Abstract
A prospecting work at the Axarquia region (Malaga, Spain) was carried out in order to identify local red grapevine cultivars preserved in ancient vineyards. A total of 11 accessions were collected in seven different plots from four municipalities and analyzed using 25 microsatellite loci for cultivar identification. The accessions analyzed were identified as eight different genotypes, seven of them corresponding to known cultivars as ‘Cabernet Sauvignon’, ‘Jaen Tinto’, ‘Molinera’, ‘Monastrell’, ‘Muscat of Alexandria’, ‘Parrel’, and ‘Romé’. In addition, one of them is referred to as the new genotype for ‘Cabriel’ cultivar. Additionally, an ampelographic characterization was carried out with 30 International Organisation of Vine and Wine (OIV) descriptors for two consecutive years for the eight accessions identified as local cultivars. This allowed the identification of a somatic variant of the ‘Muscat of Alexandria’ cultivar that affects the color of the berry and another of ‘Romé’ regarding bunch compactness.
Keywords: Vitis vinifera, autochthonous grapevine cultivar, cultivar identification, microsatellite marker, ampelographic characterization, somatic variant
1. Introduction
Within the province of Malaga, the Axarquia region is a historically recognized wine territory in Andalusia (Spain). With a mostly steep and mountainous orography, it is located in the most eastern part of the province, spreading along the coast and inland [1]. The cultivation of vines, as well as wine production and trade, have been for a long time the main foundation of the economy of this region of heroic viticulture. Like other Andalusian wine-producing areas, Axarquia has a more thousand-year-old tradition that has not been exempt from the decline that the sector suffered at the end of the last century [2]. The difficult mechanization of the vineyard, the predominance of small vineyard plots, and the low productivity have contributed to vineyard forgetfulness, and nowadays this sector remains rooted in time. Such vineyards may preserve unidentified indigenous or local varieties, which may be of interest in the current viticulture. In this respect, studying their adaptation to warm climatic conditions and their oenological potential to produce new wines could play an important role in the future [3,4]. Besides, nowadays many wine consumers demand new products, with greater diversification and personality; therein lies the growing interest of producers and consumers in ancient local cultivars [5,6].
Since the end of the 19th century, with the phylloxera (Daktulosphaira vitifoliae) arrival to Europe, genetic diversity decreased in most European vineyards [7]. In Spain, the first phylloxera outbreak was detected in Malaga (Andalusia) in 1876 [8]. This plague destroyed a large part of the vineyards in this province, which went from 112.872 ha of vineyards in 1878 to 24.180 ha in 1909 [9]. This event gave up a loss of cultivars and consequently of genetic diversity. In historical texts about the region’s viticulture, red grapevine varieties were mentioned such as ‘Cabriel’, ‘Jaén Prieto’, ‘Tempranas Negras’, ‘Alicante’ or ‘Tinto’, ‘Ubíes’, ’Corazón de Cabrito’, ‘Casiles Negras’, ‘Tinto Jaen’, ‘Teta de Negra’, or ‘Cruazno’ [10,11,12]. Actually, a large part of the Axarquia and Malaga vineyard is planted with ‘Muscat of Alexandria’ cultivar for raisin production [13]. Nevertheless, Jiménez-Cantizano et al. [14] in 2014 identified three ancient red cultivars using microsatellite markers: ‘Listán Prieto’, ‘Rome Tinto’, and ‘Jaén Tinto’ collected in vineyards in the province of Malaga.
Nuclear microsatellite markers or simple sequence repeat (SSR) have been widely used to identify and genotype grapevine cultivars [15,16,17,18,19,20,21,22]. In addition, the combination of genetic (microsatellite markers) and ampelographic methods allows the correct identification of cultivars [23]. For this purpose, in old varieties, it is a necessary activity in order to be able to preserve them as plant genetic resources in germplasm banks. Although many projects for the collection and identification of endangered cultivars have been carried out [24,25,26,27,28], there are still old vineyards, located in important wine regions, that have not been prospected. In this way, there are few works that have been developed and published regarding Andalusian ancient cultivars.
The main objective of the present study is the identification of red grapevine cultivars grown in ancient vineyards in the region of the Axarquia (Malaga, Spain). In the scope of this study, the detection of possible synonymies, denomination mistakes, and new genotypes, could contribute to an efficient preservation of old local germplasm that represents valuable genetic combinations for a new viticulture. To this end, a prospection of different ancient local red grapevine cultivars, their genetic analysis using SSR molecular markers, and their morphological description was carried out.
2. Results
As a result of the accessions genetic characterization, the presence of a new genotype, a new synonym, and three denomination mistakes were obtained. In order to confirm the identified cultivars based on the molecular results obtained, ampelographic observations were made in the vineyard over two years. In this sense, the ampelographic characterization allowed the identification of two somatic variations for ‘Muscat of Alexandria’ and ‘Romé’ cultivars.
2.1. Microsatellite Analysis
Eleven accessions were analyzed at 25 nuclear microsatellite loci resulting in eight nonredundant genotypes (Table 1).
