Abstract
Corynebacterium matruchotii may be key in tooth biofilm formation, but information about demographics, bacterial partners, and binding ligands is limited. The aims of this study were to explore C. matruchotii’s demography by age and colonization site (plaque and saliva), in vitro bacterial–bacterial interactions in coaggregation and coadhesion assays, and glycolipids as potential binding ligands in thin-layer chromatogram binding assays. C. matruchotii prevalence increased from 3 months to 18 years old, with 90% and 100% prevalence in saliva and tooth biofilm, respectively. C. matruchotii aggregated in saliva in a dose-dependent manner but lacked the ability to bind to saliva-coated hydroxyapatite. In vivo, C. matruchotii abundance paralleled that of Actinomyces naeslundii, Capnocytophaga sp. HMT 326, Fusobacterium nucleatum subsp. polymorphum, and Tannerella sp. HMT 286. In vitro, C. matruchotii bound both planktonic and surface-bound A. naeslundii, Actinomyces odontolyticus, and F. nucleatum. In addition, C. matruchotii exhibited the ability to bind glycolipids isolated from human erythrocytes (blood group O), human granulocytes, rabbit intestine, human meconium, and rat intestine. Binding assays identified candidate carbohydrate ligands as isoglobotriaosylceramide, Galα3-isoglobotriaosylceramide, lactotriaosylceramide, lactotetraosylceramide, neolactotetraosylceramide, and neolactohexaosylceramide. Thus, C. matruchotii likely uses specific plaque bacteria to adhere to the biofilm and may interact with human tissues through carbohydrate interactions.
Keywords: Corynebacterium matruchotii, demographics, aggregation, ligand, glycolipids
1. Introduction
Corynebacterium matruchotii (previously Bacterionema matruchotii) [1] is a Gram-positive actinobacterium with long filaments and a typical “wipe-handle” morphology [1]. C. matruchotii has previously been found in a morphological unit in tooth biofilms, termed “corn cobs”, formed by the filamentous organism with adherent cocci [2]. Recent studies redefined the bacterial organization in the supragingival biofilm as a hedgehog-like structure and suggested that C. matruchotii is a nucleating species in the bacterial community [3]. Morphological characterization of the hedgehog structure has revealed a network in which Streptococcus and Actinomyces species attach to the C. matruchotii filaments, with more cocci adhering to its distal filament tips [3]. Other species in the hedgehog structure belong to Porphyromonas, Haemophilus, Aggregatibacter, Neisseria, Fusobacterium, and Leptotrichia genera [3]. However, Kolenbrander and Williams [4] did not observe any interaction between C. matruchotii and Actinomyces viscosus (strains MG1, T14V, and T14AV), Actinomyces naeslundii (strains ATCC 12104, I, and W1544), or Streptococcus spp. (strains DL1, Challis, Hi, 34, and J22). Kim and Koo [5] recently confirmed that the corncob, hedgehog, and seaweed spatial architectures are common, but also documented a dome-like structure in caries-active toddlers [6].
Several 16S rRNA gene sequencing studies have found C. matruchotii among the most prevalent species in the adult human oral core microbiome, as they are present in virtually all subjects in significant abundance [7,8]. This was also confirmed in metaproteome and metatranscriptome studies [9,10]. Furthermore, C. matruchotii represents a large proportion of the protein activity in human tooth biofilms [9,10] and secretes a 50 amino acid membrane-associated proteolipid inducing calcium precipitation [11], as well as an oxidoreductase (MdbA) that catalyzes a disulfide bond formation in vitro, which may be utilized to catalyze oxidative protein folding and stabilization [12].
The relationship between C. matruchotii and oral health remains unclear. Some studies report C. matruchotii to be enumerated in caries-free children and young adults [7,13], and reduced in caries progression [13]. However, a yet unnamed Corynebacterium phylotype has been found to be among the predominant species related to caries progression in the primary and permanent dentition [14]. Similarly, some studies have found an association between C. matruchotii and periodontal health [15,16], but one study reported that C. matruchotii is more than twice as prevalent in smokers, indicating an association or co-appearance with periodontal disease, which is more common in smokers [17].
Thus, support is growing for a possible key role of C. matruchotii in oral biofilm formation, but much remains to be understood about its role in oral microbiota biology and oral health. The aims of this study were to explore C. matruchotii’s demography by age and colonization site (plaque and saliva), in vitro bacterial–bacterial interactions in coaggregation and coadhesion assays, and glycolipids as potential binding ligands in thin-layer chromatogram binding assays.
2. Materials and Methods
2.1. Study Population
Data on the prevalence and abundance of C. matruchotii and other oral bacteria were extracted from previous study cohorts: One study on age transformation of the oral microbiota [18] and one employing young adults [19]. The cohorts were described in their respective publications. Both studies with addendums were approved by the Swedish Ethical Review Authority, Sweden (Dnr 2011-90-31 M, 2012-111-31 M, 2015-389-32 M, 2017-450-31, and 2016-239-32 M) and followed the Declaration of Helsinki and the General Data Protection Regulation (GDPR). All parents and young adults were given written and verbal information before signing informed consent to participate and agreeing to the use of the information in research.
2.2. Biological Samples and Information on Medical, Lifestyle, and Living Conditions
Saliva swabs, whole chewing stimulated and parotid saliva, tooth biofilm, breast milk, and feces were sampled for microbiota characterization as described elsewhere [18,20]. Briefly, saliva swab samples were collected by swirling sterile cotton swabs in the mouth of 2-day, 3-month, 18-month, 3-year, and 5-year-old children, followed by dispersal of bacteria in 10 mM Tris-HCl, 1 mM disodium EDTA, pH 7.4. Milk was collected by the mothers after carefully cleaning the nipple with soap and water and drying with a clean towel. Feces from the children was sampled by the parents at home using tubes designed for this purpose (NC9574115, Sarstedt, Inc. SC TUBE CBS, Fisher Scientific, Gothenburg, Sweden). Written and verbal information were given to the mothers prior to sampling. Milk and feces samples were stored at −20 °C for approximately 2 weeks and then transported on ice to the lab and transferred to a −80 °C freezer until prepared for analysis.
From the young adults (18–23 years old), supragingival biofilm was collected from accessible tooth surfaces using sterile wooden toothpicks. Tooth biofilm samples were pooled by subject in buffer and the participants abstained from oral hygiene on the morning of the sampling. Whole chewing stimulated saliva was collected for 3 min in sterile test tubes.
All samples were immediately transferred to −80 °C freezers.
Information on medical and other conditions was collected via questionnaires. The young adults also reported their habitual dietary intake on a 93-question semi-quantitative food frequency questionnaire (FFQ, http://www.matval.se) as described previously [20]. Intake of energy and energy-providing nutrients were estimated as described previously [21].
For in vitro experiments, parotid saliva was collected from volunteers in ice-chilled test tubes using Lashley cups and with citric acid stimulation.
2.3. DNA Extraction and Microbiota Sequencing
Genomic DNA extraction from saliva swabs, saliva, tooth biofilm, milk, and feces samples and positive controls, PCR amplicon and library construction, Illumina sequencing, and sequence accession numbers are described in Appendix A and in the basic references [18,20].
2.4. Bacterial Cultures
C. matruchotii strains (CCUG46620 and CCUG47160) were grown on sheep blood agar plates for 48 h, washed once with 10 mM Tris-base-saline buffer (TBS) pH 7.6, and adjusted to an optical density of 1.0 at 600 nm (OD600, ~109 colony forming units (CFU)/mL) for use in coaggregation assays. For 35S-labeling, C. matruchotii was grown on blood agar plates supplemented with 35S-methionine/cysteine (NEG709A, PerkinElmer, Hägersten, Sweden), washed twice, and adjusted to an optical density of 0.2 at OD600 before use. For chromatogram binding assays, the bacteria were cultured on blood agar plates and radiolabeled by the addition of 50 μCi 35S-methionine (PerkinElmer; NEG77207MC) diluted in 0.5 mL phosphate-buffered-saline (PBS) pH 7.3 to the culture plates. After incubation for 12 h at 37 °C, the bacteria were harvested, centrifuged three times with PBS, and then suspended in PBS containing 2% (w/v) bovine serum albumin, 0.1% (w/v) NaN3, and 0.1% (w/v) Tween 20 (BSA/PBS/TWEEN) to a bacterial density of 1x108 CFU/mL. The specific activity of the suspensions was approximately 1 cpm per 100 bacteria.
