Abstract
Huntington’s disease (HD) is an inherited neurodegenerative disease. Besides the well-characterized motor symptoms, HD is marked by cognitive impairment and behavioral changes. In this study, we analyzed the blood of HD gene carries using RNA-sequencing techniques. We evaluated samples from HD gene carriers with (n = 8) and without clinically meaningful depressive symptoms (n = 8) compared with healthy controls (n = 8). Groups were age- and sex-matched. Preprocessing of data and between-group comparisons were calculated using DESeq2. The Wald test was used to generate p-values and log2 fold changes. We found 60 genes differently expressed in HD and healthy controls, of which 21 were upregulated and 39 downregulated. Within HD group, nineteen genes were differently expressed between patients with and without depression, being 6 upregulated and 13 downregulated. Several of the top differentially expressed genes are involved in nervous system development. Although preliminary, our findings corroborate the emerging view that in addition to neurodegenerative mechanisms, HD has a neurodevelopmental component. Importantly, the emergence of depression in HD might be related to these mechanisms.
Keywords: Huntington’s disease, neurodegenerative disease, RNA-seq, depression, neurodevelopment
1. Introduction
Huntington’s disease (HD) is an inherited autosomal dominant neurodegenerative disease. HD is caused by expanded CAG trinucleotide repeats in the exon 1 of the Huntingtin gene (HTT), which encodes a mutant form of the huntingtin protein (HTT) with an abnormal polyglutamine tail at the N-terminus [1,2,3]. HTT is an important protein for neurodevelopment. The knockout of the HTT in mice results in major brain abnormalities and death soon after birth [4,5,6]. In addition, depletion of wild-type HTT in the postnatal mouse brain leads to progressive neurodegeneration [7]. The presence of 40 or more CAG repeats invariably causes the disease, and longer repeats predict earlier disease onset. Mutant HTT (mHTT) is widely expressed and believed to induce neurodegeneration through different mechanisms, including abnormal interaction with other proteins, leading to cellular changes and ultimately cell death [8].
HD is marked by a triad of symptoms including motor, cognitive and psychiatric or behavioral symptoms. The clinical diagnosis of HD has been historically based on motor symptoms. However, cognitive and psychiatric symptoms are often present years prior to the onset of clinically relevant motor symptoms [9]. Psychiatric symptoms, such as anxiety, irritability, impulsivity, and depressed mood are frequent in individuals with HD and can be quite troublesome. Major depression is the most common psychiatric syndrome among premanifest HD carriers [10]. Depression may precede the onset of typical motor symptoms by 4 to 10 years, making it one of the earliest signs possibly related to HD pathophysiology. Moreover, around 40–50% of patients with HD experience depression at some point during the course of the disease [11], and many patients and their relatives consider this problem the most distressing aspect of the illness. Actually, depression seems to influence more profoundly health-related quality of life in HD than motor symptoms or cognitive dysfunction [12]. The presence of depression in HD is also an important predictor of suicidal behavior [13]. Patients with HD have been shown to commit suicide four to eight times more often than the general population [14,15,16]. The increased rate of suicide may be related to several factors, including the emotional distress of having an incurable disease alongside the elevated frequency of depression and other behavioral symptoms, such as impulsivity [13,17].
The reasons for the frequent co-occurrence of HD and depression are still unclear. The heritability nature of HD does not fully explain the high prevalence of psychiatric symptoms, especially depression, in HD [18,19]. Importantly, as the severity of depression is not associated with disease progression [20,21,22], different mechanisms are likely to affect neurons involved in mood regulation circuits and neurons involved in motor skills that are impaired in manifest or later stages of the disease. The molecular underpinnings of psychiatric symptoms in HD are poorly understood. Therefore, we designed an exploratory study to investigate peripheral blood gene expression profile through RNA-seq in HD gene carriers (with and without depression) and healthy controls. Our hypothesis is that HD gene carriers presenting with depression have a different transcriptome profile when compared with HD gene carriers without these symptoms. In addition, we assessed different genetic pathways underlying HD in comparison with healthy controls. This is the first study to evaluate gene expression profile comparing these subgroups of HD gene carriers and healthy controls. Our results advance the understanding of the biological mechanisms associated with depression in HD with the ultimate goal of identifying a more specific and effective anti-depressant strategy to improve the quality of life and decrease suicide rates of these subjects [23].
2. Results
Demographic data from HD gene carriers and controls are shown in Table 1. Groups did not differ in age, years of education, sex and BMI.
Table 1.
Demographic variables of the subjects enrolled in this study.
| Variables | Control Group (n = 8) |
HD without Depression (n = 8) | HD with Depression (n = 8) | Statistic |
|---|---|---|---|---|
| Female sex | 5 (62.5%) | 5 (62.5%) | 5 (62.5%) | χ2(1) = 0 p = 1 |
| Age | 51.91 (10.59) | 38.17 (8.44) | 48.27 (13.81) | F (2, 21) = 3.12 p = 0.067 |
| Years of education | 15.93 (4.82) | 15.57 (3.59) | 14.25 (2.96) | F (2, 21) = 0.4078 p = 0.6703 |
| BMI | 30.86 (8.09) | 29.04 (7.15) | 29.39 (6.18) | F (2, 20) = 0.3627 p = 0.7003 |
2.1. Differential Gene Expression Analysis
Sixty genes were differently expressed in HD gene carriers (n = 16) compared with healthy controls (n = 8), of which 21 were upregulated and 39 were downregulated (Table 2). When analyzing HD gene carriers with depression versus non-depression, there were 19 genes differently expressed, being 6 upregulated and 13 downregulated (Table 3).
Table 2.
Different expressed genes between HD gene carrier and healthy controls.
