Abstract
Simple Summary
The use of next generation technologies has helped to unravel the genetics of rare inherited diseases, facilitating the discovery of new susceptibility genes. Nonetheless, the sequencing of all protein-coding genes of an individual may lead to doubtful assignments of causality for non-pathological variants, so it is mandatory to perform comprehensive studies and screening of additional patients for new mutations. Here we describe a novel DNMT3A germline variant identified by whole-exome sequencing in a patient with multiple paragangliomas and papillary thyroid carcinoma. The increased methylation of DNMT3A target genes observed in the proband’s sample points towards a gain-of-function effect of the variant, contrasting with the inactivation caused by loss-of-function alterations commonly seen in other neoplasia and in patients with Tatton-Brown-Rahman syndrome. This finding stresses the diverse molecular outcomes and suggests a heterogeneous phenotypic spectrum related to DNMT3A germline variants.
Abstract
Over the past few years, next generation technologies have been applied to unravel the genetics of rare inherited diseases, facilitating the discovery of new susceptibility genes. We recently found germline DNMT3A gain-of-function variants in two patients with head and neck paragangliomas causing a characteristic hypermethylated DNA profile. Here, whole-exome sequencing identifies a novel germline DNMT3A variant (p.Gly332Arg) in a patient with bilateral carotid paragangliomas, papillary thyroid carcinoma and idiopathic intellectual disability. The variant, located in the Pro-Trp-Trp-Pro (PWWP) domain of the protein involved in chromatin targeting, affects a residue mutated in papillary thyroid tumors and located between the two residues found mutated in microcephalic dwarfism patients. Structural modelling of the variant in the DNMT3A PWWP domain predicts that the interaction with H3K36me3 will be altered. An increased methylation of DNMT3A target genes, compatible with a gain-of-function effect of the alteration, was observed in saliva DNA from the proband and in one independent acute myeloid leukemia sample carrying the same p.Gly332Arg variant. Although further studies are needed to support a causal role of DNMT3A variants in paraganglioma, the description of a new DNMT3A alteration in a patient with multiple clinical features suggests a heterogeneous phenotypic spectrum related to DNMT3A germline variants.
Keywords: paraganglioma, DNMT3A, germline variant, papillary thyroid carcinoma
1. Introduction
The drip of new susceptibility genes involved in the development of pheochromocytoma (PCC) and paraganglioma (PGL), altogether PPGLs, has not ceased since the discovery of pathogenic variants in the succinate dehydrogenase (SDH) genes in the early 2000s. Thus, up to 35–40% of PPGL patients carry a germline variant in one of the almost twenty high—or low—penetrant susceptibility genes related to the disease described so far [1]. Although the clinical characteristics associated with alterations in the major susceptibility genes are well-known, most of the recently reported genes have been found mutated only in a few patients [2,3] and sometimes even in isolated cases [4]. In order to decipher the relevance of the new additions to this growing list of genes and to unravel specific genotype-phenotype associations, the description of additional cases is necessary.
One of the last members to be incorporated to the catalogue of genes associated with hereditary PPGL is the DNA methyltransferase 3 alpha (DNMT3A) gene [2]. Apart from a catalytically inactive regulatory orthologous, DNMT3L, mammals have two catalytically active DNA methyltransferases, DNMT3A and DNMT3B, in charge of the de novo generation of DNA methylation patterns during germ cell development and in the early embryonic phase [5,6]. Germline variants in DNMT3A are associated with developmental growth disorders, while somatic variants are frequently associated with malignancy. On the one hand, germline de novo loss-of-function variants in DNMT3A cause Tatton-Brown-Rahman syndrome (TBRS), an autosomal dominant condition characterized by overgrowth, intellectual disability and distinctive facial appearance [7]. On the other hand and despite not being identified until 2010 [8], somatic variants in DNMT3A are one of the most recurrent events across blood cancers of the myeloid lineage. Up to 22% of acute myeloid leukemia (AML) patients carry a somatic variant in DNMT3A [9,10] and these alterations have been associated with poor prognosis and adverse survival outcomes for AML patients. Interestingly, there is overlap of the spectrum of DNMT3A variants in TBRS and hematological malignancies [11], with some examples of both diseases occurring in the same patient [12,13]. Moreover, a germline DNMT3A loss-of-function variant has been found causing familial AML, in this case in the absence of TBRS symptoms [14].
We recently identified missense germline variants in DNMT3A in two patients with multiple head and neck PGLs [15]. Unlike what happens in patients with TBRS and in AML and other neoplasia, in which the gene is usually inactivated by loss-of-function alterations, we demonstrated that DNMT3A-mutated PGLs exhibited a significant overall methylation, indicating an activating role of the variants. A similar activation of DNMT3A was observed later on in patients with microcephalic dwarfism [16]. This duality has been also reported for the histone methyltransferase EZH2, in which somatic gain-of-function variants are frequently found in lymphoma [17,18], while inactivating germline alterations cause the Weaver overgrowth syndrome [19]. Herein, we describe and study an additional patient developing multiple head & neck PGLs and papillary thyroid carcinoma (PTC) who carries a new gain-of-function DNMT3A germline variant.