Table 1.
Microsatellite profile of 11 grapevine accessions located in Axarquia (Malaga, Spain) analyzed at 25 microsatellite loci.
| OIV Code | Accession Code | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | M5 | M6 | M7 | M8 | M9 | M10 | M11 | |
| ssrVrZAG29 | 110 110 | 110 110 | 110 110 | 110 110 | 110 110 | 110 110 | 110 110 | 110 110 | 110 110 | 110 110 | 110 110 |
| ssrVrZAG62 | 187 203 | 185 203 | 203 203 | 187 203 | 203 203 | 187 193 | 187 195 | 187 195 | 187 203 | 187 203 | 187 195 |
| ssrVrZAG112 | 228 228 | 233 245 | 233 236 | 228 233 | 233 236 | 228 233 | 228 236 | 228 236 | 228 236 | 228 236 | 231 236 |
| ssrVrZAG67 | 130 150 | 124 124 | 137 158 | 137 137 | 137 158 | 124 137 | 130 158 | 130 158 | 137 137 | 130 158 | 124 130 |
| VVMD27 | 178 191 | 176 191 | 182 191 | 176 186 | 182 191 | 173 186 | 178 191 | 178 191 | 176 186 | 178 191 | 178 186 |
| VVMD5 | 231 235 | 224 228 | 231 237 | 222 237 | 231 237 | 228 237 | 235 237 | 235 237 | 222 231 | 235 237 | 231 237 |
| VVS2 | 135 143 | 131 148 | 133 156 | 131 150 | 133 156 | 137 150 | 135 143 | 135 143 | 131 150 | 135 143 | 131 143 |
| ssrVrZAG83 | 190 194 | 188 188 | 190 190 | 190 200 | 190 190 | 200 200 | 190 194 | 190 194 | 194 200 | 190 194 | 190 190 |
| VVMD28 | 233 257 | 243 266 | 247 259 | 243 257 | 247 259 | 233 235 | 235 257 | 235 257 | 227 257 | 235 257 | 243 247 |
| VVIh54 | 167 169 | 167 167 | 167 167 | 167 167 | 167 167 | 167 181 | 167 167 | 167 167 | 167 167 | 167 167 | 167 169 |
| VVIn73 | 264 264 | 264 264 | 256 264 | 264 264 | 256 264 | 264 268 | 264 264 | 264 264 | 264 264 | 264 264 | 264 264 |
| VMC1b11 | 185 188 | 167 185 | 188 188 | 173 188 | 188 188 | 185 185 | 185 188 | 185 188 | 173 188 | 185 188 | 185 188 |
| VVMD25 | 239 253 | 247 247 | 253 254 | 239 261 | 253 254 | 237 247 | 239 253 | 239 253 | 239 261 | 239 253 | 239 239 |
| VVIp31 | 186 190 | 188 190 | 190 190 | 180 190 | 190 190 | 190 190 | 176 190 | 176 190 | 180 196 | 176 190 | 180 192 |
| VVMD7 | 241 247 | 247 249 | 231 237 | 247 247 | 231 237 | 237 237 | 237 237 | 237 237 | 237 247 | 237 237 | 237 241 |
| VVIb01 | 290 290 | 290 294 | 290 306 | 290 290 | 290 306 | 290 290 | 290 294 | 290 294 | 290 290 | 290 294 | 290 290 |
| VVIq52 | 84 88 | 82 82 | 84 88 | 88 88 | 84 88 | 82 88 | 82 88 | 82 88 | 84 88 | 82 88 | 84 88 |
| VVMD24 | 210 210 | 212 212 | 208 208 | 208 217 | 208 208 | 208 217 | 208 208 | 208 208 | 208 208 | 208 208 | 208 208 |
| VVIp60 | 317 321 | 317 321 | 321 321 | 317 321 | 321 321 | 305 313 | 317 325 | 317 325 | 317 325 | 317 325 | 317 321 |
| VVMD32 | 250 270 | 262 270 | 248 250 | 238 254 | 248 250 | 238 238 | 254 270 | 254 270 | 238 248 | 254 270 | 254 256 |
| VVIn16 | 150 152 | 148 150 | 150 150 | 152 158 | 150 150 | 152 152 | 152 152 | 152 152 | 152 158 | 152 152 | 150 152 |
| VMC4f3.1 | 166 186 | 180 206 | 182 206 | 178 178 | 182 206 | 172 178 | 186 188 | 186 188 | 178 178 | 186 188 | 172 186 |
| ssrVrZAG79 | 244 254 | 244 252 | 244 254 | 248 258 | 244 254 | 244 244 | 244 254 | 244 254 | 240 258 | 244 254 | 240 244 |
| VVMD21 | 248 248 | 255 265 | 242 255 | 242 248 | 242 255 | 248 257 | 242 248 | 242 248 | 248 255 | 242 248 | 248 248 |
| VVIv67 | 371 375 | 375 389 | 357 375 | 357 365 | 357 375 | 365 371 | 365 375 | 365 375 | 365 365 | 365 375 | 361 365 |
On one hand, M3 and M5 accessions showed the same genotype and, on the other hand, M7, M8, and M10 (Table 2). The nonredundant genotypes obtained were compared with the Vitis International Variety Catalogue (VIVC) (www.vivc.de) [29] genotype database, Jiménez-Cantizano et al. [30] and Lacombe et al. [31] in order to detect the presence of synonymies, homonymies, and denomination mistakes. The genotypes obtained for the reference cultivars (Supplementary Table S2) were used for testing the microsatellite profiles obtained with the different databases published and comparing the relative allele sizes for the different microsatellite loci. After the comparison with the different databases, seven genotypes were identified with its prime name according to VIVC database (Table 2). Genotype III (M3 and M5 samples) has not been identified because it has not been published in the consulted databases. This genotype could be considered a new genotype, and would also correspond to the genotype of the ‘Cabriel’ cultivar identified for the first time. Additionally, the cultivar name was checked in the ampelographic section of the VIVC.