For fluorescein isothiocyanate (FITC) labeling, C. matruchotii was grown overnight on blood agar plates; washed once in PBS, supplemented with 0.1% Tween-20 (PBS-T); resuspended in 0.1 M carbonate 0.15 M NaCl2 buffer to an optical density of 1.0 at OD600; incubated with 10 mg/mL FITC (F7250, Sigma–Aldrich, Stockholm, Sweden) for 8 min, followed by three washes in PBS-T; and finally adjusted to 1.0 at OD600. Tested bacterial partner strains were grown on blood, Rogosa, or chocolate agar plates under aerobic or anaerobic conditions (Table S1). Bacteria were harvested after 48 h of growth, washed once with TBS, adjusted to an optical density of 0.5 at OD600, yielding approximately 108 CFU/mL, and frozen at −80 °C until use.
2.5. Fluid Phase Coaggregation
Saliva-mediated aggregation and bacteria–bacteria coaggregation were tested in a sedimentation assay in which equal volumes (0.1 mL) of C. matruchotii and parotid saliva or test strains were mixed in plastic round-bottom 2 mL Eppendorf tubes. The tubes were vortexed for 15 s and incubated for 10 min to allow coaggregates to form. The aggregates were scored qualitatively (++ for large, + medium, (+) small, and – no aggregates). The sedimentation assay was also used to quantify saliva-mediated aggregation by following OD600 over time in a spectrophotometer after mixing equal volumes of saliva (0.5 mL of varying concentration) and C. matruchotii (OD600 = 2.0).
2.6. Surface-Based Binding
Nitrocellulose membranes (18Ω, GE Health Care, Hybond ECL, RPN303D) were pre-wetted for 1 min, equilibrated in TBS for 10 min, and mounted in a microfiltration dot blot device (Bio-Dot Apparatus, 1706545, BioRad, Solna, Sweden). Serial dilutions of bovine serum albumin (BSA, Sigma A2153), parotid saliva, and breast milk (1:2 to 1:256 in TBS), suspensions of potential partner bacteria (50 μL, OD600 0.1 in TBS diluted from 0.1 to 0.004 in TBS, Table S1), and negative controls (TBS) were applied to assigned wells and mounted on the membrane using vacuum suction. The membranes were removed from the slot blot, rinsed, and washed (TBS, 0.1% Tween 20 (TBS-T)) before being blocked (ECL Prime blocking agent, RPN418V, GE Health Care) for 1 h. The membranes were rinsed briefly before FITC-labeled C. matruchotii was applied (50 mL, OD600 = 1.0) and allowed to bind to potential partners for 2 h at 4 °C in the dark. Finally, the membranes were rinsed and washed (3 × 5 min) in TBS-T and fluorescent signals detected using the ChemiDoc MP Imaging system (Bio-Rad).
2.7. Hydroxyapatite Adhesion and Coadhesion Assay
Adhesion of metabolically 35S-labeled C. matruchotii to hydroxyapatite beads (Bio-Rad) coated with parotid saliva, breast milk, or saliva-partner bacteria was measured as described previously [22]. The assay served as a tooth tissue model. Briefly, hydroxyapatite beads hydrated overnight (5 mg beads in 125 μL adhesion buffer (1 mM KH2PO4-K2HPO4 buffer, pH 6.5, containing 50 mM KCl, 1 mM CaCl2, and 0.1 mM MgCl2) per 96 wells) were coated with 125 μL parotid saliva or breast milk for 1 h, washed twice in adhesion buffer, and blocked with 5% BSA in adhesion buffer. Parotid saliva was pooled from up to 10 donors and serial dilutions in adhesion buffer from 1:2 to 1:256 were tested. The blocked coated beads were incubated with 35S-labeled C. matruchotii (125 μL, 108 cells/mL) for 1 h at room temperature under agitation, washed three times, resuspended in scintillation solution, and counted using MicroBeta2 (PerkinElmer, Hägersten, Sweden). The proportion of bound bacteria out of the total amount of added bacteria (percent adhesion) was calculated.
To evaluate whether bacterial partners could recruit C. matruchotii to saliva-coated hydroxyapatite surfaces, an unlabeled co-bacterial partner was bound to BSA-blocked saliva-coated hydroxyapatite beads, washed three times in adhesion buffer, and subsequently incubated for 1 h with metabolically 35S-labeled C. matruchotii (125 μL, 108 cells/mL). Finally, the beads were washed three times in adhesion buffer, resuspended in scintillation solution, and counted using MicroBeta2 (PerkinElmer). The proportion of bound bacteria out of the total amount of added bacteria (%-adhesion) was calculated and compared to C. matruchotii binding to saliva-coated beads.
2.8. Chromatogram Binding Assays
Reference glycolipids were isolated as described previously [23] and characterized by mass spectrometry [24,25] and 1H-NMR spectroscopy [26]. Thin-layer chromatography was performed on aluminum-backed silica gel 60 high performance thin-layer chromatography plates (Merck, Stockholm, Sweden). Glycosphingolipid mixtures (10–40 μg) or pure glycosphingolipids (4 μg) were applied to the plates and eluted with chloroform/methanol/water (60:35:8, by volume). Chemical detection was achieved with anisaldehyde [27].
Radiolabeled bacteria were bound to glycosphingolipids on thin-layer chromatograms as described previously [28]. Dried chromatograms were dipped in diethyl ether/n-hexane (1:5 v/v) containing 0.5% (w/v) poly(isobutyl methacrylate) for 1 min. The chromatograms were blocked with BSA/PBS/TWEEN for 2 h at room temperature before incubating the plates for 2 h at room temperature with 35S-labeled bacteria (1–5 × 106 cpm/mL) diluted in BSA/PBS/TWEEN. After washing six times with PBS and drying, the plates were autoradiographed for 12–36 h using XAR-5 x-ray films (Carestream; 8941114).
2.9. Statistical Analysis
Quantitative variables were presented as means; median with standard deviations and group differences tested by Student’s t-test. Spearman correlation coefficients were used to assess co-variations between C. matruchotii and bacteria in saliva or tooth biofilms. These analyses were performed using SPSS version 26 (IBM Corporation, Armonk, NY, USA). The tests were two-tailed and p-values < 0.05 considered significant. Taxa associated with high levels of C. matruchotii were identified in multivariate partial least squares (PLS) regression. The results are presented in a PLS loading plot with influential variables (i.e., a variable importance in projection (VIP) score >2.0). The PLS models evaluated the abundance of C. matruchotii in saliva and tooth biofilm as dependent variables (y-variable) against an independent x-block composed of all other identified bacterial species (x-variables, n = 311). All variables utilized for PLS regression analysis were auto-scaled and logarithmically transformed as needed to improve normality. SIMCA P+ version 15.0 (Umetrics AB) was used for these analyses.
3. Results
3.1. C. matruchotii Demography by Age and Sample Type
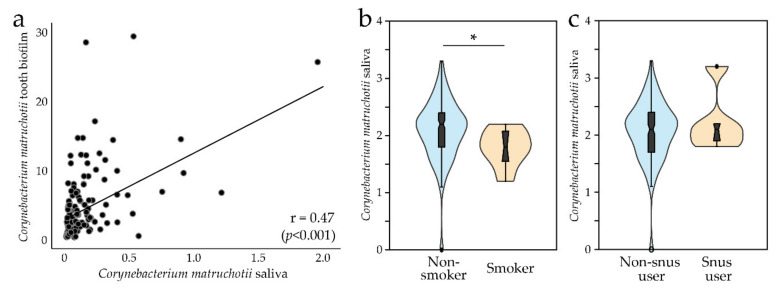
Detection of C. matruchotii in saliva was surveyed from early infancy to early adulthood (Table 1). All included participants were healthy, none had taken any antibiotic for at least 3 months before sampling, and no child was pre-term (birth weight > 2500 g) [18,20]. At 2 days and 3 months of age, C. matruchotii was virtually absent in saliva swabs (Table 1). At 18 months of age, C. matruchotii was detected in 50% of the children; thereafter, the detection prevalence increased up to 67% at 5 years of age. At 18 years of age, 98% and 100% had C. matruchotii in saliva and tooth biofilm samples, respectively, making C. matruchotii part of both the saliva and tooth biofilm core microbiome at least from 18 years of age. Though saliva detection prevalence of C. matruchotii varied by age, mean abundance was similar in carriers regardless of age (Table 1). When the infants were 3 months old, 10% of the mothers had C. matruchotii in their breast milk (Table 1). C. matruchotii was not detected in feces from 3-month or 5-year-old children (Table 1). Strikingly, the C. matruchotii abundance was 16 times higher in tooth biofilms than in saliva (i.e., 5.2% compared to 0.32%). The Spearman correlation coefficient between saliva and tooth biofilm abundance was 0.47 (p < 0.001, Figure 1a), and the linear regression coefficient with tooth biofilm abundance as the dependent and saliva abundance as the independent variable was 3.0 (95% CI 2.1, 3.8; p < 0.001).
