Abstract
Simple Summary
Anoscopus leafhoppers are a group of plant-feeding bugs that can be found in a range of grassland habitats. There are seven recognized species in the UK, some of which are difficult to tell apart. One species, Anoscopus duffieldi, has only been found at a single site, an RSPB (Royal Society for the Protection of Birds) reserve at Dungeness in Kent. As Anoscopus leafhoppers can be quite variable in colour and pattern, and in the structure of their genitalia, our aim was to establish, using DNA, whether this ‘species’ is unique or whether it is simply a variant of one of the other species. If it is unique, found nowhere else, it should be afforded special protection. Samples of all UK species, as well as another from the Czech Republic, were collected from the field, and two genes were examined. The DNA sequences showed that three species, A. duffieldi, Anoscopus albifrons and Anoscopus limicola were so closely related that they should probably be considered a single species. However, A. duffieldi are distinctive in that they live only in one area of vegetated shingle. We suggest that, until other evidence is forthcoming, A. duffieldi could be considered a locally adapted subspecies of scientific interest.
Abstract
The subfamily Aphrodinae (Hemiptera: Cicadellidae) contains ~33 species in Europe within four genera. Species in two genera in particular, Aphrodes and Anoscopus, have proved to be difficult to distinguish morphologically. Our aim was to determine the status of the putative species Anoscopus duffieldi, found only on the RSPB Nature Reserve at Dungeness, Kent, a possible rare UK endemic. DNA from samples of all seven UK Anoscopus species (plus Anoscopus alpinus from the Czech Republic) were sequenced using parts of the mitochondrial cytochrome oxidase I and 16S rRNA genes. Bayesian inference phylogenies were created. Specimens of each species clustered into monophyletic groups, except for Anoscopus albifrons, A. duffieldi and Anoscopus limicola. Two A. albifrons specimens grouped with A. duffieldi repeatedly with strong support, and the remaining A. albifrons clustered within A. limicola. Genetic distances suggest that A. albifrons and A. limicola are a single interbreeding population (0% divergence), while A. albifrons and A. duffieldi diverged by only 0.28%. Shared haplotypes between A. albifrons, A. limicola and A. duffieldi strongly suggest interbreeding, although misidentification may also explain these topologies. However, all A. duffieldi clustered together in the trees. A conservative approach might be to treat A. duffieldi, until other evidence is forthcoming, as a possible endemic subspecies.
Keywords: Anoscopus duffieldi, Aphrodinae, Cicadellidae, endemic species, genetic distance, molecular separation
1. Introduction
Conventional methods of species identification and separation rely on morphological features to distinguish taxa [1]. Erroneous identification can undermine taxonomy, ecological research, conservation efforts and ecosystem management [2,3,4]. Serious problems can arise when type specimens are involved [5]. Differences between morphologically similar species, and their phylogenetic relationships, can often be resolved using genetic evidence [6,7,8,9].
Auchenorrhyncha are within the fifth most diverse insect order, Hemiptera [10,11], and comprise ~43,000 species worldwide, including leafhoppers, planthoppers, treehoppers, froghoppers (spittlebugs) and cicadas [12,13]. These herbivorous insects variously feed on xylem or phloem sap or mesophyll contents [14]. Many leafhoppers are plant pathogen vectors [15,16] or are studied as part of conservation efforts [14,17] or evaluation of community structure [18]. Species separation within many leafhopper genera is seriously understudied and hampered by the presence of morphologically cryptic species and biotypes.
The subfamily Aphrodinae (Hemiptera: Cicadellidae) contains 33 species in Europe within four genera: Stroggylocephalus Flor, 1861; Planaphrodes Hamilton, 1975; Aphrodes Curtis, 1833; and Anoscopus Kirschbaum, 1868 [19,20], of which 15 species and all genera occur in the UK [21]. Species in Aphrodes, Planaphrodes and Anoscopus are morphologically similar, to the extent that all Anoscopus were previously regarded as Aphrodes [22,23,24,25]. The current split between Anoscopus and Aphrodes dates back to Hamilton [24]. External characters were used to distinguish between leafhopper species until the late 1930s, when the aedeagus became the primary discriminator [26]. Some Aphrodes and Anoscopus species have proved difficult to distinguish morphologically, the differences being mainly based upon the details of the male aedeagus, such as its shape and positions of spines [9,20,27]. However, these characters are subject to intraspecific variability and interspecific overlap, and females and nymphs cannot be separated reliably [9]. More accurate morphological identification of male Aphrodes and Anoscopus is generally possible using a combination of aedeagus and external morphometric measurements [9,20,28].
A possibly new species of Aphrodes was reported by Duffield [29] from Dungeness, Kent. Duffield noted banded elytra on three males, similar to Aphrodes assimilis (now Anoscopus assimilis (Signoret, 1879)), which had not been recorded in Britain at the time. Additional morphological characters of A. assimilis, published by Ribaut [22], supported Duffield’s initial identification. Subsequently, Duffield sent specimens to Le Quesne, who established it as a new species, Aphrodes duffieldi (now Anoscopus duffieldi (Le Quesne, 1964)), possibly confined to Kent, based upon aedeagus characters [30]. Le Quesne [23] later considered that A. duffieldi could be synonymous with Anoscopus alpinus (Wagner, 1955), with this continental species regarded as conspecific with A. assimilis by Nast [31], Hamilton [24] and, with question marks, Remane and Fröhlich [27]. Guglielmino and Buckle [20] treated A. assimilis and A. alpinus as separate species, based on differences in the forewing shape, colour and aedeagus size and structure. They also studied the morphology of nine male specimens of A. duffieldi from the type locality and noticed a large variability in the aedeagus morphology and external similarity to another species, A. albifrons (Linnaeus, 1758), suggesting that A. duffieldi specimens may represent hybrids between A. albifrons and A. alpinus or A. assimilis, but concluded that the problem needed further research.
Britain has very few endemic taxa, and therefore species (and subspecies) that are found to be endemic are often given high levels of protection (e.g., designated Sites of Special Scientific Interest) [32]. The present study independently tests the findings of previous morphologically-based work by using molecular data. Our aim was to separate species of Anoscopus by analysis of DNA sequences in order to resolve the status of A. duffieldi at its only known location in the UK and, as far as is known, the world [21] and some other taxonomic uncertainties within the Anoscopus genus. Accurate species separation is an essential precursor to meaningful ecological research and conservation planning in this genus.
2. Materials and Methods
2.1. Specimen Collection
Anoscopus specimens from the UK were mostly collected by suction sampling between 2011 and 2015 (Figure 1). Samples of A. alpinus were acquired in the Czech Republic in 2015. Details of the collection sites and preservation methods are described in Table 1. The material was initially identified based on morphology using the keys by Le Quesne [23], Biedermann and Niedringhaus [25] and Wilson et al. [21]. Anoscopus specimens from Dungeness were attributed to A. duffieldi or A. albifrons based on the aedeagal characters used by Le Quesne [23] and Guglielmino and Buckle [20], although the published differences between these taxa are slight and some specimens displayed characters that appeared intermediate. No other Anoscopus species were collected from this site. Photographs and drawings of the Anoscopus species have been published previously [20,25,33].