Table 2.
Genotypes identified for the 11 grapevine accessions characterized at 25 microsatellite loci.
| Genotype | Code Accession | Local Name | Prime Name * |
|---|---|---|---|
| I | M1 | Casiles Negra | MOLINERA |
| II | M2 | Moscatel de Alejandría Tinta | MUSCAT OF ALEXANDRIA |
| III | M3, M5 | Unknown/Cabriel | - |
| IV | M4 | Romé | MONASTRELL |
| V | M6 | Romé | CABERNET SAUVIGNON |
| VI | M7, M8, M10 | Romé | ROMÉ |
| VII | M9 | Romé | PARREL |
| VIII | M11 | Jaén Tinto | JAEN TINTO |
* Prime name according to VIVC (www.vivc.de).
Three denomination mistakes were detected for samples M4, M6, and M9, known locally as ‘Romé’, but identified as ‘Monastrell’, ‘Cabernet Sauvignon’, and ‘Parrel’, respectively (Table 2). Furthermore, ‘Casiles Negra’ accession presented a similar genotype of ‘Molinera’ and also, the name ‘Casiles Negra’, is not included in the VIVC database. Therefore, ‘Casiles Negra’ should be considered a new synonym of ‘Molinera’ cultivar.
2.2. Ampelographic Characterization
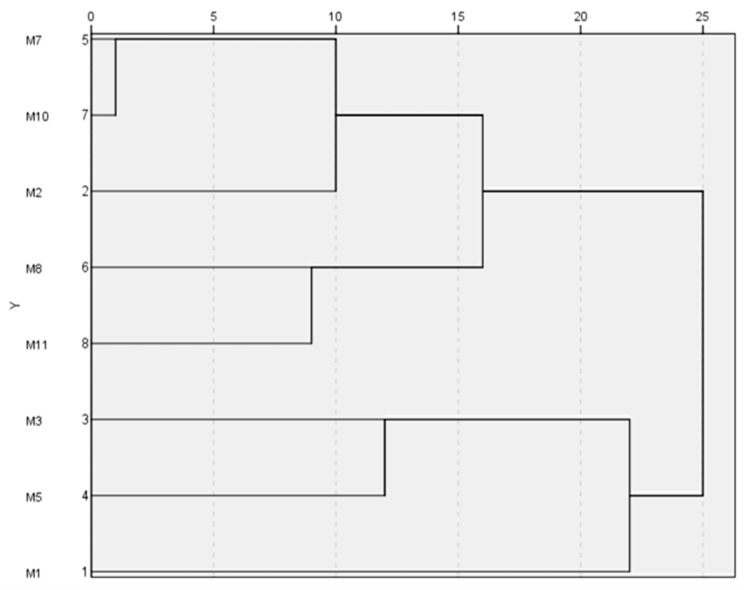
Table 3 shows the results of the morphological characterization of the identified accessions considered as minor Andalusian cultivars. Each accession presented a different phenotype for the 30 evaluated descriptors OIV, except for the accessions M7 and M10 that showed the same phenotype (Table 4, Figure 1) and genotype (Table 1 and Table 2). Nevertheless, M8 accession presented identical genotype at 25 microsatellite loci with M7 and M10 but different phenotype (Table 1). Both accessions are clearly different in the expression of six OIV descriptors (OIV 204, OIV 206, OIV 208, OIV 209, OIV 222, and OIV 238). M7 and M10 have loose bunch and M8 showed very dense bunch. These phenotypic differences detected are those that could allow the establishment of somatic variants or clones in the same cultivar.
Table 3.