Table 1.
Corynebacterium matruchotii detection in saliva, breast milk, and feces by age, and tooth biofilm samples at 18 years of age.
| Age at Sampling | ||||||
|---|---|---|---|---|---|---|
| 2 Days | 3 Months | 18 Months | 3 Years | 5 Years | 18–23 Years | |
| Saliva | ||||||
| Numbers (% female) | 206 (47) | 155 (46) | 132 (42) | 140 (45) | 116 (49) | 175 (51) |
| Smoker, number | 8 | |||||
| Prevalence carrier, % | 1.9 | 0 | 50 | 61.4 | 67.2 | 98.3 |
| Relative abundance, median; mean | 3.0; 4.1 | 0.0; 0.0 | 0.26; 0.46 | 0.19; 0.34 | 0.08; 0.20 | 0.32; 0.54 |
| Tooth Biofilm | ||||||
| Numbers (% female) | 45 (60) | |||||
| Smoker, number | 4 | |||||
| Prevalence carrier, % | 100 | |||||
| Relative abundance, median; mean | 5.2; 6.5 | |||||
| Breast Milk | ||||||
| Numbers (% female) | 113 (46) | |||||
| Prevalence carrier, % | 10.1 (12) | |||||
| Relative abundance, median; mean | 0.10; 0.10 | |||||
| Feces | ||||||
| Numbers (% female) | 135 (61) | 52 (48) | ||||
| Prevalence carrier, % | 0 | 0 | ||||
| Relative abundance, median; mean | 0 | 0 | ||||
Relative abundances are given as median; mean. Relative abundance of C. matruchotii is given as mean values (i.e., % of all reads in the participants in whom C. matruchotii was present).
Figure 1.
(a) Scatter plot of C. matruchotii abundance in saliva versus tooth biofilm. (b) Violin plots with a box plot comparing C. matruchotii abundance in saliva by smoking and (c) use of Swedish snus (snuff). * p < 0.05.
3.2. C. matruchotii and Lifestyle Factors
We found no association between C. matruchotii abundance and early infancy feeding mode (breast feeding or not) or birth mode (vaginally or by caesarian section), use of pacifier, or the way the parents cleaned the pacifier (all evaluated at 18 months of age) (Table S2). Young adults who smoked had significantly lower abundance of C. matruchotii in saliva (mean abundance (log value) 1.77 versus 2.09, median 1.79 versus 2.16, p = 0.032, Figure 1b). The levels did not differ between those who used Swedish snus (snuff) and those who did not (Figure 1c). Differences by tobacco use could not be tested for tooth biofilm samples because too few participants with tooth biofilm samples used tobacco. Furthermore, no association was found between C. matruchotii abundance and number of teeth, saliva flow rate, caries scores, or intake of sugar or any other food or dietary component in the young adults (Table S2).
3.3. Saliva and Breast Milk Mediated Fluid Phase Aggregation and Solid Phase Adhesion of C. matruchotii
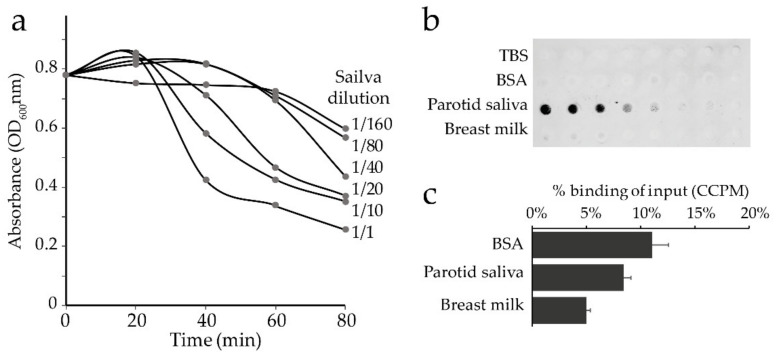
C. matruchotii (CCUG46620) aggregation in parotid saliva was evaluated using a sedimentation assay, and saliva- or breast milk-mediated adhesion was evaluated by a surface overlay assay and by binding to saliva- or breast milk-coated hydroxyapatite. Saliva aggregated C. matruchotii in a dose-dependent manner (Figure 2a). C. matruchotii also bound to saliva-coated nitrocellulose membranes (Figure 2b) but had a limited ability to bind to saliva coated on hydroxyapatite (Figure 2c). Breast milk did not mediate binding of C. matruchotii when coated on nitrocellulose membranes (Figure 2b) or hydroxyapatite beads (Figure 2c).
Figure 2.
Saliva and breast milk aggregation and adhesion of C. matruchotii. (a) Sedimentation assay of aggregation in parotid saliva by time and (b) binding of Tris-base-saline buffer (TBS) (Ctrl), bovine serum albumin (BSA, 2.5 mg to 0.020 mg per mL TBS), pooled parotid saliva (diluted 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, 1:128, 1:256 in TBS), and pooled fat-free breast milk (diluted 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, 1:128, 1:256 in TBS). (c) Proportion (%) S35-labeled C. matruchotii bound to BSA-, saliva-, or breast milk-coated hydroxyapatite beads. CCPM; Corrected counts per minute.
3.4. In Vivo Associations between C. matruchotii and Other Oral Bacteria
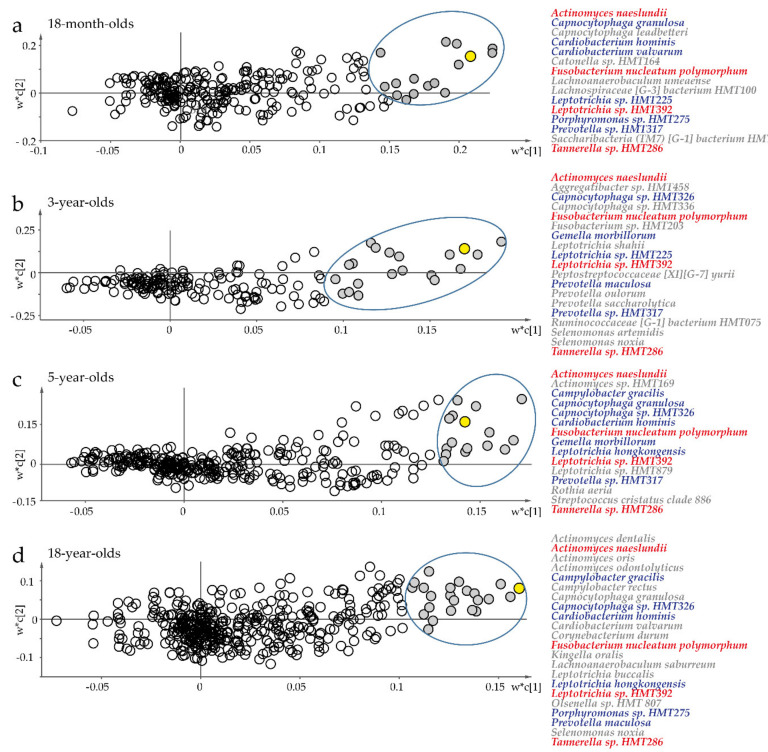
To identify potential biofilm bacteria partners for C. matruchotii in biofilm formation, multivariate PLS modeling was performed by age strata with C. matruchotii abundance as the dependent variable. All models were very strong, with an explanatory power (R2) > 74% and predicted power (Q2) > 53%. Four bacterial species were strongly associated (significant VIP values > 2.0) with saliva abundance of C. matruchotii at all ages (Figure 3a–d; Table S3): A. naeslundii, F. nucleatum subsp. polymorphum, Leptotrichia sp. HMT 392, and Tannerella sp. HMT 286. However, single species were similarly strongly associated with C. matruchotii abundance at two or three separate ages (Figure 3a–d; Table S3). Similar to saliva, very strong associations (VIP > 2.0) were seen between C. matruchotii abundance and A. naeslundii and Tannerella sp. HMT 286 in tooth biofilm samples from young adults, and with Actinomyces massiliensis, Actinomyces odontolyticus (sp. HMT 178), Actinomyces sp. HMT 171, Campylobacter gracilis Capnocytophaga sp. HMT 336, Cardiobacterium hominis, Cardiobacterium valvarum, Lachnoanaerobaculum saburreum, Leptotrichia hofstadii, Leptotrichia wadei, and Selenomonas noxia in saliva samples (Table S3). Univariate Spearman correlation analyses confirmed the associations identified by PLS in saliva and tooth biofilm.