| Gene ID | Gene Name | log2FoldChange | Stat | p Value | padj |
|---|---|---|---|---|---|
| ENSG00000109956 | B3GAT1 | 1.484 | 4.332 | 0.000 | 0.019415 |
| ENSG00000205336 | ADGRG1 | 1.293 | 4.605 | 0.000 | 0.019415 |
| ENSG00000165568 | AKR1E2 | −1.007 | −4.185 | 0.000 | 0.026784 |
| ENSG00000206172 | HBA1 | −1.208 | −3.890 | 0.000 | 0.038152 |
| ENSG00000110203 | FOLR3 | −1.687 | −3.265 | 0.001 | 0.059707 |
| ENSG00000234389 | AC007278.1 | −1.262 | −3.162 | 0.002 | 0.068408 |
| ENSG00000137267 | TUBB2A | −1.591 | −2.213 | 0.027 | 0.192338 |
| ENSG00000196565 | HBG2 | −1.006 | −2.166 | 0.030 | 0.201445 |
| ENSG00000239839 | DEFA3 | 1.101 | 2.017 | 0.044 | 0.234289 |
| ENSG00000237973 | MTCO1P12 | 2.565 | 3.763 | 0.000 | NA |
| ENSG00000248527 | MTATP6P1 | 1.036 | 3.042 | 0.002 | NA |
| ENSG00000162631 | NTNG1 | −1.102 | −2.618 | 0.009 | NA |
| ENSG00000196539 | OR2T3 | −1.329 | −3.064 | 0.002 | NA |
| ENSG00000200488 | RN7SKP203 | −1.059 | −2.842 | 0.004 | NA |
| ENSG00000071909 | MYO3B | 1.142 | 2.789 | 0.005 | NA |
| ENSG00000144485 | HES6 | −1.141 | −3.027 | 0.002 | NA |
| ENSG00000144908 | ALDH1L1 | −1.288 | −2.189 | 0.029 | NA |
| ENSG00000145362 | ANK2 | −1.017 | −1.961 | 0.050 | NA |
| ENSG00000178636 | AC092656.1 | −1.233 | −3.359 | 0.001 | NA |
| ENSG00000247199 | AC091948.1 | −1.128 | −4.107 | 0.000 | NA |
| ENSG00000230202 | AL450405.1 | 2.670 | 4.467 | 0.000 | NA |
| ENSG00000260997 | AC004847.1 | 1.032 | 3.541 | 0.000 | NA |
| ENSG00000075213 | SEMA3A | −1.025 | −2.605 | 0.009 | NA |
| ENSG00000279483 | AC090498.1 | −1.257 | −2.763 | 0.006 | NA |
| ENSG00000276819 | TRBV15 | −1.360 | −3.558 | 0.000 | NA |
| ENSG00000201098 | RNY1 | −1.164 | −2.702 | 0.007 | NA |
| ENSG00000234449 | FAM239A | −2.310 | −2.901 | 0.004 | NA |
| ENSG00000215374 | FAM66B | −1.026 | −4.054 | 0.000 | NA |
| ENSG00000240905 | RN7SL798P | 1.118 | 3.315 | 0.001 | NA |
| ENSG00000184350 | MRGPRE | −1.929 | −2.127 | 0.033 | NA |
| ENSG00000006071 | ABCC8 | −1.368 | −2.151 | 0.031 | NA |
| ENSG00000170959 | DCDC1 | −1.127 | −2.440 | 0.015 | NA |
| ENSG00000156113 | KCNMA1 | −1.153 | −2.707 | 0.007 | NA |
| ENSG00000235602 | POU5F1P3 | 1.022 | 3.920 | 0.000 | NA |
| ENSG00000225231 | LINC02470 | −1.765 | −2.060 | 0.039 | NA |
| ENSG00000177359 | AC024940.2 | −1.687 | −3.254 | 0.001 | NA |
| ENSG00000273824 | AC008033.3 | 1.148 | 2.862 | 0.004 | NA |
| ENSG00000123201 | GUCY1B2 | −1.532 | −3.138 | 0.002 | NA |
| ENSG00000102837 | OLFM4 | 1.081 | 2.248 | 0.025 | NA |
| ENSG00000139926 | FRMD6 | −1.146 | −3.784 | 0.000 | NA |
| ENSG00000021645 | NRXN3 | −1.020 | −2.433 | 0.015 | NA |
| ENSG00000189419 | SPATA41 | −1.037 | −3.232 | 0.001 | NA |
| ENSG00000205918 | PDPK2P | 1.111 | 3.006 | 0.003 | NA |
| ENSG00000261245 | AC093520.2 | 1.096 | 2.797 | 0.005 | NA |
| ENSG00000270124 | AC092127.2 | 1.040 | 3.505 | 0.000 | NA |
| ENSG00000262074 | SNORD3B-2 | −1.154 | −2.194 | 0.028 | NA |
| ENSG00000276241 | AC243829.2 | 1.579 | 3.073 | 0.002 | NA |
| ENSG00000274512 | TBC1D3L | 1.021 | 2.304 | 0.021 | NA |
| ENSG00000142449 | FBN3 | −1.148 | −1.982 | 0.047 | NA |
| ENSG00000187244 | BCAM | −1.063 | −2.163 | 0.031 | NA |
| ENSG00000262874 | C19orf84 | 1.224 | 3.628 | 0.000 | NA |
| ENSG00000233493 | TMEM238 | −1.064 | −4.849 | 0.000 | NA |
| ENSG00000179954 | SSC5D | −1.192 | −2.558 | 0.011 | NA |
| ENSG00000196263 | ZNF471 | −1.153 | −3.938 | 0.000 | NA |
| ENSG00000211659 | IGLV3-25 | −1.369 | −3.483 | 0.000 | NA |
| ENSG00000264063 | MIR3687-2 | −1.068 | −2.784 | 0.005 | NA |
| ENSG00000215533 | LINC00189 | 1.417 | 2.738 | 0.006 | NA |
| ENSG00000236056 | GAPDHP14 | 1.401 | 2.836 | 0.005 | NA |
| ENSG00000255568 | BRWD1-AS2 | −1.119 | −3.732 | 0.000 | NA |
| ENSG00000210049 | MT-TF | 1.037 | 2.638 | 0.008 | NA |
Stat: statistics; padj: p value adjustable.
Table 3.
Different expressed genes between HD gene carrier with depression and HD gene carrier without depression.
| Gene ID | Gene Name | log2FoldChange | Stat | p Value | padj |
|---|---|---|---|---|---|
| ENSG00000130202 | NECTIN2 | −1.20871551 | −2.27773 | 0.022743 | 0.999462 |
| ENSG00000235169 | SMIM1 | 1.286216213 | 2.0662 | 0.03881 | 0.999462 |
| ENSG00000163646 | CLRN1 | −1.417542862 | −2.08074 | 0.037458 | 0.999462 |
| ENSG00000233058 | LINC00884 | −1.037747878 | −2.89202 | 0.003828 | 0.999462 |
| ENSG00000010030 | ETV7 | 1.070004254 | 2.301148 | 0.021383 | 0.999462 |
| ENSG00000215018 | COL28A1 | −1.217119591 | −2.50457 | 0.01226 | 0.999462 |
| ENSG00000175445 | LPL | 1.308483772 | 2.189831 | 0.028537 | 0.999462 |
| ENSG00000178860 | MSC | −1.100681038 | −2.98191 | 0.002865 | 0.999462 |
| ENSG00000159247 | TUBBP5 | −1.267966389 | −2.35356 | 0.018595 | 0.999462 |
| ENSG00000196565 | HBG2 | −1.350354678 | −2.52451 | 0.011586 | 0.999462 |
| ENSG00000251381 | LINC00958 | 2.26201877 | 2.3048 | 0.021178 | 0.999462 |
| ENSG00000254789 | AC073172.1 | −1.332994545 | −2.7906 | 0.005261 | 0.999462 |
| ENSG00000255508 | AP002990.1 | −1.058759746 | −3.49951 | 0.000466 | 0.999462 |
| ENSG00000078114 | NEBL | 2.623958542 | 3.216931 | 0.001296 | 0.999462 |
| ENSG00000200830 | RN7SKP134 | −1.036440243 | −2.74592 | 0.006034 | 0.999462 |
| ENSG00000135116 | HRK | −1.007746896 | −2.79251 | 0.00523 | 0.999462 |
| ENSG00000124107 | SLPI | −1.250074721 | −2.10266 | 0.035496 | 0.999462 |
| ENSG00000226025 | AC005515.1 | 1.025633489 | 2.438544 | 0.014747 | 0.999462 |
| ENSG00000160233 | LRRC3 | −1.09935177 | −2.93576 | 0.003327 | 0.999462 |
Stat: statistics; padj: p value adjustable.