Ampelographic characteristics of 30 OIV descriptors on grapevine accessions located in Axarquia (Malaga, Spain).
| Accession Code | ||||||||
|---|---|---|---|---|---|---|---|---|
| OIV Code | M1 | M2 | M3 | M5 | M7 | M8 | M10 | M11 |
| OIV 065 | 9 | 5 | 7 | 5 | 5 | 5 | 5 | 5 |
| OIV 067 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| OIV 068 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 |
| OIV 070 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| OIV 071 | 1 | 2 | 1 | 2 | 1 | 1 | 1 | 1 |
| OIV 072 | 1 | 5 | 5 | 1 | 3 | 3 | 3 | 1 |
| OIV 074 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 1 |
| OIV 076 | 5 | 2 | 5 | 5 | 3 | 3 | 3 | 2 |
| OIV 079 | 3 | 7 | 3 | 3 | 7 | 7 | 7 | 3 |
| OIV 080 | 2 | 3 | 3 | 3 | 2 | 2 | 2 | 1 |
| OIV 081-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| OIV 081-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| OIV 082 | 4 | 1 | 1 | 1 | 3 | 3 | 3 | 1 |
| OIV 083-1 | 2 | 3 | 3 | 3 | 2 | 2 | 2 | 3 |
| OIV 083-2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| OIV 084 | 1 | 3 | 1 | 1 | 7 | 7 | 7 | 7 |
| OIV 085 | 1 | 3 | 1 | 1 | 3 | 3 | 3 | 3 |
| OIV 202 | 7 | 5 | 7 | 5 | 5 | 5 | 5 | 7 |
| OIV 203 | 7 | 3 | 5 | 3 | 5 | 5 | 5 | 7 |
| OIV 204 | 3 | 3 | 1 | 5 | 3 | 9 | 3 | 9 |
| OIV 206 | 5 | 3 | 7 | 5 | 5 | 3 | 5 | 5 |
| OIV 208 | 2 | 2 | 1 | 1 | 2 | 1 | 2 | 2 |
| OIV 209 | 3 | 3 | 3 | 2 | 2 | 1 | 2 | 2 |
| OIV 220 | 9 | 7 | 5 | 5 | 5 | 5 | 5 | 5 |
| OIV 221 | 5 | 5 | 3 | 3 | 5 | 5 | 5 | 5 |
| OIV 222 | 7 | 1 | 1 | 2 | 1 | 2 | 1 | 2 |
| OIV 223 | 3 | 3 | 3 | 3 | 2 | 2 | 2 | 2 |
| OIV 225 | 5 | 3 | 5 | 5 | 5 | 5 | 5 | 5 |
| OIV 238 | 5 | 5 | 5 | 7 | 7 | 5 | 7 | 5 |
| OIV 241 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
Table 4.
Number of different observations between different accessions characterized with 30 OIV descriptors.
| M1 | M2 | M3 | M5 | M7 | M8 | M10 | |
|---|---|---|---|---|---|---|---|
| M2 | 16 | ||||||
| M3 | 12 | 14 | |||||
| M5 | 16 | 14 | 12 | ||||
| M7 | 14 | 14 | 17 | 17 | |||
| M8 | 16 | 15 | 16 | 18 | 6 | ||
| M10 | 14 | 14 | 17 | 17 | 0 | 6 | |
| M11 | 13 | 15 | 15 | 15 | 12 | 9 | 12 |
Figure 1.
Dendrogram representing the differences among the different studied accessions based on hierarchical cluster analysis (HCA) of ampelographic characterization employing an average link between groups and re-scaled distance cluster combination.
‘Moscatel de Alejandría Tinta’ (M2) accession showed the same microsatellite profile with ‘Muscat of Alexandria’, but different berry color; thus, it could be concluded that ‘Moscatel de Alejandría Tinta’ is a red somatic variant for berry color of ‘Muscat of Alexandria’.
3. Discussion
During the last 30 years, in Europe, the interest of grapevine growers and wine producers for old and autochthonous cultivars has increased and, therefore, it has become necessary to correctly identify the different cultivars [24]. There are still diverse grapevine synonymies (the same cultivar known under different names) and homonymies (different cultivars known under the same name) to clarify, that alongside with the existence of unnamed accessions, are a source of misidentification and confusion regarding grapevine cultivars designations [27,32,33]. Of the eight genotypes identified in this research work (Table 2), only five correspond to minor Andalusian cultivars (‘Molinera’, ‘Muscat of Alexandria’, ‘Romé’, ’Cabriel’, ‘Jaén Tinto’). These cultivars were cultivated in the province of Malaga at the beginning of the XIX century according Clemente y Rubio [11]. This work has allowed to identify the genotype of the ‘Cabriel’ cultivar for the first time. This genotype is not included in VIVC database which aims to virtually assemble all accessions maintained in the existing collections worldwide [34]. In addition, this cultivar is only conserved in Axarquia’s vineyards; accordingly, VIVC (www.vivc.de) is not preserved in the different holding institutions.
Additionally, ‘Casiles Negra’ accession presented a similar genotype of ‘Molinera’ cultivar. ‘Casiles’ name is not listed in the VIVC database. Nevertheless, García de la Leña [12] cites the ‘Casiles Negras’ cultivar in 1792 among the grapevine cultivars grown in the province of Malaga. Clemente y Rubio [11], in 1807, cited ‘Casiles de Málaga’ cultivar. This result suggests that ‘Casiles Negras’ should be considered as a new synonym of ‘Molinera’.
