Abstract
The molecular circadian clock is a self-regulating transcription/translation cycle of positive (Bmal1, Clock/Npas2) and negative (Per1,2,3, Cry1,2) regulatory components. While the molecular clock has been well-characterized in the body’s master circadian pacemaker, the hypothalamic suprachiasmatic nucleus (SCN), only a few studies have examined both the positive and negative clock components in extra-SCN brain tissue. Furthermore, there has yet to be a direct comparison of male and female clock gene expression in the brain. This comparison is warranted, as there are sex differences in circadian functioning and disorders associated with disrupted clock gene expression. This study examined basal clock gene expression (Per1, Per2, Bmal1 mRNA) in the SCN, prefrontal cortex (PFC), rostral agranular insula, hypothalamic paraventricular nucleus (PVN), amygdala, and hippocampus of male and female rats at 4-h intervals throughout a 12:12h light:dark cycle. There was a significant rhythm of Per1, Per2, and Bmal1 in the SCN, PFC, insula, PVN, subregions of the hippocampus, and amygdala with a 24-h period, suggesting the importance of an oscillating molecular clock in extra-SCN brain regions. There were three distinct clock gene expression profiles across the brain regions, indicative of diversity amongst brain clocks. Although generally the clock gene expression profiles were similar between male and female rats, there were some sex differences in the robustness of clock gene expression (e.g., females had less robust rhythms in the medial PFC, more robust rhythms in the hippocampus, and a greater mesor in the medial amygdala). Furthermore, females with a regular estrous cycle had attenuated aggregate rhythms in clock gene expression in the PFC compared to non-cycling females. This suggests that gonadal hormones may modulate the expression of the molecular clock.
Keywords: clock genes, medial prefrontal cortex, amygdala, hippocampus, paraventricular nucleus of the hypothalamus, sex differences, extra-SCN, molecular clock
Introduction
The existence and characteristics of intrinsic molecular circadian clocks in brain regions outside of the body’s master circadian pacemaker, the suprachiasmatic nucleus of the hypothalamus (SCN), require further exploration, as these intrinsic clocks, if operational, may contribute to the normal functioning of these extra-SCN brain regions. Furthermore, there are sex differences in circadian rhythms (Bailey & Silver, 2014), yet there has been virtually no direct comparison of basal clock gene expression at the mRNA level in the brains of male and female subjects. The approximately 24-hour rhythmic expression of clock genes in the SCN comprises a molecular clock that is necessary for the expression of circadian rhythms throughout the body (Reppert & Weaver, 2002; Takahashi et al., 2008). This molecular clock consists of counter-regulatory and oscillatory transcription/translation interactions between positive (Bmal1, Clock or Npas2) and negative (Per1, Per2, Per3, Cry1, Cry2) clock gene encoded transcription regulators (Darlington et al., 1998; Dunlap, 1999; Gekakis, 1998; Reppert & Weaver, 2002). Expression of both the positive and negative clock gene components are thought to comprise a functional molecular clock, as genetic knockdown of either the positive or negative components results in behavioral arrhythmicity in rodents (Bae et al., 2001; Bunger et al., 2000).
The SCN exhibits rhythmic expression of Bmal1, Per1, Per2, Cry1, and Cry2 mRNA with a genetically determined period that approximates 24-h (Bae et al., 2001). These genes have been found to be rhythmically expressed in many other mammalian brain and peripheral tissues, raising the prospect that extra-SCN clock gene expression may directly contribute to neural function throughout the brain (Abe et al., 2002; Sakamoto et al., 1998). In rodents, rhythmic clock gene expression has been found in a variety of peripheral tissues, and their rhythmic expression contributes to the optimal functioning of these tissues (Balsalobre, 2000; Hastings et al., 2007; Marcheva et al., 2013; Yamamoto et al., 2004; Yamazaki et al., 2000). Rhythmic clock gene expression has also been observed in some extra-SCN brain regions (Amir et al., 2004; Ángeles-Castellanos et al., 2007; Feillet et al., 2008; Girotti et al., 2009; Guilding & Piggins, 2007; Harbour et al., 2013, 2014; Lamont et al., 2005; Masubuchi et al., 2000; Perrin et al., 2006; Rath et al., 2013; Rath et al., 2014; Reick, 2001; Yamamoto et al., 2004; but see Abe et al., 2002). Most reports of clock gene expression in the brain have provided limited insight into the nature of extra-SCN clocks because those reports focused on only one or a few clock genes and brain regions (Guilding & Piggins, 2007). Only more recently has both the positive (Bmal1) and negative (Per1/Per2) components of the molecular clock been well-examined within a few extra-SCN brain regions (PVN, central amygdala, hippocampus, neocortex, and cerebellum) (Girotti et al., 2009; Harbour et al., 2014; Jilg et al., 2010; Rath et al., 2013). Thus, it is not possible to discern from the extant literature whether there is evidence for oscillatory expression of positive and negative regulatory clock components within most brain regions, whether intrinsic clock gene expression is ubiquitous throughout the brain, and whether the expression rhythms (phase relationships and amplitude) are similar across brain regions.
Normal clock gene expression has been implicated in overall mental health (Bunney et al., 2015; Bunney & Bunney, 2000; Etain et al., 2011; Johansson et al., 2002; Lamont et al., 2009; Li et al., 2013; McCarthy & Welsh, 2012; McClung, 2007; Partonen et al., 2007). In mice, mutation or knockdown of Clock or Per1/Per2 are associated with manic-like behavior and increased anxiety (Dzirasa et al., 2011; Mukherjee et al., 2010; Spencer et al., 2012). The prevalence of mood disorders associated with clock gene disruption (e.g., depression, anxiety, and post-traumatic stress disorder) is greater in women compared to men. There also appears to be a role for gonadal hormones in circadian function (Bailey & Silver, 2014; Morin, 1980; Thomas & Armstrong, 1989) and clock gene expression in female rodents (He et al., 2007; Nakamura et al., 2001;2005; 2010; Perrin et al., 2006; Smith et al., 2010). However, there has yet to be a study to directly compare in males and females the expression of positive and negative clock genes.
Consequently, the objective of our first experiment was to examine in male and female rats clock gene expression of both the positive (Bmal1) and negative (Per1, Per2) components of the molecular clock in brain regions important in the regulation of emotion, mood, and stress responsivity (prefrontal cortex (PFC), rostral agranular insula, amygdala, and hippocampus). In a follow-up experiment we examined whether clock gene expression profiles varied in female rats depending on their estrous cycle parameters (presence or absence of cyclicity). For comparison purposes we also examined clock gene mRNA expression in the SCN and PVN, brain regions we previously demonstrated have robust rhythmic expression of each of these clock genes (Girotti et al., 2009). The dysfunction of the medial PFC (mPFC), hippocampus, and amygdala are implicated in the same mood and anxiety disorders associated with disrupted clock gene expression (Adhikari, 2014; Del Casale et al., 2013; Drevets et al., 2008; Koenigs & Grafman, 2009; Mayberg et al., 2005). The mPFC, amygdala, and hippocampus are also critical components of fear conditioning and extinction of fear conditioning, both of which exhibit diurnal fluctuations in behavioral expression (Chaudhury & Colwell, 2002; Eckel-Mahan et al., 2008; Smarr et al., 2014; Valentinuzzi et al., 2001; Woodruff et al., 2015). Characterization of basal clock gene expression in these brain regions may contribute to a better understanding of circadian function in mental health.
MATERIALS AND METHODS
Animals
Sprague Dawley rats (Harlan, Indianapolis, IN), aged approximately 2.5 months, were used in both experiments. Rats were allowed 2 weeks to acclimate to the facility, cage mates, and light:dark cycle. Half of the rats were maintained on a normal 12:12 h light:dark cycle (lights on at 0500 h or 0530 h) and the other half were maintained on a reverse 12:12 h light:dark cycle (lights on at 1700 h or 1730 h). Rats were divided evenly between four individual rooms that were temperature and humidity-controlled. Rats were pair-housed and had free access to food and water. Procedures for the ethical treatment of the animals were conducted in accordance to the guidelines found within the Guide for the Care and Use of Laboratory Animals (DHHS Publication No. (NIH) 80-23, revised 2010 8th edition) and were approved by the University of Colorado’s Institutional Animal Care and Use Committee.
Experiment 1: Basal clock gene expression throughout the brain of male and female rats
In order to assess basal clock gene expression, two cohorts of rats were used, with sex counterbalanced across cohorts (total of 48 males and 49 females; 8-9 rats of each sex at each time point). Females and males were age-matched and housed in separate rooms. Female estrous cycle was not tracked for this first experiment. Thus, all female data from this experiment were pooled and compiled regardless of estrous cycle stage.
Experiment 2: Basal clock gene expression throughout the brain of normally cycling versus non-cycling female rats
74 female rats were used (12-13 rats at each time point). Estrous cycle phase was assessed by daily vaginal lavage for 2 weeks. The rats on the reverse light:dark schedule were lavaged daily at zeitgeber time 1 (ZT1 – ZT refers to the number of hours after the light phase onset), and the rats on the normal light:dark schedule were lavaged daily at ZT8. Lavages began 2 days after arrival and after the light:dark schedule was set. For lavage, a blunt tip glass eye dropper filled with ~0.5mL of sterile saline was inserted into the vagina. The saline was expelled into the vagina two to three times before being taken back up by the eye dropper in order to gently wash off vaginal cells. A drop of the sample was placed on a microscope slide and immediately examined under 40x light microscopy to assess estrous cycle stage. Rats were considered cyclers if they had at least three different estrous cycle stages within the last five days before sacrifice (Goldman et al., 2007; Marcondes et al., 2002).
Tissue Collection
For both experiments, rats were removed from their home cage and immediately decapitated at six evenly spaced times across the 24 hour day (ZT0, 4, 8, 12, 16, 20). Procedures were performed under red light conditions for rats killed during the dark phase (ZT12, 16, 20). Trunk blood was collected into EDTA-coated tubes and centrifuged at 4,000 rpms for 10 minutes at 4°C. Plasma was then aliquoted and stored at −70°C until subsequent use. Brains were extracted and flash frozen in −25 ± 5°C isopentane chilled with dry ice. Brains were then cut (12 μm thick coronal sections) with a cryostat (Leica CM 1850) at the level of the mPFC (Bregma ~ 2.2-3.2 mm anterior to Bregma), SCN (Bregma ~ 1.3 to 1.4 mm posterior to Bregma), PVN (Bregma ~ 1.8 to 1.88 mm posterior to Bregma), and hippocampus/amygdala (Bregma ~ 2.5 to 2.8 mm posterior to Bregma) according to Paxinos & Watson (4th edition). Brain slices were thaw-mounted onto Colorfrost Plus microscope slides, and stored at −70°C until subsequent use.