2. Results
2.1. Whole Exome Sequencing Analysis
The filtering process resulted in 541 variants, six of which were found in the public archive ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) (Table S1) and only one was found in a gene previously associated to PPGLs, DNMT3A (c.994G > A, p.Gly332Arg; VAF = 0.48). Interestingly, the only submission found in ClinVar regarding this variant was a de novo alteration identified in a patient with intellectual disability. The variant, subsequently validated by Sanger sequencing, was not found in 1942 population-matched unrelated Spanish exomes (http://csvs.babelomics.org/) and appeared in 3 of >140,000 exomes (allele frequency = 0.00001194) in the Genome Aggregation Consortium (gnomAD) database (http://gnomad.broadinstitute.org/). This frequency is below the frequency found for pathogenic variants in SDHB, a gene whose alterations have the lowest penetrance amongst those found in the known PPGL susceptibility genes. The Gly332 residue is located within the Pro-Trp-Trp-Pro (PWWP) domain of DNMT3A, described to function as a chromatin methylation reader by recognizing both DNA and histone methylated lysines. There are 31 missense variants in gnomAD, including p.Gly332Arg, located within the PWWP domain of DNMT3A, all of them with frequencies bellow 6.4 × 10−5 (Table S2).
2.2. Presence of DNMT3A p.Gly332Arg in Databases and In Silico Predictions
The PredictSNP1 consensus classifier predicted the change to be deleterious (87% expected accuracy), Varsome (https://varsome.com) considered it as likely pathogenic and its frequency in gnomAD was lower than the threshold for DNMT3A. Three different missense somatic variants have been reported in five independent samples in COSMIC (https://cancer.sanger.ac.uk/cosmic) or cBioPortal (https://www.cbioportal.org/) databases affecting the Gly332 residue (Figure 1a): p.Gly332Glu (n = 1; found in one clear cell renal cell carcinoma), p.Gly332Arg (n = 3; found in PTC, AML and essential thrombocythemia) and p.Gly332Val (n = 1; found in one PTC); although the causality of these variants has not been assessed.
Figure 1.
Missense variants and conservation of the PWWP domain of DNMT3A. (a) Representation of DNMT3A missense variants affecting the PWWP domain. Upper part, gain-of-function germline variants (denoted with a grey box) reported in patients with PPGL or microcephalic dwarfism. Bottom part, somatic variants reported in COSMIC (https://cancer.sanger.ac.uk/cosmic) and/or cBioPortal (www.cbioportal.org) databases affecting the same amino acids found mutated in the mentioned patients and/or affecting other residues involved in the aromatic cage [16,20] (denoted in red letters). (b) Multiple-sequence alignment of PWWP domains performed with Clustal Omega (European Bioinformatics Institute, Cambridge, UK). The alignment includes at least a member of each of the six classes of human PWWP domains since DNMT3A and DNMT3B share the same class (DNMT3B related) and PWWP3A and HDGF are also included in the same class (HDGF related) [20]. The position of DNMT3A-Gly332 is denoted with a green box.
Gly332 is conserved in four of the six classes of PWWP domains and the different residues found in this position have a weak similarity between them (Figure 1b). In addition, Gly332 is located in between Trp330 and Asp333, two pivotal residues for the aromatic cage of DNMT3A that recognizes tri-methylated lysine 36 of histone H3 (H3K36me3) (Figure 2). In the predicted structure of the PWWP domain of the DNMT3A Gly332Arg mutant, the position of the Arg lies opposite to the Phe303 residue and could form an additional side to the conserved aromatic cage formed by the amino acids Phe303, Trp306, Trp330 and Asp333. Thus, it is possible that an Arg residue in the 332 position could promote the stabilization of the binding to the trimethyl lysine present in histone H3.
Figure 2.
DNMT3A Gly332Arg mutant structure predicted with Phyre2. Trymethyl lysine (in yellow) is modelled from the DNMT3B structure (PDB code ID: 5CIU). The Arg332 mutant residue is colored in orange and the pivotal residues involved in the aromatic cage are colored in green and labeled.
2.3. Methylation Profiling
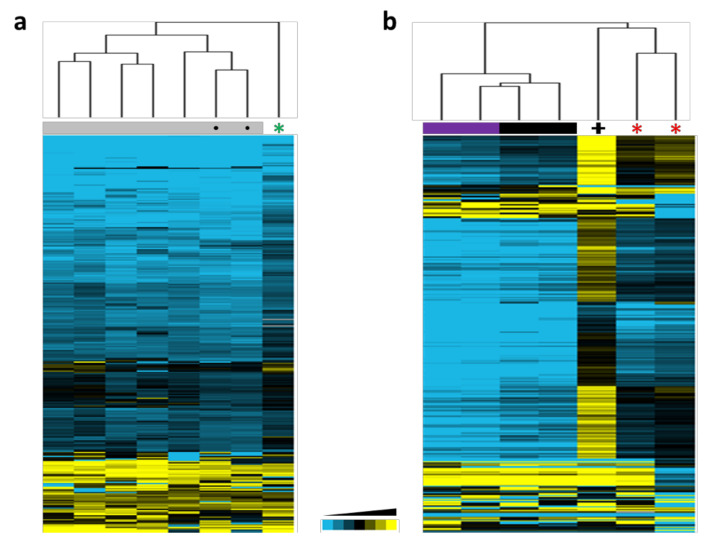
In order to obtain a representative methylation profile of DNMT3A-mutated tissues, we first got a list of 307 probes differentially methylated in DNMT3A-mutated tissues (Table S3). Then, we used this list to interrogate Illumina methylation data obtained from DNA saliva samples coming from the proband, two age/gender-matched controls and five healthy individuals. Hierarchical clustering grouped all the controls together and separated from the proband’s sample that showed a recognizable methylated profile (Figure 3a), suggesting that the p.Gly332Arg DNMT3A variant produces a similar alteration in germline global methylation than the observed for the previously reported PPGL patients.