As for the ampelographic description, this is the methodology that enables the identification of variants or clones in a cultivar [35]. This work has allowed the identification of several somatic variants of local cultivars as they are considered ‘Romé’ and ’Muscat of Alexandria’. In the case of ‘Romé’, the differences found between the accessions studied mainly affect bunch compactness. Grapevine bunch compactness is an economically important trait since it affects several major components of fruit quality. Foremost, compact clusters are more susceptible to pests and diseases [36]. Another somatic variant was detected for ‘Muscat de Alexandria’ cultivar, it is known with the local name ‘Moscatel de Alejandría Tinta’ because it presents red berries. Traditionally, when clones or somatic variants of the same variety have the same phenotypes different enough to be grown for the production of different wines, they are grouped in different cultivars [37] that could keep the name of the progenitor variety [38]. This somatic variant for the berry color of ‘Muscat of Alexandría’ was previously identified by De Lorenzis et al. [39]. They characterized ‘Zibbibo’ (synonymy of ‘Muscat of Alesandría’) and ‘Zibbibo Nero’ and determined that the color locus structure of ‘Zibibbo’ and its putative parents suggested that ‘Zibibbo Nero’ is a berry color revertant of ‘Zibibbo’. In this case, ‘Moscatel de Alejandría Tinta’ and ‘Zibibbo Nero’ would be different names for the same clone. However, the fact that ‘Moscatel de Alejandria Tinta’ and ‘Zibibbo Nero’ have black berries does not mean that they are the same clone but that they can be two different clones with black berries. Another somatic variant for the berry shape has also been described in Andalusia for a ‘Muscat of Alexandria’ accession collected in an ancient vineyard [40].
All these autochthonous cultivars and somatic variants located in the Axarquia region should be studied in order to generate knowledge to make new type of wines. Additionally, it could help to develop strategies to adapt viticulture in different regions to diverse models and markets that nowadays require to ensure the sustainability of the crop. According to Sancho-Galán et al. [41], in order to promote the cultivation of old and autochthonous cultivars, it would be necessary to apply for their inclusion in the Official Register of Authorized Varieties.
4. Materials and Methods
4.1. Plant Material
A set of 11 grapevine accessions located in seven vineyards of the Axarquia (Malaga, Spain) were studied. All studied accessions were collected and labelled with local names, except the sample accession M3 named as unknown (Supplementary Table S1, Supplementary Figure S1). Six of the accessions (M4, M6, M7, M8, M9, and M10) were named with the same local name, but were located in different vineyard plots. These accessions were analyzed with microsatellite markers and morphologic descriptors. Supplementary Table S1 and Supplemental Figure S1 show the code, location, and the local name accession. Furthermore, six reference grapevine cultivars (‘Airén’, ‘Cabernet Sauvignon’, ‘Chardonnay’, ‘Garnacha’, ‘Pinot noir’, and ‘Syrah’) were also included to test for microsatellite profiles obtained with the different database published [29,30,31].
The morphological descriptions were performed for the eight accessions (M1, M2, M3, M5, M7, M8, M10, and M11) identified as minor Andalusian cultivars.
4.2. DNA Extraction and Microsatellite Analysis
Two independent samples were analyzed for each accession. DNA was extracted from wood material using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. A total of 25 nuclear microsatellite loci were employed to perform the varietal identification following the methodology proposed by Urrestarazu et al. (2015) [42]. PCR mix was carried out in GeneAMP 9700 (Applied Biosystems, Foster City, CA, USA), and the amplified products were separated by capillary electrophoresis, using an automated sequencer ABI PRISM 3130 (Applied Biosystems, Foster City, CA, USA). Fluorescent labelled fragments (6-FAM, VIC, PET, and NED) were detected and sized using GeneMapper v. 3.7, and fragment lengths were assessed with the help of internal standards GeneScan-500 LIZTM (Applied Biosystems, Foster City, CA, USA). The comparison of the SSR obtained was performed using a microsatellite toolkit v. 9.0 software [43]. Lastly, the microsatellite genotypes obtained after the analysis were compared to the genetic profiles given by Jiménez-Cantizano et al. [30] and Lacombe et al. [31], and to the data contained in European grapevine database of microsatellite profiles VIVC [21].
4.3. Ampelographic Characterization
A total of 30 OIV descriptors were studied, 17 for mature leaves, six for bunches, and seven for berries (Supplementary Table S3). The morphological characterization was carried out during two consecutive years (2018 and 2019) in field and using a set of 30 descriptors selected from the International Organization of Vine and Wine’s descriptor list [44], including both qualitative and quantitative characteristics, observed or measured in 10 leaves, bunches, and berries. The ampelographic characterization was performed by three different ampelographers, over two years and the modal value is expressed following Benito et al. [45] criteria.