Figure 3.
Multivariate partial least squares (PLS) regression of saliva microbiota to target potential partner bacteria for C. matruchotii. Scatter plots of PLS loading based on a model with C. matruchotii abundance as the dependent variable (Y, yellow filled symbol) and abundance of all other detected species in saliva swab samples as the independent variable block (X) for (a) 18-month-old infants, (b) 3-year-old children, (c) 5-year-old children, and (d) young adults. Unfilled symbols represent species with variable importance in projection (VIP) <2.0. Red text indicates species that were found in saliva at all ages, and blue indicates those found at two or three ages. w* describes the PLS weights from the combination of the original variables in the X-swarm; c is the component number. Species with a significant VIP score >2.0 are indicated.
3.5. Fluid Phase C. matruchotii Coaggregation with Other Bacterial Species
C. matruchotii was present in all tooth biofilm samples but did not present strong binding to saliva-coated hydroxyapatite; therefore, we hypothesized that C. matruchotii utilizes one or several partner bacteria to manifest in the tooth biofilm. A set of species were selected from the in vivo multivariate PLS screening (Figure 3) and previously reported interactions with C. matruchotii [3]. The first step was to test a panel of potential candidate species (83 strains in 47 species) for coaggregation with planktonic C. matruchotii (Table S4).
Coaggregation occurred between C. matruchotii and strains of A. naeslundii, Actinomyces odontolyticus, Corynebacterium durum, Fusobacterium nucleatum subsp. nucleatum, Fusobacterium nucleatum subsp. polymorphum, Fusobacterium periodonticum, and Porphyromonas gingivalis; weak coaggregation was seen for Aggregatibacter actinomycetemcomitans, Gemella haemolysans, Streptococcus oralis, and Veillonella dispar (Table S4). Other species in Actinomyces, Bifidobacterium, Campylobacter, Filifactor, Haemophilus, Lachnoanaerobaculum, Lactobacillus, Leptotrichia, Neisseria, Prevotella, Rothia, Selenomonas, Streptococcus, Tannerella, Veillonella, or Scardovia did not coaggregate with C. matruchotii in this assay (Table S4).
3.6. C. matruchotii Coadhesion Partners in Surface-Based Assays
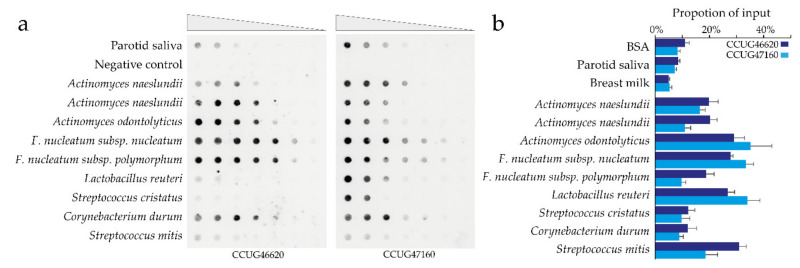
Next, the same set of species were tested as potential coadhesion partners for C. matruchotii using an overlay assay. Coadhesion occurred for A. naeslundii, A. odontolyticus, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, C. durum, L. reuteri, S. cristatus, and S. mitis (Figure S1). A few more species (i.e., A. actinomycetemcomitans, Rothia dentocariosa) exhibited weak coadhesion, but the majority had limited signs of coadhesion (Figure S1). Serial dilution of the potential coadhesion partners A. naeslundii, A. odontolyticus, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, L. reuteri, S. cristatus, C. durum, and S. mitis exhibited interactions, though with some C. matruchotii strain variation for strains CCUG46620 and CCUG47160 (Figure 4a; Table S4).
Figure 4.
Surface-based C. matruchotii coadhesion partners. (a) Serial dilution of potential coadhesion partners for C. matruchotii (50 μL of OD600 from 0.5 to 0.004) were evaluated for binding of FITC-labeled C. matruchotii. The signal represents the binding of labeled C. matruchotii to filters coated with potential partner bacteria. (b) Proportion 35S-labeled C. matruchotii binding to saliva-coated or saliva-partner bacteria-coated hydroxyapatite beads. The bars represent mean and SD of three independent experiments.
3.7. Actinomyces spp. Recruitment of C. matruchotii to Hydroxyapatite
The next step was to examine whether C. matruchotii could utilize bacterial coadhesion partners to bind to tooth surfaces using a saliva and partner bacteria-coated hydroxyapatite bead assay. The test strains were selected from our surface overlay assay. A. naeslundii, A. odontolyticus, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, L. reuteri, and S. mitis exhibited a significant ability to recruit C. matruchotii to the saliva-coated hydroxyapatite bead. C. durum and S. cristatus species could not mediate C. matruchotii recruitment. The recruitment pattern was largely similar for the two tested C. matruchotii strains (CCUG46620 and CCUG47160) (Figure 4b; Table S4).
3.8. Carbohydrate Mediated Fluid Phase Coaggregation between C. matruchotii and Partner Bacteria
To understand the planktonic bacteria–bacteria interaction mode between C. matruchotii and A. naeslundii, A. odontolyticus, F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, F. peridonticum, or P. gingivalis, heat treatment was used to denature surface proteins and differentiate between protein–protein or protein–carbohydrate interactions. Heat treatment of C. matruchotii disrupted the coaggregation with A. naeslundii and A. odontolyticus (Table S5), and heat treatment of F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, F. peridonticum, or P. gingivalis abolished aggregation with C. matruchotii. This suggests that C. matruchotii may utilize a protein to bind a carbohydrate structure on Actinomyces spp. and a carbohydrate epitope to interact with a protein on F. nucleatum subsp. nucleatum, F. nucleatum subsp. polymorphum, F. peridonticum, and P. gingivalis.
3.9. Glycolipid Binding
To map potential carbohydrate ligands for C. matruchotii adhesion, a panel of monosaccharides (fructose, glucose, galactose) and disaccharides (sucrose, maltose, lactose) were screened for inhibition of coaggregation. None of the tested monosaccharides or disaccharides had a visual effect on the coaggregation between C. matruchotii and A. naeslundii or A. odontolyticus (data not shown).
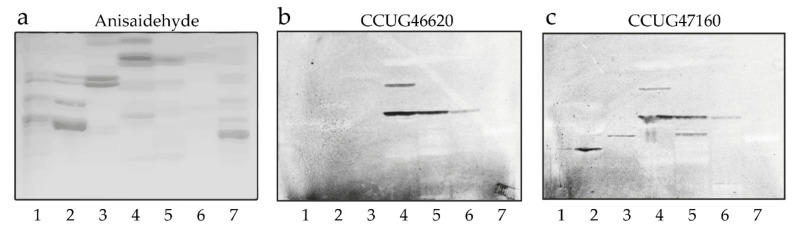
To further evaluate potential carbohydrate ligands for C. matruchotii, a number of glycolipid binding experiments were performed. In the first set of binding experiments, we used mixtures of glycolipids from different sources in order to expose the bacteria to a large number of variant carbohydrate sequences. We obtained selective binding to compounds that, on thin-layer chromatograms, migrated in the triaosylceramide to hexaosylceramide regions in the non-acid fractions from human blood group O erythrocytes, granulocytes, and meconium, as well as rabbit and rat intestines (Figure 5, lanes 2–6). There was also occasional binding to fast-migrating compounds in the total non-acid glycolipid fractions. However, this was judged to be a non-specific interaction caused by the large amounts of these compounds on the chromatograms because no binding to any monoglycosylceramides or diglycosylceramides was obtained when using defined amounts of pure glycolipids (see below). In addition, no binding was observed when binding of the two C. matruchotii strains to acid glycolipids was tested (data not shown).
Figure 5.