2.2. Gene Ontology Analysis
Comparing HD gene carriers and healthy controls, 109 enriched pathways were identified (Table 4). Among the enriched pathways, many of them contain the top differentially expressed gene ADGRG1, such as GO:0010573~vascular endothelial growth factor production, GO:0021801~cerebral cortex radial glia guided migration, GO:0021796~cerebral cortex regionalization GO:0021819~layer formation in cerebral cortex. Noteworthy, there were other enriched pathways with different genes involved in neurodevelopment, such as the GO:2001224~positive regulation of neuron migration and GO:0007155~cell adhesion.
Table 4.
The 20 most significant different pathways between HD gene carriers and healthy control.
| Genes | Process_Name | Significant_ Genes_Count |
Total_Genes_ Group_Count |
Percent_ Significant_Genes |
p-Value | padj-Value |
|---|---|---|---|---|---|---|
| HBG2; HBA1; | GO:0015671~oxygen transport | 2 | 14 | 14.286 | 0.00013 | 0.013186 |
| ANK2; ABCC8; | GO:0043268~positive regulation of potassium ion transport | 2 | 10 | 20.000 | 0.00007 | 0.013186 |
| ANK2; SEMA3A; | GO:0002027~regulation of heart rate | 2 | 31 | 6.452 | 0.00058 | 0.026314 |
| FRMD6; | GO:0003383~apical constriction | 1 | 3 | 33.333 | 0.00427 | 0.026314 |
| ABCC8; KCNMA1; | GO:0006813~potassium ion transport | 2 | 78 | 2.564 | 0.00337 | 0.026314 |
| SSC5D; IGLV3-25; HBA1; | GO:0006898~receptor-mediated endocytosis | 3 | 185 | 1.622 | 0.00110 | 0.026314 |
| ALDH1L1; | GO:0009258~10-formyltetrahydrofolate catabolic process | 1 | 2 | 50.000 | 0.00321 | 0.026314 |
| ADGRG1; | GO:0010573~vascular endothelial growth factor production | 1 | 3 | 33.333 | 0.00427 | 0.026314 |
| ADGRG1; | GO:0021801~cerebral cortex radial glia guided migration | 1 | 2 | 50.000 | 0.00321 | 0.026314 |
| SEMA3A; | GO:0021828~gonadotrophin-releasing hormone neuronal migration to the hypothalamus | 1 | 2 | 50.000 | 0.00321 | 0.026314 |
| FRMD6; | GO:0032970~regulation of actin filament-based process | 1 | 2 | 50.000 | 0.00321 | 0.026314 |
| ANK2; | GO:0033292~T-tubule organization | 1 | 3 | 33.333 | 0.00427 | 0.026314 |
| KCNMA1; | GO:0034465~response to carbon monoxide | 1 | 3 | 33.333 | 0.00427 | 0.026314 |
| ANK2; FRMD6; | GO:0034613~cellular protein localization | 2 | 40 | 5.000 | 0.00094 | 0.026314 |
| ANK2; | GO:0036309~protein localization to M-band | 1 | 2 | 50.000 | 0.00321 | 0.026314 |
| ANK2; | GO:0036371~protein localization to T-tubule | 1 | 1 | 100.000 | 0.00214 | 0.026314 |
| SEMA3A; | GO:0036486~ventral trunk neural crest cell migration | 1 | 3 | 33.333 | 0.00427 | 0.026314 |
| SSC5D; | GO:0042494~detection of bacterial lipoprotein | 1 | 1 | 100.000 | 0.00214 | 0.026314 |
| SEMA3A; | GO:0048880~sensory system development | 1 | 3 | 33.333 | 0.00427 | 0.026314 |
| SSC5D; DEFA3; | GO:0050830~defense response to Gram-positive bacterium | 2 | 66 | 3.030 | 0.00245 | 0.026314 |
padj-value: p value adjustable.
In the comparison among HD gene carriers, 61 enriched pathways were identified (Table 5). Several pathways have the NECTIN2 gene, as GO:0002891~positive regulation of immunoglobulin mediated immune response, GO:0034332~adherens junction organization, GO:0007157~heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules.
Table 5.
The 20 most significant different pathways between HD gene carries with depression and HD gene carriers without depression.
| Genes | Process_Name | Significant_ Genes_Count |
Total_Genes_ Group_Count |
Percent_ Significant_Genes |
p-Value | padj-Value |
|---|---|---|---|---|---|---|
| NECTIN2; | GO:0002891~positive regulation of immunoglobulin mediated immune response | 1 | 3 | 33.3333 | 0.00136 | 0.008475 |
| SLPI; COL28A1; | GO:0010951~negative regulation of endopeptidase activity | 2 | 124 | 1.6129 | 0.00084 | 0.008475 |
| MSC; | GO:0014707~branchiomeric skeletal muscle development | 1 | 3 | 33.3333 | 0.00136 | 0.008475 |
| NECTIN2; | GO:0030382~sperm mitochondrion organization | 1 | 2 | 50.0000 | 0.00102 | 0.008475 |
| NECTIN2; | GO:0032990~cell part morphogenesis | 1 | 1 | 100.0000 | 0.00068 | 0.008475 |
| NECTIN2; | GO:0033005~positive regulation of mast cell activation | 1 | 2 | 50.0000 | 0.00102 | 0.008475 |
| LPL; | GO:0034371~chylomicron remodeling | 1 | 3 | 33.3333 | 0.00136 | 0.008475 |
| NECTIN2; | GO:0044406~adhesion of symbiont to host | 3 | 33.3333 | 0.00136 | 0.008475 | |
| NECTIN2; | GO:0046814~coreceptor-mediated virion attachment to host cell | 1 | 1 | 100.0000 | 0.00068 | 0.008475 |
| NECTIN2; | GO:0051654~establishment of mitochondrion localization | 1 | 2 | 50.0000 | 0.00102 | 0.008475 |
| NECTIN2; | GO:0060370~susceptibility to T cell mediated cytotoxicity | 1 | 3 | 33.3333 | 0.00136 | 0.008475 |
| NEBL; | GO:0071691~cardiac muscle thin filament assembly | 1 | 1 | 100.0000 | 0.00068 | 0.008475 |
| NECTIN2; | GO:0042271~susceptibility to natural killer cell mediated cytotoxicity | 1 | 4 | 25.0000 | 0.00169 | 0.009079 |
| NECTIN2; | GO:0046596~regulation of viral entry into host cell | 1 | 4 | 25.0000 | 0.00169 | 0.009079 |
| NECTIN2; | GO:0002860~positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target | 1 | 7 | 14.2857 | 0.00271 | 0.009770 |
| NECTIN2; | GO:0007289~spermatid nucleus differentiation | 1 | 8 | 12.5000 | 0.00305 | 0.009770 |
| LPL; | GO:0010886~positive regulation of cholesterol storage | 1 | 7 | 14.2857 | 0.00271 | 0.009770 |
| LPL; | GO:0010890~positive regulation of sequestering of triglyceride | 1 | 7 | 14.2857 | 0.00271 | 0.009770 |
| NECTIN2; | GO:0019064~fusion of virus membrane with host plasma membrane | 1 | 8 | 12.5000 | 0.00305 | 0.009770 |
| HRK; | GO:0032464~positive regulation of protein homooligomerization | 1 | 8 | 12.5000 | 0.00305 | 0.009770 |
padj-value: p value adjustable.
