interleukin-6 Stimulates Defective Angiogenesis. Cancer Res 75, 3098–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CD, and Olson LK (2011). Modulation of palmitate-induced endoplasmic reticulum stress and apoptosis in pancreatic beta-cells by stearoyl-CoA desaturase and Elovl6. American journal of physiology. Endocrinology and metabolism 300, E640–649. [DOI] [PubMed] [Google Scholar]
- Halbleib K, Pesek K, Covino R, Hofbauer HF, Wunnicke D, Hanelt I, Hummer G, and Ernst R. (2017). Activation of the Unfolded Protein Response by Lipid Bilayer Stress Mol Cell. [DOI] [PubMed] [Google Scholar]
- Haldar AVa.D. (1994). Purification and Characterization of Glycerophosphate Acyltransferase from Rat Liver Mitochondria. J. Biol. Chem n44 269, 27209–27215. [PubMed] [Google Scholar]
- Han GS, and Carman GM (2010). Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem 285, 14628–14638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SP, Wu MS, Shun CT, Wang HP, Lin MT, Kuo ML, and Lin JT (2004). Interleukin-6 increases vascular endothelial growth factor and angiogenesis in gastric carcinoma. J Biomed Sci 11, 517–527. [DOI] [PubMed] [Google Scholar]
- Kamada F, Aoki Y, Narisawa A, Abe Y, Komatsuzaki S, Kikuchi A, Kanno J, Niihori T, Ono M, Ishii N, et al. (2011). A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet 56, 34–40. [DOI] [PubMed] [Google Scholar]
- Kampmann M, Bassik MC, and Weissman JS (2013). Integrated platform for genome-wide screening and construction of high-density genetic interaction maps in mammalian cells. Proc Natl Acad Sci U S A 110, E2317–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M, Bassik MC, and Weissman JS (2014). Functional genomics platform for pooled screening and generation of mammalian genetic interaction maps. Nat Protoc 9, 1825–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst AM, Gao K, Nelson CC, and Li G. (2009). Nuclear factor kappa B subunit p50 promotes melanoma angiogenesis by upregulating interleukin-6 expression. Int J Cancer 124, 494–501. [DOI] [PubMed] [Google Scholar]
- Kitai Y, Ariyama H, Kono N, Oikawa D, Iwawaki T, and Arai H. (2013). Membrane lipid saturation activates IRE1alpha without inducing clustering. Genes Cells 18, 798–809. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Matsuda Y, Hitomi T, Okuda H, Shioi H, Matsuda T, Imai H, Sone M, Taura D, Harada KH, et al. (2015). Biochemical and Functional Characterization of RNF213 (Mysterin) R4810K, a Susceptibility Mutation of Moyamoya Disease, in Angiogenesis In Vitro and In Vivo. Journal of the American Heart Association 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Yamazaki S, Takashima S, Liu W, Okuda H, Yan J, Fujii Y, Hitomi T, Harada KH, Habu T, et al. (2013). Ablation of Rnf213 retards progression of diabetes in the Akita mouse. Biochem Biophys Res Commun 432, 519–525. [DOI] [PubMed] [Google Scholar]
- Kotani Y, Morito D, Yamazaki S, Ogino K, Kawakami K, Takashima S, Hirata H, and Nagata K. (2015). Neuromuscular regulation in zebrafish by a large AAA+ ATPase/ubiquitin ligase, mysterin/RNF213. Scientific reports 5, 16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lê S, Josse J, and Husson F. (2008). FactoMineR: An R package for multivariate analysis. Journal of Statistical Software 25, 1–18. [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, and Schaffer JE (2003). Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proceedings of the National Academy of Sciences 100, 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, McLaren DG, Chen D, Kan Y, Stout SJ, Shen X, Murphy BA, Forrest G, Karanam B, Sonatore L, et al. (2015). Potential mechanism of enhanced postprandial glucagon-like peptide-1 release following treatment with a diacylglycerol acyltransferase 1 inhibitor. Pharmacol Res Perspect 3, e00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Hitomi T, Kobayashi H, Harada KH, and Koizumi A. (2012). Distribution of Moyamoya Disease Susceptibility Polymorphism p.R4810K in RNF213 in East and Southeast Asian Populations. Neurologia medico-chirurgica 52, 299–303. [DOI] [PubMed] [Google Scholar]