A hierarchical clustering analysis (HCA) using Ward method and the Euclidean square distance was performed, using the statistical software SPSS 24.0 (SPSS Inc., Chicago, IL, USA) to classify clusters according to samples similarity and dissimilarity.
5. Conclusions
The plant material that was localized and identified for the first time in this work is a source of interest for the wine sector. Molecular microsatellite analysis allowed the correct identification of the different red grapevine accessions located in ancient vineyards in the Axarquia region. A total eight cultivars were identified in this work of which only five correspond to Andalusian minor local cultivars. In addition, a new genotype was identified for ‘Cabriel’ cultivar. Ampelographic description of the minor local cultivars has contributed to detecting two somatic variants or clones, one for ‘Muscat of Alexandria’ and another one for ‘Romé’.
Acknowledgments
The authors would like to acknowledge the support received from Axarquia region private vineyards.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/9/11/1572/s1, Figure S1. Vineyard map location. Table S1. Code and local name accession, location, and municipality of the vineyard in Malaga (Spain). Table S2. Microsatellite profile of six reference cultivars analyzed at 25 microsatellite loci. Table S3. Code, description, and scale of the 30 OIV descriptors selected for the ampelographic characterization.
Author Contributions
Conceptualization, A.J.-C., A.A.-A., P.S.-G. and V.P.; methodology, A.J.-C., A.M.-M., A.A.-A. and P.S.-G.; software, A.J.-C.; validation, A.J.-C., A.A.-A., P.S.-G. and V.P.; formal analysis, A.J.-C. and A.M.-M.; investigation, A.J.-C., A.A.-A. and P.S.-G.; resources, A.J.-C. and A.M.-M.; data curation, A.J.-C. and P.S.-G.; writing—original draft preparation, A.J.-C., A.A.-A. and P.S.-G.; writing—review and editing, A.J.-C., A.A.-A., P.S.-G. and V.P.; visualization, A.J.-C., A.M.-M., A.A.-A. and P.S.-G.; supervision, A.J.-C., A.A.-A. and P.S.-G. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Benabent M. La Axarquía, un paisaje en proceso de transformación. Rev. PH Inst. Andal. Patrim Histórico. 2009;71:44–51. [Google Scholar]
- 2.Quintana-Toret F.J. Origen histórico de la viticultura malagueña. Baetica Estud. Arte Geogr. Hist. 1985;8:393–404. [Google Scholar]
- 3.Jiménez-Cantizano A., Amores-Arrocha A., Gutiérrez-Escobar R., Palacios V. Short communication: Identification and relationship of the autochthonous ‘Romé’ and ‘Rome Tinto’ grapevine cultivars. Span. J. Agric. Res. 2018;16:e07SC02. doi: 10.5424/sjar/2018164-13142. [DOI] [Google Scholar]
- 4.Sancho-Galán P., Amores-Arrocha A., Palacios V., Jiménez-Cantizano A. Genetical, morphological and physicochemical characterization of the autochthonous cultivar ‘Uva Rey’ (Vitis vinifera L.) Agronomy. 2019;9:563. doi: 10.3390/agronomy9090563. [DOI] [Google Scholar]
- 5.García-Muñoz S., Muñoz-Organero G., De Andrés M.T., Cabello F. Ampelography: An old technique with future uses: The case of minor varieties of Vitis vinifera L. from the Balearic Islands. J. Inter. Sci. Vigne Vin. 2011;45:125–137. doi: 10.20870/oeno-one.2011.45.3.1497. [DOI] [Google Scholar]
- 6.Zinelabidine L.H., Cunha J., Eiras-Diaz J.E., Cabello F., Martínez-Zapater J.M., Ibánez J. Pedigree analysis of the Spanish grapevine cultivar ‘Heben’. Vitis. 2015;54:81–86. [Google Scholar]
- 7.Martínez-Zapater J.M., Carmona M.J., Díaz-Riquelme J., Fernández L., Lijavetzky D. Grapevine genetics after the genome sequence: Challenges and limitations. Aust. J. Grape Wine Res. 2010;16:33–46. doi: 10.1111/j.1755-0238.2009.00073.x. [DOI] [Google Scholar]
- 8.De los Salmones G. La Invasión Filoxérica en España y las Cepas Americanas. 1st ed. Tipolitografía de Luis Tasso; Barcelona, Spain: 1893. [Google Scholar]
- 9.Carnero T. Expansión Vinícola y Atraso Agrario 1870–1900. La Viticultiura Española Durante la Gran Depression. 2nd ed. Ministerio de Agricultura; Madrid, Spain: 1980. [Google Scholar]
- 10.Ghiara B. La Vinificación Mediante el Exclusivo Empleo de la Asepsia Industrial. 1st ed. Escuela Tipográfica Salesiana; Malaga, Spain: 1918. [Google Scholar]
- 11.Clemente y Rubio S.D.R. Ensayo Sobre las Variedades de la vid Común que Vegetan en Andalucía. Imprenta de Villalpando; Madrid, Spain: 1807. [Google Scholar]