Screening for C. matruchotii carbohydrate recognition by binding to glycolipids on thin-layer chromatograms. (a) Chemical detection by anisaldehyde. (b) Autoradiograms obtained by binding of C. matruchotii strain CCUG46620 and (c) C. matruchotii strain CCUG47160, followed by autoradiography for 12 h. The solvent system used was chloroform/methanol/water (60:35:8, by volume). Lane 1, non-acid glycosphingolipids of human blood group AB erythrocytes, 40 μg; lane 2, non-acid glycosphingolipids of human blood group O erythrocytes, 40 μg; lane 3, non-acid glycosphingolipids of human granulocytes, 40 μg; lane 4, non-acid glycosphingolipids of rabbit intestine, 40 μg; lane 5, non-acid glycosphingolipids of human meconium, 40 μg; lane 6, non-acid glycosphingolipids of rat intestine, 40 μg; lane 7, non-acid glycosphingolipids of sheep intestine, 40 μg.
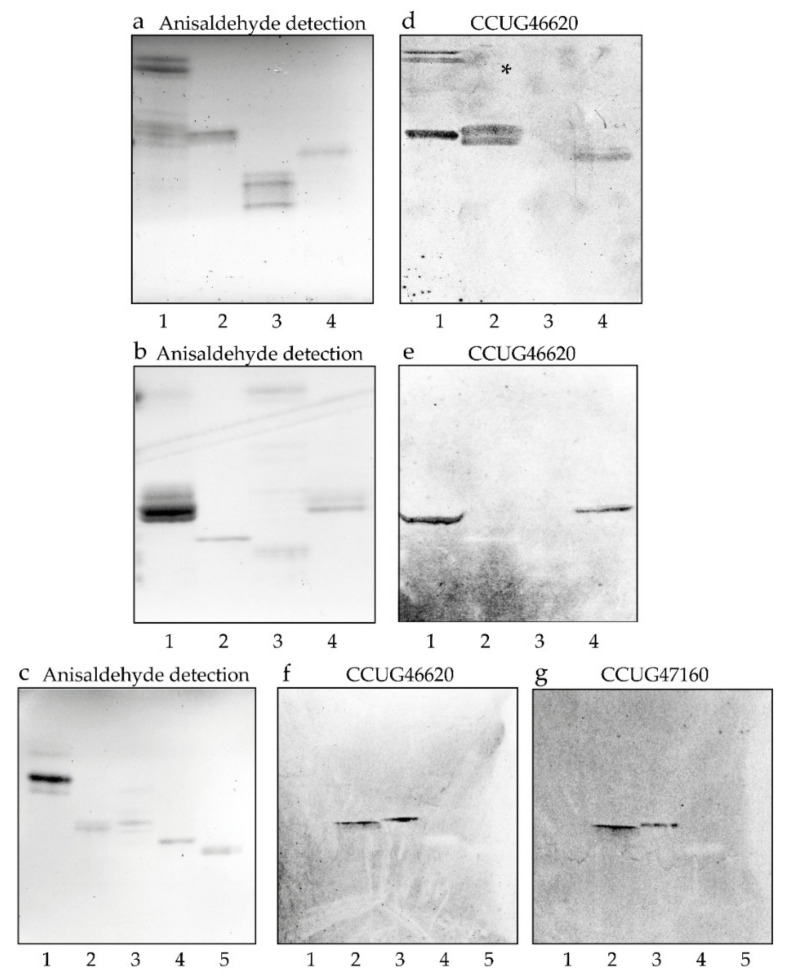
As the glycolipid mixtures have been characterized in previous studies [29,30,31,32,33], we examined the binding of C. matruchotii to a number of representative, structurally characterized reference compounds from our glycolipid library. The results from these binding assays are exemplified in Figure 6 and summarized in Table 2. Two triglycosylceramides, isoglobotriaosylceramide (Figure 6d, lane 2) and lactotriaosylceramide (Figure 6e, lane 4), were recognized by the bacteria (see Table 2 for glycolipid structures). In the case of isoglobotriaosylceramide, a substitution of the terminal Gal by Galα in the 3-position was tolerated with retained binding of the bacteria (Galα3-isoglobotriaosylceramide; Figure 6d, lane 4), whereas a substitution with GalNAcβ in the 3-position abrogated the binding (isoglobotetraosylceramide; Table 2, No. 9). No binding to other glycolipids with terminal Galα3 was observed, such as the Galili penta- and heptaosylceramides (Figure 6d, lane 3), demonstrating that a terminal Galα3 was not enough to support binding of the bacteria.
Figure 6.
Binding of C. matruchotii to reference glycosphingolipids on thin-layer chromatograms. Chemical detection by anisaldehyde (a–c), and autoradiograms obtained by binding of C. matruchotii strain CCUG46620 (d–f) and C. matruchotii strain CCUG47160 (g), followed by autoradiography for 12 h. The solvent system used was chloroform/methanol/water (60:35:8, by volume). The lanes on (a,d) are: Lane 1, non-acid glycosphingolipids of rat intestine, 40 μg; lane 2, isoglobotriaosylceramide (Galα3Galβ4Glcβ1Cer), 4 μg; lane 3, Galili pentaosylceramide (Galα3Galβ4GlcNAcβ3Galβ4Glcβ1Cer) and Galili heptaosylceramide (Galα3Galβ4GlcNAcβ3Galβ4GlcNAcβ3Galβ4Glcβ1Cer), 4 μg; lane 4, Galα3-isoglobotriaosylceramide (Galα3Galα3Galβ4Glcβ1Cer), 4 μg. The lanes on (b,e) are: Lane 1, triglycosylceramides of human meconium (globotriaosylceramide, Galα4Galβ4Glcβ1Cer and lactotriaosylceramide, GlcNAcβ3Galβ4Glcβ1Cer, mixture), 10 μg; lane 2, H type 1 pentaosylceramide (Fucα2Galβ3GlcNAcβ3Galβ4Glcβ1Cer), 4 μg; lane 3, non-acid glycosphingolipids of porcine intestine, 20 μg; lane 4, lactotriaosylceramide (GlcNAcβ3Galβ4Glcβ1Cer), 4 μg. The lanes on (c,f,g) are: Lane 1, globotriaosylceramide (Galα4Galβ4Glcβ1Cer), 8 μg; lane 2, lactotetraosylceramide (Galβ3GlcNAcβ3Galβ4Glcβ1Cer), 4 μg; lane 3, neolactotetraosylceramide (Galβ4GlcNAcβ3Galβ4Glcβ1Cer), 4 μg; lane 4, H type 2 pentaosylceramide (Fucα2Galβ4GlcNAcβ3Galβ4Glcβ1Cer), 4 μg; lane 5, Lex pentaosylceramide (Galβ4(Fucα3)GlcNAcβ3Galβ4Glcβ1Cer), 4 μg.
Table 2.
Binding of C. matruchotii CCUG46620 and CCUG47160 to glycolipids on thin-layer chromatograms.
| No | Trivial Name | Structure | Binding a |
|---|---|---|---|
| 1. | Galactosylceramide | Galβ1Cer | - |
| 2. | Sulfatide | SO3-Galβ1Cer | - |
| 3. | LacCer | Galβ4Glcβ1Cer | - |
| 4. | Neu5Gc-GM3 | Neu5Gcα3Galβ4Glcβ1Cer | - |
| 5. | Globotri | Galα4Galβ4Glcβ1Cer | - |
| 6. | Isoglobotri | Galα3Galβ4Glcβ1Cer | + |
| 7. | Galα3-isoglobotri | Galα3Galα3Galβ4Glcβ1Cer | + |
| 8. | Globotetra | GalNAcβ3Galα4Galβ4Glcβ1Cer | - |
| 9. | Isoglobotetra | GalNAcβ3Galα3Galβ4Glcβ1Cer | - |
| 10. | Galili penta | Galα3Galβ4GlcNAcβ3Galβ4Glcβ1Cer | - |
| 11. | Galili hepta | Galα3Galβ4GlcNAcβ3Galβ4GlcNAcβ3Galβ4Glcβ1Cer | - |
| 12. | Forssman | GalNAcα3GalNAcβ3Galα4Galβ4Glcβ1Cer | - |
| 13. | Lactotri | GlcNAcβ3Galβ4Glcβ1Cer | + |
| 14. | Lactotetra | Galβ3GlcNAcβ3Galβ4Glcβ1Cer | + |
| 15. | Neolactotetra | Galβ4GlcNAcβ3Galβ4Glcβ1Cer | + |
| 16. | Neolactohexa | Galβ4GlcNAcβ3Galβ4GlcNAcβ3Galβ4Glcβ1Cer | + |
| 17. | H type 1 penta | Fucα2Galβ3GlcNAcβ3Galβ4Glcβ1Cer | - |
| 18. | H type 2 penta | Fucα2Galβ4GlcNAcβ3Galβ4Glcβ1Cer | - |
| 19. | Lex penta | Galβ4(Fucα3)GlcNAcβ3Galβ4Glcβ1Cer | - |
| 20. | A type 1 hexa | GalNAcα3(Fucα2)Galβ3GlcNAcβ3Galβ4Glcβ1Cer | - |
| 21. | Leb hexa | Fucα2Galβ3(Fucα4)GlcNAcβ3Galβ4Glcβ1Cer | - |
a + denotes a binding when 4 μg of the glycolipid was applied on the thin-layer chromatogram, whereas - denotes no binding, even at 4 mg.