Corticosterone (CORT) Hormone Assays
Plasma samples were assayed in duplicate using an ELISA kit according to the manufacturer’s instructions (Cat #ADI-901-097 Enzo Life Sciences, Plymouth Meeting, PA or Cat #K014-H1 Arbor Assays, Ann Arbor, MI). Sensitivity for CORT was 18.6 pg/mL (Enzo Life Sciences) or 27.0 pg/mL (Arbor Assays) according to the manufacturer. Plasma was diluted 1:50 in assay buffer and heat inactivated for 60 minutes at 65°C in order to denature corticosteroid binding globulin. Intra-assay coefficient of variation averaged 8.9%, and the inter-assay coefficient of variation was 12.8%.
In situ Hybridization
In situ hybridization for Per1, Per2, and Bmal1 mRNA was performed as previously described (Ginsberg et al.,2003; Girotti et al., 2009) with slight modifications as follows. Hybridization was performed in a 50% formamide humidified atmosphere at 54°C for 16-18 hours. Slides were then treated with RNase A (Cat #R5503, Sigma, St. Louis, MO) at 37°C for an hour, washed in decreasing concentrations of standard saline citrate solution (SSC), incubated in 0.1X SSC at 65°C for an hour, then dehydrated through a series of ethanol washes. Dried slides were then exposed to X-ray film for 2-4 weeks, after which films were digitized by use of Northern Light lightbox model B95 (Imaging Res. Inc, St. Catharines, Ontario, CAN), a Sony CCD video camera model XC-ST70 fitted with a Navitar 7000 zoom lens (Rochester, NY) connected to a LG3-01 frame grabber (Scion Corp., Frederick, MD) inside a Dell Dimension 500, and captured with Scion Image beta rel. 4.0.2.
Densitometry
Digitized brain images were analyzed using ImageJ64 (NIH) to quantify the mean optical density (OD) of regions of interest (ROI). ROIs were hand drawn using visible anatomical landmarks with aid of the Paxinos & Watson Brain Atlas. An experimenter blind to the treatment group assignments generated ROIs for both hemispheres. PFC and rostral agranular insula (insula) measures were based on 6 coronal slices per brain, and all other measures were based on 4 coronal slices per brain. The mean gray values of all ROIs were converted into uncalibrated ODs using ImageJ (NIH). Densitometry was performed on films in which the gray level signal fell within the linear range of the gray level to OD relationship. These ODs were then averaged to get the mean OD for each animal. The mean OD for each rat was normalized by converting the mean OD values to a percent of mean value for the male ZT0 group in Experiment 1, or a percent of the mean value for the overall ZT0 group for Experiment 2.
Statistical Analysis
Data were analyzed by two-way analysis of variance (ANOVA) in order to assess clock gene variation across time of day and sex in Experiment 1, or clock gene variation across time of day and estrous cyclicity in Experiment 2. Rats in Experiment 2 were divided into non-cyclers or cyclers, rather than stage of estrous cycle, in order to increase power of analysis due to a limited number of subjects in each estrous stage at each ZT. For significant main effects or interactions, Fischer’s least significant difference (FLSD) post-hoc test was used to assess significant sex or cyclicity differences at a specific time of day. P-values < 0.05 were considered to indicate significant differences between groups. Statistical Package for Social Sciences (SPSS, Mac version 21, 2012) was used for ANOVA and post-hoc analysis. For most clock gene measures in most brain regions there was minimal, but occasional missing data due to the absence within that assay of tissue sections for a particular brain that included the appropriate region of interest. Those missing data are reflected in the small variations in denominator degrees of freedom for the ANOVA analyses. Due to technical difficulties, there were not enough tissue sections to measure Per2 gene expression for the first cohort of rats in Experiment 1 at the level of the SCN. Consequently, that measure is based on group sizes of 4-5 rats. All line graphs showing relative clock gene expression over the course of the day have double-plotted ZT0/ZT24 data for aid in visualization of the underlying rhythm.
Data with a significant time of day effect were further analyzed using least-squares rhythmometry method with ChronoLab 3.0.3 (Mojón et al., 1992) and separated out by male or female (Experiment 1), and by cycling versus non-cycling (Experiment 2). Data were tested for fit to a cosine curve constrained to a 24-h period and considered to have a significant 24-h period if the p-value was < 0.05. In cases where there was a significant 24-h period, estimates of amplitude and acrophase, along with their corresponding 95% confidence intervals (CIs), were generated. Amplitude refers to the distance from the MESOR (rhythm adjusted mean) to the peak of the rhythm, and is reported in units of normalized percent OD values in each experiment. Acrophase, the phase angle where the peak of the fitted curve falls, was converted to ZT time.
RESULTS
Experiment 1: Basal clock gene expression throughout the brain of male and female rats
CORT hormone levels
There was a robust diurnal rhythm of basal CORT levels, with an acrophase centered around the onset of the dark phase (male acrophase = 12.7±0.9, 95% confidence interval (CI); female acrophase = 13.8±1.5, 95% CI) [Supplemental Fig 1]. Two-way ANOVA found significant main effects of ZT and sex. Post-hoc tests (FLSD) revealed that females had greater levels of CORT compared to males at ZT8-20 (i.e., during all times except the diurnal nadir) [Table 1, Supplemental Fig 1].
Table 1.
Experiment 1: Statistical analysis of basal Per1, Per2, and Bmal1 mRNA throughout the brain.
| Cosinor Analysis (× = p<0.05) | ||||||
|---|---|---|---|---|---|---|
| ZT | Sex | Sex × ZT | Male | Female | ||
| Per1 | SCN | F(5,82) = 20.9** | F(1,82) = 1.7 | F(5,82) = 0.0 | × | × |
| PVN | F(5,84) = 12.8** | F(1,84) = 0.0 | F(5,84) = 0.4 | × | × | |
| AC | F(5,84) = 7.7** | F(1,84) = 0.5 | F(5,84) = 0.5 | × | × | |
| PL | F(5,84) = 3.0* | F(1,84) = 0.0 | F(5,84) = 0.4 | × | ||
| IL | F(5,82) = 3.0* | F(1,82) = 0.0 | F(5,82) = 0.9 | × | ||
| VO | F(5,84) = 11.5** | F(1,84) = 0.4 | F(5,84) = 0.7 | × | × | |
| Insula | F(5,84) = 8.9** | F(1,84) = 0.0 | F(5,84) = 0.5 | × | × | |
| CA1 | F(5,85) = 4.5* | F(1,85) = 0.1 | F(5,85) = 0.7 | × | ||
| CA3 | F(5,85) = 4.2* | F(1,85) = 0.9 | F(5.85) = 1.2 | 0.07 | × | |
| supraDG | F(5,85) = 2.0 | F(1,85) = 0.4 | F(5,85) = 0.8 | |||
| infraDG | F(5,85) = 1.2 | F(1,85) = 0.4 | F(5,85) = 1.2 | |||
| CEA | F(5,85) = 5.1** | F(1,85) = 0.3 | F(5,85) = 1.5 | |||
| BLA | F(5,85) = 6.7** | F(1,85) = 0.1 | F(5,85) = 2.2 | × | × | |
| MEA | F(5,85) = 2.9* | F(1,85) = 0.1 | F(5,85) = 0.2 | |||
| Bmal | SCN | F(5,82) = 22.1** | F(1,82) = 0.0 | F(5,82) = 0.6 | × | × |
| PVN | F(5,83) = 32.2** | F(1,83) = 0.2 | F(5,83) = 1.5 | × | × | |
| AC | F(5,83) = 10.5** | F(1,83) = 0.0 | F(5,83) = 1.8 | × | × | |
| PL | F(5,83) = 8.3** | F(1,83) = 0.2 | F(5,83) = 1.0 | × | × | |
| IL | F(5,83) = 5.8** | F(1,83) = 0.1 | F(5,83) = 1.1 | × | × | |
| VO | F(5,83) = 10.2** | F(1,83) = 0.0 | F(5,83) = 0.6 | × | × | |
| Insula | F(5,83) = 13.4** | F(1,83) = 0.0 | F(5,83) = 1.0 | × | × | |
| CA1 | F(5,85) = 3.4* | F(1,85) = 0.0 | F(5,85) = 0.8 | 0.05 | × | |
| CA3 | F(5,85) = 3.2* | F(1,85) = 0.9 | F(5,85) = 0.4 | 0.07 | × | |
| supraDG | F(5,85) = 1.9 | F(1,85) = 0.9 | F(5,85) = 0.7 | |||
| infraDG | F(5,85) = 1.8 | F(1,85) = 0.2 | F(5,85) = 0.7 | |||
| CEA | F(5,85) = 9.5** | F(1,85) = 12.2* | F(5,85) = 1.4 | × | × | |
| BLA | F(5,85) = 7.6** | F(1,85) = 1.5 | F(5,85) = 0.9 | × | × | |
| MEA | F(5,85) = 7.0** | F(1,85) = 20.7* | F(5,85) = 0.5 | × | × | |
| Per2 | SCN | F(5,36) = 23.4** | F(1,36) = 2.4 | F(5,36) = 1.1 | × | × |
| PVN | F(5,78) = 14.3** | F(1,78) = 0.4 | F(5,78) = 0.8 | × | × | |
| AC | F(5,84) = 12.0** | F(1,84) = 0.3 | F(5,84) = 1.3 | × | × | |
| PL | F(5,84) = 6.7** | F(1,84) = 0.1 | F(5,84) = 1.3 | × | × | |
| IL | F(5,81) = 4.0* | F(1,81) = 0.0 | F(5,81) = 1.0 | × | ||
| VO | F(5,84) = 17.9** | F(1,84) = 0.3 | F(5,84) = 1.0 | × | × | |
| Insula | F(5,84) = 18.0** | F(1,84) = 0.0 | F(5,84) = 1.3 | × | × | |
| CA1 | F(5,85) = 10.6** | F(1,85) = 0.1 | F(5,85) = 1.2 | × | × | |
| CA3 | F(5,85) = 11.0** | F(1,85) = 0.0 | F(5,85) = 1.7 | × | × | |
| supraDG | F(5,85) = 3.3* | F(1,85) = 0.1 | F(5,85) = 1.2 | × | ||
| infraDG | F(5,85) = 1.7 | F(1,85) = 0.2 | F(5,85) = 0.8 | |||
| CEA | F(5,85) = 5.1** | F(1,85) = 0.9 | F(5,85) = 0.5 | 0.06 | × | |
| BLA | F(5,85) = 7.9** | F(1,85) = 0.1 | F(5,85) = 2.1 | × | × | |
| MEA | F(5,85) = 2.8* | F(1,85) = 0.5 | F(5,85) = 0.7 | |||
| CORT | F(5,85) = 10.2** | F(1,85) = 29.4* | F(5,85) = 2.0 | × | × | |
Two-way ANOVA results for effect of time of day (ZT), sex, and their interaction,
p < 0.05,
p < 0.001.