Figure 3.
Hierarchical clusterings using methylation data from 307 probes found significantly differentially methylated between DNMT3A-mutated and non-mutated tissues in patients with PPGL [15]. (a) Unsupervised profiling using saliva DNA from the index patient (denoted with a green asterisk), in-home healthy controls (denoted with grey boxes and a dark circle) and healthy donors obtained from Gene Expression Omnibus (GEO) (denoted with grey boxes). (b) Profiling using methylation data from blood DNA obtained from two patients with PPGL (denoted with red asterisks) [15] compared to methylation data (GSE120428) [16] corresponding to: one patient with microcephalic dwarfism (denoted with a black cross), two patients with TBRS (denoted with purple boxes) and two controls (indicated with black boxes).
We also speculated that the methylated profile observed upon alteration of DNMT3A in PPGL patients could be similar to the one described for patients with microcephalic dwarfism harboring germline gain-of-function DNMT3A variants. To explore this, we investigated the methylation status of the 307 differentially methylated probes in lymphocyte DNAs from the previously reported PGL DNMT3A variant carriers [15] and in blood samples from patients with either overgrowth or dwarfism due to DNMT3A germline variants [16]. Thus, we found that the blood DNA from PGL patients showed a similar profile and clustered together with the one obtained from the patient with microcephalic dwarfism (Figure 3b). The higher methylation observed in the patient with dwarfism suggests a higher impact of this variant in the activity of DNMT3A. Moreover, the mild differences observed between controls and the overgrowth patients suggest that the increased methylation caused by the gain-of-function probably affects non-canonical DNMT3A target regions unaltered in the presence of loss-of-function variants.
In addition, we were able to collect an AML sample carrying the p.Gly332Arg DNMT3A variant and investigated whether this somatic substitution could also be causing a gain-of-function in AML. To explore this, we profiled one AML sample carrying the p.Gly332Arg DNMT3A variant (this sample also carries another DNMT3A variant and two frameshift variants in TET2), with AML samples extracted from TCGA and carriers of DNMT3A somatic variants known to cause global methylation alterations (i.e., the bona fide p.Arg882 DNMT3A loss-of-function alteration). We also included three samples carrying additional frameshift TET2 variants. The unsupervised clustering, using the aforementioned methylation signature, revealed that the p.Gly332Arg mutated AML showed a completely different profile to the one observed for the samples carrying the recurrent p.Arg882 DNMT3A inactivating variant (Figure S1). By including TET2 mutated samples in the comparison, we discarded that this gene had an effect in the profiling. Moreover, we discarded a potential batch effect by performing a consensus clustering analysis with randomly selected sets of probes. Overall, these results suggest that the p.Gly332Arg DNMT3A variant led, in germline and also in AML, to a characteristic methylation profile, distinct to the profile caused by well-known DNMT3A inactivating variants. Although the sample carrying the p.Gly332Arg variant harbors an additional variant in the MTase domain of DNMT3A (p.Arg720Cys), its low allele frequency together with the fact that variants in this domain likely disrupt the catalytic activity of the enzyme, supports that the observed methylated profile is caused by the PWWP variant.
3. Discussion
Description of additional patients mutated in novel disease genes and comprehensive studies are required to advance in their clinical characterization. The link between global DNA hypermethylation and PPGL was first described in 2008, with the identification of a CpG island methylator phenotype (CIMP) in abdominal paragangliomas [21]. Later it was demonstrated that this was a consequence of oncometabolite accumulation, caused by pathogenic variants in Krebs cycle genes. More recently, disrupting variants in ATRX and in other chromatin-remodeling genes, including post-zygotic H3F3A gain-of-function alterations, have also been found recurrently in PPGLs [22,23]. The identification of DNMT3A activating variants leading to a specific CIMP further supported the association between epigenetic modifications and PPGL development.
In addition to the variants reported in patients with PGLs, germline gain-of-function DNMT3A variants located in the PWWP domain which cause widespread DNA hypermethylation at polycomb-regulated regions (with the H3K27me3 mark) have been found in patients with microcephalic dwarfism, an extreme growth disorder [16]. A similar growth deficiency has been observed in mice carrying a DNMT3A PWWP germline variant in the Asp329 residue (corresponding to the human Asp333). This variant causes a gain-of-function phenotype associated with the recruitment of DNMT3A towards H3K27me3-marked chromatin. This results in DNA hypermethylation and de-repression of developmental regulatory genes that manifests phenotypically as dominant postnatal growth retardation [24]. In the same way, the murine equivalent to the human Lys299Ile found in patients with PGL [15], disrupts both DNA and H3K36me2/3 binding by altering the aromatic cage conformation of the PWWP domain of DNMT3A, finally leading to disruption of the sub-nuclear localization of DNMT3A [25]. In PGL patients, we demonstrated that germline DNMT3A variants in residues within the PWWP domain caused significant hypermethylation of homeobox-containing genes involved in early embryonic development. However, long-term enforced expression of transgenic DNMT3A did not cause global hypermethylation of the genome [26]. This contrasts with the widespread hypomethylation observed in TBRS patients carrying germline DNMT3A variants, especially enriched at genes involved in morphogenesis, development and malignancy predisposition pathways [27].