In the case of lactotriaosylceramide, C. matruchotii also bound when the terminal GlcNAc was substituted with Galβ in the 3-position, creating lactotetraosylceramide (Figure 6f,g, lane 2), or in the 4-position, creating neolactotetraosylceramide (Figure 6f,g, lane 3). Neolactohexaosylceramide (Table 2, No. 16) was also recognized by the bacteria. Further elongation of the terminal Gal of lactotetraosylceramide or neolactotetraosylceramide by Fucα in the 2-position (H type 1 pentaosylceramide, Figure 6e, lane 2; and the H type 2 pentaosylceramide, Figure 6f,g, lane 4) eliminated binding of C. matruchotii. Substitution of the GlcNAc of neolactotetraosylceramide with Fucα in the 3-position also blocked bacterial binding (Lex pentaosylceramide; Figure 6f,g, lane 5).
4. Discussion
The present study surveyed the presence of C. matruchotii in saliva and tooth biofilm from birth to young adulthood and explored phenotypical characteristics of C. matruchotii related to its presence in saliva and establishment in tooth biofilms. The main findings are that the species establishes after 3 months of age and then reaches nearly 100% by early adulthood. C. matruchotii is enriched in tooth biofilms compared to saliva but does not bind directly to the tooth saliva pellicle and may need an adjuvant component to establish itself. C. matruchotii recognition of lactotriaosylceramide, lactotetraosylceramide, and neolactotetraosylceramide may mediate binding to human tissues.
Our findings also support that C. matruchotii is part of the core tooth microbiota in humans. Due to the relatively late colonization, establishment may be associated with tooth eruption, but it is not evidently associated with the number of teeth in early adulthood. Saliva binding in the planktonic phase, but with a limited ability to bind saliva-coated tooth surfaces, suggests binding to a non-tooth absorbing protein or glycoprotein within the saliva, or hidden epitopes after absorption to the tooth surface and that the oral biofilm establishment may build on partner bacteria-dependent recruitment. This would be in line with the prerequisite of tooth eruption and formation of a tooth biofilm before C. matruchotii establishment. Theoretically, binding may also be mediated by a structurally modified tooth pellicle component, but this was not tested in the present study.
C. matruchotii has been indicated to be in close proximity with Actinomyces spp. at the tooth surface [3]. In vivo, A. naeslundii was associated with the abundance of C. matruchotii at all ages and, in vitro, A. naeslundii bound C. matruchotii in both planktonic and overlay assays and possessed the ability to recruit C. matruchotii to the tooth surface. We can hypothesize that A. naeslundii, which is an initial tooth colonizer [34], binds to the saliva pellicle [35] and forms a layer that provides C. matruchotii attachment sites. In addition to A. naeslundii, C. matruchotii associated with F. nucleatum subsp. polymorphum, Leptotrichia sp. HMT 392, and Tannerella sp. HMT 286 in in vivo and in vitro binding assays suggests that Fusobacterium, Porphyromonas, Aggregatibacter, and Streptococcus may directly interact with C. matruchotii. To some extent, this is in line with in vivo localization in which the neighbors along the C. matruchotii filament consisted of Fusobacterium, Leptotrichia, and Capnocytophaga, and at its distal end Streptococcus and Porphyromonas, with frequently observed Haemophilus-Aggregatibacter and Neisseriaceae [3]. Notably, Capnocytophaga spp. were associated with the in vivo abundance of C. matruchotii at all ages but did not directly bind to C. matruchotii. The binding properties of C. matruchotii suggest both protein and carbohydrate interactions with its bacterial partners. Similar dual mechanisms have been observed for A. naeslundii [36], but further studies are required to dissect the C. matruchotii bacterial interaction network.
Glycolipid binding assays demonstrated selective binding of C. matruchotii to isoglobotriaosylceramide, Galα3-isoglobotriaosylceramide, lactotriaosylceramide, lactotetraosylceramide, and neolactotetraosylceramide. Isoglobotriaosylceramide is not likely to be a physiologically relevant ligand for C. matruchotii in humans because this glycolipid has not been found in human tissues [37]. Galα3-isoglobotriaosylceramide has been characterized in cat intestine [38], but not yet in humans. C. matruchotii ligands in humans are most likely lactotriaosylceramide, lactotetraosylceramide, and neolactotetraosylceramide. Lactotetraosylceramide has been identified in the human gastrointestinal tract [39,40,41], whereas neolactotetraosylceramide is widely distributed in human tissues and has been characterized in erythrocytes [42], neutrophils [33], brain [43], and stomach [40]. Lactotriaosylceramide is the precursor of both lactotetraosylceramide and neolactotetraosylceramide and should also be present in these tissues. Furthermore, the terminal Galβ4GlcNAcβ sequence of neolactotetraosylceramide is a common core structure in the carbohydrate chains of glycoproteins [44].
The current study has some strengths and limitations that should be recognized and considered when evaluating the results. The strengths include a combination of in vivo clinical data and in vitro experimental data to support the conclusions and the wide age range from earliest infancy to early adulthood. In addition, two different C. matruchotii strains were used to cover some within-species genetic variations. The two strains mainly presented overlapping results, but also some phenotypic variation. In addition, for the majority of tested C. matruchotii bacterial binding partners, a limited number of strains within in each species were evaluated, limiting the coverage of intra-species variations.
The limitations include a relatively low number of participants with milk and feces samples; however, as the results were consistent among the samples analyzed, these samples rendered conclusive results on the low detection prevalence and abundance. In addition, the use of next generation sequences (NGS) for the identification of microorganisms is potentially associated with errors at various steps, including PCR amplification, sequencing, and bioinformatics. Furthermore, NGS identification includes dead bacteria, and, finally, self-reported information is prone to bias [45].
C. matruchotii colonizes the oral cavity, the skin [46], and likely the nose and nasopharynx, but is not detected in the gastrointestinal tract or human milk, but the association between C. matruchotii and health is still unclear. Interestingly, abundance of the Corynebacterium genus was negatively associated with head and neck squamous cell cancer risk, which also parallels less smoking [47]. We also found smoking to be associated with reduced levels of C. matruchotii, which also coincides with some previous studies [12,15], but not others [17,48]. Notably, C. matruchotii has been suggested to be a low immunogenic species compared to other oral bacteria [49], and the finding that C. matruchotii possesses the ability to bind glycolipids isolated from tissues is of interest for future studies. Genetic tools are required to further understand C. matruchotii [12,50]. Such tools were valuable when the C. matruchotii MdbA gene was characterized, indicating that C. matruchotii possesses the ability to catalyze disulfide bond formation in vitro, and has a similar function as the Corynebacterium diphtheriae MdbA gene, which is required to maintain normal cell growth and morphology, toxin production, and pilus assembly [12].
In conclusion, the present study shows that C. matruchotii establishes after 3 months of age and has reached nearly 100% at early adulthood. C. matruchotii oral biofilm establishment is likely to require both tooth eruption and formation of a pre-existing bacterial biofilm, potentially by A. naeslundii. In addition, C. matruchotii has the ability to interact with glycolipids present on human cells and exhibits specific binding to lactotriaosylceramide, lactotetraosylceramide, neolactotetraosylceramide, isoglobotriaosylceramide, and Galα3-isoglobotriaosylceramide.
Acknowledgments
Agnetha Rönnlund is acknowledged for skillful laboratory work.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/11/1780/s1. Figure S1: Surface-based C. matruchotii coadhesion partners, Table S1: List of bacterial species and strains with culture medium and conditions for use in aggregation and adhesion experiments, Table S2: C. matruchotii in saliva by various lifestyle factors, Table S3: Candidate partner bacteria for C. matruchotii in oral biofilm formation, Table S4: Ability of bacterial partners to coaggregate, coadhere, and recruit C. matruchotii to the tooth surface, Table S5: Bacterial–bacterial binding properties.