Figure 1.
United Kingdom locations where Anoscopus were sampled (site details given in Table 1) with enlarged inset for SE England. Scale bars indicate distances in kilometers.
Table 1.
Collection site, year and storage method for the Anoscopus specimens sequenced.
| Anoscopus Species | Collection Site | Ordnance Survey + | Co-Ordinates (N, E) | Year Collected | Storage |
|---|---|---|---|---|---|
| albifrons | Dungeness, Kent | TR076190 | 50.933074, 0.95310771 | 2013 | 100% Ethanol * |
| Newtimber Hill, Sussex | TQ268119 | 50.892674, −0.19846861 | 2015 | 100% Ethanol | |
| albiger | Wartling, Sussex | TQ666085 | 50.852027, 0.36542503 | 2014/15 | Dried/100% Ethanol |
| alpinus | Mt Kralicky Sneznik, Czech Rep. | 50.206401, 16.849404 | 2015 | 100% Ethanol | |
| duffieldi | Dungeness, Kent | TR076190 | 50.933074, 0.95310771 | 2014/15 | 100% Ethanol |
| flavostriatus | Winding Bottom, Sussex | TQ191087 | 50.865548, −0.30893968 | 2013 | Dried |
| 2014/15 | 100% Ethanol | ||||
| limicola | Colne Point, Essex | TM108124 | 51.770566, 1.0538762 | 2011 | 100% Ethanol * |
| Malacleit, Outer Hebrides | NF790730 | 56.632199, −7.3805002 | 2012 | Dried | |
| serratulae | Rye Harbour, Sussex | TQ931192 | 50.939904, 0.74711850 | 2015 | 100% Ethanol |
| histrionicus | Merthyr Common, Wales | SO071058 | 51.743173, −3.3469341 | 2015 | Ethanol § |
+ Ordinance Survey is the National mapping agency for Great Britain and is widely used for determining co-ordinates. * specimens were frozen prior to transfer into 100% ethanol. § specimen killed with ethyl acetate, frozen and dried prior to transfer into molecular-grade ethanol.
2.2. Choice of Molecular Markers
The mitochondrial cytochrome c oxidase subunit 1 (CO1) gene in particular has been used to resolve species-level separation and relationships in animal taxa, including insects, due to its relatively rapid mutation rate [34,35], lack of recombination and highly conserved regions for relatively easy amplification from small or degraded specimens [7,36]. As such, there is a wide range of primers designed for this region [8]. Mitochondrial ribosomal genes (e.g., 16S) can also be useful for barcoding and phylogenetics of closely related species [8] but in many taxa are more conserved than the CO1 barcoding region. Mitochondrial DNA is also suitable for calculating genetic distances within and between species [37]; however, due to maternal inheritance, hybridisation between species may occur, altering phylogenetic results. For this and other reasons, parallel nuclear gene analysis has become increasingly used to ensure correct relationships. The nuclear 28S ribosomal gene has 12 divergent domains (D1-D12) within five fragments, differing in variability [38]. Some domains have been previously used in leafhopper phylogenetic studies [15,38,39]. We initially chose therefore to target regions of the CO1, 16S and 28S genes to facilitate species separation.
2.3. DNA Extraction and PCR Amplification
Qiagen’s DNeasy® Blood and Tissue kit (Qiagen, Hilden, Germany) was used to extract DNA from all Anoscopus specimens following the manufacturer’s protocol.
PCR reaction mixtures for both CO1 and 16S amplification consisted of 5 μL Multiplex master mix (Qiagen, Hilden, Germany), 3.6 μL RNase free water, 0.2 μL of each primer (10 pmol/μL) and 1 μL extracted DNA with a final volume of 10 μL. All PCRs had an initial 15 min denaturation step at 95 °C. General invertebrate primers, LCO1490 and HCO2198, targeting the mt CO1 gene [40] (Table 2), used a PCR protocol with 42 cycles as follows: 30 s at 94 °C, 90 s at 50 °C and 90 s at 72 °C and a final 10 min elongation step at 72 °C. 16S rRNA primers LR-J-12887 and LR-N-13398 [41] (Table 2) were used with 35 cycles of the following: 30 s at 94 °C, 90 s at 51 °C and 90 s at 72 °C, prior to 10 min elongation at 72 °C. Successful PCR products were purified using 1.25 μL Multicore 10X Buffer, 0.5 μL TSAP and 0.25 μL EXO1 (Thermofisher Scientific, Newport, Wales, UK) in a final volume of 2 μL with one thermal cycle of 30 min at 37 °C, 15 min at 80 °C and 5 min at 12 °C, before submission to Eurofins MWG Operon (Ebersburg, Germany) for sequencing. Amplification of nuclear 28S ribosomal DNA was also attempted using two sets of primer pairs. The first pair was originally designed by Hillis and Dixon [42] with modifications by Zahniser [43]; 28SP & 28SM2 were used to target Fragment I (D2-D3) and the second pair from Dietrich et al. [38]; 28SIIF and 28SIIR amplified Fragment II (D3-D6). PCR mixes were described as above for CO1 and 16S with thermal conditions of 15 min at 95 °C and 30 cycles of 1 min at 94 °C, 1 min at 51 °C, 2 min at 72 °C and a final elongation of 7 min at 72 °C.
Table 2.
Forward (top) and reverse (bottom) primer names, sequences (5′ to 3′) and reference for each primer pair for the mitochondrial CO1 (Cytochrome Oxidase 1) gene, the mitochondrial ribosomal gene16S rRNA and the nuclear 28S ribosomal RNA gene (rRNA).
| Locus | Primer Name | Primer Sequence (5′–3′) | Reference |
|---|---|---|---|
| CO1 | LCO1490 | GGTCAACAAATCATAAAGATATTGG | Folmer et al., 1994 [40] |
| HCO2198 | TAAACTTCAGGGTGACCAAAAAATCA | ||
| 16S | LR-J-12887 | CCGGTYTGAACTCARATCAWGT | Fu et al., 2014 [41] |
| LR-N-13398 | CTGTTTAWCAAAAACATTTC | ||
| 28S | 28SP | AGTCGKGTTGCTTGAKAGTGCAG | Zahniser, 2008 [43] |
| 28SM2 | TTCGGGTCCCAACGTGTACG | ||
| 28SII’(F) | GGGACCCGTCTTGAAACAC | Dietrich et al., 2001 [38] | |
| 28SII’(R) | ACCCTCCTACTCGTCAAGG |
2.4. Sequencing Analysis
All sequences obtained were confirmed to be of the mitochondrial CO1 gene, because no stop codons were found and the nucleotide sequences corresponded to the expected amino acids of the first 600 bp of the CO1 gene, and this was confirmed by a Blast search. Chromatograms were analysed using Sequencher v4.9 (Gene Codes, Ann Arbor, MI, USA.), resolving sequence ambiguities and producing consensus sequences with final lengths of 600, 420 and 1358 bp for the CO1, 16S rRNA and 28S genes, respectively. Contigs were created in Sequencher, with 28S fragments I and II separately sequenced and concatenated to generate contigs before being aligned in ClustalX v2.1 [44].