Cosinor analysis test for 24 h rhythmicity was analyzed separately for each sex. SCN = suprachiasmatic nucleus, PVN = paraventricular nucleus of the hypothalamus, AC = anterior cingulate cortex, PL = prelimbic cortex, IL = infralimbic cortex, VO = ventral orbital cortex, Insula = rostral agranular insular cortex, CA1 (of hippocampus), CA3 (of hippocampus), supraDG = superior blade of the dentate gyrus, infraDG = inferior blade of the dentate gyrus, CEA = central amygdala, BLA = basolateral amygdala, MEA = medial amygdala.
Clock Gene Analysis – Sex Comparison of Time of Day (ZT) and 24-h Rhythmic Expression
As expected, the SCN had robust, 24-h rhythmic Per1, Per2, and Bmal1 mRNA expression (significant main effect of ZT; significant cosinor analysis) [Table 1, Fig 1]. There was no main effect of sex or sex by ZT interaction for Per1, Per2, and Bmal1 mRNA in the SCN. There were also no sex differences in acrophase or amplitude for all clock genes examined (cosinor analysis). This general profile of 24-h rhythmic expression of each of the clock genes in both male and female rats was observed in many of the other brain regions of interest. Thus, there was also robust 24-h rhythmic Per1, Per2, and Bmal1 mRNA expression for both sexes in the PVN, anterior cingulate cortex (AC), ventral orbital cortex (VO), rostral agranular insula (insula), and basal lateral amygdala (BLA) [Table 1, Fig 1, Fig 2, Fig 3, Fig 4, Supplemental Fig 2]. Although there was not a significant sex by ZT interaction for clock gene expression in any of the brain regions examined, cosinor analysis indicated that males but not females had a robust 24-h rhythm of Per1 (prelimbic and infralimbic cortex) and Per2 (infralimbic cortex) mRNA in subregions of the medial prefrontal cortex [Table 1]. On the other hand, females but not males had a robust 24-h rhythm of Per1 (CA1 and CA3), Bmal1 (CA1 and CA3), and Per2 (supra blade of the dentate gyrus) mRNA in subregions of the hippocampus, and a more robust 24-h rhythm of Per2 mRNA in the central nucleus of the amygdala (CEA) [Table 1]. The only main effect of sex observed was for Bmal1 mRNA expression in the CEA and medial amygdala (MEA) [Table 1]. Post-hoc analysis revealed that this sex effect is due to greater Bmal1 mRNA expression in females compared to males at ZT0 and ZT20 in the CEA (greater peak levels in females), and ZT0, ZT8, and ZT20 in the MEA (greater peak and trough levels in females).
Figure 1.

A) 24-h rhythmic expression of Per1, Per2, and Bmal1 mRNA in the SCN and PVN. There was an overall significant ZT effect (two-way ANOVA, p < 0.05) and 24-h rhythm (cosinor analysis, p < 0.05) for all clock genes in both the SCN and PVN of both males and females (n=8 for each sex and ZT time, except n = 4 for SCN Per2 mRNA). Female profiles are denoted by filled circles connected by a solid line; male profiles are denoted by open squares connected by a dashed line. B) Representative autoradiographic images were taken from the diurnal peak and trough of each clock gene’s rhythmic expression in the SCN and PVN (brain regions of interest denoted within black rectangle).
Figure 2.

24-h rhythmic expression of Per1, Per2, and Bmal1 mRNA in the anterior cingulate (AC), prelimbic (PL), infralimbic (IL), and ventral orbital (VO) subregions of the prefrontal cortex, as well as the rostral agranular insula. There was an overall significant ZT effect for all clock genes across all subregions of the PFC and insula (two-way ANOVA, p < 0.05; n=8 for each sex and ZT time). Although there was a significant 24-h rhythm of each clock gene for males in all brain regions (cosinor analysis, p < 0.05), females failed to have a significant rhythm for Per1 in the PL and IL subregions, and for Per2 in the IL. Female profiles are denoted by filled circles connected by a solid line; male profiles are denoted by open squares connected by a dashed line.
Figure 3.
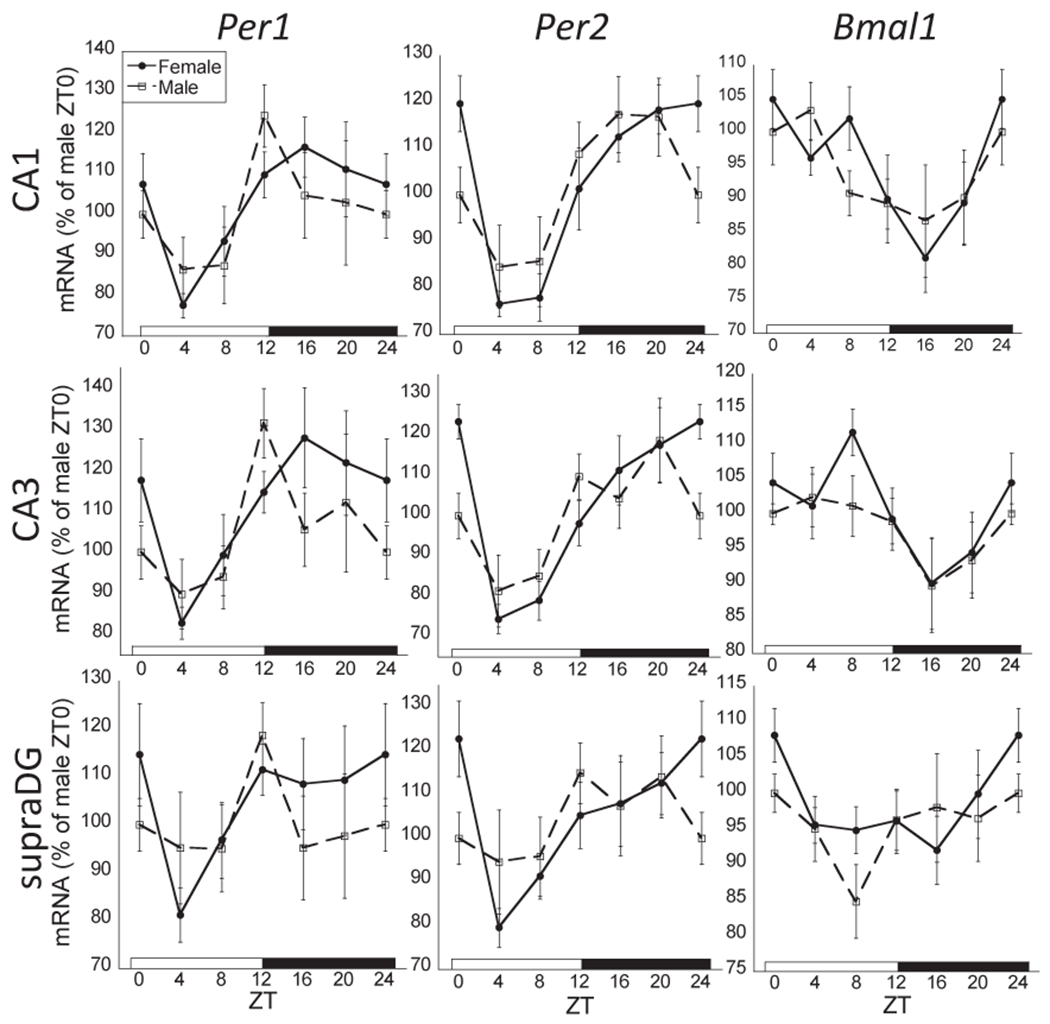
24-h rhythmic expression of Per1, Per2, and Bmal1 mRNA in the CA1 and CA3 subregions of the hippocampus in male and female rats. There was an overall significant ZT effect for all clock genes in the CA1 and CA3 (two-way ANOVA, p < 0.05; n=8 for each sex and ZT time). Whereas females had significant rhythmic expression of each clock gene in both CA1 and CA3 (cosinor analysis, p < 0.05), males failed to have significant rhythmic Per1 and Bmal1 mRNA expression in the CA1 and CA3. All clock genes failed to exhibit rhythmic expression in the inferior (not pictured) and superior blades of the dentate gyrus (ZT effect, 2-way ANOVA, p > 0.05), however, Per2 mRNA expression was rhythmic in the supra DG for females (cosinor analysis, p < 0.05). Female profiles are denoted by filled circles connected by a solid line; male profiles are denoted by open squares connected by a dashed line.
Figure 4.

24-h rhythmic expression of Per1, Per2, and Bmal1 mRNA in the amygdala. There was an overall significant ZT effect for all clock genes in the BLA, MEA, and CEA (two-way ANOVA, p < 0.05; n=8 for each sex and ZT time). There was also a significant sex effect for Bmal1 mRNA in the CEA and MEA (two-way ANOVA, p < 0.05; *p < 0.05, FLSD). Cosinor analysis found all clock genes to be rhythmic in the BLA for both males and females (p < 0.05). In the CEA, cosinor analysis found Bmal1 mRNA rhythmic for both males and females (p < 0.05), and Per2 mRNA rhythmic in females (p < 0.05) with males trending toward rhythmic expression (p = 0.06). In the MEA, only Bmal1 mRNA was rhythmic for both males and females (p < 0.05). Female profiles are denoted by filled circles connected by a solid line; male profiles are denoted by open squares connected by a dashed line.
Some brain regions examined lacked a significant 24-h rhythm of clock gene expression for either sex [Table 1]. Specifically, there was an absence of rhythmic Per1 and Bmal1 mRNA in the dentate gyrus (DG). Although there was a significant ZT effect for Per1 mRNA in all three amygdala subdivisions, only in the BLA did that expression have a significant 24-h rhythm. Similarly, there was a significant ZT effect for Per1 and Per2 mRNA in the MEA, but that expression did not have a significant 24-h rhythm.
Clock Gene Analysis – Acrophase Comparison Within and Across Brain Regions
The 24-h rhythmic expression of Bmal1, Per1, and Per2 mRNA in the brain regions examined revealed three distinct composite profiles, suggesting that the molecular clock varies in its overall phase relationship between different brain regions. The three main clock profiles are: 1) Bmal1 mRNA acrophase in the dark phase with Per1/Per2 mRNA antiphasic to Bmal1 mRNA; 2) Bmal1 mRNA acrophase in the light phase with Per1/Per2 mRNA antiphasic to Bmal1 mRNA; 3) Bmal1 mRNA acrophase around the transition from dark to light phase with Per1/Per2 mRNA acrophase in the early/mid dark phase [Fig. 5].
Figure 5.