Appendix A
Genomic DNA was extracted from saliva, saliva swabs, tooth biofilm, milk, and feces samples, and positive controls created using the GenElute™ Bacterial Genomic DNA Kit (Sigma–Aldrich, St. Louis, MO, USA) with the addition of lysozyme and mutanolysin, RNase, and proteinase K to ensure harvest from both Gram-positive and Gram-negative bacteria. Bacterial 16S rRNA gene amplicons were generated from the v3–v4 (357F-forward TACGGGAGGCAGCAG and 800R-reverse CCAGGGTATCTAATCC primers) variable regions of the 16S rRNA gene, sequenced on an Illumina MiSeq platform, and demultiplexed using deML [51]. Amplicon primers were removed from the obtained sequences and quality checked in QIIME 2 (qiime2.org) [52]. Length trimming, denoising, and chimera and PhiX removal were achieved using DADA2 [53] within QIIME2, resulting in identification of amplicon sequence variants (ASVs) per sample [54]. Default parameters in DADA2 were used with left trimming of 13 bp for both forward and reversed reads, and right trimming at 230 bp for the reversed sequences and 268 bp for the forward sequences. ASVs with only one read were filtered, and species or phylotypes represented by at least 25 reads with 98.5% identity in the extended Human Oral Microbiome Database (eHOMD, http://www.homd.org) [55,56] were retained. For simplicity, both named and unnamed phylotypes are referred to as species in the text. Mixtures of known mock bacteria were used as positive controls and sterile water as the negative control.
Author Contributions
A.E. and I.J. conceptualized the study; P.L.H. and L.E. contributed clinical data; A.E., I.J., P.L.H. and S.T. contributed financial support; A.E. and A.B. performed laboratory analyses, including NGS sequencing; A.E., S.T., A.B., and I.J. performed formal data analyses; A.E., I.J., and S.T. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Patent Revenue Fund for Research in Preventive Odontology (grant number 2018-03), the County Council of Västerbotten Spjutspetsmedel (grant number RV 581231), the County Council of Västerbotten Internal Research Foundation (grant number VLL-831231), and the Swedish Cancer Foundation (grant number 18 0760). None of the funding bodies had any influence on the design, data collection, analysis, interpretation, or writing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gilmour M.N. The classification of organisms termed Leptotrichia (Leptothrix) buccalis. II. Reproduction of Bacterionema matruchotii. Bacteriol. Rev. 1961;25:142–151. doi: 10.1128/MMBR.25.2.142-151.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S.J. A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch. Oral Biol. 1972;17:613–616. doi: 10.1016/0003-9969(72)90081-7. [DOI] [PubMed] [Google Scholar]
- 3.Mark Welch J.L., Rossetti B.J., Rieken C.W., Dewhirst F.E., Borisy G.G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolenbrander P.E., Williams B.L. Prevalence of viridans streptococci exhibiting lactose-inhibitable coaggregation with oral actinomycetes. Infect. Immun. 1983;41:449–452. doi: 10.1128/IAI.41.2.449-452.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D., Barraza J.P., Arthur R.A., Hara A., Lewis K., Liu Y., Scisci E.L., Hajishengallis E., Whiteley M., Koo H. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc. Natl. Acad. Sci. USA. 2020;117:12375–12386. doi: 10.1073/pnas.1919099117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mark Welch J.L., Dewhirst F.E., Borisy G.G. Biogeography of the Oral Microbiome: The Site-Specialist Hypothesis. Ann. Rev. Microbiol. 2019;73:335–358. doi: 10.1146/annurev-micro-090817-062503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksson L., Lif Holgerson P., Johansson I. Saliva and tooth biofilm bacterial microbiota in adolescents in a low caries community. Sci. Rep. 2017;7:5861. doi: 10.1038/s41598-017-06221-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fakhruddin K.S., Ngo H.C., Samaranayake L.P. Cariogenic microbiome and microbiota of the early primary dentition: A contemporary overview. Oral Dis. 2019;25:982–995. doi: 10.1111/odi.12932. [DOI] [PubMed] [Google Scholar]
- 9.Belda-Ferre P., Williamson J., Simon-Soro A., Artacho A., Jensen O.N., Mira A. The human oral metaproteome reveals potential biomarkers for caries disease. Proteomics. 2015;15:3497–3507. doi: 10.1002/pmic.201400600. [DOI] [PubMed] [Google Scholar]
- 10.Benitez-Paez A., Belda-Ferre P., Simon-Soro A., Mira A. Microbiota diversity and gene expression dynamics in human oral biofilms. BMC Genom. 2014;15:311. doi: 10.1186/1471-2164-15-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Dijk S., Dean D.D., Liu Y., Zhao Y., Chirgwin J.M., Schwartz Z., Boyan B.D. Purification, amino acid sequence, and cDNA sequence of a novel calcium-precipitating proteolipid involved in calcification of corynebacterium matruchotii. Calcif. Tissue Int. 1998;62:350–358. doi: 10.1007/s002239900443. [DOI] [PubMed] [Google Scholar]
- 12.Luong T.T., Tirgar R., Reardon-Robinson M.E., Joachimiak A., Osipiuk J., Ton-That H. Structural Basis of a Thiol-Disulfide Oxidoreductase in the Hedgehog-Forming Actinobacterium Corynebacterium matruchotii. J. Bacteriol. 2018;200 doi: 10.1128/JB.00783-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross E.L., Leys E.J., Gasparovich S.R., Firestone N.D., Schwartzbaum J.A., Janies D.A., Asnani K., Griffen A.L. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J. Clin. Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aas J.A., Griffen A.L., Dardis S.R., Lee A.M., Olsen I., Dewhirst F.E., Leys E.J., Paster B.J. Bacteria of dental caries in primary and permanent teeth in children and young adults. J. Clin. Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Acharya A., Chen T., Chan Y., Watt R.M., Jin L., Mattheos N. Species-Level Salivary Microbial Indicators of Well-Resolved Periodontitis: A Preliminary Investigation. Front. Cell Infect. Microbiol. 2019;9:347. doi: 10.3389/fcimb.2019.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nibali L., Sousa V., Davrandi M., Spratt D., Alyahya Q., Dopico J., Donos N. Differences in the periodontal microbiome of successfully treated and persistent aggressive periodontitis. J. Clin. Periodontol. 2020;47:980–990. doi: 10.1111/jcpe.13330. [DOI] [PubMed] [Google Scholar]
- 17.Moon J.H., Lee J.H., Lee J.Y. Subgingival microbiome in smokers and non-smokers in Korean chronic periodontitis patients. Mol. Oral Microbiol. 2015;30:227–241. doi: 10.1111/omi.12086. [DOI] [PubMed] [Google Scholar]
- 18.Lif Holgerson P., Esberg A., Sjodin A., West C.E., Johansson I. A longitudinal study of the development of the saliva microbiome in infants 2 days to 5 years compared to the microbiome in adolescents. Sci. Rep. 2020;10:9629. doi: 10.1038/s41598-020-66658-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esberg A., Haworth S., Kuja-Halkola R., Magnusson P.K.E., Johansson I. Heritability of Oral Microbiota and Immune Responses to Oral Bacteria. Microorganisms. 2020;8:1126. doi: 10.3390/microorganisms8081126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esberg A., Haworth S., Hasslof P., Lif Holgerson P., Johansson I. Oral Microbiota Profile Associates with Sugar Intake and Taste Preference Genes. Nutrients. 2020;12:681. doi: 10.3390/nu12030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eriksson L., Esberg A., Haworth S., Holgerson P.L., Johansson I. Allelic Variation in Taste Genes Is Associated with Taste and Diet Preferences and Dental Caries. Nutrients. 2019;11:1491. doi: 10.3390/nu11071491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibbons R.J., Hay D.I. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces viscosus LY7 to apatitic surfaces. Infect. Immun. 1988;56:439–445. doi: 10.1128/IAI.56.2.439-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlsson K.A. Preparation of total nonacid glycolipids for overlay analysis of receptors for bacteria and viruses and for other studies. Methods Enzymol. 1987;138:212–220. doi: 10.1016/0076-6879(87)38018-8. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson H., Halim A., Teneberg S. Differentiation of glycosphingolipid-derived glycan structural isomers by liquid chromatography/mass spectrometry. Glycobiology. 2010;20:1103–1116. doi: 10.1093/glycob/cwq070. [DOI] [PubMed] [Google Scholar]