2.3. Validation of Microarray Data by Real-Time qPCR
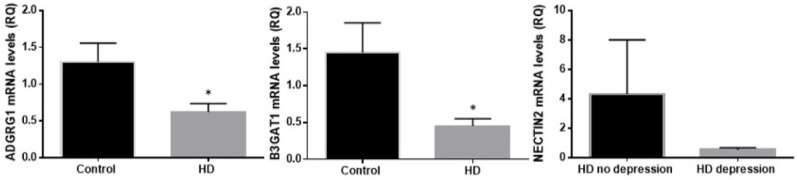
We validated the RNA-seq results by determining mRNA levels of the top three differentially expressed genes (Figure 1). Regarding ADGRG1 and B3GAT1, real-time qPCR confirmed RNAseq results. For NECTIN2, the results showed the same tendency, but without reaching statistical significance.
Figure 1.
mRNA levels of the top three differentially expressed genes. *: p < 0.05.
3. Discussion
In this study, we examined RNA-seq gene expression in the blood of HD gene carriers with and without depressive symptoms and healthy controls. Differences between HD gene carriers and controls were marked, with several genes related to neurodevelopmental pathways. Conversely, differences among HD gene carriers with and without depression were less pronounced. As several of the genes and enriched pathways are involved in the development of the nervous system, our preliminary findings corroborate the emerging view that in addition to being a neurodegenerative disease, HD has a neurodevelopmental component.
The gene B3GAT1 was among the top differentially expressed genes between HD gene carriers and controls. This gene is the key enzyme during the biosynthesis of the carbohydrate epitope HNK-1. The HNK-1 epitope is mainly present in the brain, more specifically in a number of cell adhesion molecules (CAM) important for neuronal cell adhesion, synaptic plasticity, and, therefore, neurodevelopment [24,25]. B3GAT1 has also been implicated in major psychiatric disorders, as schizophrenia and schizoaffective disorders [26]. Mice deficient for B3GAT1 exhibited normal development of gross anatomical features, but had impairments of high-order brain functions, including learning, and memory [25].
Another gene differentially expressed between HD gene carriers and healthy controls was ADGRG1 (or GPR56). This gene encodes a member of the G protein-coupled receptor family. ADGRG1 plays critical roles in the development of several organs, including the brain, with extensive implications for human diseases and their treatment [27,28,29,30,31]. Mutations in ADGRG1 cause a severe human brain malformation called bilateral frontoparietal polymicrogyria, characterized by cortical lamination defects, cerebellar hypoplasia, and central nervous system hypomyelination [32,33]. In addition, ADGRG1 regulates embryonic brain development and postnatal myelination [34,35].
Both B3GAT1 and ADGRG1 are involved in neurodevelopment, corroborating recent studies that propose the conceptualization of HD as a neurodevelopmental disease [36,37]. It has been proposed that CAG repeat expansion may be phylogenetically relevant for brain development [38]. Animal models, in vitro studies and molecular research studies have shown that the protein HTT mediates a variety of developmental processes in the central nervous system [36,39]. Accordingly, the presence of mHTT may influence neuronal homeostasis throughout development, ultimately leading to premature cell death and, hence, neurodegeneration from otherwise nonlethal stressors. Importantly, prior to neuronal death, mHTT may cause subclinical neurodevelopmental abnormalities [40,41]. In humans, a study evaluating fractal dimension (FD), a sensitive measure of cortical neurodevelopment, showed that the premanisfest HD subjects differed from healthy controls in the amount of cortical folding in temporal regions, and in motor and visual areas. This spatial pattern of FD differs from what has been observed in well-defined neurodevelopmental disorders such as autism. In this latter case, higher cortical folding was seen in frontal, temporal and parietal regions, and more pronounced in children than in young adults [42]. Altogether these findings suggest that HTT gene expression may be a factor that contributes to cortical development, especially those regions that differ between patients and controls [43].
Comparing HD gene carriers with depression and without depression, one of the top differentially expressed gene was NECTIN2. NECTIN2 is downregulated in HD gene carriers with depression. The correspondent protein is a CAM, i.e., an important structural substrate required for synaptic plasticity and synaptogenesis. NECTIN2 is part of the nectin family of four structurally similar type-I membrane glycoproteins belonging to the immunoglobulin superfamily (IgSF) [44]. CAMs are trans-synaptic anchors and mediators of experience-dependent signaling, dynamically modulating synaptic activity and plasticity [45,46]. These proteins participate in synaptogenesis, neural growth, synaptic maturation, and modulate synaptic function through interactions with other synaptic proteins and receptors [45,47]. Nectins are also able to interact and activate membrane receptors of different growth factors, such as fibroblast growth factor, platelet-derived growth factor and the vascular endothelial growth factor, regulating proliferation, differentiation, and cell survival [48]. These interactions are relevant for depression given the role played by growth factors in its pathophysiology [49].
CAM dysfunction has been associated with several neuropsychiatric conditions [50,51,52,53]. In addition, chronic stress can affect the expression of CAMs, with the chronic restraint model of depression leading to decreased expression of CAMs in the hippocampus [52]. This observation goes in line with the concept that neurodevelopmental impairment and/or dysfunction increase the vulnerability to psychiatric disorders by altering the developmental programming of brain regions that are associated with affective and cognitive processing [54,55,56]. Indeed, the presence of neurodevelopmental abnormalities, notably involving cortical regions, is associated with an increased lifetime risk for depression [54]. The establishment of cortical thickness depends on different processes such as cell death, synaptogenesis, synaptic pruning, and myelination during the first two decades of life, with a dynamic synaptic reorganization modulated by environmental influences [57,58,59,60]. Abnormal brain volume has been consistently shown in depression, and a recent meta-analysis including over 10,000 subjects showed cortical abnormalities in adults and adolescents with major depression [61].
We also performed gene ontology (GO) analysis to identify the pathways that are enriched by differently expressed genes. The pathway ‘cerebral cortex radial glia guided migration’ has the presence of ADGRG1. This pathway is involved in neuronal radial migration in the developing cerebral cortex. The cerebral cortex has a well-organized six-layered architecture. The establishment of cortical layers in the mammal developing cortex requires an elaborate control of multiple processes, such as cell proliferation, differentiation, apoptosis, and neuronal migration [62]. Studies have revealed that radial glia are present during corticogenesis and their processes span the full thickness of the cortical wall [63]. Defects in radial glial cells could lead to cortical heterotopias, suggesting that normal radial glial cells are critical for cerebral cortex development [64,65]. One of the pathways containing NECTIN2 is called ‘cell part morphogenesis’ that refers to how structures of a cell are generated and organized. Altogether, these results implicate the involvement of neurodevelopmental pathways in HD and, more specifically, in the emergence of depression.