- 12.De la Leña C.G. Disertación en Recomendación y Defensa del Famoso vino Malagueño Pero Ximen y Modo de Formarlo. 1st ed. Servicio de Publicaciones y Divulgación Científica de la Universidad de Málaga; Malaga, Spain: 1792. [Google Scholar]
- 13.Sánchez M.T., Gómez M.D., López M.I., López M.J. El viñedo en la comarca de «La Axarquía» (Málaga). Situación actual y futuro. Alimentaria. 1997;225:65–71. [Google Scholar]
- 14.Jiménez-Cantizano A. Ph.D. Thesis. Universidad de Cadiz; Cadiz, Spain: 2014. Caracterización Molecular del Banco de Germoplasma de vid del Rancho de la Merced. [Google Scholar]
- 15.Marsal G., Méndez J.J., Mateo J.M., Ferrer S., Canals J.M., Zamora F., Fort F. Molecular characterization of Vitis vinifera L. local cultivars from volcanic areas (Canary Islands and Madeira) using SSR markers. Oeno One. 2019;4:667–680. doi: 10.20870/oeno-one.2019.53.4.2404. [DOI] [Google Scholar]
- 16.Cipriani G., Spadotto A., Jurnam I., Di Gaspero G., Crespan M., Meneghetti S., Frare E., Vignani R., Cresti M., Morgante M., et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages, and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010;121:1569–1585. doi: 10.1007/s00122-010-1411-9. [DOI] [PubMed] [Google Scholar]
- 17.Cho K.H., Bae K.M., Noh J.H., Shin I.S., Kim S.H., Kim J.H., Kim D.Y., Hwang H.S. Genetic diversity and identification of Korean grapevine cultivars using SSR markers. Korean. J. Breed. Sci. 2011;43:422–429. [Google Scholar]
- 18.Basheer-Salimia R., Lorenzi S., Batarseh F., Moreno-Sanz P., Emanuelli F., Grando M.S. Molecular identification and genetic relationships of Palestinian grapevine cultivars. Mol. Biotechnol. 2014;56:546–556. doi: 10.1007/s12033-013-9728-7. [DOI] [PubMed] [Google Scholar]
- 19.Losada E.D., Salgado A.T., Ramos-Cabrer A.M., Segade S.R., Diéguez S.C., Pereira-Lorenzo S. Twenty microsatellites (SSRs) reveal two main origins of variability in grapevine cultivars from Northwestern Spain. Vitis. 2010;49:55–62. [Google Scholar]
- 20.Hizarci Y., Ercisli S., Yuksel C., Ergul A. Genetic characterization and relatedness among autochthonous grapevine cultivars from Northeast Turkey by Simple Sequence Repeats (SSR) J. Appl. Bot. Food Qual. 2013;85:224–228. [Google Scholar]
- 21.Dong Z., Liu W., Li X., Tan W., Zhao Q., Wang M., Ren R., Ma X., Tang X. Genetic relationships of 34 grapevine varieties and construction of molecular fingerprints by SSR markers. Biotechnol. Biotechnol. Equip. 2017;32:942–950. doi: 10.1080/13102818.2018.1450162. [DOI] [Google Scholar]
- 22.Eyduran S.P., Ercisli S., Akin M., Eyduran E. Genetic characterization of autochthonous grapevine cultivars from Eastern Turkkey by simple sequence repeats (SSRs) Biotechnol. Biotechnol. Equip. 2016;30:26–31. doi: 10.1080/13102818.2015.1105726. [DOI] [Google Scholar]
- 23.Lopes M.S., Sefc K.M., Eiras-Dias E., Steinkellner H., Câmara Laimer M.L., Câmara Machado A. The use of microsatellites for germplasm management in a Portuguese grapevine collection. Theor. Appl. Genet. 1999;99:733–739. doi: 10.1007/s001220051291. [DOI] [PubMed] [Google Scholar]
- 24.González-Andrés F., Martín J.P., Yuste J., Rubio J.A., Arranz C., Ortiz J.M. Identification and molecular biodiversity of autochthonous grapevine cultivars in the “Comarca del Bierzo”, León, Spain. Vitis. 2007;46:71–76. [Google Scholar]
- 25.Boccacci P., Torello-Marinoni D., Gambino G., Botta R., Schneider A. Genetic Characterization of Endangered Grape Cultivars of Reggio Emilia Province. Am. J. Enol. Vitic. 2005;56:411–416. [Google Scholar]
- 26.Casanova J., Mozas P., Ortiz J.M. Ampelography and microsatellite DNA analysis of autochthonous and endangered grapevine cultivars in the province of Huesca (Spain) Span. J. Agric. Res. 2011;9:790–800. doi: 10.5424/sjar/20110903-431-10. [DOI] [Google Scholar]
- 27.Balda P., Ibáñez J., Sancha J.C., de Toda F.M. Characterization and identification of minority red grape varieties recovered in Rioja, Spain. Am. J. Enol. Vitic. 2014;65:148–152. doi: 10.5344/ajev.2013.13050. [DOI] [Google Scholar]
- 28.Ferreira V., Pinto-Carnide O., Mota T., Martín J.P., Ortíz J.M., Castro I. Identification of minority grapevine cultivars from Vinhos Verdes Portuguese DOC Region. Vitis. 2015;54:53–58. [Google Scholar]
- 29.Vitis International Variety Catalogue. [(accessed on 23 October 2020)]; Available online: www.vivc.de.