- 25.Samuelsson B.E., Pimlott W., Karlsson K.A. Mass spectrometry of mixtures of intact glycosphingolipids. Methods Enzymol. 1990;193:623–646. doi: 10.1016/0076-6879(90)93442-n. [DOI] [PubMed] [Google Scholar]
- 26.Koerner T.A., Jr., Prestegard J.H., Demou P.C., Yu R.K. High-resolution proton NMR studies of gangliosides. 1. Use of homonuclear two-dimensional spin-echo J-correlated spectroscopy for determination of residue composition and anomeric configurations. Biochemistry. 1983;22:2676–2687. doi: 10.1021/bi00280a014. [DOI] [PubMed] [Google Scholar]
- 27.Waldi D. Sprühreagentien für die dünnschicht-chromatographie. In: Stahl E., editor. Dünnschicht-Chromatographie. Springer; Berlin/Heidelberg, Germany: 1962. pp. 496–515. [Google Scholar]
- 28.Roche N., Angstrom J., Hurtig M., Larsson T., Boren T., Teneberg S. Helicobacter pylori and complex gangliosides. Infect. Immun. 2004;72:1519–1529. doi: 10.1128/IAI.72.3.1519-1529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breimer M.E., Hansson G.C., Karlsson K.A., Leffler H. Glycosphingolipids of rat tissues. Different composition of epithelial and nonepithelial cells of small intestine. J. Biol. Chem. 1982;257:557–568. [PubMed] [Google Scholar]
- 30.Breimer M.E., Hansson G.C., Karlsson K.A., Leffler H., Pimlott W., Samuelsson B.E. Selected ion monitoring of glycospingolipid mixtures. Identification of several blood group type glycolipids in the small intestine of an individual rabbit. Biomed. Mass Spectrom. 1979;6:231–241. doi: 10.1002/bms.1200060603. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson K.A., Larson G. Molecular characterization of cell-surface antigens of human fetal tissue: Meconium, a rich source of epithelial blood-group glycolipids. FEBS Lett. 1978;87:283–287. doi: 10.1016/0014-5793(78)80352-4. [DOI] [PubMed] [Google Scholar]
- 32.Koscielak J., Miller-Podraza H., Krauze R., Cedergren B. Glycolipid composition of blood group P erythrocytes. FEBS Lett. 1976;66:250–253. doi: 10.1016/0014-5793(76)80515-7. [DOI] [PubMed] [Google Scholar]
- 33.Macher B.A., Klock J.C. Isolation and chemical characterization of neutral glycosphingolipids of human neutrophils. J. Biol. Chem. 1980;255:2092–2096. [PubMed] [Google Scholar]
- 34.Dige I., Raarup M.K., Nyengaard J.R., Kilian M., Nyvad B. Actinomyces naeslundii in initial dental biofilm formation. Microbiology. 2009;155:2116–2126. doi: 10.1099/mic.0.027706-0. [DOI] [PubMed] [Google Scholar]
- 35.Hawkins B.W., Cannon R.D., Jenkinson H.F. Interactions of Actinomyces naeslundii strains T14V and ATCC 12104 with saliva, collagen and fibrinogen. Arch. Oral Biol. 1993;38:533–535. doi: 10.1016/0003-9969(93)90191-N. [DOI] [PubMed] [Google Scholar]
- 36.Cisar J.O., Kolenbrander P.E., McIntire F.C. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 1979;24:742–752. doi: 10.1128/IAI.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Speak A.O., Salio M., Neville D.C., Fontaine J., Priestman D.A., Platt N., Heare T., Butters T.D., Dwek R.A., Trottein F., et al. Implications for invariant natural killer T cell ligands due to the restricted presence of isoglobotrihexosylceramide in mammals. Proc. Natl. Acad. Sci. USA. 2007;104:5971–5976. doi: 10.1073/pnas.0607285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teneberg S., Angstrom J., Ljungh A. Carbohydrate recognition by enterohemorrhagic Escherichia coli: Characterization of a novel glycosphingolipid from cat small intestine. Glycobiology. 2004;14:187–196. doi: 10.1093/glycob/cwh015. [DOI] [PubMed] [Google Scholar]
- 39.Bjork S., Breimer M.E., Hansson G.C., Karlsson K.A., Leffler H. Structures of blood group glycosphingolipids of human small intestine. A relation between the expression of fucolipids of epithelial cells and the ABO, Le and Se phenotype of the donor. J. Biol. Chem. 1987;262:6758–6765. [PubMed] [Google Scholar]
- 40.Jin C., Barone A., Boren T., Teneberg S. Helicobacter pylori-binding nonacid glycosphingolipids in the human stomach. J. Biol. Chem. 2018;293:17248–17266. doi: 10.1074/jbc.RA118.004854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teneberg S., Leonardsson I., Karlsson H., Jovall P.A., Angstrom J., Danielsson D., Naslund I., Ljungh A., Wadstrom T., Karlsson K.A. Lactotetraosylceramide, a novel glycosphingolipid receptor for Helicobacter pylori, present in human gastric epithelium. J. Biol. Chem. 2002;277:19709–19719. doi: 10.1074/jbc.M201113200. [DOI] [PubMed] [Google Scholar]
- 42.Siddiqui B., Hakomori S. A ceramide tetrasaccharide of human erythrocyte membrane reacting with anti-type XIV pneumococcal polysaccharide antiserum. Biochim. Biophys. Acta. 1973;330:147–155. doi: 10.1016/0005-2736(73)90219-8. [DOI] [PubMed] [Google Scholar]
- 43.Ishikawa Y., Gasa S., Minami R., Makita A. Characterization of neutral glycosphingolipids from fetal human brain: Evidence for stage-specific expression of the globo, ganglio, and neolacto series in the central nervous system. J. Biochem. 1987;101:1369–1375. doi: 10.1093/oxfordjournals.jbchem.a122005. [DOI] [PubMed] [Google Scholar]
- 44.Stanley P., Cummings R.D. Structures Common to Different Glycans. In: Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etzler M.E., editors. Essentials of Glycobiology. 3rd ed. Cold Spring Harbor; New York, NY, USA: 2009. [PubMed] [Google Scholar]
- 45.Johansson I., Hallmans G., Wikman A., Biessy C., Riboli E., Kaaks R. Validation and calibration of food-frequency questionnaire measurements in the Northern Sweden Health and Disease cohort. Public Health Nutr. 2002;5:487–496. doi: 10.1079/PHN2001315. [DOI] [PubMed] [Google Scholar]
- 46.Park S.Y., Kim H.S., Lee S.H., Kim S. Characterization and Analysis of the Skin Microbiota in Acne: Impact of Systemic Antibiotics. J. Clin. Med. 2020;9:168. doi: 10.3390/jcm9010168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayes R.B., Ahn J., Fan X., Peters B.A., Ma Y., Yang L., Agalliu I., Burk R.D., Ganly I., Purdue M.P., et al. Association of Oral Microbiome With Risk for Incident Head and Neck Squamous Cell Cancer. JAMA Oncol. 2018;4:358–365. doi: 10.1001/jamaoncol.2017.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Boeck I., Wittouck S., Wuyts S., Oerlemans E.F.M., van den Broek M.F.L., Vandenheuvel D., Vanderveken O., Lebeer S. Comparing the Healthy Nose and Nasopharynx Microbiota Reveals Continuity As Well As Niche-Specificity. Front. Microbiol. 2017;8:2372. doi: 10.3389/fmicb.2017.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Esberg A., Johansson A., Claesson R., Johansson I. 43-Year Temporal Trends in Immune Response to Oral Bacteria in a Swedish Population. Pathogens. 2020;9:544. doi: 10.3390/pathogens9070544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang C., Nguyen M.T., Ton-That H. Genetic Manipulation of Corynebacterium diphtheriae and Other Corynebacterium Species. Curr. Protoc. Microbiol. 2020;58:e111. doi: 10.1002/cpmc.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Renaud G., Stenzel U., Maricic T., Wiebe V., Kelso J. deML: Robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics. 2015;31:770–772. doi: 10.1093/bioinformatics/btu719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Callahan B.J., McMurdie P.J., Holmes S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017;11:2639–2643. doi: 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen T., Yu W.H., Izard J., Baranova O.V., Lakshmanan A., Dewhirst F.E. The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Escapa I.F., Chen T., Huang Y., Gajare P., Dewhirst F.E., Lemon K.P. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): A Resource for the Microbiome of the Human Aerodigestive Tract. mSystems. 2018;3 doi: 10.1128/mSystems.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.