Interestingly, another enriched pathway in the comparison among HD carriers was the positive regulation of ‘immunoglobulin mediated immune response’ in subjects with depression. This pathway has NECTIN2 and is involved in processes that activate or increase the frequency, rate, or extent of an immunoglobulin-mediated immune response. The immune system is a relevant player in the pathological cascade of neurodegenerative diseases triggered by misfolded proteins, including HD [66]. In addition, immune changes have been associated with the pathophysiology of depression [67].
There are limitations in this study that must be considered. First, this is a cross-sectional study with a small sample size. The analyses were performed in the blood and might not accurately reflect alterations in the central nervous system. Despite these shortcomings, this is the first study to compare peripheral (i.e., blood) gene expression profile between HD gene carriers and healthy controls.
In summary, we found distinctive gene expression related to neurodevelopmental pathways in HD. These findings are in line with recent studies suggesting that HD is not only a neurodegenerative disease but also has a neurodevelopmental component. Our findings also suggest that this neurodevelopmental component can contribute to the increased rates of depression in HD. Future directions include analyses of the identified genes in larger cohorts of HD carriers aiming to better understand the pathophysiology and risk factors associated with depression (and other behavioral correlates) in HD.
4. Material and Methods
4.1. Subjects and Clinical Assessments
This study included 16 patients with a genetic diagnosis of HD (9 premanifest and 7 manifest HD, i.e., patients with a clinical diagnosis of HD), being eight HD gene carriers with symptoms of depression and eight patients without, and a group of 8 healthy controls. HD gene carriers were recruited from the Huntington Disease Society of America (HDSA) Center of Excellence at University of Texas Health Science Center at Houston (UTHealth). Controls were recruited from the local community, comprising a group of people with no history of neurological or psychiatric disorders. Genetic diagnosis was confirmed by a genotype CAG allele ≥ 36. A movement disorders specialist evaluated all patients and the clinical diagnosis of HD was based on established motor signs, i.e., a Diagnostic Confidence Level (DCL) set at 4 in the Unified HD Rating Scale (UHDRS) (1996). All subjects provided written informed consent before admission to the study (Approval Number: HSC-MS-17-0234, and issue on 10 May 2017). The Research Ethics Committees of UTHealth approved this study.
The clinical evaluation included a questionnaire about socio-demographic information alongside motor, cognitive and behavioral assessments. HD gene carriers were subjected to motor function evaluation with the UHDRS (1996). Behavioral symptoms were evaluated through the short version of the Problem Behaviors Assessment (PBS-s). The PBA-s is a semi-structured interview containing 11 items, each designed to measure the severity and frequency of different behavioral symptoms in HD (McNally G et al., 2015). Patients with a score zero or < 2 were considered without depression symptoms. Patients had moderate to severe symptoms of depression.
4.2. Blood Sampling
Peripheral blood was collected from participants by venipuncture into PAXgene Blood RNA Tubes (PreAnalytix, QIAGEN, Inc., Germantown, MD, USA) on the same day of clinical assessment. RNA was isolated with the PreAnalytix kit (QIAGEN, Inc., Germantown, MD, USA) according to the manufacturer’s instructions. Total RNA samples were quantified and sent to a core facility to perform RNA-sequencing.
4.3. Gene Expression Analysis
RNA samples were quantified upon receipt using Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA) and RNA integrity was checked with 4200 TapeStation (Agilent Technologies, Palo Alto, CA, USA). rRNA depletion along with globin depletion was performed using Globin Zero Gold kit (Illumina, San Diego, CA, USA).
RNA sequencing library preparation used NEBNext Ultra RNA Library Prep Kit for Illumina by following the manufacturer’s recommendations (NEB, Ipswich, MA, USA). Briefly, enriched RNAs were fragmented for 15 minutes at 94 °C. First strand and second strand cDNA were subsequently synthesized. cDNA fragments were end repaired and adenylated at 3’ends, and universal adapter was ligated to cDNA fragments, followed by index addition and library enrichment with limited cycle PCR. Sequencing libraries were validated using the Agilent Tapestation 4200 (Agilent Technologies, Palo Alto, CA, USA), and quantified by using Qubit 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) as well as by quantitative PCR (Applied Biosystems, Carlsbad, CA, USA).
The sequencing libraries were multiplexed and clustered on two lanes of a flow cell and loaded on the Illumina HiSeq instrument according to manufacturer’s instructions. The samples were sequenced using a 2 × 150 Paired End (PE) configuration. Image analysis and base calling were conducted by the HiSeq Control Software (HCS). Raw sequence data (.bcl files) generated from Illumina HiSeq were converted into fastq files and de-multiplexed using Illumina’s bcl2fastq 2.17 software. One mismatch was allowed for index sequence identification.
4.4. Data Analysis
After demultiplexing, sequence data were checked for overall quality and yield. Then, sequence reads were trimmed to remove possible adapter sequences and nucleotides with poor quality using Trimmomatic v.0.36. The trimmed reads were mapped to the Homo sapiens reference genome GRCh38 available on ENSEMBL using the STAR aligner v.2.5.2b. The STAR aligner is a splice aware aligner that detects splice junctions and incorporates them to help align the entire read sequences. BAM files were generated as a result of this step. Unique gene hit counts were calculated by using feature Counts from the Subread package v.1.5.2. Only unique reads within exon regions were counted.
After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis. Using DESeq2, a comparison of gene expression between the groups of samples was performed. The Wald test was used to generate p-values and Log2 fold changes. Genes with adjusted p-values < 0.05 and absolute log2 fold changes > 1 were called as differentially expressed genes for each comparison. A gene ontology analysis was performed on the statistically significant set of genes by implementing the software GeneSCF v1.1. The GO list was used to cluster the set of genes based on their biological process and determine their statistical significance. A PCA analysis was performed using the "plotPCA" function within the DESeq2 R package. The plot shows the samples in a 2D plane spanned by their first two principal components. The top 500 genes, selected by highest row variance, were used to generate the plot.
4.5. Gene Ontology Analysis
A gene ontology analysis was performed on the statistically significant set of genes by implementing the software GeneSCF v.1.1-p2. The goa_human GO list was used to cluster the set of genes based on their biological processes and determine their statistical significance. A list of genes clustered based on their gene ontologies was generated, adjusted p-value less than 0.05 in the differentially expressed gene sets.