Varsome classifies as likely pathogenic 74% (23/31) of the missense variants found in gnomAD within the PWWP domain of DNMT3A (Table S2). None of them appeared in TBRS patients and 58% (18/31) are found as somatic events in the COSMIC database, mostly affecting hematopoietic and lymphoid tissues. The conservation of Gly332 within the PWWP domain and its location between Trp330 and Asp333, the two residues altered in patients with microcephalic dwarfism [16], highly suggest a disruptive role of the variant. The substitution of the wild-type Gly332 by an Arg could stabilize the link with trimethylated histone H3 causing, as occurred with the previously reported variants found in PGL patients, a gain of alternative activities by DNMT3A.
Herein we have demonstrated that blood DNA from a patient with microcephalic dwarfism shows a similar profile to the observed for PGL patients, suggesting that a comparable activation of DNMT3A is occurring in both phenotypes. Moreover, overall alterations of DNA methylation have been also observed in blood cells from TET2 (hypermethylation) or EZH2 (hypomethylation) germline pathogenic loss-of-function variant carriers [28,29]. Thus, the methylated signature found in saliva DNA from the PGL patient described herein further supports the activating role of the new germline alteration found within the PWWP domain of DNMT3A. However and although the patient described herein showed idiopathic intellectual disability and short stature, there is no clear phenotypic overlap between PGL and microcephalic dwarfism. Interestingly, an AML pedigree carrying a germline DNMT3A loss-of-function mutation has no symptoms of TBRS and only three patients with TBRS have been reported to develop AML [14]. This could be suggestive of specific genotype-phenotype associations for some DNMT3A germline variants but there are other possible explanations such as the presence of additional modifier variants influencing the phenotypic outcome of these individuals.
Somatic alterations in the epigenetic regulators TET2 and DNMT3A represent a frequent cause of clonal hematopoiesis, an aging-related mechanism in which somatic variants in early blood cell progenitors confer an advantage to mutated clones. Albeit asymptomatic, this mechanism is associated with a higher risk of cardiovascular disease and hematologic malignancies [30,31,32,33]. Interestingly, several variants affecting the Gly332 residue (including p.Gly332Arg and p.Gly332Glu) have been found in healthy individuals with clonal hematopoietic mutations [32,34,35]. In fact, the Gly332Arg variant is included in a list of leukemogenic driver mutations affecting genes known to promote clonal expansion of hematopoietic stem cells (Table S2). It has been suggested that the presence of some pathogenic TBRS variants amongst the general population is due to age-related clonal hematopoiesis, questioning the utility of databases such as gnomAD in DNMT3A variant pathogenicity stratification [13]. The methylated profile observed in the AML sample carrying the p.Gly332Arg variant further suggests that alteration of specific residues within the PWWP domain of DNMT3A leading to activation of its methylation capacities, may have oncogenic potential to both endocrine malignancies (PPGL) and AML.
Thyroid tumors show the second highest missense mutation rate (0.23; number of missense variants/number samples) affecting the DNMT3A PWWP domain amongst all cancers included in COSMIC, even higher than AML (0.13) (Table 1). Two recent studies described DNMT3A pathogenic variants occurring at a frequency of 1.2% (12/1013) in PTC [36] and associated with adverse clinical outcomes [37]. Interestingly thyroid toxic adenomas have been recently found in all mutation carriers belonging to a TET2 pedigree [38]. Both, adenomas and microcarcinomas, are precursor neoplasms of the thyroid suggesting a role of these epigenetic alterations in malignancy initiation [39]. Moreover, COSMIC reports only one DNMT3A variant affecting the microcephalic dwarfism-mutated residue Asp333 and it affects an anaplastic thyroid carcinoma [40], further supporting the relevance of the PWWP domain in thyroid cancer. All these findings suggest that DNMT3A alteration and the subsequent overall change in DNA methylation, plays an important role in thyroid pathogenesis as well, though the activating or inactivating nature of the alterations is still unknown. To note, AML, thyroid cancer and PPGL show the lowest tumor mutational burden amongst all neoplasia (n = 10–14) [41], something that indicates a low number of passenger variants and stresses the relevance for tumor development of mutated genes, such as DNMT3A.
Table 1.
Variants affecting the PWWP domain in cancers included in the COSMIC database.
| Tissue | Number Samples Mutated | Samples Tested | % Samples with Mutations |
|---|---|---|---|
| Endometrium | 7 | 958 | 0.730688935 |
| Thyroid | 5 | 2151 | 0.232450023 |
| Large intestine | 8 | 4480 | 0.178571429 |
| Urinary tract | 2 | 1204 | 0.166112957 |
| Bone | 1 | 724 | 0.138121547 |
| AML | 18 | 13,565 | 0.132694434 |
| Skin | 2 | 1780 | 0.112359551 |
| Kidney | 3 | 2790 | 0.107526882 |
| Hematopoietic and lymphoid (≠ AML) | 13 | 14,357 | 0.090548165 |
| Lung | 4 | 4931 | 0.081119448 |
| Breast | 4 | 5131 | 0.077957513 |
| Biliary tract | 1 | 1474 | 0.067842605 |
| Central nervous system | 2 | 3246 | 0.061614295 |
| Esophagus | 1 | 1668 | 0.059952038 |
| Upper aerodigestive tract | 1 | 1795 | 0.055710306 |
| Liver | 1 | 2370 | 0.042194093 |
| Prostate | 1 | 2869 | 0.03485535 |
In summary, we describe and perform methylome analysis for a novel DNMT3A activating variant in a patient with multiple clinical features. Although the causal role of DNMT3A variants in PPGL has not been demonstrated, our results stress the heterogeneous phenotypic spectrum related to DNMT3A germline variants and support a dual nature of this gene in disease.