2.5. Phylogenetic Analyses
A likelihood ratio test as implemented in jModelTest v2.1.7 [45,46] was used to determine the best-fit model of DNA substitution under the Akaike Information Criterion (AIC). Additional parameters such as base frequencies, the shape parameter of the gamma distribution [47,48] and the proportion of invariable sites (I) were also estimated. This model was subsequently used in Bayesian Inference as implemented in MrBayes v3.2 [49] and then used to calculate distances. Four chains were run for 5 × 106 generations using random starting trees and flat priors. Trees and parameters were recorded every 100th generation, and two runs were performed simultaneously. Split frequencies were compared every 100th generation, and chain convergence was evaluated in Tracer v1.6 [50]. All runs used the default heating and swap parameters. In addition, FigTree v1.4.2 [51] was used to view the Bayesian trees with posterior probabilities. Three phylogenies were produced based on the mt CO1 gene, 16S rRNA gene and a concatenated dataset, with Aphrodes bicincta (Schrank, 1776) as the closely related outgroup which suitably resolved the ingroup taxa. The CO1 and 28S sequences for A. bicincta were downloaded from GenBank (accession numbers KR042069.1 for CO1 and AF304579.1 for 28S), and an archived DNA extract was sequenced for the 16S rRNA outgroup as this sequence was not present in GenBank.
2.6. Population-Level Analyses
Diversity indices such as haplotype diversity (the probability that two randomly chosen sequences are different in the sample) [52] and nucleotide diversity, π (the average number of nucleotide differences per site between two sequences) [53], were calculated for each phylogenetic lineage as identified in the Bayesian tree using DnaSP version 6 [54,55]. Within- and between-group pairwise estimates of nucleotide sequence divergence were generated in MEGA v6.0 [56] (Table 3 and Table 4) by implementing a correction factor as described in Nei and Li [37].
Table 3.
Within-group (intraspecific) mean pairwise distances (d) with corresponding standard errors (generated by MEGA v6.0; [56] for CO1and 16S, for Anoscopus species. Anoscopus histrionicus was not included as there was only one individual available. d = divergence, S.E. = standard error.
| Species | CO1 | 16S | ||
|---|---|---|---|---|
| d (%) | S.E. (%) | d (%) | S.E. (%) | |
| albifrons | 0.8 | 0.2 | 0.4 | 0.2 |
| albiger | 2.4 | 0.4 | 0.7 | 0.3 |
| alpinus | 0.6 | 0.2 | 0.3 | 0.3 |
| duffieldi | 0.3 | 0.2 | 0.3 | 0.3 |
| flavostriatus | 0.3 | 0.2 | 0.3 | 0.3 |
| limicola | 0.3 | 0.1 | 0.8 | 0.5 |
| serratulae | 0.4 | 0.2 | 0.5 | 0.3 |
Table 4.
Mitochondrial cytochrome oxidase 1 and 16S rRNA per cent haplotype and nucleotide diversity as identified by the network and phylogenetic analyses within Anoscopus.
| Species | CO1 | 16S rRNA | ||
|---|---|---|---|---|
| Haplotype Diversity | Nucleotide Diversity | Haplotype Diversity | Nucleotide Diversity | |
| A. albifrons | 0.81 | 0.004 | 0.5 | 0.001 |
| A. albiger | 1.00 | 0.024 | 0.46 | 0.003 |
| A. alpinus | 0.98 | 0.006 | 0.29 | 0.0007 |
| A. duffieldi | 0.68 | 0.003 | 0.44 | 0.001 |
| A. flavostriatus | 0.65 | 0.001 | 0.33 | 0.0009 |
| A. limicola | 0.38 | 0.001 | 0.11 | 0.0009 |
| A. serratulae | 0.81 | 0.004 | 0.29 | 0.001 |
Haplotype networks for both the CO1 and 16S rRNA genes were constructed showing the minimum mutational steps between different haplotypes using TCS (Templeton Crandall Singh network) with 95% confidence limits [57]. The haplotype networks, in conjunction with frequencies and geographic distribution of different haplotypes, were used to depict geographical and potential ancestor–descendant relationships among the identified sequences.
3. Results
3.1. Nucleotide and Haplotype Diversity
Nucleotide and haplotype diversity for species (CO1 gene) varied from 0.001–0.024 and 0.38–1.00, respectively. Nucleotide and haplotype diversity for the 16S rRNA gene was lower (values varied from 0.0009 to 0.003 and 0.11 to 0.5, respectively). These rather low nucleotide diversity values for both gene regions are indicative of shallow divergences [58]. Species Anoscopus albiger (Germar, 1821), A. albifrons, A. alpinus and A. serratulae (Fabricius, 1775) were characterised by high haplotypic diversity values, indicating the high incidence of locality-specific haplotypes (Table 4).
3.2. Phylogenetic Analyses
The best fit General Time Reversible (GTR + G (0.089) + I (0.449)) and Hasegawa–Kishino–Yano 85 (HKY85 + G (0.024) models of substitution were applied in all phylogenetic analyses for the CO1 and 16S rRNA genes, respectively.
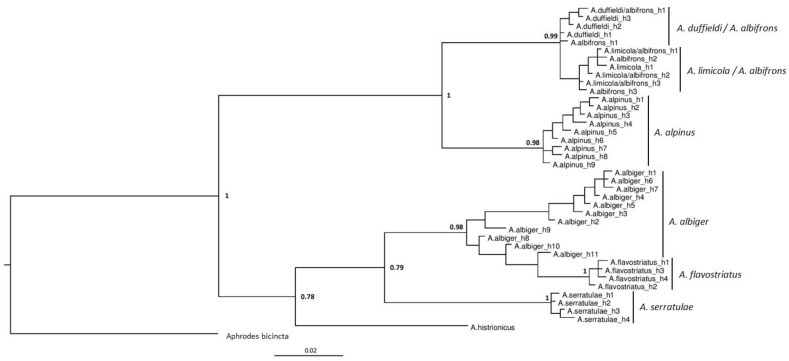
Three phylogenies were generated based on the CO1 gene (Figure 2), 16S rRNA gene (Figure 3) and the combined dataset including both the CO1 and 16S genes (Figure 4). Although sequencing was successful for 28S, this region failed to resolve closely related species as sequences were identical, hence these results are not shown. Phylogenies based on the CO1 gene and the 16S rRNA gene, in general, reflect the same topologies and mainly separated taxa monophyletically, with a few exceptions. Each tree separated the entire Anoscopus genus into two subgroups; one containing A. albifrons, A. limicola (Edwards, 1908), A. duffieldi and A. alpinus and the other A. albiger, A. flavostriatus (Donovan, 1799), A. serratulae and A. histrionicus (Fabricius, 1794), with strong support in each tree (posterior probabilities of 1) (Figure 2, Figure 3 and Figure 4). This was also reflected in the networks (Figure 5 and Figure 6). Anoscopus albifrons was clustered within both A. limicola and A. duffieldi, with two A. albifrons specimens grouping with A. duffieldi sequences and a number of A. albifrons and A. limicola specimens clustering together. There were shared haplotypes between A. duffieldi and A. albifrons and also between A. limicola and A. albifrons, indicating identical sequences across these taxa. The fourth species within this subgroup, A. alpinus, was clearly separated from the A. duffieldi, A. limicola and A. albifrons aggregate.