4.6. Real-Time Quantitative PCR
Three top differentially expressed genes between groups were selected for validation using real time quantitative PCR. Briefly, RNA samples (300 ng) were initially converted into cDNA using the High Capacity cDNA Synthesis Kit (Life Technologies, Carlsbad, CA) and later diluted 2 times for the PCR reactions. Amplifications of Adhesion G Protein-Coupled Receptor G1 (ADGRG1), Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1 (B3GAT1) and Nectin Cell Adhesion Molecule 2 (NECTIN2) were performed in 12 μL-reactions using inventoried FAM-MGB-labeled TaqMan Gene Expression Assays (Hs00938474_m1, Hs01024500_m1, Hs01071562_m1 for ADGRG1, B3GAT1 and NECTIN2, respectively) and the VIC-MGB_PL-labelled beta-2-microglobulin (B2M) as endogenous control (Hs00187842_m1). PCR reactions were run on a QuantStudio 7 Flex Real-Time PCR System (Life Technologies, Massachusetts, USA) with each sample assayed in triplicate. Data were analyzed by the 2(-Delta Delta C(T)) method.
4.7. Statistical Analysis
After extraction of gene hit counts, the gene hit counts table was used for downstream differential expression analysis. Preprocessing of data and between-group comparisons were calculated using DESeq2. The Wald test was used to generate p-values and log2 fold changes. Genes with a p-value < 0.05 and absolute log2 fold change > 1 were called as differentially expressed genes. In addition, we clustered differentially expressed genes by their gene ontology (GO) using GeneSCF and the enrichment of GO terms was tested by Fisher exact test. In the PCR results we used t-tests with a significance level of p < 0.05 were used to evaluate for possible group differences.
Acknowledgments
The Neuropsychiatry Program is partly supported by the Department of Psychiatry and Behavioral Sciences, UT Health Houston. The UT Health Center of Excellence in Huntington’s disease is partly funded by the Huntington’s disease Society of America.
Author Contributions
G.D.C. did the genetic analyses and drafted the first version of the manuscript; N.P.R. and E.F.S. recruited all the subjects and contributed to the statistical analysis and writing of the final version of the manuscript; A.L.T. designed the study, oversaw all research steps, and wrote the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Department of Psychiatry and Behavioral Sciences, UT Health Houston and Huntington’s disease Society of America.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.MacDonald M.E., Barnes G., Srinidhi J., Duyao M.P., Ambrose C.M., Myers R.H., Gray J., Conneally P.M., Young A., Penney J., et al. Gametic but not somatic instability of CAG repeat length in Huntington’s disease. J. Med. Genet. 1993;30:982–986. doi: 10.1136/jmg.30.12.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorsey E.R., Beck C.A., Darwin K., Nichols P., Brocht A.F., Biglan K.M., Shoulson I., Huntington Study Group C.I. Natural history of Huntington disease. JAMA Neurol. 2013;70:1520–1530. doi: 10.1001/jamaneurol.2013.4408. [DOI] [PubMed] [Google Scholar]
- 3.Testa C.M., Jankovic J. Huntington disease: A quarter century of progress since the gene discovery. J. Neurol. Sci. 2019;396:52–68. doi: 10.1016/j.jns.2018.09.022. [DOI] [PubMed] [Google Scholar]
- 4.Nasir J., Floresco S.B., O’Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich P., Shanmugasundaram R., Shuyu E., Dragatsis I. Congenital hydrocephalus associated with abnormal subcommissural organ in mice lacking huntingtin in Wnt1 cell lineages. Hum. Mol. Genet. 2009;18:142–150. doi: 10.1093/hmg/ddn324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taran A.S., Shuvalova L.D., Lagarkova M.A., Alieva I.B. Huntington’s Disease-An Outlook on the Interplay of the HTT Protein, Microtubules and Actin Cytoskeletal Components. Cells. 2020;9:1514. doi: 10.3390/cells9061514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dragatsis I., Levine M.S., Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 8.Zuccato C., Valenza M., Cattaneo E. Molecular mechanisms and potential therapeutical targets in Huntington’s disease. Physiol. Rev. 2010;90:905–981. doi: 10.1152/physrev.00041.2009. [DOI] [PubMed] [Google Scholar]
- 9.Ross C.A., Reilmann R., Cardoso F., McCusker E.A., Testa C.M., Stout J.C., Leavitt B.R., Pei Z., Landwehrmeyer B., Martinez A., et al. Movement Disorder Society Task Force Viewpoint: Huntington’s Disease Diagnostic Categories. Mov. Disord. Clin. Pract. 2019;6:541–546. doi: 10.1002/mdc3.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reedeker W., van der Mast R.C., Giltay E.J., Kooistra T.A., Roos R.A., van Duijn E. Psychiatric disorders in Huntington’s disease: A 2-year follow-up study. Psychosomatics. 2012;53:220–229. doi: 10.1016/j.psym.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 11.Duff K., Paulsen J.S., Beglinger L.J., Langbehn D.R., Stout J.C., Predict H.D.I.o.t.H.S.G. Psychiatric symptoms in Huntington’s disease before diagnosis: The predict-HD study. Biol. Psychiatry. 2007;62:1341–1346. doi: 10.1016/j.biopsych.2006.11.034. [DOI] [PubMed] [Google Scholar]
- 12.Ho A.K., Gilbert A.S., Mason S.L., Goodman A.O., Barker R.A. Health-related quality of life in Huntington’s disease: Which factors matter most? Mov. Disord. Off. J. Mov. Disord. Soc. 2009;24:574–578. doi: 10.1002/mds.22412. [DOI] [PubMed] [Google Scholar]
- 13.Wetzel H.H., Gehl C.R., Dellefave-Castillo L., Schiffman J.F., Shannon K.M., Paulsen J.S., Huntington Study Group Suicidal ideation in Huntington disease: The role of comorbidity. Psychiatry Res. 2011;188:372–376. doi: 10.1016/j.psychres.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrer L.A. Suicide and attempted suicide in Huntington disease: Implications for preclinical testing of persons at risk. Am. J. Med Genet. 1986;24:305–311. doi: 10.1002/ajmg.1320240211. [DOI] [PubMed] [Google Scholar]
- 15.Robins Wahlin T.B., Backman L., Lundin A., Haegermark A., Winblad B., Anvret M. High suicidal ideation in persons testing for Huntington’s disease. Acta Neurol. Scand. 2000;102:150–161. doi: 10.1034/j.1600-0404.2000.102003150.x. [DOI] [PubMed] [Google Scholar]
- 16.Schoenfeld M., Myers R.H., Cupples L.A., Berkman B., Sax D.S., Clark E. Increased rate of suicide among patients with Huntington’s disease. J. Neurol. Neurosurg. Psychiatry. 1984;47:1283–1287. doi: 10.1136/jnnp.47.12.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiedorowicz J.G., Mills J.A., Ruggle A., Langbehn D., Paulsen J.S., Group P.-H.I.o.t.H.S. Suicidal behavior in prodromal Huntington disease. Neuro Degener. Dis. 2011;8:483–490. doi: 10.1159/000327754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almqvist E.W., Brinkman R.R., Wiggins S., Hayden M.R. Canadian Collaborative Study of Predictive, T. Psychological consequences and predictors of adverse events in the first 5 years after predictive testing for Huntington’s disease. Clin. Genet. 2003;64:300–309. doi: 10.1034/j.1399-0004.2003.00157.x. [DOI] [PubMed] [Google Scholar]