4. Materials and Methods
4.1. Patients and Samples
A 38-year-old woman with idiopathic intellectual disability was remitted to the hospital with a cervical mass of three months of evolution. The initial diagnosis was multinodular goiter with cysts. Two years later, she underwent total thyroidectomy with a diagnosis of nodular hyperplasia and a papillary thyroid microcarcinoma of 0.5 mm and stage I (T1, N0, M0) that was subsequently treated with radioactive iodine (I-131). One year later, during the follow-up, two PGLs located in the right carotid (3 × 3.5 cm) and in the left vagus nerve were diagnosed by imaging and underwent surgical removal. Thirteen years later, a left carotid PGL (2.5 × 2.2 cm) was detected by computed tomography and echography. The tumor was not operated and, since then, the patient, who had no family history of thyroid cancer or PPGLs, has been subject to close follow-up of the established lesion. DNA from saliva was obtained from the patient and from two age- and gender-matched healthy controls, whereas tumor samples were not available. An informed consent was obtained from her legal guardian.
Tumor DNA from a patient with AML and carrying two somatic variants in DNMT3A (p.Gly332Arg and p.Arg720Cys) and two in TET2 (p.Arg1216Ter and p.Arg544Ter) was used for methylation profiling.
4.2. Targeted Next-Generation Sequencing
Whole-exome sequencing was carried out at the National Centre for Genomic Analysis (CNAG) on DNA extracted from a saliva sample from the patient by OrageneTM self-collection kit (DNA GenoteK, Ontario, Canada). Exome capture was performed using the Roche NimbleGen exome capture kit (Roche NimbleGen, Madison, WI, USA) and exome sequencing (mean coverage > 80×) was performed by 2 × 100 base paired-end technology using a HiSeq2000 sequencer (Illumina Inc., San Diego, CA, USA). Alignment of the reads was performed by the GEM5 and BFAST programs and the SAMtools program was used (http://samtools.sourceforge.net) to identify single nucleotide substitutions and small insertions and deletions. Filtering was applied to exclude variants present in gnomAD with AF > 0.00005, variants present in internal exomes or in the CIBERER Spanish Variant Server (http://csvs.babelomics.org/), noncoding substitutions and variants in genomic regions with low mappability, with low depth readings or quality or with the alternative allele present in < 20% of reads.
4.3. In Silico Predictions
The PredictSNP1 consensus classifier [42] was used to predict the effect of the only nucleotide substitution affecting a gene previously associated with PPGL and that passed all filtering steps. The Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/) was used to generate an alignment of the six classes of human PWWP domains. The amino acid sequence of the DNMT3A PWWP domain (in which Gly332Arg mutant is located) was submitted to the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2) for structure prediction [43]. Twenty templates were selected to model the protein based on heuristics to maximize confidence, percentage identity and alignment coverage. The H3K36me3 peptide was modelled into the DNMT3A PWWP Gly332Arg mutant structure obtained using the DNMT3B PWWP domain structure bound to H3K36me3 peptide (PDB code 5CIU) [44] as a reference to ensure no clashes with the methylated lysine.
4.4. DNA Methylation Array
Bisulfite conversion of the AML tumor and the saliva DNAs from the proband and two age-and gender-matched controls was performed using the EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) and genome-wide DNA methylation was assayed using the Infinium MethylationEPIC BeadChip (Illumina) at the Centro Nacional de Genotipado (CEGEN-ISCIII) (www.cegen.org), as previously described [45]. This BeadChip interrogates over 850,000 methylation sites per sample at single-nucleotide resolution. M values were used for statistical analyses.
For methylation analyses, first, supervised DNA analyses (using data from Reference [15]) were performed to obtain a list of differentially methylated probes (n = 307; false discovery rate (FDR) < 0.15) between DNMT3A-mutated tissues (blood and tumors) from patients with PGL and healthy controls (Figure S2). Following, we used this list of probes for profiling MethylationEPIC data obtained from the DNA saliva samples from the proband, the two age- gender-matched controls and five additional saliva samples of healthy donors obtained from Gene Expression Omnibus (GEO) (GSE111631). In a subsequent analysis, we used the same list of 307 probes to profile methylation data from blood of patients with microcephalic dwarfism (n = 1) or overgrowth syndrome (n = 2) (GSE120428) [16] and compare them to controls (n = 2) and to PGLs carrying DNMT3A variants (n = 2) [15]. Finally, we extracted methylation data from 12 AML samples available from The Cancer Genome Atlas (TCGA), corresponding to 213 out of the 307 probes aforementioned for hierarchical clustering with one additional AML sample carrying the p.Gly332Arg variant, another DNMT3A variant (p.Arg720Cys) and two frameshift TET2 mutations. The TCGA samples were all carriers of mutations in DNMT3A known to cause global methylation alterations (i.e., p.Arg882Cys/His) and three of them also harbored TET2 frameshift mutations. The reliability of the clusters was verified by consensus clustering, a robust clustering method which obtains the consensus across multiple runs of a clustering algorithm and assess the stability of the discovered clusters by using resampling techniques [46]. Hierarchical clustering of methylation data was performed using GeneCluster 2.0 [47].