Figure 2.
Bayesian phylogeny based on mitochondrial cytochrome c oxidase subunit 1 (CO1) DNA data. Haplotypes were identified in DnaSP v5.10.01 [55] from individual sequences, with species names separated by ‘/’ indicating shared haplotypes across two different taxa. Bayesian posterior probabilities are labelled at each node with the whole tree rooted by using the closely related Aphrodes bicincta as an outgroup.
Figure 3.
Bayesian phylogeny based on mitochondrial 16S rRNA data. Haplotypes were identified in DnaSP v5.10.01 [55] from individual sequences, with species names separated by ‘/’ indicating shared haplotypes across two different species. Bayesian posterior probabilities are labelled at each node with the whole tree rooted by using the closely related Aphrodes bicincta as an outgroup.
Figure 4.
Bayesian phylogeny based on the concatenated mitochondrial CO1 and 16S rRNA data. Haplotypes were identified in DnaSP v5.10.01 [55] from individual sequences, with species names separated by ‘/’ indicating shared haplotypes across two different species. Bayesian posterior probabilities are labelled at each node with the whole tree rooted by using the closely related Aphrodes bicincta as an outgroup.
Figure 5.
TCS network based on the mitochondrial CO1 gene. Each circle represents a unique haplotype, and the size of each circle is proportional to the number of samples. Cross-hatching along branches represents the number of mutational steps. (TCS only connects alleles with a 95% confidence limit, i.e., 10 steps). The colours represent localities, and the circles are labelled with the name of the haplotype as seen in Figure 2, Figure 3 and Figure 4. Stars indicate ancestral haplotypes.
Figure 6.
TCS network based on the mitochondrial 16S rRNA gene. Each circle represents a unique haplotype, and the size of each circle is proportional to the number of samples. Cross-hatching along branches represents the number of mutational steps. (TCS only connects alleles with a 95% confidence limit, i.e., 10 steps). The colours represent localities, and the circles are labelled with the name of the haplotype as seen in Figure 2, Figure 3 and Figure 4. Stars indicate ancestral haplotypes.
The other major clade that included A. albiger, A. flavostriatus, A. serratulae and A. histrionicus was resolved somewhat differently, but all species were clearly separated in the 16S tree (Figure 3), concatenated dataset (Figure 4) and network (Figure 6).
3.3. Population Level Analyses
Within-species sequence divergences were low in all species for the mitochondrial CO1 gene (divergences < 0.8%, CO1) except A. albiger, which harboured more within-species diversity (2.4%, CO1, Table 3). Genetic distances between A. duffieldi, A. limicola and A. albifrons were low, especially between A. albifrons and A. limicola, where a sequence divergence value of 0.05% was recorded for the CO1 gene and 0% for both the ribosomal 16S rRNA gene and the combined dataset. Sequence divergence values between A. albifrons and A. duffieldi were 0.35% (CO1), 0.15% (16S) and 0.35% (CO1 + 16S). Likewise, divergences between A. limicola and A. duffieldi were low at 0.8% (CO1), 0.15% (16S) and 0.55% (16S + CO1) (Table 5). These distances between species were well below the within-species genetic distances for A. albiger. However, it connects with the three-species aggregate when using 16S gene sequences (Figure 6). Haplotype diversity between species (groups identified in the TCS network) ranged from 0.38 to 1 and 0.11 to 0.50 for the CO1 and 16S, respectively. The suggested heterogeneity within A. albiger is mirrored by the high haplotype and nucleotide diversities recorded for this species (Table 4).
Table 5.
Mean pairwise distances (%) (corrected for intra-specific distances) between Anoscopus taxa for CO1 and 16S in bold below each diagonal with standard error values (%) above (generated by MEGA v6.0; [56] Tamura et al., 2013). Anoscopus histrionicus was not included as there was only one individual available.
| Loci and Species Name | Mean Genetic Distance/Standard Error (%) | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| CO1 | ||||||
| 1. albifrons | 1 | 0.9 | 0.3 | 1.2 | 0.2 | |
| 2. albiger | 10.2 | 1.2 | 1 | 0.7 | 1.1 | |
| 3. alpinus | 6.2 | 10.6 | 0.9 | 1.3 | 0.9 | |
| 4. duffieldi | 0.35 | 10.05 | 6.35 | 1.2 | 0.4 | |
| 5. flavostriatus | 13.25 | 4.15 | 13.25 | 13.1 | 1.2 | |
| 6. limicola | 0.05 | 10.65 | 6.55 | 0.8 | 13.7 | |
| 7. serratulae | 11.4 | 6.7 | 11.7 | 11.15 | 8.25 | 11.85 |
| 16S | ||||||
| 1. albifrons | 1.2 | 0.7 | 0.2 | 1 | 0.1 | |
| 2. albiger | 5.25 | 1.1 | 1.2 | 0.8 | 1.2 | |
| 3. alpinus | 1.95 | 5.2 | 0.6 | 1 | 0.6 | |
| 4. duffieldi | 0.15 | 5.2 | 1.9 | 1 | 0.3 | |
| 5. flavostriatus | 5.25 | 3.6 | 4.9 | 5.2 | 1 | |
| 6. limicola | 0 | 5.35 | 2.15 | 0.15 | 5.35 | |
| 7. serratulae | 5.05 | 4.1 | 5.2 | 4.9 | 2.6 | 5.05 |
The geographic distribution of CO1 and 16S diversity within the genus Anoscopus are illustrated in Figure 5 and Figure 6, respectively. Using CO1 mitochondrial DNA sequences, the three well-defined lineages as indicated in the phylogenetic analyses (Figure 2, Figure 3 and Figure 4) were also geographically well defined (Figure 5). Anoscopus alpinus haplotypes were only recorded from the Czech Republic, A. serratulae haplotypes from Rye Habour and A. albiger haplotypes from Wartling. These networks could not be connected with 95% confidence, which indicates that these groups represent good biological species. Individuals representing A. flavostriatus and the remainder of A. albiger were connected with no shared haplotypes between these two species. All A. flavostriatus haplotypes were recorded from Winding Bottom and A. albiger individuals from Wartling. Individuals within A. albiger were separated from each other by up to 10 mutational steps, indicating higher levels of sequence variation within this species, and some of the A. albiger haplotypes could not be connected to each other and were closer to some of the A. flavostriatus haplotypes. However, there were no shared haplotypes between these two species. All those A. flavostriatus haplotypes were recorded from Winding Bottom and A. albiger individuals from Wartling. The last network included individuals of A. albifrons, A. limicola and A. duffieldi, with haplotypes being shared between species and localities. The results from the 16S rRNA networks (Figure 6) showed a similar pattern, with the exception that A. alpinus could be connected to the A. albifrons, A. limicola and A. duffieldi network. In addition, when using 16S rRNA sequences, A. albiger haplotypes were connected within one network.