- 19.Van Duijn E., Kingma E.M., Timman R., Zitman F.G., Tibben A., Roos R.A., van der Mast R.C. Cross-sectional study on prevalences of psychiatric disorders in mutation carriers of Huntington’s disease compared with mutation-negative first-degree relatives. J. Clin. Psychiatry. 2008;69:1804–1810. doi: 10.4088/JCP.v69n1116. [DOI] [PubMed] [Google Scholar]
- 20.Berrios G.E., Wagle A.C., Markova I.S., Wagle S.A., Ho L.W., Rubinsztein D.C., Whittaker J., Ffrench-Constant C., Kershaw A., Rosser A., et al. Psychiatric symptoms and CAG repeats in neurologically asymptomatic Huntington’s disease gene carriers. Psychiatry Res. 2001;102:217–225. doi: 10.1016/S0165-1781(01)00257-8. [DOI] [PubMed] [Google Scholar]
- 21.Craufurd D., Thompson J.C., Snowden J.S. Behavioral changes in Huntington Disease. Neuropsychiatry Neuropsychol. Behav. Neurol. 2001;14:219–226. [PubMed] [Google Scholar]
- 22.Kingma E.M., van Duijn E., Timman R., van der Mast R.C., Roos R.A. Behavioural problems in Huntington’s disease using the Problem Behaviours Assessment. Gen. Hosp. Psychiatry. 2008;30:155–161. doi: 10.1016/j.genhosppsych.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Pla P., Orvoen S., Saudou F., David D.J., Humbert S. Mood disorders in Huntington’s disease: From behavior to cellular and molecular mechanisms. Front. Behav. Neurosci. 2014;8:135. doi: 10.3389/fnbeh.2014.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleene R., Schachner M. Glycans and neural cell interactions. Nat. Rev. Neurosci. 2004;5:195–208. doi: 10.1038/nrn1349. [DOI] [PubMed] [Google Scholar]
- 25.Morita I., Kizuka Y., Kakuda S., Oka S. Expression and function of the HNK-1 carbohydrate. J. Biochem. 2008;143:719–724. doi: 10.1093/jb/mvm221. [DOI] [PubMed] [Google Scholar]
- 26.Jeffries A.R., Mungall A.J., Dawson E., Halls K., Langford C.F., Murray R.M., Dunham I., Powell J.F. beta-1,3-Glucuronyltransferase-1 gene implicated as a candidate for a schizophrenia-like psychosis through molecular analysis of a balanced translocation. Mol. Psychiatry. 2003;8:654–663. doi: 10.1038/sj.mp.4001382. [DOI] [PubMed] [Google Scholar]
- 27.Hamann J., Aust G., Arac D., Engel F.B., Formstone C., Fredriksson R., Hall R.A., Harty B.L., Kirchhoff C., Knapp B., et al. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol. Rev. 2015;67:338–367. doi: 10.1124/pr.114.009647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno M., Pedrosa L., Pare L., Pineda E., Bejarano L., Martinez J., Balasubramaniyan V., Ezhilarasan R., Kallarackal N., Kim S.H., et al. GPR56/ADGRG1 Inhibits Mesenchymal Differentiation and Radioresistance in Glioblastoma. Cell Rep. 2017;21:2183–2197. doi: 10.1016/j.celrep.2017.10.083. [DOI] [PubMed] [Google Scholar]
- 29.Bahi-Buisson N., Poirier K., Boddaert N., Fallet-Bianco C., Specchio N., Bertini E., Caglayan O., Lascelles K., Elie C., Rambaud J., et al. GPR56-related bilateral frontoparietal polymicrogyria: Further evidence for an overlap with the cobblestone complex. Brain A J. Neurol. 2010;133:3194–3209. doi: 10.1093/brain/awq259. [DOI] [PubMed] [Google Scholar]
- 30.Bae B.I., Tietjen I., Atabay K.D., Evrony G.D., Johnson M.B., Asare E., Wang P.P., Murayama A.Y., Im K., Lisgo S.N., et al. Evolutionarily dynamic alternative splicing of GPR56 regulates regional cerebral cortical patterning. Science. 2014;343:764–768. doi: 10.1126/science.1244392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu B., Luo R., Jin P., Li T., Oak H.C., Giera S., Monk K.R., Lak P., Shoichet B.K., Piao X. GAIN domain-mediated cleavage is required for activation of G protein-coupled receptor 56 (GPR56) by its natural ligands and a small-molecule agonist. J. Biol. Chem. 2019;294:19246–19254. doi: 10.1074/jbc.RA119.008234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piao X., Hill R.S., Bodell A., Chang B.S., Basel-Vanagaite L., Straussberg R., Dobyns W.B., Qasrawi B., Winter R.M., Innes A.M., et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 33.Piao X., Chang B.S., Bodell A., Woods K., Benzeev B., Topcu M., Guerrini R., Goldberg-Stern H., Sztriha L., Dobyns W.B., et al. Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann. Neurol. 2005;58:680–687. doi: 10.1002/ana.20616. [DOI] [PubMed] [Google Scholar]
- 34.Giera S., Deng Y., Luo R., Ackerman S.D., Mogha A., Monk K.R., Ying Y., Jeong S.J., Makinodan M., Bialas A.R., et al. The adhesion G protein-coupled receptor GPR56 is a cell-autonomous regulator of oligodendrocyte development. Nat. Commun. 2015;6:6121. doi: 10.1038/ncomms7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giera S., Luo R., Ying Y., Ackerman S.D., Jeong S.J., Stoveken H.M., Folts C.J., Welsh C.A., Tall G.G., Stevens B., et al. Microglial transglutaminase-2 drives myelination and myelin repair via GPR56/ADGRG1 in oligodendrocyte precursor cells. Elife. 2018;7:e33385. doi: 10.7554/eLife.33385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiatr K., Szlachcic W.J., Trzeciak M., Figlerowicz M., Figiel M. Huntington Disease as a Neurodevelopmental Disorder and Early Signs of the Disease in Stem Cells. Mol. Neurobiol. 2018;55:3351–3371. doi: 10.1007/s12035-017-0477-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van der Plas E., Langbehn D.R., Conrad A.L., Koscik T.R., Tereshchenko A., Epping E.A., Magnotta V.A., Nopoulos P.C. Abnormal brain development in child and adolescent carriers of mutant huntingtin. Neurology. 2019;93:e1021–e1030. doi: 10.1212/WNL.0000000000008066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cattaneo E., Zuccato C., Tartari M. Normal huntingtin function: An alternative approach to Huntington’s disease. Nat. Rev. Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 39.Reiner A., Dragatsis I., Zeitlin S., Goldowitz D. Wild-type huntingtin plays a role in brain development and neuronal survival. Mol. Neurobiol. 2003;28:259–276. doi: 10.1385/MN:28:3:259. [DOI] [PubMed] [Google Scholar]
- 40.Nopoulos P.C. Huntington disease: A single-gene degenerative disorder of the striatum. Dialogues Clin. Neurosci. 2016;18:91–98. doi: 10.31887/DCNS.2016.18.1/pnopoulos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mehler M.F., Gokhan S. Mechanisms underlying neural cell death in neurodegenerative diseases: Alterations of a developmentally-mediated cellular rheostat. Trends Neurosci. 2000;23:599–605. doi: 10.1016/S0166-2236(00)01705-7. [DOI] [PubMed] [Google Scholar]