5. Conclusions
We describe a novel germline DNMT3A variant (p.Gly332Arg) identified by whole-exome sequencing in a patient with multiple clinical features: bilateral carotid PGLs, PTC and idiopathic intellectual disability. The variant affects a residue found mutated in papillary thyroid tumors and the structural modelling of the PWWP domain predicts the substitution to alter the interaction of DNMT3A with H3K36me3. Moreover, we observed an increased methylation of DNMT3A target genes in saliva DNA from the proband and in one independent AML sample carrying the same p.Gly332Arg variant, compatible with a gain-of-function effect of the alteration. Although the causal role of DNMT3A variants in PPGL has not been demonstrated, our finding stresses the heterogeneous phenotypic spectrum related to DNMT3A germline variants.
Acknowledgments
The Human Genotyping Unit is a member of the Centro Nacional de Genotipado–Plataforma de Recursos Biomoleculares (CeGen-PRB3) and is supported by grant PT17/0019 of the PE I+D+i 2013–2016, funded by ISCIII and ERDF.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/11/3304/s1, Figure S1: Hierarchical clustering performed with one AML sample carrying the p.Gly332Arg DNMT3A mutation (black circle) and 12 AML samples from TCGA [48] carrying mutations known to cause global methylation alterations: loss-of-function p.Arg882Cys/His DNMT3A mutations (blue boxes) and TET2 frameshift mutations (purple boxes). Profiling was based on methylation data from the 213 probes, out of the 307 significantly differentially methylated between DNMT3A-mutated and non-mutated tissues in patients with PPGL, contained in the Infinium HumanMethylation450 BeadChip, Figure S2: Schematic representation of the methylation analysis pipeline, Table S1: Filtered exome variants found in ClinVar, Table S2: Variants affecting the PWWP domain found in gnomAD, Table S3: List of differentially methylated CpGs between DNMT3A-mutated and non-mutated samples.
Author Contributions
Conceptualization A.C.; methodology S.M., J.C., A.C., B.C., J.M.R.-R.; software J.C., J.L., Á.M.M.-M.; validation, S.M., A.C., M.S.; formal analysis S.M., A.C.; investigation S.M., M.M., A.C.; resources J.M.-L., R.A., X.M.-G.; data curation A.C.; writing—original draft preparation S.M., A.C.; writing—review and editing S.M., L.J.L.-G., C.R.-A., C.M.-C., M.R., A.C.; visualization A.C.; supervision M.R., A.C.; project administration, A.C.; funding acquisition, A.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Instituto de Salud Carlos III (ISCIII), through the “Acción Estratégica en Salud” (AES) (projects PI18/00454 and PI17/01796 to A.C. and M.R., repectively), cofounded by the European Regional Development Fund (ERDF).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fishbein L. Pheochromocytoma/Paraganglioma: Is This a Genetic Disorder? Curr. Cardiol. Rep. 2019;21:104. doi: 10.1007/s11886-019-1184-y. [DOI] [PubMed] [Google Scholar]
- 2.Remacha L., Pirman D., Mahoney C.E., Coloma J., Calsina B., Curras-Freixes M., Leton R., Torres-Perez R., Richter S., Pita G., et al. Recurrent Germline DLST Mutations in Individuals with Multiple Pheochromocytomas and Paragangliomas. Am. J. Hum. Genet. 2019;104:651–664. doi: 10.1016/j.ajhg.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buffet A., Morin A., Castro-Vega L.J., Habarou F., Lussey-Lepoutre C., Letouze E., Lefebvre H., Guilhem I., Haissaguerre M., Raingeard I., et al. Germline Mutations in the Mitochondrial 2-Oxoglutarate/Malate Carrier SLC25A11 Gene Confer a Predisposition to Metastatic Paragangliomas. Cancer Res. 2018;78:1914–1922. doi: 10.1158/0008-5472.CAN-17-2463. [DOI] [PubMed] [Google Scholar]
- 4.Remacha L., Comino-Mendez I., Richter S., Contreras L., Curras-Freixes M., Pita G., Leton R., Galarreta A., Torres-Perez R., Honrado E., et al. Targeted exome sequencing of Krebs cycle genes reveals candidate cancer–predisposing mutations in pheochromocytomas and paragangliomas. Clin. Cancer Res. 2017;23:6315–6325. doi: 10.1158/1078-0432.CCR-16-2250. [DOI] [PubMed] [Google Scholar]
- 5.Gowher H., Jeltsch A. Mammalian DNA methyltransferases: New discoveries and open questions. Biochem. Soc. Trans. 2018;46:1191–1202. doi: 10.1042/BST20170574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yagi M., Kabata M., Tanaka A., Ukai T., Ohta S., Nakabayashi K., Shimizu M., Hata K., Meissner A., Yamamoto T., et al. Identification of distinct loci for de novo DNA methylation by DNMT3A and DNMT3B during mammalian development. Nat. Commun. 2020;11:3199. doi: 10.1038/s41467-020-16989-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatton-Brown K., Seal S., Ruark E., Harmer J., Ramsay E., Del Vecchio Duarte S., Zachariou A., Hanks S., O’Brien E., Aksglaede L., et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E., Kandoth C., Payton J.E., Baty J., Welch J., et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L., Rau R., Goodell M.A. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudry S.F., Chevassut T.J. Epigenetic Guardian: A Review of the DNA Methyltransferase DNMT3A in Acute Myeloid Leukaemia and Clonal Haematopoiesis. BioMed Res. Int. 2017;2017:5473197. doi: 10.1155/2017/5473197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen W., Heeley J.M., Carlston C.M., Acuna-Hidalgo R., Nillesen W.M., Dent K.M., Douglas G.V., Levine K.L., Bayrak-Toydemir P., Marcelis C.L., et al. The spectrum of DNMT3A variants in Tatton-Brown-Rahman syndrome overlaps with that in hematologic malignancies. Am. J. Med. Genet. A. 2017;173:3022–3028. doi: 10.1002/ajmg.a.38485. [DOI] [PubMed] [Google Scholar]