4. Discussion
Species separation in Aphrodes and some of Anoscopus (e.g., A. duffieldi and A. albifrons) has hitherto been by aedeagus morphology. However, these characters alone have proved to be unreliable, although when combined with external morphometrics have been shown to improve species separation, at least for male Aphrodes [9,28]. It is likely that sexual vibrational communication signals in Anoscopus would provide additional evidence that may be diagnostic, as shown in Aphrodes [28], but this requires specialist equipment and expertise that is not widely available. Some Canadian Anoscopus spp. have been barcoded previously based on specimens collected in Canada and Corsica [59,60], but here we used DNA barcoding for the first time to separate all the known species of Anoscopus in the UK, with unexpected results.
The phylogenetic trees showed a major, deep division within the Anoscopus genus, with one subgroup comprising A. albifrons, A. limicola, A. duffieldi and A. alpinus (albifrons subgroup) and the other including A. albiger, A. flavostriatus, A. serratulae and A. histrionicus (albiger subgroup). There appear to be no obvious morphological differences between these subgroups that might warrant further taxonomic recognition. However, our analysis lacked several additional Anoscopus species and subspecies described from continental Europe, Asia and the Canary Islands [20,61], which would be needed to fully understand the phylogeny of the genus. However, it should be remembered that our aim here was primarily to separate species from the UK and determine the status of A. duffieldi, and not to generate a complete phylogeny.
4.1. Anoscopus duffieldi and Related Taxa (Albifrons Subgroup)
The phylogenetic trees (Figure 2, Figure 3 and Figure 4) and analysis of genetic distances (Table 3 and Table 5) clearly show that A. duffieldi is not conspecific with A. alpinus (based on specimens from the Czech Republic), as proposed by Le Quesne [23]. Besides differences in mitochondrial DNA sequences, both taxa also differ in habitat preferences and have allopatric distributions. Anoscopus duffieldi has only been recorded from vegetated coastal shingle at Dungeness, Kent [21], while Anoscopus alpinus is restricted to heaths, bogs and subalpine grasslands at high elevations (between 880 and 2970 m a.s.l.) of central and eastern European mountains: the Alps, Hercynian mountains, the Balkans and probably also the Carpathians [12,20,62]. However, we cannot eliminate the possibility that A. duffieldi is synonymous with another continental species, A. assimilis, to which it is also morphologically similar [12,24,27], because the latter species was missing in our molecular dataset. Anoscopus assimilis has been reported from meadows, pastures and undergrowth of mixed forests at low to montane elevations of the western Mediterranean region, and its distribution seems to extend in western France as far north as to Brittany [20,22,63]. We were able to download and examine two identical CO1 sequences thought to be A. assimilis collected in Corsica [60] (GenBank accession numbers MK816310 and MK188564), but these were acquired from females and hence, the authors acknowledge, impossible to accurately identify morphologically. They shared an identical haplotype with both A. duffieldi and A. albifrons (h1 in Figure 2).
Anoscopus duffieldi was found sympatrically with A. albifrons at Dungeness. Anoscopus duffieldi cluster together in all trees (Figure 5 and Figure 6). However, in the same cluster with A. duffieldi are specimens of A. albifrons, including a haplotype that is found in both species. Possible reasons for this include misidentification caused by intermediate aedeagal characters. Alternatively, there may be uni-directional hybridisation where male individuals of A. albifrons mate with female individuals of A. duffieldi to produce morphologically A. albifrons individuals but with A. duffieldi mitochondria. Hybridisation may also result in mixed characters [20], hindering correct identification. The term ‘hybridisation’ of course is not entirely correct for crosses between taxa that are subspecies or ecotypes.
Anoscopus duffieldi specimens are also closely related to a mixed cluster of A. albifrons and A. limicola. For the mitochondrial genes studied, there is no evidence that these are separate species. Sequence divergence values between A. albifrons and A. limicola was estimated at 0–0.05% (Table 5). There were several shared haplotypes between these two taxa, suggesting that they are one interbreeding population. Interestingly, a shared haplotype between A. limicola and A. albifrons is found at both Newtimber Hill on the south coast and Malacleit in the Outer Hebrides (morphologically identified as A. albifrons at Newtimber Hill and A. limicola in Malacleit). Divergence values between A. duffieldi and both A. albifrons and A. limicola were lower than would be expected for different species, and much lower than between other species of Anoscopus (Table 5). Anoscopus albifrons and A. limicola differ mainly in the general size, subtle details of aedeagus shape and ecology. While the former is a quite eurytopic and widely distributed grassland species, the latter has been considered to be a salt marshes specialist, particularly on the grass species Puccinellia maritima, and restricted to western European coasts [12,20,33,63].
4.2. Other Anoscopus Species (Albiger Subgroup)
All of the other species separated well, forming monophyletic groups with low intraspecific genetic diversity, with the exception of A. albiger. Some haplotypes of this highly genetically diverse species show affinities with A. flavostriatus in the CO1 tree (Figure 2), but these two species are resolved into monophyletic sister groups in the 16S and combined (CO1 + 16S) trees (Figure 3 and Figure 4). All the A. albiger specimens came from the same location (Wartling), yet each of the individuals harboured a unique haplotype. This strongly suggests high levels of genetic diversity within this species. Anoscopus albiger was clearly different from all the other Anoscopus in having far greater intraspecific diversity (e.g., Table 3, 2.4% at CO1, compared with <0.8% for all other groups). A possible explanation for this intraspecific diversity within A. albiger is that it has had a very different history in the UK compared with the other Anoscopus species. One possibility is that this is a relict species that managed to survive in the UK through the last ice age, retaining high levels of genetic diversity. The low levels of genetic diversity shown in all of the other Anoscopus species may indicate that they went through genetic bottlenecks during post-glacial recolonization. More sampling from other parts of the UK, Ireland and continental Europe may help to resolve this question.