Plot of acrophases (± 95% CI) for Per1, Bmal1, and Per2 mRNA in all brain regions examined in males and females separately (A-C) and males and females combined (D-F). There were 3 distinct molecular clock profiles: 1) Bmal1 mRNA acrophase in the dark phase with Per1/Per2 mRNA antiphasic to Bmal1 mRNA (A,D), 2) Bmal1 mRNA acrophase in the light phase with Per1/Per2 mRNA antiphasic to Bmal1 mRNA (B, E), and 3) Bmal1 mRNA acrophase around the transition from dark to light phase with Per1/Per2 mRNA acrophase in the early/mid dark phase (C,F). The Bmal1 mRNA acrophase clusters for each of the 3 distinct profiles are highlighted within black rectangles. Acrophase estimates are for rhythms with a significant 24-h period, p < 0.05; cosinor analysis (NS = not significant).
Bmal1 mRNA acrophase in the dark phase with Per1/Per2 mRNA antiphasic to Bmal1 mRNA
The SCN and the CEA both had Bmal1 mRNA acrophase at ~ZT18. In the SCN, the acrophase of Per1 mRNA was antiphasic to Bmal1 mRNA and occurred at ~ZT5 [Fig 5]. The acrophase of Per2 mRNA expression occurred at ~ZT9, between that of Per1 and Bmal1 mRNA. This is the only brain region examined in which the acrophase of Per2 mRNA was significantly different than that of Per1 mRNA (no overlap of 95% CIs). In the CEA, Per1 mRNA failed to be rhythmic, but Per2 mRNA was rhythmic and antiphasic (acrophase ~ZT4) to Bmal1 mRNA expression. Considered together, both the SCN and CEA had a Bmal1 mRNA acrophase in the dark phase and an antiphasic acrophase of a negative component of the molecular clock (either Per1 or Per2 mRNA).
Bmal1 mRNA acrophase in the light phase with Per1/Per2 mRNA antiphasic to Bmal1 mRNA
The PVN, insula, VO, and the CA1 and CA3 subregions of the hippocampus had a Bmal1 mRNA acrophase in the beginning of the light phase [Fig 5]. In these brain areas, both Per1 and Per2 mRNA expression had an acrophase (~ZT13-18) that was approximately antiphasic to the Bmal1 mRNA acrophase (~ZT0-5). It is noteworthy that the acrophases for Bmal1 and Per1 mRNA in these brain areas were also anti-phasic to their respective acrophases in the SCN, demonstrating distinct patterns of rhythmicity between the two hypothalamic nuclei. In these brain regions, the acrophases for Per1 and Per2 mRNA were similar to each other.
Bmal1 mRNA acrophase at the transition from dark to light with Per1/Per2 mRNA acrophase during the early-mid dark phase
Subregions of the medial PFC (AC, PL, IL), the BLA, and the MEA had an acrophase of Bmal1 mRNA around the late dark phase (ZT20-24), and an acrophase of Per1 and Per2 mRNA during the early to mid dark phase (ZT11-16) [Fig 5]. The acrophases of Per1 and Per2 mRNA of these brain regions were similar to the Per1 and Per2 mRNA acrophases seen in the PVN, insula, VO, and the CA1 and CA3 subregions of the hippocampus. The exception is the MEA, which lacked 24-h rhythmic expression of both Per1 and Per2.
Throughout nearly all brain regions examined, there was a distinct trend for the acrophase of each of the three clock genes to be slightly phase-delayed in females compared to males, although within any one brain region this sex difference was not statistically significant [Fig 5].
Rhythmic Clock Gene Amplitude Comparison Across Brain Regions
There was robust Per1, Per2, and Bmal1 mRNA expression in the SCN and PVN, with relatively large amplitudes for each 24-h rhythm [Fig 6]. Consistent with other reports (Harbour et al., 2013, 2014; Girotti et al., 2009), in these two brain regions there was a larger amplitude of Per1 and Per2 mRNA 24-h rhythms than there was for Bmal1 mRNA. Notably, the amplitudes of the 24-h rhythms of Bmal1 and Per2 mRNA were lower in all other brain regions examined. The amplitude of Per1 mRNA 24-h rhythm was somewhat lower in the PFC subregions and insula compared to the SCN and PVN, and was distinctly lower in the hippocampus and amygdala.
Figure 6.

Plot of amplitudes (± 95% CI) for the rhythmic expression of Per1, Bmal1, and Per2 mRNA in all brain regions examined in males and females combined. The SCN and PVN had the greatest amplitudes for Per1, Bmal1 and Per2 mRNA rhythmic expression compared to all other brain regions examined. In all cases where there was a significant 24 h rhythm for both sexes (Table 1, Fig. 6), there was no significant sex difference in amplitude. Brain regions of interest are arranged in rostral to caudal order on the x-axis. Instances where there was not a significant rhythm have been marked as not significant (NS).
Experiment 2: Basal clock gene expression throughout the brain of normally cycling versus non-cycling female rats
Estrous Cyclicity and CORT levels
A large percentage of female rats (43%) in this experiment were considered non-cyclers. This lack of cycling could be due to some cases of vaginal lavage-induced pseudopregnancy (Becker et al., 2005), but perhaps largely due to the Lee-Boot effect, as females were pair-housed and in rooms with only females (Van der Lee & Boot, 1955; 1956). Similar to Experiment 1, females had a large diurnal rhythm in basal plasma CORT levels (two-way ANOVA and cosinor analysis, p < 0.05) with an acrophase of 11.2±1.7 95% CI. There was no effect of cyclicity or ZT by cyclicity interaction.
Clock gene expression
Two-way ANOVA and cosinor analysis found similar acrophases and amplitudes for 24-h rhythms of Bmal1 and Per2 mRNA expression as in Experiment 1 [Table 2 & Supplemental Table 1]. Per1 mRNA failed to have a significant main effect of ZT, with the SCN and PVN as the exceptions. Similar to the profile for females in Experiment 1, Per1 and Per2 expression in the prelimibc cortex (PL) and infralimbic cortex (IL) failed to be rhythmic, suggestive of an attenuated amplitude in females compared to males. There was a significant cyclicity × ZT interaction for Bmal1 mRNA expression in the AC (F(5,62) = 2.8, p = 0.02), PL (F(5,62) = 2.8, p = 0.02), IL (F(5,62) = 2.6, p = 0.03), and VO (F (5,62) = 2.6, p = 0.03), where non-cyclers had more robust 24-h Bmal1 expression compared to cyclers [Fig 7]. Bmal1 mRNA expression in the subregions of the PFC were the only instances where there was a significant cyclicity by ZT interaction [Table 2]. Examination of cosinor analysis revealed that while both non-cyclers and cyclers had rhythmic Bmal1 mRNA expression in the PFC, cyclers had a blunted rhythm (smaller amplitude) compared to non-cyclers. In addition to a blunted amplitude, cyclers also had a significant difference (cosinor analysis) in acrophase of Bmal1 mRNA rhythm in the PL and IL subregions. When including all of these brain regions in an ANOVA analysis that treated brain region as a repeated measure factor, there was a significant cyclicity by ZT interaction (F5,62 = 2.9; p = 0.02) [Fig 7].
Table 2.
Experiment 2: Statistical analysis of basal Per1, Per2, and Bmal1 mRNA throughout the brain.
| Cosinor Analysis (× = p<0.05) | ||||||
|---|---|---|---|---|---|---|
| ZT | Cyclicity | Cyclicity × ZT | Acyclic | Cyclic | ||
| per1 | SCN | F(5,63) = 30.5** | F(1,63) = 0.4 | F(5,63) = 2.3 | × | × |
| PVN | F(5,63) = 5.1* | F(1,63) = 2.8 | F(5,63) = 0.6 | × | × | |
| AC | F(5,62) = 0.6 | F(1,62) = 0.3 | F(5,62) = 1.4 | |||
| PL | F(5,62) = 0.5 | F(1,62) = 0.9 | F(5,62) = 1.6 | |||
| IL | F(5,62) = 0.6 | F(1,62) = 0.7 | F(5,62) = 1.8 | |||
| VO | F(5,62) = 0.9 | F(1,62) = 1.0 | F(5,62) = 1.5 | × | ||
| Insula | F(5,62) =1.2 | F(1,62) = 0.1 | F(5,62) = 1.4 | × | ||
| CA1 | F(5,63) = 0.8 | F(1,63) = 0.4 | F(5,63) = 1.8 | |||
| CA3 | F(5,63) = 0.6 | F(1,63) = 0.3 | F(5,63) = 1.2 | |||
| supraDG | F(5,63) =1.0 | F(1,63) = 0.9 | F(5,63) = 1.4 | |||
| infraDG | F(5,63) = 1.1 | F(1,63) = 1.7 | F(5,63) = 1.5 | |||
| CEA | F(5,62) = 0.8 | F(1,62) = 0.0 | F(5.62) = 1.4 | |||
| BLA | F(5,62) = 2.2 | F(1,62) = 0.0 | F(5,62) = 1.9 | × | ||
| MEA | F(5,62) = 1.5 | F(1,62) = 0.0 | F(5,62) = 1.5 | |||
| Bmal | SCN | F(5,63) = 4.6* | F(1,63) = 1.3 | F(5,63) = 1.4 | 0.07 | × |
| PVN | F(5,63) = 3.6* | F(1,63) = 0.1 | F(5,63) = 1.2 | × | ||
| AC | F(5,62) = 7.0** | F(1,62) = 4.6* | F(5,62) = 2.8* | × | × | |
| PL | F(5,62) = 4.7* | F(1,62) = 2.3 | F(5,62) = 2.8* | × | × | |
| IL | F(5,62) = 2.1 | F(1,62) = 4.3* | F(5,62) = 2.6* | × | × | |
| VO | F(5,62) = 6.3** | F(1,62) = 2.4 | F(5,62) = 2.6* | × | × | |
| Insula | F(5,62) = 10.97** | F(1,62) = 4.1* | F(5,62) = 2.1 | × | × | |
| CA1 | F(5,63) = 1.9 | F(1,63) = 0.7 | F(5,63) = 0.5 | |||
| CA3 | F(5,63) = 2.2 | F(1,63) = 0.1 | F(5,63) = 0.5 | |||
| supraDG | F(5,63) = 1.2 | F(1,63) = 0.3 | F(5,63) = 0.2 | |||
| infraDG | F(5,63) = 0.8 | F(1,63) = 0.0 | F(5,63) = 0.3 | |||
| CEA | F(5,62) = 2.6 | F(1,62) = 0.5 | F(5,62) = 0.4 | × | ||
| BLA | F(5,62) = 44* | F(1,62) = 0.8 | F(5,62) = 0.4 | × | ||
| MEA | F(5,62) = 6.0** | F(1,62) = 0.1 | F(5,62) = 1.5 | × | ||
| Per2 | SCN | F(5,63) = 34.6** | F(1,63) = 0.1 | F(5,63) = 0.7 | × | × |
| PVN | F(5,63) = 10.1** | F(1,63) = 0.1 | F(5,63) = 1.1 | × | × | |
| AC | F(5,62) = 4.0* | F(1,62) = 1.0 | F(5,62) = 0.3 | × | × | |
| PL | F(5,62) = 0.7 | F(1,62) = 0.4 | F(5,62) = 0.3 | |||
| IL | F(5,62) = 0.8 | F(1,62) = 0.8 | F(5,62) = 0.4 | |||
| VO | F(5,62) = 5.7** | F(1,62) = 0.5 | F(5,62) = 0.4 | × | × | |
| Insula | F(5,62) = 7.2 | F(1,62) = 0.5 | F(5,62) = 0.3 | × | × | |
| CA1 | F(5,62) =1.8 | F(1,62) = 0.0 | F(5,62) = 1.5 | |||
| CA3 | F(5,62) =1.6 | F(1,62) = 0.3 | F(5,62) = 1.4 | × | ||
| supraDG | F(5,62) =0.7 | F(1,62) = 0.0 | F(5,62) = 1.9 | |||
| infraDG | F(5,62) =1.3 | F(1,62) = 0.0 | F(5,62) = 2.0 | |||
| CEA | F(5,62) = 3.9* | F(1,62) = 0.3 | F(5,62) = 1.5 | × | ||
| BLA | F(5,62) = 0.7 | F(1,62) = 0.2 | F(5,62) = 0.5 | |||
| MEA | F(5,62) = 0.5 | F(1,62) = 0.0 | F(5,62) = 1.0 | |||
| CORT | F(5,63) = 5.5** | F(1,63) = 0.0 | F(5,63) = 0.4 | × | × | |
Two-way ANOVA results for effect of time of day (ZT), estrous cyclicity, and their interaction,
p < 0.05,
p < 0.001.