- 12.Hollink I., van den Ouweland A.M.W., Beverloo H.B., Arentsen-Peters S., Zwaan C.M., Wagner A. Acute myeloid leukaemia in a case with Tatton-Brown-Rahman syndrome: The peculiar DNMT3A R882 mutation. J. Med. Genet. 2017;54:805–808. doi: 10.1136/jmedgenet-2017-104574. [DOI] [PubMed] [Google Scholar]
- 13.Tatton-Brown K., Zachariou A., Loveday C., Renwick A., Mahamdallie S., Aksglaede L., Baralle D., Barge-Schaapveld D., Blyth M., Bouma M., et al. The Tatton-Brown-Rahman Syndrome: A clinical study of 55 individuals with de novo constitutive DNMT3A variants. Wellcome Open Res. 2018;3:46. doi: 10.12688/wellcomeopenres.14430.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiNardo C.D., Beird H.C., Estecio M., Hardikar S., Takahashi K., Bannon S.A., Borthakur G., Jabbour E., Gumbs C., Khoury J.D., et al. Germline DNMT3A mutation in familial acute myeloid leukaemia. Epigenetics. 2020 doi: 10.1080/15592294.2020.1809871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remacha L., Curras-Freixes M., Torres-Ruiz R., Schiavi F., Torres-Perez R., Calsina B., Leton R., Comino-Mendez I., Roldan-Romero J.M., Montero-Conde C., et al. Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet. Med. 2018;20:1644–1651. doi: 10.1038/s41436-018-0003-y. [DOI] [PubMed] [Google Scholar]
- 16.Heyn P., Logan C.V., Fluteau A., Challis R.C., Auchynnikava T., Martin C.A., Marsh J.A., Taglini F., Kilanowski F., Parry D.A., et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat. Genet. 2019;51:96–105. doi: 10.1038/s41588-018-0274-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yap D.B., Chu J., Berg T., Schapira M., Cheng S.W., Moradian A., Morin R.D., Mungall A.J., Meissner B., Boyle M., et al. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katoh M. Mutation spectra of histone methyltransferases with canonical SET domains and EZH2-targeted therapy. Epigenomics. 2016;8:285–305. doi: 10.2217/epi.15.89. [DOI] [PubMed] [Google Scholar]
- 19.Tatton-Brown K., Hanks S., Ruark E., Zachariou A., Duarte Sdel V., Ramsay E., Snape K., Murray A., Perdeaux E.R., Seal S., et al. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–1133. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu H., Zeng H., Lam R., Tempel W., Amaya M.F., Xu C., Dombrovski L., Qiu W., Wang Y., Min J. Structural and histone binding ability characterizations of human PWWP domains. PLoS ONE. 2011;6:e18919. doi: 10.1371/journal.pone.0018919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geli J., Kiss N., Karimi M., Lee J.J., Backdahl M., Ekstrom T.J., Larsson C. Global and regional CpG methylation in pheochromocytomas and abdominal paragangliomas: Association to malignant behavior. Clin. Cancer Res. 2008;14:2551–2559. doi: 10.1158/1078-0432.CCR-07-1867. [DOI] [PubMed] [Google Scholar]
- 22.Toledo R.A., Qin Y., Cheng Z.M., Gao Q., Iwata S., Silva G.M., Prasad M.L., Ocal I.T., Rao S., Aronin N., et al. Recurrent Mutations of Chromatin-Remodeling Genes and Kinase Receptors in Pheochromocytomas and Paragangliomas. Clin. Cancer Res. 2016;22:2301–2310. doi: 10.1158/1078-0432.CCR-15-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fishbein L., Khare S., Wubbenhorst B., DeSloover D., D’Andrea K., Merrill S., Cho N.W., Greenberg R.A., Else T., Montone K., et al. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nat. Commun. 2015;6:6140. doi: 10.1038/ncomms7140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sendzikaite G., Hanna C.W., Stewart-Morgan K.R., Ivanova E., Kelsey G. A DNMT3A PWWP mutation leads to methylation of bivalent chromatin and growth retardation in mice. Nat. Commun. 2019;10:1884. doi: 10.1038/s41467-019-09713-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dukatz M., Holzer K., Choudalakis M., Emperle M., Lungu C., Bashtrykov P., Jeltsch A. H3K36me2/3 Binding and DNA Binding of the DNA Methyltransferase DNMT3A PWWP Domain Both Contribute to its Chromatin Interaction. J. Mol. Biol. 2019;431:5063–5074. doi: 10.1016/j.jmb.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Ketkar S., Verdoni A.M., Smith A.M., Bangert C.V., Leight E.R., Chen D.Y., Brune M.K., Helton N.M., Hoock M., George D.R., et al. Remethylation of Dnmt3a (-/-) hematopoietic cells is associated with partial correction of gene dysregulation and reduced myeloid skewing. Proc. Natl. Acad. Sci. USA. 2020;117:3123–3134. doi: 10.1073/pnas.1918611117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeffries A.R., Maroofian R., Salter C.G., Chioza B.A., Cross H.E., Patton M.A., Dempster E., Temple I.K., Mackay D.J.G., Rezwan F.I., et al. Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging. Genome Res. 2019;29:1057–1066. doi: 10.1101/gr.243584.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choufani S., Gibson W.T., Turinsky A.L., Chung B.H.Y., Wang T., Garg K., Vitriolo A., Cohen A.S.A., Cyrus S., Goodman S., et al. DNA Methylation Signature for EZH2 Functionally Classifies Sequence Variants in Three PRC2 Complex Genes. Am. J. Hum. Genet. 2020;106:596–610. doi: 10.1016/j.ajhg.2020.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaasinen E., Kuismin O., Rajamaki K., Ristolainen H., Aavikko M., Kondelin J., Saarinen S., Berta D.G., Katainen R., Hirvonen E.A.M., et al. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat. Commun. 2019;10:1252. doi: 10.1038/s41467-019-09198-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaner J., Desai P., Mencia-Trinchant N., Guzman M.L., Roboz G.J., Hassane D.C. Clonal Hematopoiesis and Premalignant Diseases. Cold Spring Harb. Perspect. Med. 2019;10:a035675. doi: 10.1101/cshperspect.a035675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Genovese G., Kahler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izzo F., Lee S.C., Poran A., Chaligne R., Gaiti F., Gross B., Murali R.R., Deochand S.D., Ang C., Jones P.W., et al. DNA methylation disruption reshapes the hematopoietic differentiation landscape. Nat. Genet. 2020;52:378–387. doi: 10.1038/s41588-020-0595-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young A.L., Tong R.S., Birmann B.M., Druley T.E. Clonal hematopoiesis and risk of acute myeloid leukemia. Haematologica. 2019;104:2410–2417. doi: 10.3324/haematol.2018.215269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watson C.J., Papula A.L., Poon G.Y.P., Wong W.H., Young A.L., Druley T.E., Fisher D.S., Blundell J.R. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science. 2020;367:1449–1454. doi: 10.1126/science.aay9333. [DOI] [PubMed] [Google Scholar]
- 36.Siraj A.K., Pratheeshkumar P., Parvathareddy S.K., Bu R., Masoodi T., Iqbal K., Al-Rasheed M., Al-Dayel F., Al-Sobhi S.S., Alzahrani A.S., et al. Prognostic significance of DNMT3A alterations in Middle Eastern papillary thyroid carcinoma. Eur. J. Cancer. 2019;117:133–144. doi: 10.1016/j.ejca.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 37.Guo L.C., Zhu W.D., Ma X.Y., Ni H., Zhong E.J., Shao Y.W., Yu J., Gu D.M., Ji S.D., Xu H.D., et al. Mutations of genes including DNMT3A detected by next-generation sequencing in thyroid cancer. Cancer Biol. Ther. 2019;20:240–246. doi: 10.1080/15384047.2018.1523856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duployez N., Goursaud L., Fenwarth L., Bories C., Marceau-Renaut A., Boyer T., Fournier E., Nibourel O., Roche-Lestienne C., Huet G., et al. Familial myeloid malignancies with germline TET2 mutation. Leukemia. 2020;34:1450–1453. doi: 10.1038/s41375-019-0675-6. [DOI] [PubMed] [Google Scholar]
- 39.Chmielik E., Rusinek D., Oczko-Wojciechowska M., Jarzab M., Krajewska J., Czarniecka A., Jarzab B. Heterogeneity of Thyroid Cancer. Pathobiology. 2018;85:117–129. doi: 10.1159/000486422. [DOI] [PubMed] [Google Scholar]
- 40.Pozdeyev N., Gay L.M., Sokol E.S., Hartmaier R., Deaver K.E., Davis S., French J.D., Borre P.V., LaBarbera D.V., Tan A.C., et al. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin. Cancer Res. 2018;24:3059–3068. doi: 10.1158/1078-0432.CCR-18-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y., Liu Z.X., Jiang Z.H., Wang X.S. Identification of genomic features associated with immunotherapy response in gastrointestinal cancers. World J. Gastrointest. Oncol. 2019;11:270–280. doi: 10.4251/wjgo.v11.i4.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E.D., Zendulka J., Brezovsky J., Damborsky J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014;10:e1003440. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rondelet G., Dal Maso T., Willems L., Wouters J. Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B. J. Struct. Biol. 2016;194:357–367. doi: 10.1016/j.jsb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Bibikova M., Le J., Barnes B., Saedinia-Melnyk S., Zhou L., Shen R., Gunderson K.L. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics. 2009;1:177–200. doi: 10.2217/epi.09.14. [DOI] [PubMed] [Google Scholar]
- 46.Monti S., Tamayo P., Mesirov J., Golub T. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Mach. Learn. 2003;52:91–118. doi: 10.1023/A:1023949509487. [DOI] [Google Scholar]
- 47.Reich M., Ohm K., Angelo M., Tamayo P., Mesirov J.P. GeneCluster 2.0: An advanced toolset for bioarray analysis. Bioinformatics. 2004;20:1797–1798. doi: 10.1093/bioinformatics/bth138. [DOI] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research Network. Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.