5. Conclusions
Five out of eight Anoscopus taxa studied were clearly separated through mtDNA barcoding, and based on both morphological and molecular evidence, they represent distinct species. For the remaining three taxa (A. duffieldi, A. albifrons and A. limicola), there is little support for their status as separate species based on our molecular evidence. Pairwise genetic distances among these three taxa were very low, ranging from 0% to 0.55% (CO1 + 16S). In contrast, pairwise comparisons between all other species, and between these other species and A. duffieldi, A. albifrons and A. limicola, ranged from 4.05% to 10.45%. There is little support therefore for A. duffieldi as a separate species. However, specimens of A. duffieldi did cluster together in the trees, so it would be prudent to protect this population until other evidence is forthcoming and in the meantime treat A. duffieldi provisionally as a subspecies with a unique morphotype or simply a different ecotype or possibly host race of A. albifrons. More research would be needed to establish which term would be most appropriate. The only habitat and site on which they have been found is dry shingle, dominated by the grass Anthoxanthum odoratum L. (Figure 7). It might be appropriate to attribute a similar status (subspecies, ecotype or host race of A. albifrons) to A. limicola. All the A. limicola specimens came from saltmarsh dominated by Puccinellia maritima (Hudson), which is consistent with previously published data on the ecology of this taxon [12]. Anoscopus albifrons has been considered a generalist species found in a wide range of habitats. Guglielmino and Bückle [20] recently described another distinct morphotype as a subspecies from southern Europe, A. albifrons mappus. Anoscopus albifrons in its broad sense may turn out to represent a single polymorphic species or a complex of incipient species undergoing a process of speciation, but this suggestion clearly requires more research.
Figure 7.
Vegetated shingle at Dungeness dominated by the grass Anthoxanthum odoratum. Dungeness is the only known site where Anoscopus duffieldi has been recorded. Photo taken by Alan Stewart.
Further work could include sequencing of nuclear genes that are less conserved than 28S. This might shed light on possible cases of hybridisation. Other studies have successfully amplified the nuclear protein-coding genes Histone 3 and Wingless from leafhoppers to resolve relationships between species [64]. The hypervariable mitochondrial D-loop region could possibly resolve the relationships further. Microsatellites have been developed for the closely related Aphrodes [65], and these should be tested to see whether they work on Anoscopus. If not, more specific microsatellites could be developed. Another option is to include other characteristics such as vibrational signals [9] and ecological information such as habitat and food plant data [20]. Future studies should also aim to include material of A. assimilis and other continental taxa which are missing from our analysis.
Britain has few endemic species, and these have historically been afforded priority status by conservationists within the Biodiversity Acton Plan process [32]. Significantly, in the context of this study, however, Britain has several distinct subspecific varieties or forms of invertebrates that are endemic, often differing from their continental counterparts ecologically as well as morphologically. Furthermore, and probably because of its unique habitat for invertebrates (Figure 7) ([66]), Dungeness harbours a significant number of these endemic variants [32], of which A. duffieldi may be one.
Acknowledgments
Thanks to Robert Elmer for extracting the DNA during his final year undergraduate project at Cardiff University.
Author Contributions
The laboratory work was mainly conducted by J.R. and the paper arose from her Professional Training Year report. Analysis of the sequence data was conducted by J.R. and I.-R.M.R., both of whom generated the graphics. Assistance in the laboratory and the analysis of the data were provided by J.E.S. and R.J.M.-G. Specimens of Anoscopus were identified morphologically and provided by A.J.A.S., M.R.W., and I.M. The project was supervised throughout by W.O.C.S. The paper was mainly drafted by J.R., W.O.C.S., and I.-R.M.R., but all authors contributed to, or approved, the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
We thank Mark Gurney from the Royal Society for the Protection of Birds for providing William Symondson with the funds to undertake this work and for permission to collect material from the Dungeness reserve.
Conflicts of Interest
The authors declare no conflict of interest.
Availability of Data and Material
GenBank accession numbers for all Anoscopus species and haplotypes, plus haplotypes shared between species, plus the 16S sequence for A. bicincta: Anoscopus albifrons COI MW204907, MW204910, MW204913 16S MW228389, MW228390; Anoscopus albiger COI MW204884-MW204894 16S MW228379-MW228381; Anoscopus alpinus COI MW204914-MW204922 16S MW228393, MW228394; Anoscopus duffieldi COI MW204904-MW204906 16S MW228392; Anoscopus flavostriatus COI MW204895-MW204898 16S MW228382, MW228383; Anoscopus histrionicus COI MW204923 16S MW228386; Anoscopus limicola COI MW204911 16S MW228391; Anoscopus serratulae COI MW204899-MW204902 16S W228384, MW228385; Anoscopus limicola/albifrons haplotype COI MW204908, MW204909, MW204912 16S MW228387; Anoscopus duffieldi/albifrons haplotype COI MW204903 16S MW228388; Aphrodes bicincta 16S MW228395.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Austin J.J., Melville J. Incorporating historical museum specimens into molecular systematic and conservation genetics research. Mol. Ecol. Notes. 2006;6:1089–1092. doi: 10.1111/j.1471-8286.2006.01443.x. [DOI] [Google Scholar]
- 2.Hebert P.D.N., Gregory T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 2005;54:852–859. doi: 10.1080/10635150500354886. [DOI] [PubMed] [Google Scholar]
- 3.Dexter K.G., Pennington T.D., Cunningham C.W. Using DNA to assess errors in tropical tree identifications: How often are ecologists wrong and when does it matter? Ecol. Monogr. 2010;80:267–286. doi: 10.1890/09-0267.1. [DOI] [Google Scholar]
- 4.Lis B., Lis B., Ziaja D.J. In BOLD we trust? A commentary on the reliability of specimen identification for DNA barcoding: A case study on burrower bugs (Hemiptera: Heteroptera: Cydnidae) Zootaxa. 2016;4114:83–86. doi: 10.11646/zootaxa.4114.1.6. [DOI] [PubMed] [Google Scholar]