Cosinor analysis test for 24 h rhythmicity was analyzed separately for female rats that displayed a regular estrous cycle (Cyclic; n=5-10 per ZT time; N=42) and those that did not (Acyclic; n=2-8, per ZT time; N=32).
Figure 7.

Comparison of Per1, Per2 and Bmal1 mRNA in the SCN, PL, and IL of female rats separated according to estrous cyclicity status. Note that there was a blunted rhythm and significant phase-advance for Bmal1 mRNA in the PL and IL, but not SCN, of female rats that had regular estrous cycles (cyclic; n=5-10 per ZT time; N=42) compared to female rats with less regular estrous cycles (acyclic; n=2-8 per ZT time; N=32) (Significant cyclicity status by ZT interaction, two-way ANOVA, p < 0.05). There were also significant differences of cyclicity status on Bmal1 mRNA at ZT0 (PL, IL) and ZT4 (IL) (two-way ANOVA, FLSD, *p < 0.05). Cyclicity status had no significant effect on clock gene expression in the SCN. Per1 and Per2 mRNA were not rhythmic in the PL and IL (p > 0.05). Acyclic profiles are denoted by filled circles connected by a solid line; cyclic profiles are denoted by open squares connected by a dashed line.
DISCUSSION
We found a 24-h rhythmic expression profile for both positive (Bmal1) and negative (Per1, Per2) regulatory clock genes across a range of brain structures that are important for emotion-related learning and control. The phase relationship between the expression of these clock genes varied within and between brain regions with three distinct profiles evident. Overall, these rhythms were similar between male and female rats. However, in female rats, the acrophase of all clock genes in nearly all brain regions examined was slightly phase-delayed compared to male rats. There were also some brain region specific sex differences in the robustness of rhythmic clock gene expression. In addition, the PFC had less robust rhythmic clock gene expression in the composite data of female rats at various phases of the estrous cycle compared to females that were not cycling.
Three Distinct Molecular Clock Profiles
The molecular clock in the SCN represents one of the distinct molecular clocks found in this study – Bmal1 mRNA acrophase in the dark phase with antiphasic expression of Per1/Per2 mRNA. These acrophases matched that of previous studies seen in the SCN (Dunlap, 1999; Girotti et al., 2009, Harbour et al., 2014). This was the only tissue examined where there was a significant difference in the acrophase of Per1 and Per2 mRNA with a ~4-h phase-delay of Per2 mRNA relative to Per1 mRNA, as previously reported in several different rodent species (Albrecht et al., 1997; Vosko et al., 2009). The CEA was the only extra-SCN brain region examined in this study that had a phase relationship for the positive and negative clock gene components similar to the SCN. Per2 mRNA was rhythmic in the CEA and its acrophase was antiphasic to the acrophase of Bmal1 mRNA. Our results are consistent with the previous finding that PER2 protein diurnal expression in the CEA has a similar, although somewhat advanced phase relationship with PER2 protein expression in the SCN, which differs from other brain regions (BLA, DG) (Harbour et al., 2013; Lamont et al., 2005). Our results are also in close agreement with the recent finding that Bmal1 and Per2 mRNA in the CEA and SCN have similar phase-relationships, with acrophase estimates very close to ours (Harbour et al., 2014). The CEA integrates neural input to produce expression or inhibition of fear (LeDoux et al., 1988; Wilensky et al., 2006). Circadian variations have been observed in conditioned fear expression and conditioned fear extinction memory (Chaudhury & Colwell, 2002; Eckel-Mahan et al., 2008; Valentinuzzi et al., 2001; Woodruff et al, 2015; Pace-Schott et al., 2013). Whether the shared phase-relationship of clock gene expression between the SCN and CEA has a functional significance for circadian modulation of conditioned fear remains to be determined. The SCN and CEA are comprised predominantly of GABAergic neurons (Sun & Cassell, 1993; Sun et al., 1994; Wagner et al., 1997), which may contribute to the somewhat unique circadian clock relationship of those two brain regions.
The second distinct molecular clock observed was characterized by a Bmal1 mRNA acrophase during the light phase with anti-phasic expression of Per1/Per2 mRNA. This was evident in the PVN, the CA1 and CA3 subregions of the hippocampus, the VO, and the rostral insula. In the PVN there were no sex differences in acrophase or amplitude of any of the clock genes despite a robust sex difference seen in the amplitude of diurnal CORT levels. In the hippocampal DG, the only significant clock gene expression rhythm that we observed was in female rats (Per2 mRNA), consistent with the more robust CA1 and CA3 rhythms evident in female rats compared to males [Fig 3 & 5]. Despite the overall lack of statistical significance, the general diurnal expression profile for each clock gene in the DG was similar to that seen in the CA1 and CA3 [Fig 3]. Previous studies have shown rhythmic clock gene expression in rodent DG (Feillet et al., 2008; Harbour et al., 2014; Lamont et al., 2005; Gilhooley et al., 2011), whereas others failed to see rhythmic Per1 or Per2 mRNA in DG or whole hippocampus of rodents (Shieh et al., 2005). Wang et al. (2009) observed rhythmic but blunted amplitude of Per2 mRNA and PER2 protein in the DG compared to CA1, CA2, and CA3 of mouse hippocampus. Consequently, there may be some rhythmic clock gene expression in the DG with an expression profile similar, but less robust, to the profile in the CA1 and CA3. Performance on hippocampal-dependent learning and memory tasks has been shown to have diurnal differences (Eckel-Mahan et al., 2008; Smarr et al., 2014), possibly reflecting rhythmic clock gene expression in the hippocampus.
The third distinct molecular clock observed had an acrophase of Bmal1 mRNA that occurred at the transition from the dark to the light phase and acrophases of Per1 and Per2 mRNA that occurred during the early to mid dark phase. Subregions of the medial PFC (AC, PL, IL) and BLA shared this molecular clock. This is the first study to show rhythmic expression of these clock genes throughout the mPFC. A key feature of this third molecular clock profile is the lack of fully anti-phasic rhythms between Bmal1 and Per1/Per2 mRNA due to the distinct acrophase of Bmal1 compared to other brain regions. This lack of anti-phasic expression may be due to phenotypic variations in clock gene mRNA or protein half-lives, resulting in an anti-phasic molecular clock evident primarily at the protein level. Additionally, CRY in these tissues may play a bigger role in the negative regulation of Bmal1 mRNA expression than PER1 or PER2 (Shearman, 2000). It should also be noted that in the SCN, only the rhythmic expression of Per1 mRNA, not Per2 mRNA, is fully anti-phasic to Bmal1 mRNA.
Possible Factors Contributing to Tissue Differences in the Molecular Clock
The 24-h rhythmic expression of Per1 and Per2 mRNA in the brain regions examined had two distinct acrophases: in the mid-light phase or in the early to mid-dark phase. The timing/entrainment of these rhythms may be related to the presence of a glucocorticoid response element (GRE) and cAMP response element (CRE) in the promoter region of both genes (Colwell, 2011; So et al., 2009; Yamamoto et al., 2005). In the SCN, light increases activation of cAMP response element binding protein (CREB), which results in rapid increase of Per1, and somewhat delayed Per2 transcription (Miyake et al., 2000; Shearman, 2000; Tischkau, 2002; Zylka et al., 1998). Thus, it may be expected that the acrophase of Per1 and Per2 expression occurs during the light phase in the SCN. In all other brain regions examined except the CEA, the acrophase of Per1 and Per2 mRNA occurred during the first half of the dark phase. The increased nocturnal physical activity and general arousal during the dark phase is associated with increased neural activation, which could increase Per1 and Per2 expression via CREB activation. Interestingly, in humans and a diurnal rodent (degu) Per1/2 gene expression in extra-SCN brain regions has been found to have an acrophase during the light phase (Li et al., 2013; Vosko et al., 2009).
Glucocorticoids have been shown to rapidly induce Per1 mRNA in fibroblasts, liver, and hippocampus, and Per2 mRNA in mesenchymal stem cells, likely via the GRE associated with these genes’ promoter region (Balsalobre, 2000; Conway-Campbell et al., 2010; So et al., 2009; T. Yamamoto et al., 2005). Peak Per1 and Per2 mRNA levels seen during the early dark phase in extra-SCN brain regions could be influenced by the daily peak in circulating CORT levels present at the onset of the dark phase. The diurnal CORT peak may contribute to daily GRE-mediated induction of Per1 and Per2 mRNA expression and act as an entrainment factor for extra-SCN clocks (Balsalobre et al., 2000; Pezük et al., 2012; Segall et al. 2006; 2009). There is very little GR expression in the rodent SCN (Balsalobre et al., 2000; Rosenfeld et al., 1988; Rosenfeld et al., 1992). Thus, Per1 and Per2 mRNA expression in the SCN would not be subject to this daily entraining influence.