- 30.Jiménez-Cantizano A., García de Luján A., Arroyo-García R. Molecular characterization of table grape varieties preserved in the Rancho de la Merced Grapevine Germplasm Bank (Spain) Vitis. 2018;57:93–101. [Google Scholar]
- 31.Lacombe T., Boursiquot J.M., Laucou V., Di Vecchi-Staraz M., Péros J.P., This P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.) Theor. Appl. Genet. 2013;126:401–414. doi: 10.1007/s00122-012-1988-2. [DOI] [PubMed] [Google Scholar]
- 32.Popescu C.F., Crespan M. Combining Microsatellite Markers and Ampelography for Better Management of Romanian Grapevine Germplasm Collections. Not. Sci. Biol. 2018;10:193–198. doi: 10.15835/nsb10210297. [DOI] [Google Scholar]
- 33.Martín J.P., Arranz C., Castro I.D., Yuste J., Rubio J.A., Pinto-Carnide O., Ortiz J.M. Prospection and identification of grapevine varieties cultivated in north Portugal and northwest Spain. Vitis. 2011;50:29–33. [Google Scholar]
- 34.Maul E., Töpfer R. BIO Web of Conferences, Proceedings of the 38th World Congress of Vine and Wine (Part 1), Mainz, Germany, 5–10 July 2015. EDP Sciences; Les Ulis, France: 2015. Vitis International Variety Catalogue (VIVC): A cultivar database referenced by genetic profiles and morphology; p. 01009. [Google Scholar]
- 35.Sancho-Galán P., Amores-Arrocha A., Palacios V., Jiménez-Cantizano A. Preliminary Study of Somatic Variants of Palomino Fino (Vitis vinifera L.) Grown in a Warm Climate Region (Andalusia, Spain) Agronomy. 2020;10:654. doi: 10.3390/agronomy10050654. [DOI] [Google Scholar]
- 36.Tello J., Ibáñez J. What do we know about grapevine bunch compactness? A state-of-the-art review. Aust. J. Grape Wine Res. 2018;24:6–23. doi: 10.1111/ajgw.12310. [DOI] [Google Scholar]
- 37.This P., Boursiquot J.M. Essai de définition du cépage. Prog. Agric. Vitic. 1999;116:359–361. [Google Scholar]
- 38.Carbonell-Bejerano P., Royo C., Mauri N., Ibáñez J., Zapater J.M. Somatic variation and cultivar innovation in grapevine. In: Morata A., Loira I., editors. Advances in Grape and Wine Biotechnology. 1st ed. IntechOpen; London, UK: 2016. [Google Scholar]
- 39.De Lorenzis G., Squadrito M., Brancadoro L., Scienza A. Zibibbo Nero characterization, a red-wine grape revertant of Muscat of Alexandria. Mol. Biotechnol. 2015;57:265–274. doi: 10.1007/s12033-014-9820-7. [DOI] [PubMed] [Google Scholar]
- 40.Jiménez- Cantizano A., Lara M., Ocete M.E., Ocete R. Short communication: Characterization of the relic Almuñécar grapevine cultivar. Span. J. Agric. Res. 2012;10:454–460. doi: 10.5424/sjar/2012102-549-11. [DOI] [Google Scholar]
- 41.Sancho-Galán P., Amores-Arrocha A., Palacios V., Jiménez-Cantizano A. Identification and characterization of white grape varieties autochthonous of a warm climate region (Andalusia, Spain) Agronomy. 2020;10:205. doi: 10.3390/agronomy10020205. [DOI] [Google Scholar]
- 42.Urrestarazu J., Royo J., Santesteban L.G., Miranda C. Evaluating the Influence of the Microsatellite Marker Set on the Genetic Structure Inferred in Pyrus communis L. PLoS ONE. 2015;10:e0138417. doi: 10.1371/journal.pone.0138417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S.D.E. Ph.D. Thesis. University of Dublin; Dublin, Ireland: 2001. Trypanotolerance in West African Cattle and the Population Genetic Effects of Selection. [Google Scholar]
- 44.Organisation Internationale de la Vigne et du Vin (OIV) OIV Descriptor List for Grape Varieties and Vitis Species. 2nd ed. OIV; Paris, France: 2009. [Google Scholar]
- 45.Benito A., Muñoz-Organero G., de Andrés M.T., Ocete R., García-Muñoz S., López M.A., Arroyo-García R., Cabello F. Ex situ ampelographical characterisation of wild Vitis vinifera from fifty-one Spanish populations. Aust. J. Grape Wine Res. 2017;23:143–152. doi: 10.1111/ajgw.12250. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.