- 5.Bluemel J.K., King R., Virant-Doberlet M., Symondson W.O.C. Primers for identification of type and other archived specimens of Aphrodes leafhoppers (Hemiptera, Cicadellidae) Mol. Ecol. Resour. 2011;11:770–774. doi: 10.1111/j.1755-0998.2011.03008.x. [DOI] [PubMed] [Google Scholar]
- 6.Seabra S.G., Pina-Martins F., Marabuto E., Yurtsever S., Halkka O., Quartau J.A., Paulo O.S. Molecular phylogeny and DNA barcoding in the meadow-spittlebug Philaenus spumarius (Hemiptera, Cercopidae) and its related species. Mol. Phylogenet. Evol. 2010;56:462–467. doi: 10.1016/j.ympev.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 7.Aly S.M. Reliability of long vs short COI markers in identification of forensically important flies. Croat. Med. J. 2014;55:19–26. doi: 10.3325/cmj.2014.55.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Mandal S., Chhakchhuak L., Gurusubramanian G., Kumar N.S. Mitochondrial markers for identification and phylogenetic studies in insects—A review. DNA Barcodes. 2014;2:1–9. doi: 10.2478/dna-2014-0001. [DOI] [Google Scholar]
- 9.Bluemel J.K., Derlink M., Pavlovčič P., Russo I.-R.M., King R.A., Corbett E., Sherrard-Smith E., Blejec A., Wilson M.R., Stewart A.J.A., et al. Integrating vibrational signals, mitochondrial DNA and morphology for species determination in the genus Aphrodes (Hemiptera: Cicadellidae) Syst. Entomol. 2014;39:304–324. doi: 10.1111/syen.12056. [DOI] [Google Scholar]
- 10.Kirby P., Stewart A.J., Wilson M.R. True bugs, leaf-and planthoppers, and their allies. In: Hawksworth D.L., editor. The Changing Wildlife of Great Britain and Ireland. Volume 17. CRC Press; Boco Raton, FL, USA: 2001. pp. 262–299. [Google Scholar]
- 11.Forero D. The systematics of the Hemiptera. Rev. Colomb. Entomol. 2008;34:1–21. [Google Scholar]
- 12.Nickel H. The Leafhoppers and Planthoppers of Germany (Hemiptera: Auchenorrhyncha): Patterns and Strategies in a Highly Diverse Group of Phytophagous Insects. Pensoft; Sofia, Bulgaria: Goecke & Evers; Keltern, Germany: 2003. [Google Scholar]
- 13.Bartlett C.R., Deitz L.L., Dmitriev D.A., Sanborn A.F., Soulier-Perkins A., Wallace M.S. The diversity of the true hoppers (Hemiptera: Auchenorrhyncha) In: Foottit R.G., Adler P.H., editors. Insect Biodiversity: Science and Society. Volume 2. John Wiley & Sons; Chichester, UK: 2018. pp. 501–590. [Google Scholar]
- 14.Biedermann R., Achtziger R., Nickel H., Stewart A.J.A. Conservation of grassland leafhoppers: A brief review. J. Insect Conserv. 2005;9:229–243. doi: 10.1007/s10841-005-0531-z. [DOI] [Google Scholar]
- 15.Zahniser J.N., Dietrich C.H. Phylogeny of the leafhopper subfamily Deltocephalinae (Insecta: Auchenorrhyncha: Cicadellidae) and related subfamilies based on morphology. Syst. Biodivers. 2008;6:1–24. doi: 10.1017/S1477200007002617. [DOI] [Google Scholar]
- 16.Dietrich C.H. Overview of the phylogeny, taxonomy and diversity of the leafhopper (Hemiptera: Auchenorrhyncha: Cicadomorpha: Membracoidea: Cicadellidae) vectors of plant pathogens. In: Chang C.-I., Lee C.-Y., Hsien-Tzung Shih H.-T., editors. Proceedings of the 2013 International Symposium on Insect Vectors and Insect-Borne Diseases, Taichung, Taiwan, August 2013. Volume 173. Special Publication of TARI; Taiwan Agricultural Research Institute; Taichung, Taiwan: 2013. pp. 47–70. [Google Scholar]
- 17.Rothenbücher J., Schaefer M. Conservation of leafhoppers in floodplain grasslands—Trade-off between diversity and naturalness in a northern German national park. J. Insect Cons. 2005;9:335–349. doi: 10.1007/s10841-005-0514-0. [DOI] [Google Scholar]
- 18.Schuch S., Wesche K., Schaefer M. Long-term decline in the abundance of leafhoppers and planthoppers (Auchenorrhyncha) in Central European protected dry grasslands. Biol. Conserv. 2012;149:75–83. doi: 10.1016/j.biocon.2012.02.006. [DOI] [Google Scholar]
- 19.Endrestøl A., Elven H. Two species of Aphrodinae (Hemiptera, Cicadellidae) new to the Norwegian fauna. Norweg. J. Entomol. 2009;56:24–27. [Google Scholar]
- 20.Guglielmino A., Bückle C. Revision of Errhomeninae and Aphrodinae (Hemiptera, Cicadomorpha) in Italy with remarks on their variability and distribution in adjacent regions and description of three new taxa. Zootaxa. 2015;3906:1–66. doi: 10.11646/zootaxa.3906.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Wilson M.R., Stewart A., Biedermann R., Nickel H., Niedringhaus R. The Planthoppers and Leafhoppers of Britain and Ireland. Identification Keys to All Families and Genera and all British and Irish Species not Recorded from Germany. Cicadina, Supplement 2. WABV Fründ; Scheeßel, Germany: 2015. [Google Scholar]
- 22.Ribaut H. Homoptères Auchénorhinques. II. (Jassidae) Volume 57. Faune de France; Paris, France: 1952. p. 474. [Google Scholar]
- 23.Le Quesne W.J. Hemiptera (Cicadomorpha), excluding Deltocephalinae and Typhlocybinae. Handbooks for the Identification of British Insects. Royal Entomological Society; London, UK: 1965. [Google Scholar]
- 24.Hamilton K.G.A. A review of the northern hemisphere aphrodina (Rhynchota: Homoptera: Cicadellidae), with special reference to the nearctic fauna. Can. Entomol. 1975;107:1009–1027. doi: 10.4039/Ent1071009-10. [DOI] [Google Scholar]
- 25.Biedermann R., Niedringhaus R. The Plant- and Leafhoppers of Germany: Identification Key to All Species. WABV Fründ; Scheeßel, Germany: 2009. [Google Scholar]
- 26.Barnett D.E. Some new preparation techniques used in leafhopper identification. Fla. Entomol. 1976;59:321–323. doi: 10.2307/3494272. [DOI] [Google Scholar]
- 27.Remane R., Fröhlich W. Beiträge zur Chorologie einiger Zikaden-Arten (Homoptera Auchenorrhyncha) in der Westpaläarktis. Marbg. Entomol. Publik. 1994;2:131–188. [Google Scholar]
- 28.Tishechkin D.Y. On the taxonomy and distribution of Aphrodes bicincta (Schrank, 1776) species group (Homoptera: Auchenorrhyncha: Cicadellidae: Aphrodinae) in Eastern Palaearctic. Zootaxa. 2017;4318:167–176. doi: 10.11646/zootaxa.4318.1.9. [DOI] [Google Scholar]
- 29.Duffield C.A.W. The genus aphrodes (Homoptera; Auchenorhyncha) Trans. Kent Field Club. 1963;1:155–160. [Google Scholar]
- 30.Le Quesne W.J. Some taxonomic changes and additions in the British Cicadellidae (Hemiptera) including a new species and subspecies. Proc. R. Entomol. Soc. B. 1964;33:73–116. doi: 10.1111/j.1365-3113.1964.tb01616.x. [DOI] [Google Scholar]
- 31.Nast J. Palaearctic Auchenorrhyncha (Homoptera) an Annotated Check List. Polish Scientific Publishers; Warsaw, Poland: 1992. [Google Scholar]
- 32.Key R.S., Drake C.M., Sheppard D.A. Conservation of Invertebrates in England: A Review and Framework. English Nature; Peterborough, UK: 2000. English Nature Science Report 35. [Google Scholar]