While there were two distinct acrophases for Per1 and Per2 mRNA evident in the brain regions examined, there were three distinct acrophases for Bmal1 mRNA. This suggests that the phase relationship of rhythmic Bmal1 mRNA is not solely determined by Per1/Per2 mRNA profiles. Retinoid-related orphan receptor (ROR) is a protein that positively induces Bmal1 transcription (Sato et al., 2004). There are tissue variations in the expressed isoforms of ROR that may differentially modulate the oscillatory relationship between Bmal1 gene expression and the other molecular clock elements (Emery & Reppert, 2004).
Tissue Comparison of the Molecular Clock Amplitude
Overall, there was a robust amplitude of rhythmic Per1, Per2, and Bmal1 mRNA in the SCN and PVN of male and female rats, with no differences between the sexes. Clock gene expression in all other brain regions, while still rhythmic, had attenuated amplitudes compared to the SCN and PVN. These other brain regions may have less intercellular synchronization compared to the SCN and PVN. Increased synchronization could be due to a greater shared phenotypic response to the entraining influence of local neural, paracrine or hormonal factors. It is also possible that clock gene rhythmic expression patterns (phase, amplitude) may differ between cell types within some brain regions, resulting in a blunted composite amplitude at the tissue level.
Gonadal Steroid Modulation of the Molecular Clock
There were small sex differences in the robustness of rhythmic clock gene expression in certain extra-SCN brain regions. In females, there were less robust Per1 and Per2 rhythms in the PL and IL, brain areas important in stress adaptation, fear extinction, and emotional control (Dalley et al., 2004; Jones et al., 2011; Quirk et al., 2006). This may reflect individual rat variations in rhythm parameters due to fluctuating gonadal hormone levels as data for female rats in Experiment 1 were pooled regardless of estrous cycle status. Perrin et al. (2006) demonstrated that PER2 protein amplitude changes depending on estrous cycle stage in the CEA and oval nucleus of the bed nucleus of the stria terminalis. There are sex differences in mPFC-modulated behaviors that rely on gonadal hormone status (Farrell et al., 2013; Fenton et al., 2014; Sutcliffe et al., 2007), and diurnal variations in performance of these tasks has also been shown in male rodents (Chaudhury & Colwell, 2002; Eckel-Mahan et al., 2008; Smarr et al., 2014; Woodruff et al., 2015). In contrast to the PFC, female rats exhibited more robust rhythmic Per1 and Bmal1 mRNA in the CA1 and CA3 hippocampal subregions compared to males, which in this brain region may be related to the greater amplitude of diurnal CORT levels in females [Supplemental Fig 1] (Atkinson & Waddell, 1997; Goel et al., 2014; Viau, 2002).
There was a distinct trend for the acrophase of female rats to be slightly phase-delayed compared to males for all clock genes in nearly all brain regions [Fig. 5], which may be due to the influence of gonadal hormones. Previous studies have found that administration of estradiol or progesterone to ovariectomized rats, whether acute or chronic, alters the acrophase of clock gene expression in some peripheral tissues and the SCN (He et al., 2007; Nakamura et al., 2001; 2005; 2010; Smith et al., 2010). In addition, ovariectomy has been found to increase Per1 transcriptional synchrony between cells in the SCN and decrease synchrony in some peripheral tissues (Murphy et al., 2013). We also observed significant sex differences in Bmal1 mRNA expression within the CEA and MEA. Females had greater peak Bmal1 mRNA expression in the CEA, and overall greater Bmal1 mRNA levels (mesor) in the MEA compared to males. The MEA has high concentrations of gonadal hormone receptors (Gray & Bingaman, 1996; Li et al., 1993; Merchenthaler et al., 2004; Simerly et al., 1990), and gonadal hormones may regulate overall Bmal1 mRNA expression levels in the amygdala subregions.
There was a trend for blunted aggregate clock gene rhythmic expression in females that have robust cycling of the estrous cycle compared to non-cyclers. This difference was predominantly evident for Bmal1 mRNA expression in the PFC where, in addition to a blunted amplitude, cycling females also had a slightly phase-advanced acrophase of Bmal1 mRNA compared to noncycling females [Fig 7]. This influence of an active estrous cycle may indicate that fluctuating gonadal hormone levels in female rats contributes to varying clock gene expression, resulting in blunted amplitude at the composite level. Our results are consistent with a study that found robust diurnal rhythm of PER2 protein in the CEA and the oval nucleus of the bed nucleus of the stria terminalis of ovariectomized female rats, whereas in non-ovariectomized rats, the amplitude of PER2 protein rhythm varied with estrous cycle phase (Perrin et al., 2006).
Concluding Summary
We found 24-h rhythmic expression of both positive and negative regulatory clock genes within a network of brain regions important for emotional regulation in male and female rats. However, there was diversity in acrophase of these molecular clocks that varied depending on brain region. In addition, females had a consistent and distinct trend to have acrophases slightly phase-delayed compared to males. There were also small differences in the robustness of rhythmic clock gene expression that depended on sex and estrous cycle status, which may be due to activational effects of gonadal hormones. Further research is necessary to determine the basis and functional significance of the brain region variations in molecular clock phase-relationships and how gonadal hormones may influence these molecular clocks. This knowledge may lead to a better understanding of the neurobiological basis of sex differences in risk for certain psychological disorders.
Supplementary Material
References
- Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, & Block GD. (2002). Circadian rhythms in isolated brain regions. Journal of Neuroscience, 22(1), 350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A (2014). Distributed circuits underlying anxiety. Frontiers in Behavioral Neuroscience, 8:112. doi: 10.3389/fnbeh.2014.00112/abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Sun ZS, Eichele G, & Lee CC. (1997). A differential response of two putative mammalian circadian regulators, mPer1 and mPer2, to light. Cell, 91(7), 1055–1064. [DOI] [PubMed] [Google Scholar]
- Amir S, Waddington Lamont E, Robinson B, & Stewart J. (2004). A Circadian Rhythm in the Expression of PERIOD2 Protein Reveals a Novel SCN-Controlled Oscillator in the Oval Nucleus of the Bed Nucleus of the Stria Terminalis. Journal of Neuroscience, 24(4), 781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson HC, & Waddell BJ. (1997). Circadian variation in basal plasma corticosterone and adrenocorticotropin in the rat: sexual dimorphism and changes across the estrous cycle. Endocrinology, 138(9), 3842–3848. [DOI] [PubMed] [Google Scholar]
- Ángeles-Castellanos M, Mendoza J, & Escobar C. (2007). Restricted feeding schedules phase shift daily rhythms of c-Fos and protein Per1 immunoreactivity in corticolimbic regions in rats. Neuroscience, 144(1), 344–355. [DOI] [PubMed] [Google Scholar]
- Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, & Weaver DR. (2001). Differential Functions of mPer1, mPer2, and mPer3 in the SCN Circadian Clock. Neuron, 30(2), 525–536. [DOI] [PubMed] [Google Scholar]
- Bailey M, & Silver R. (2014). Sex differences in circadian timing systems: implications for disease. Frontiers in Neuroendocrinology, 35(1), 111–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A (2000). Resetting of Circadian Time in Peripheral Tissues by Glucocorticoid Signaling. Science, 289(5488), 2344–2347. [DOI] [PubMed] [Google Scholar]
- Becker JB, Arnold AP, Berkley KJ, Blaustein JD, Eckel LA, Hampson E, Herman JP, Marts S, Sadee W, Steiner M, Taylor J, & Young E. (2005). Strategies and Methods for Research on Sex Differences in Brain and Behavior. Endocrinology, 146(4), 1650–1673. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, & Bradfield CA. (2000). Mop3 Is an Essential Component of the Master Circadian Pacemaker in Mammals. Cell, 103(7), 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney BG, Li JZ, Walsh DM, Stein R, Vawter MP, Cartagena P, Barchas JD, Schatzberg AF, Myers RM, Watson SJ, Akil H, Bunney WE. (2015). Circadian dysregulation of clock genes: clues to rapid treatments in major depressive disorder. Molecular Psychiatry, 20(1), 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney WE, & Bunney BG. (2000). Molecular clock genes in man and lower animals: possible implications for circadian abnormalities in depression. Neuropsychopharmacology, 22(4), 335–345. [DOI] [PubMed] [Google Scholar]
- Chaudhury D, & Colwell CS. (2002). Circadian modulation of learning and memory in fear-conditioned mice. Behavioural Brain Research, 133(1), 95–108. [DOI] [PubMed] [Google Scholar]
- Christianson JP, & Greenwood BN. (2014). Stress-protective neural circuits: not all roads lead through the prefrontal cortex. Stress, 17(1), 1–12. [DOI] [PubMed] [Google Scholar]
- Colwell CS. (2011). Linking neural activity and molecular oscillations in the SCN. Nature Reviews Neuroscience, 12(10), 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway-Campbell BL, Sarabdjitsingh RA, McKenna MA, Pooley JR, Kershaw YM, Meijer OC, de Kloet ER, & Lightman SL. (2010). Glucocorticoid Ultradian Rhythmicity Directs Cyclical Gene Pulsing of the Clock Gene Period 1 in Rat Hippocampus. Journal of Neuroendocrinology, 22(10), 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalley JW, Cardinal RN, & Robbins TW. (2004). Prefrontal executive and cognitive functions in rodents: neural and neurochemical substrates. Neuroscience & Biobehavioral Reviews, 28(7), 771–784. [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA. (1998). Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science (New York, NY), 280(5369), 1599–1603. [DOI] [PubMed] [Google Scholar]
- Del Casale A, Serata D, Rapinesi C, Kotzalidis GD, Angeletti G, Tatarelli R, Ferracuti S, Girardi P. (2013). Structural neuroimaging in patients with panic disorder: findings and limitations of recent studies. Psychiatria Danubina, 25(2), 108–114. [PubMed] [Google Scholar]