- 42.Awate S.P., Win L., Yushkevich P., Schultz R.T., Gee J.C. 3D cerebral cortical morphometry in autism: Increased folding in children and adolescents in frontal, parietal, and temporal lobes. Med. Image Comput. Comput. Assist. Interv. 2008;11:559–567. doi: 10.1007/978-3-540-85988-8_67. [DOI] [PubMed] [Google Scholar]
- 43.Kubera K.M., Schmitgen M.M., Hirjak D., Wolf R.C., Orth M. Cortical neurodevelopment in pre-manifest Huntington’s disease. Neuroimage Clin. 2019;23:101913. doi: 10.1016/j.nicl.2019.101913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrison O.J., Vendome J., Brasch J., Jin X., Hong S., Katsamba P.S., Ahlsen G., Troyanovsky R.B., Troyanovsky S.M., Honig B., et al. Nectin ectodomain structures reveal a canonical adhesive interface. Nat. Struct. Mol. Biol. 2012;19:906–915. doi: 10.1038/nsmb.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalva M.B., McClelland A.C., Kayser M.S. Cell adhesion molecules: Signalling functions at the synapse. Nat. Rev. Neurosci. 2007;8:206–220. doi: 10.1038/nrn2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shapiro L. Self-recognition at the atomic level: Understanding the astonishing molecular diversity of homophilic Dscams. Neuron. 2007;56:10–13. doi: 10.1016/j.neuron.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 47.Parrish J.Z., Emoto K., Kim M.D., Jan Y.N. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annu. Rev. Neurosci. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- 48.Mizutani K., Takai Y. Nectin spot: A novel type of nectin-mediated cell adhesion apparatus. Biochem. J. 2016;473:2691–2715. doi: 10.1042/BCJ20160235. [DOI] [PubMed] [Google Scholar]
- 49.Audet M.C., Anisman H. Interplay between pro-inflammatory cytokines and growth factors in depressive illnesses. Front. Cell. Neurosci. 2013;7:68. doi: 10.3389/fncel.2013.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jamain S., Quach H., Betancur C., Rastam M., Colineaux C., Gillberg I.C., Soderstrom H., Giros B., Leboyer M., Gillberg C., et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat. Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirov G., Gumus D., Chen W., Norton N., Georgieva L., Sari M., O’Donovan M.C., Erdogan F., Owen M.J., Ropers H.H., et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 52.Sandi C. Stress, cognitive impairment and cell adhesion molecules. Nat. Rev. Neurosci. 2004;5:917–930. doi: 10.1038/nrn1555. [DOI] [PubMed] [Google Scholar]
- 53.Yan J., Oliveira G., Coutinho A., Yang C., Feng J., Katz C., Sram J., Bockholt A., Jones I.R., Craddock N., et al. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol. Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- 54.Peterson B.S., Wang Z., Horga G., Warner V., Rutherford B., Klahr K.W., Graniello B., Wickramaratne P., Garcia F., Yu S., et al. Discriminating risk and resilience endophenotypes from lifetime illness effects in familial major depressive disorder. JAMA Psychiatry. 2014;71:136–148. doi: 10.1001/jamapsychiatry.2013.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gee D.G., Casey B.J. The Impact of Developmental Timing for Stress and Recovery. Neurobiol. Stress. 2015;1:184–194. doi: 10.1016/j.ynstr.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLaughlin K.A., Koenen K.C., Bromet E.J., Karam E.G., Liu H., Petukhova M., Ruscio A.M., Sampson N.A., Stein D.J., Aguilar-Gaxiola S., et al. Childhood adversities and post-traumatic stress disorder: Evidence for stress sensitisation in the World Mental Health Surveys. Br. J. Psychiatry J. Ment. Sci. 2017;211:280–288. doi: 10.1192/bjp.bp.116.197640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirjak D., Huber M., Kirchler E., Kubera K.M., Karner M., Sambataro F., Freudenmann R.W., Wolf R.C. Cortical features of distinct developmental trajectories in patients with delusional infestation. Prog. Neuro Psychopharmacol. Biol. Psychiatry. 2017;76:72–79. doi: 10.1016/j.pnpbp.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 58.Oostermeijer S., Whittle S., Suo C., Allen N.B., Simmons J.G., Vijayakumar N., van de Ven P.M., Jansen L.M., Yucel M., Popma A. Trajectories of adolescent conduct problems in relation to cortical thickness development: A longitudinal MRI study. Transl. Psychiatry. 2016;6:e899. doi: 10.1038/tp.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vijayakumar N., Allen N.B., Youssef G., Dennison M., Yucel M., Simmons J.G., Whittle S. Brain development during adolescence: A mixed-longitudinal investigation of cortical thickness, surface area, and volume. Hum. Brain Mapp. 2016;37:2027–2038. doi: 10.1002/hbm.23154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zilles K., Palomero-Gallagher N., Amunts K. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 2013;36:275–284. doi: 10.1016/j.tins.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 61.Schmaal L., Hibar D.P., Samann P.G., Hall G.B., Baune B.T., Jahanshad N., Cheung J.W., van Erp T.G.M., Bos D., Ikram M.A., et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol. Psychiatry. 2017;22:900–909. doi: 10.1038/mp.2016.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barkovich A.J., Guerrini R., Kuzniecky R.I., Jackson G.D., Dobyns W.B. A developmental and genetic classification for malformations of cortical development: Update 2012. Brain A J. Neurol. 2012;135:1348–1369. doi: 10.1093/brain/aws019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Misson J.P., Austin C.P., Takahashi T., Cepko C.L., Caviness V.S., Jr. The alignment of migrating neural cells in relation to the murine neopallial radial glial fiber system. Cereb. Cortex. 1991;1:221–229. doi: 10.1093/cercor/1.3.221. [DOI] [PubMed] [Google Scholar]
- 64.Jossin Y., Lee M., Klezovitch O., Kon E., Cossard A., Lien W.H., Fernandez T.E., Cooper J.A., Vasioukhin V. Llgl1 Connects Cell Polarity with Cell-Cell Adhesion in Embryonic Neural Stem Cells. Dev. Cell. 2017;41:481–495. e485. doi: 10.1016/j.devcel.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Beattie R., Hippenmeyer S. Mechanisms of radial glia progenitor cell lineage progression. FEBS Lett. 2017;591:3993–4008. doi: 10.1002/1873-3468.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramsingh A.I., Manley K., Rong Y., Reilly A., Messer A. Transcriptional dysregulation of inflammatory/immune pathways after active vaccination against Huntington’s disease. Hum. Mol. Genet. 2015;24:6186–6197. doi: 10.1093/hmg/ddv335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colpo G.D., Leboyer M., Dantzer R., Trivedi M.H., Teixeira A.L. Immune-based strategies for mood disorders: Facts and challenges. Expert Rev. Neurother. 2017;18:139–152. doi: 10.1080/14737175.2018.1407242. [DOI] [PMC free article] [PubMed] [Google Scholar]