- Liu W, Morito D, Takashima S, Mineharu Y, Kobayashi H, Hitomi T, Hashikata H, Matsuura N, Yamazaki S, Toyoda A, et al. (2011). Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS One 6, e22542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Miyazaki-Anzai S, Keenan AL, Okamura K, Kendrick J, Chonchol M, Offermanns S, Ntambi JM, Kuro OM, and Miyazaki M. (2015). Saturated phosphatidic acids mediate saturated fatty acid-induced vascular calcification and lipotoxicity. J Clin Invest 125, 4544–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, Behlke MA, Ory DS, and Schaffer JE (2011). Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell metabolism 14, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle CA, Klett EL, and Coleman RA (2009). Hepatic triacylglycerol accumulation and insulin resistance. J Lipid Res 50 Suppl, S74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaez-Rivas M, and Zhang Q. (2016). Comprehensive untargeted lipidomic analysis using core-shell C30 particle column and high field orbitrap mass spectrometer. J Chromatogr A 1440, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton CM, and Ntambi JM (2009). Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab 297, E28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumen MB, Ishida Y, Muramatsu M, Yamamoto M, and Honjo T. (1997). Inhibition of carnitine palmitoyltransferase I augments sphingolipid synthesis and palmitate-induced apoptosis. J Biol Chem 272, 3324–3329. [DOI] [PubMed] [Google Scholar]
- Prasad SS, Garg A, and Agarwal AK (2011). Enzymatic activities of the human AGPAT isoform 3 and isoform 5: localization of AGPAT5 to mitochondria. J Lipid Res 52, 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robblee MM, Kim CC, Porter Abate J, Valdearcos M, Sandlund KL, Shenoy MK, Volmer R, Iwawaki T, and Koliwad SK (2016). Saturated Fatty Acids Engage an IRE1alpha-Dependent Pathway to Activate the NLRP3 Inflammasome in Myeloid Cells. Cell reports 14, 2611–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro M, Zhou YT, Levi M, and Unger RH (1998). Fatty acid-induced beta cell apoptosis: A link between obesity and diabetes. Proc. Natl. Acad. Sci. USA Vol 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindou H, Hishikawa D, Harayama T, Eto M, and Shimizu T. (2013). Generation of membrane diversity by lysophospholipid acyltransferases. J Biochem 154, 21–28. [DOI] [PubMed] [Google Scholar]
- Strittmatter P, Spatz L, Corcoran D, Rogers MJ, Setlow B, and Redline R. (1974). Purification and properties of rat liver microsomal stearyl coenzyme A desaturase. Proceedings of the National Academy of Sciences of the United States of America 71, 4565–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi R, and Ishikawa M. (2010). Precise and global identification of phospholipid molecular species by an Orbitrap mass spectrometer and automated search engine Lipid Search. J Chromatogr A 1217, 4229–4239. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, and Reue K. (2009). Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. American journal of physiology. Endocrinology and metabolism 296, E1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin SM, Lancaster GI, Darby I, Febbraio MA, and Watt MJ (2006). Apoptosis in skeletal muscle myotubes is induced by ceramides and is positively related to insulin resistance. Am J Physiol Endocrinol Metab 291, E1341–1350. [DOI] [PubMed] [Google Scholar]
- Udeshi ND, Mertins P, Svinkina T, and Carr SA (2013). Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc 8, 1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, and Ron D. (2013). Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci U S A 110, 4628–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Briggs MR, Hua X, Yokoyama C, Goldstein JL, and Brown MS (1993). Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J Biol Chem 268, 14497–14504. [PubMed] [Google Scholar]
- Wei Y, Wang D, Topczewski F, and Pagliassotti MJ (2006). Saturated fatty acids induce endoplasmic reticulum stress and apoptosis independently of ceramide in liver cells. Am J Physiol Endocrinol Metab 291, E275–281. [DOI] [PubMed] [Google Scholar]
- Zhu XGSN, Shen Y, La K, Klompstra D, Gultekin Y, Fidelin J, Molina H, Hang H, Min W, and Birsoy K. CHP1 activates microsomal GPATs and is necessary for glycerolipid synthesis. Molecular Cell. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T, Uchikata T, Sakamoto S, Yokoi Y, Fukusaki E, and Bamba T. (2013). Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J Chromatogr A 1292, 211–218. [DOI] [PubMed] [Google Scholar]
- Yamashita A, Hayashi Y, Matsumoto N, Nemoto-Sasaki Y, Oka S, Tanikawa T, and Sugiura T. (2014). Glycerophosphate/Acylglycerophosphate acyltransferases. Biology (Basel) 3, 801–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CL, Monetti M, Burri BJ, and Farese RV Jr. (2005). The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J Lipid Res 46, 1502–1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Screen result table, Related to Figure 3.
Table S4. Proteomics data, Related to Figure 6.
Table S5. List of primers, Related to STAR Methods.