- Donnelly NA, Holtzman T, Rich PD, Nevado-Holgado AJ, Fernando ABP, Van Dijck G, Holzhammer T, Paul O, Ruther P, Paulsen O, Robbins TW, Dalley JW. (2014). Oscillatory activity in the medial prefrontal cortex and nucleus accumbens correlates with impulsivity and reward outcome. PLoS ONE, 9(10), e111300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevets WC, Savitz J, & Trimble M. (2008). The subgenual anterior cingulate cortex in mood disorders. CNS Spectrums, 13(8), 663–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC. (1999). Molecular bases for circadian clocks. Cell, 96(2), 271–290. [DOI] [PubMed] [Google Scholar]
- Dzirasa K, McGarity DL, Bhattacharya A, Kumar S, Takahashi JS, Dunson D, McClung CA, & Nicolelis MAL. (2011). Impaired Limbic Gamma Oscillatory Synchrony during Anxiety-Related Behavior in a Genetic Mouse Model of Bipolar Mania. Journal of Neuroscience, 31(17), 6449–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Phan T, Han S, Wang H, Chan GCK, Scheiner ZS, & Storm DR. (2008). Circadian oscillation of hippocampal MAPK activity and cAmp: implications for memory persistence. Nature Neuroscience, 11(9), 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P & Reppert SM. (2004). A Rhythmic Ror. Neuron, 43(4), 443–446. [DOI] [PubMed] [Google Scholar]
- Etain B, Milhiet V, Bellivier F, & Leboyer M. (2011). Genetics of circadian rhythms and mood spectrum disorders. European Neuropsychopharmacology, 21, S676–S682. [DOI] [PubMed] [Google Scholar]
- Farrell MR, Sengelaub DR, & Wellman CL. (2013). Sex differences and chronic stress effects on the neural circuitry underlying fear conditioning and extinction. Physiology & Behavior, 122, 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feillet CA, Mendoza J, Albrecht U, Pévet P, & Challet E. (2008). Forebrain oscillators ticking with different clock hands. Molecular and Cellular Neuroscience, 37(2), 209–221. [DOI] [PubMed] [Google Scholar]
- Fenton GE, Pollard AK, Halliday DM, Mason R, Bredy TW, & Stevenson CW. (2014). Persistent prelimbic cortex activity contributes to enhanced learned fear expression in females. Learning & Memory, 21(2), 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, & Weitz CJ. (1998). Role of the CLOCK Protein in the Mammalian Circadian Mechanism. Science, 280(5369), 1564–1569. [DOI] [PubMed] [Google Scholar]
- Gilhooley MJ, Pinnock SB, & Herbert J. (2011). Rhythmic expression of Per1 in the dentate gyrus is Supplementalressed by corticosterone: Implications for neurogenesis. Neuroscience Letters, 489(3), 177–181. [DOI] [PubMed] [Google Scholar]
- Ginsberg AB, Campeau S, Day HE, & Spencer RL. (2003). Acute glucocorticoid pretreatment Supplementalresses stress-induced hypothalamic-pituitary-adrenal axis hormone secretion and expression of corticotropin-releasing hormone hnRNA but does not affect c-fos mRNA or fos protein expression in the paraventricular nucleus of the hypothalamus. Journal of Neuroendocrinology, 15(11), 1075–1083. [DOI] [PubMed] [Google Scholar]
- Girotti M, Weinberg MS, & Spencer RL. (2009). Diurnal expression of functional and clock-related genes throughout the rat HPA axis: system-wide shifts in response to a restricted feeding schedule. AJP: Endocrinology and Metabolism, 296(4), E888–E897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel N, Workman JL, Lee TT, Innala L, & Viau V. (2014). Sex Differences in the HPA Axis. Compr Physiol, 4(3). 1121–1155. [DOI] [PubMed] [Google Scholar]
- Goldman JM, Murr AS, & Cooper RL. (2007). The rodent estrous cycle: characterization of vaginal cytology and its utility in toxicological studies. Birth Defects Research Part B: Developmental and Reproductive Toxicology, 80(2), 84–97. [DOI] [PubMed] [Google Scholar]
- Gray TS, & Bingaman EW. (1996). The amygdala: corticotropin-releasing factor, steroids, and stress. Critical Reviews in Neurobiology, 10(2), 155–168. [DOI] [PubMed] [Google Scholar]
- Guilding C, & Piggins HD. (2007). Challenging the omnipotence of the suprachiasmatic timekeeper: are circadian oscillators present throughout the mammalian brain? European Journal of Neuroscience, 25(11), 3195–3216. [DOI] [PubMed] [Google Scholar]
- Harbour VL, Weigl Y, Robinson B, & Amir S. (2013). Comprehensive mapping of regional expression of the clock protein PERIOD2 in the rat forebrain across the 24-h day. PLoS ONE, 8(10), e76391.doi: 10.1371/journal.pone.0076391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour VL, Weigl Y, Robinson B, & Amir S. (2014). Phase Differences in Expression of Circadian Clock Genes in the Central Nucleus of the Amygdala, Dentate Gyrus, and Suprachiasmatic Nucleus in the Rat. PLoS ONE, 9(7), e103309. doi: 10.1371/journal.pone.0103309.t001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings M, O'Neill JS, & Maywood ES. (2007). Circadian clocks: regulators of endocrine and metabolic rhythms. Journal of Endocrinology, 195(2), 187–198. [DOI] [PubMed] [Google Scholar]
- He PJ, Hirata M, Yamauchi N, & Hattori MA. (2007). Up-regulation of Per1 expression by estradiol and progesterone in the rat uterus. Journal of Endocrinology, 194(3), 511–519. [DOI] [PubMed] [Google Scholar]
- Jilg A, Lesny S, Peruzki N, Schwegler H, Selbach O, Dehghani F, & Stehle JH. (2010). Temporal dynamics of mouse hippocampal clock gene expression Supplementalort memory processing. Hippocampus, 20(3), 377–388. [DOI] [PubMed] [Google Scholar]
- Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, Kieseppä T, Lichtermann D, Praschak-Rieder N, Neumeister A, Nilsson L-G, Kasper S, Peltonen L, Adolfsson R, Schalling M, Partonen T. (2002). Circadian Clock-Related Polymorphisms in Seasonal Affective Disorder and their Relevance to Diurnal Preference. Neuropsychopharmacology, 28(4), 734–739. [DOI] [PubMed] [Google Scholar]
- Jones KR, Myers B, & Herman JP. (2011). Stimulation of the prelimbic cortex differentially modulates neuroendocrine responses to psychogenic and systemic stressors. Physiology & Behavior, 104(2), 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenigs M, & Grafman J. (2009). Posttraumatic Stress Disorder: The Role of Medial Prefrontal Cortex and Amygdala. The Neuroscientist, 15(5), 540–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont EW, Coutu DL, Cermakian N, & Boivin DB. (2009). Circadian rhythms and clock genes in psychotic disorders. The Israel Journal of Psychiatry and Related Sciences, 47(1), 27–35. [PubMed] [Google Scholar]
- Lamont EW, Robinson B, Stewart J, & Amir S. (2005). The central and basolateral nuclei of the amygdala exhibit opposite diurnal rhythms of expression of the clock protein Period2. Proceedings of the National Academy of Sciences, 102(11), 4180–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDoux JE, Iwata J, Cicchetti P, & Reis DJ. (1988). Different projections of the central amygdaloid nucleus mediate autonomic and behavioral correlates of conditioned fear. The Journal of Neuroscience, 8(7), 2517–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Blaustein JD, De Vries GJ, & Wade GN. (1993). Estrogen-receptor immunoreactivity in hamster brain: preoptic area, hypothalamus and amygdala. Brain Research, 631(2), 304–312. [DOI] [PubMed] [Google Scholar]
- Li JZ, Bunney BG, Meng F, Hagenauer MH, Walsh DM, Vawter MP, Evans SJ, Coudary PV, Cartagena P, Carchas JD, Schatzberg AF, Jones EG, Myers RM, Watson SJ, Akil H, Bunney WE. (2013). Circadian patterns of gene expression in the human brain and disruption in major depressive disorder. Proceedings of the National Academy of Sciences of the United States of America, 110(24), 9950–9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Peek CB, Affinati A, Maury E, & Bass J. (2013). Circadian Clocks and Metabolism. Handbook of Experimental Pharmacology, 217, 127–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcondes FK, Bianchi FJ, & Tanno AP. (2002). Determination of the estrous cycle phases of rats: some helpful considerations. Brazilian Journal of Biology, 62(4A), 609–614. [DOI] [PubMed] [Google Scholar]
- Masubuchi S, Honma S, Abe H, Ishizaki K, Namihira M, Ikeda M, & Honma K-I. (2000). Clock genes outside the suprachiasmatic nucleus involved in manifestation of locomotor activity rhythm in rats. European Journal of Neuroscience, 12(12), 4206–4214. [PubMed] [Google Scholar]
- Mayberg HS, Lozano AM, Voon V, McNeely HE, Seminowicz D, Hamani C, Schwalb JM, & Kennedy SH. (2005). Deep Brain Stimulation for Treatment-Resistant Depression. Neuron, 45(5), 651–660. [DOI] [PubMed] [Google Scholar]
- McCarthy MJ, & Welsh DK. (2012). Cellular Circadian Clocks in Mood Disorders. Journal of Biological Rhythms, 27(5), 339–352. [DOI] [PubMed] [Google Scholar]
- McClung CA. (2007). Circadian genes, rhythms and the biology of mood disorders. Pharmacology & Therapeutics, 114(2), 222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh JL. (2004). The Amygdala Modulates the Consolidation of Memories of Emotionally Arousing Experiences. Annual Review of Neuroscience, 27(1), 1–28. [DOI] [PubMed] [Google Scholar]
- Merchenthaler I, Lane MV, Numan S, & Dellovade TL. (2004). Distribution of estrogen receptor α and β in the mouse central nervous system: In vivo autoradiographic and immunocytochemical analyses. Journal of Comparative Neurology, 473(2), 270–291. [DOI] [PubMed] [Google Scholar]
- Miyake S, Sumi Y, Yan L, Takekida S, Fukuyama T, Ishida Y, Yamaguchi S, Yagita K, & Okamura H. (2000). Phase-dependent responses of Per1 and Per2 genes to a light-stimulus in the suprachiasmatic nucleus of the rat. Neuroscience Letters, 294(1), 41–44. [DOI] [PubMed] [Google Scholar]
- Mojón A, Fernández JR, & Hermida RC. (1992). Chronolab: An Interactive Software Package for Chronobiologic Time Series Analysis Written for the Macintosh Computer. Chronobiology Int, 9(6), 403–412. [DOI] [PubMed] [Google Scholar]
- Morin LP. (1980) Effect of ovarian hormones on synchrony of hamster circadian rhythms. Physiol Behav, 24(4), 741–749. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Coque L, Cao J-L, Kumar J, Chakravarty S, Asaithamby A, Graham A, Gordon E, Enwright JF 3rd, DiLeone RJ, Birnbaum SG, Cooper DC, & McClung CA. (2010). Knockdown of Clock in the Ventral Tegmental Area Through RNA Interference Results in a Mixed State of Mania and Depression-Like Behavior. Biological Psychiatry, 68(6), 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy ZC, Pezük P, Menaker M, & Sellix MT. (2013). Effects of ovarian hormones on internal circadian organization in rats. Biol of Reprod, 89(2), 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TJ, Moriya T, Inoue S, Shimazoe T, Watanabe S, Ebihara S, & Shinohara K. (2005). Estrogen differentially regulates expression ofPer1 andPer2 genes between central and peripheral clocks and between reproductive and nonreproductive tissues in female rats. Journal of Neuroscience Research, 82(5), 622–630. [DOI] [PubMed] [Google Scholar]