- 33.Ossiannilsson F. Fauna Entomologica Scandinavica—Part 2. Volume 7 Scandinavian Science Press; Klampenborg, Denmark: 1981. The auchenorrhyncha (homoptera) of fennoscandia and Denmark. [Google Scholar]
- 34.Pentinsaari M., Salmela H., Mutanen M., Roslin T. Molecular evolution of a widely-adopted taxonomic marker (COI) across the animal tree of life. Sci. Rep. 2016;6:35275. doi: 10.1038/srep35275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Percy D.M. Making the most of your host: The Metrosideros-feeding psyllids (Hemiptera, Psylloidea) of the Hawaiian Islands. ZooKeys. 2007;649:1–163. doi: 10.3897/zookeys.649.10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebert P.D., Ratnasingham S., De Waard J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nei M., Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dietrich C., Rakitov R., Holmes J., Black W. Phylogeny of the major lineages of membracoidea (Insecta: Hemiptera: Cicadomorpha) based on 28S rDNA sequences. Mol. Phylogenet. Evol. 2001;18:293–305. doi: 10.1006/mpev.2000.0873. [DOI] [PubMed] [Google Scholar]
- 39.Dai R.H., Chen X.X., Li Z.Z. Phylogeny of Deltocephalinae (Hemiptera: Cicadellidae) from China based on partial 16S rDNA and 28S rDNA D2 sequences combined with morphological characters. Acta Entomol. Sin. 2008;51:1055–1064. [Google Scholar]
- 40.Folmer O., Black M., Hoeh W., Lutz R., Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 41.Fu J.-Y., Han B.-Y., Xiao Q. Mitochondrial COI and 16sRNA evidence for a single species hypothesis of E. vitis, J. formosana and E. onukii in East Asia. PLoS ONE. 2014;9:e115259. doi: 10.1371/journal.pone.0115259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hillis D.M., Dixon M.T. Ribosomal DNA: Molecular evolution and phylogenetic inference. Q. Rev. Biol. 1991;66:411–453. doi: 10.1086/417338. [DOI] [PubMed] [Google Scholar]
- 43.Zahniser J.N. Ph.D Thesis. University of Illinois; Champaign, IL, USA: 2008. [(accessed on 28 October 2020)]. Systematics of the Leafhopper Subfamily Deltocephalinae (Hemiptera: Cicadellidae) and the Tribe Chiasmini: Phylogeny, Classification, and Biogeography. Available online: http://hdl.handle.net/2142/86469. [Google Scholar]
- 44.Larkin M., Blackshields G., Brown N., Chenna R., Mcgettigan P., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 45.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 46.Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z., Goldman N., Friday A. Comparison of models from nucleotide substitution used in maximum likelihood phylogenetic estimation. Mol. Biol. Evol. 1994;11:316–324. doi: 10.1093/oxfordjournals.molbev.a040112. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 1996;144:1941–1950. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- 49.Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rambaut A., Suchard M., Xie W., Drummond A. Tracer v. 1.6; Institute of Evolutionary Biology, University of Edinburgh. [(accessed on 28 October 2020)];2014 Available online: https://github.com/beast-dev/tracer/releases/tag/v1.6.
- 51.Rambaut A. FigTree 1.4. 2; University of Edinburgh: Edinburgh, Scotland. [(accessed on 28 October 2020)];2014 Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 52.Nei M., Tajima F. Genetic drift and estimation of effective population size. Genetics. 1981;98:625–640. doi: 10.1093/genetics/98.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York, NY, USA: 1987. p. 512. [Google Scholar]
- 54.Rozas J., Rozas R. DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- 55.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 56.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clement M., Posada D., Crandall K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 58.Avise J.C. Phylogeography: The History and Formation of Species. Harvard University Press; Cambridge, UK: 2000. [Google Scholar]
- 59.Gwiazdowski R.A., Foottit R.G., Maw H.E.L., Hebert P.D.N. The Hemiptera (Insecta) of Canada: Constructing a reference library of DNA barcodes. PLoS ONE. 2015;10:e0125635. doi: 10.1371/journal.pone.0125635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Albre J., Gibernau M. Diversity and temporal variations of the Hemiptera Auchenorrhyncha fauna in the Ajaccio region (France, Corsica) Ann. Soc. Entomol. Fr. 2019;55:497–508. doi: 10.1080/00379271.2019.1688189. [DOI] [Google Scholar]
- 61.Lindberg H. Hemiptera insularum canariensium. systematik, ökologie und verbreitung der kanarischen heteropteren und cicadinen. Comment. Biol. 1954;14:1–304. [Google Scholar]
- 62.Malenovský I. New records of Auchenorrhyncha (Hemiptera) for the Czech Republic. Acta Musei Morav. Scient. Boil. 2013;98:235–263. [Google Scholar]
- 63.Della Giustina W. Faune de France, 73. Volume 3 Fédération Française des Sociétés de Sciences Naturelles et INRA; Paris, France: 1989. Homoptères Cicadellidae. Compléments aux ouvrages d’Henri Ribaut. [Google Scholar]
- 64.Bennett G.M., O’Grady P.M. Review of the native Hawaiian leafhopper genus Nesophrosyne (Hemiptera: Cicadellidae: Deltocephalinae) with description of eight new species associated with Broussaisia arguta (Hydrangeaceae) Zootaxa. 2011;2805:1–25. doi: 10.11646/zootaxa.2805.1.1. [DOI] [Google Scholar]
- 65.Derlink M., Pipan B., Pavlovčič P., Jones L., Meglič V., Symondson W.O.C., Virant-Doberlet M. Characterization of eleven polymorphic microsatellite markers for leafhoppers of the genus Aphrodes (Hemiptera: Cicadellidae) Conserv. Genet. Resour. 2014;6:933–935. doi: 10.1007/s12686-014-0245-1. [DOI] [Google Scholar]
- 66.Morris R.K.A., Parsons M.S. JNCC Report 77. Joint Nature Conservation Committee; Peterborough, UK: 1992. A survey of invertebrate communities on the shingle of Dungeness, Rye Harbour and Orford Ness. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GenBank accession numbers for all Anoscopus species and haplotypes, plus haplotypes shared between species, plus the 16S sequence for A. bicincta: Anoscopus albifrons COI MW204907, MW204910, MW204913 16S MW228389, MW228390; Anoscopus albiger COI MW204884-MW204894 16S MW228379-MW228381; Anoscopus alpinus COI MW204914-MW204922 16S MW228393, MW228394; Anoscopus duffieldi COI MW204904-MW204906 16S MW228392; Anoscopus flavostriatus COI MW204895-MW204898 16S MW228382, MW228383; Anoscopus histrionicus COI MW204923 16S MW228386; Anoscopus limicola COI MW204911 16S MW228391; Anoscopus serratulae COI MW204899-MW204902 16S W228384, MW228385; Anoscopus limicola/albifrons haplotype COI MW204908, MW204909, MW204912 16S MW228387; Anoscopus duffieldi/albifrons haplotype COI MW204903 16S MW228388; Aphrodes bicincta 16S MW228395.