- Nakamura TJ, Sellix MT, Kudo T, Nakao N, Yoshimura T, Ebihara S, Colwell CS, & Block GD. (2010). Influence of the estrous cycle on clock gene expression in reproductive tissues: Effects of fluctuating ovarian steroid hormone levels. Steroids, 75(3), 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TJ, Shinohara K, Funabashi T, & Kimura F. (2001). Effect of estrogen on the expression of Cry1 and Cry2 mRNAs in the suprachiasmatic nucleus of female rats. Neuroscience, 41(3), 251–255. [DOI] [PubMed] [Google Scholar]
- Pace-Schott EF, Spencer RMC, Vijayakumar S, Ahmed NAK, Verga PW, Orr SP, Pitman RK, & Milad MR. (2013). Extinction of conditioned fear is better learned and recalled in the morning than in the evening. Journal of Psychiatric Research, 47(11), 1776–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partonen T, Treutlein J, Alpman A, Frank J, Johansson C, Depner M, Aron L, Rietschel M, Wellek S, Soronen P, Paunio T, Koch A, Chen P, Lathrop M, Adolfsson R, Persson M-L, Kasper S, Schalling M, Peltonen L, & Schumann G. (2007). Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Annals of Medicine, 39(3), 229–238. [DOI] [PubMed] [Google Scholar]
- Perrin JS, Segall LA, Harbour VL, Woodside B, & Amir S. (2006). The expression of the clock protein PER2 in the limbic forebrain is modulated by the estrous cycle. Proceedings of the National Academy of Sciences, 103(14), 5591–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezük P, Mohawk JA, Wang LA, & Menaker M. (2012). Glucocorticoids as entraining signals for peripheral circadian oscillators. Endocrinology, 153(10), 4775–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirk GJ, Garcia R, & González-Lima F. (2006). Prefrontal Mechanisms in Extinction of Conditioned Fear. Biological Psychiatry, 60(4), 337–343. [DOI] [PubMed] [Google Scholar]
- Rath MF, Rohde K, Fahrenkrug J, & Møller M. (2013). Circadian clock components in the rat neocortex: daily dynamics, localization and regulation. Brain Structure and Function, 218(2), 551–562. [DOI] [PubMed] [Google Scholar]
- Rath MF, Rovsing L, & Møller M. (2014). Circadian oscillators in the mouse brain: molecular clock components in the neocortex and cerebellar cortex. Cell and Tissue Research, 357(3), 743–755. [DOI] [PubMed] [Google Scholar]
- Reick M (2001). NPAS2: An Analog of Clock Operative in the Mammalian Forebrain. Science, 293(5529), 506–509. [DOI] [PubMed] [Google Scholar]
- Reppert SM, & Weaver DR. (2002). Coordination of circadian timing in mammals. Nature, 418(6901), 935–941. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P, Van Eekelen JA, Levine S, & De Kloet ER. (1988). Ontogeny of the type 2 glucocorticoid receptor in discrete rat brain regions: an immunocytochemical study. Brain Res, 470(1), 119–127. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P, Van Eekelen JA, Levine S, & De Kloet ER. (1993). Ontogeny of corticosteroid receptors in the brain. Cell Mol Neurobiol, 13(4), 295–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, & Ishida N. (1998). Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. The Journal of Biological Chemistry, 273(42), 27039–27042. [DOI] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, GitzGerald GA, Kay SA, & Hogenesch JB. (2004). A Functional Genomics Strategy Reveals Rora as a Component of the Mammalian Circadian Clock. Neuron, 43(4), 527–537. [DOI] [PubMed] [Google Scholar]
- Segall LA, Perrin JS, Walker CD, Stewart J, & Amir S. (2006). Glucocorticoid rhythms control the rhythm of expression of the clock protein, Period2, in the oval nucleus of the stria terminalis and central nucleus of the amygdala in rats. Neurosci Res, 140(3), 753–757. [DOI] [PubMed] [Google Scholar]
- Segall LA, Milet A, Tronche F, & Amir S. (2009). Brain glucocorticoid receptors are necessary for the rhythmic expression of the clock protein, PERIOD2, in the central extended amygdala in mice. Neurosci Letters, 457(1), 58–60. [DOI] [PubMed] [Google Scholar]
- Shearman LP. (2000). Interacting Molecular Loops in the Mammalian Circadian Clock. Science, 288(5468), 1013–1019. [DOI] [PubMed] [Google Scholar]
- Shieh KR, Yang S-C, Lu X-Y, Akil H, & Watson SJ. (2005). Diurnal rhythmic expression of the rhythm-related genes, rPeriod1, rPeriod2, and rClock, in the rat brain. Journal of Biomedical Science, 12, 209–217. [DOI] [PubMed] [Google Scholar]
- Simerly RB, Chang C, Muramatsu M, & Swanson LW. (1990). Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: an in situ hybridization study. The Journal of Comparative Neurology, 294(1), 76–95. [DOI] [PubMed] [Google Scholar]
- Smarr BL, Jennings KJ, Driscoll JR, & Kriegsfeld LJ. (2014). A time to remember: The role of circadian clocks in learning and memory. Behavioral Neuroscience, 128(3), 283–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BJ, Sutton GM, Wu X, Yu G, Goh BC, Hebert T, Pelled G, Gazit Z, Butler AA, & Gimble JM. (2010). Ovariectomy and genes encoding core circadian regulatory proteins in murine bone. Osteoporosis International, 22(5), 1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So AYL, Bernal TU, Pillsbury ML, Yamamoto KR, & Feldman BJ. (2009). Glucocorticoid regulation of the circadian clock modulates glucose homeostasis. Proceedings of the National Academy of Sciences, 106(41), 17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S, Falcon E, Kumar J, Krishnan V, Mukherjee S, Birnbaum SG, & McClung CA. (2012). Circadian genes Period 1and Period 2in the nucleus accumbens regulate anxiety-related behavior. European Journal of Neuroscience, 37(2), 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, & Cassell MD. (1993). Intrinsic GABAergic neurons in the rat central extended amygdala. Journal of Comparative Neurology, 330(3), 381–404. [DOI] [PubMed] [Google Scholar]
- Sun N, Yi H, & Cassell MD. (1994). Evidence for a GABAergic interface between cortical afferents and brainstem projection neurons in the rat central extended amygdala. The Journal of Comparative Neurology, 340(1), 43–64. [DOI] [PubMed] [Google Scholar]
- Sutcliffe JS, Marshall KM, & Neill JC. (2007). Influence of gender on working and spatial memory in the novel object recognition task in the rat. Behavioural Brain Research, 177(1), 117–125. [DOI] [PubMed] [Google Scholar]
- Takahashi JS, Hong H-K, Ko CH, & McDearmon EL. (2008). The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nature Reviews Genetics, 9(10), 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, Sakakida Y, Okumura K, Takashima N, & Okamura H. (1998). A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells, 3(3), 167–176. [DOI] [PubMed] [Google Scholar]
- Thomas EM, & Armstrong SM. (1989). Effect of ovariectomy and estradiol on unity of female rat circadian rhythms. The American Journal of Physiology, 257(5 Pt 2), R1241–50. [DOI] [PubMed] [Google Scholar]
- Tischkau SA, Mitchell JW, Tyan S-H, Buchanan GF, & Gillette MU. (2002). Ca2+/cAMP Response Element-binding Protein (CREB)-dependent Activation of Per1 Is Required for Light-induced Signaling in the Suprachiasmatic Nucleus Circadian Clock. Journal of Biological Chemistry, 278(2), 718–723. [DOI] [PubMed] [Google Scholar]
- Valentinuzzi VS, Kolker DE, Vitaterna MH, Ferrari EA, Takahashi JS, & Turek FW. (2001). Effect of circadian phase on context and cued fear conditioning in C57BL/6J mice. Animal Learning & Behavior, 29(2), 133–142. [Google Scholar]
- Van der Lee S, & Boot LM. (1955). Spontaneous pseudopregnancy in mice. Acta Physiologica Et Pharmacologica Neerlandica, 4(3), 442. [PubMed] [Google Scholar]
- Van der Lee S, & Boot LM. (1956). Spontaneous pseudopregnancy in mice. II. Acta Physiologica Et Pharmacologica Neerlandica, 5(2), 213–215. [PubMed] [Google Scholar]
- Viau V (2002). Functional cross‐talk between the hypothalamic‐pituitary‐gonadal and‐adrenal axes. Journal of Neuroendocrinology, 14(6), 506–513. [DOI] [PubMed] [Google Scholar]
- Vosko AM, Hagenauer MH, Hummer DL, & Lee TM. (2009). Period gene expression in the diurnal degu (Octodon degus) differs fro the nocturnal laboratory rat (Rattus norvegicus). Am J Physiol Regul Integr Comp Physiol, 296(2), R353–R361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Castel M, Gainer H, & Yarom Y. (1997). GABA in the mammalian suprachiasmatic nucleus and its role in diurnal rhythmicity. Nature, 387(6633), 598–603. [DOI] [PubMed] [Google Scholar]
- Wang LM, Dragich JM, Kudo T, Odom IH, Welsh DK, O'Dell TJ, & Colwell CS. (2009). Expression of the circadian clock gene Period2 in the hippocampus: possible implications for synaptic plasticity and learned behaviour. ASN Neuro, 1(3), pii: e00012. doi: 10.1042/AN20090020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilensky AE, Schafe GE, Kristensen MP, & LeDoux JE. (2006). Rethinking the Fear Circuit: The Central Nucleus of the Amygdala Is Required for the Acquisition, Consolidation, and Expression of Pavlovian Fear Conditioning. Journal of Neuroscience, 26(48), 12387–12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff ER, Greenwood BN, Chun LE, Fardi S, Hinds LR, & Spencer RL. (2015). Adrenal-dependent diurnal modulation of conditioned fear extinction learning. Behavioural Brain Research, 286, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Soma H, Akashi M, Mamine T, & Takumi T. (2004). Transcriptional oscillation of canonical clock genes in mouse peripheral tissues. BMC Molecular Biology, 5(1), 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Nakahata Y, Tanaka M, Yoshida M, Soma H, Shinohara K, Yasuda A, Mamine T, & Takumi T. (2005). Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. Journal of Biological Chemistry, 280(51), 42036–42043. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, & Tei H. (2000). Resetting central and peripheral circadian oscillators in transgenic rats. Science (New York, NY), 288(5466), 682–685. [DOI] [PubMed] [Google Scholar]
- Yang S-T, Shi Y, Wang Q, Peng J-Y, & Li B-M (2014). Neuronal representation of working memory in the medial prefrontal cortex of rats. Molecular Brain, 7, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Shearman LP, Weaver DR, & Reppert SM. (1998). Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron, 20(6), 1103–1110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


